Abstract
Fluorescent quantum dots are emerging as an important tool for imaging cells and tissues, and their unique optical and physical properties have captured the attention of the research community. The most common types of commercially available quantum dots consist of a nanocrystalline semiconductor core composed of cadmium selenide with a zinc sulfide capping layer and an outer polymer layer to facilitate conjugation to targeting biomolecules such as immunoglobulins. They exhibit high fluorescent quantum yields and have large absorption cross-sections, possess excellent photostability, and can be synthesized so that their narrow-band fluorescence emission can occur in a wide spectrum of colors. These properties make them excellent candidates for serving as multiplexing molecular beacons using a variety of imaging modalities including highly correlated microscopies. Whereas much attention has been focused on quantum-dot applications for live-cell imaging, we have sought to characterize and exploit their utility for enabling simultaneous multiprotein immunolabeling in fixed cells and tissues. Considerations for their application to immunolabeling for correlated light and electron microscopic analysis are discussed.
Keywords: Quantum dot, light microscopy, fluorescence microscopy, electron microscopy, immunolabeling, multiphoton microscopy
INTRODUCTION
Toxicologic pathology stands to benefit enormously from continuing advances in both fluorescence microscopy technologies and methodologies. These advances include confocal and multiphoton microscopy (Denk et al., 1990), deconvolution (Chen et al., 1995), total internal reflection fluorescence microscopy (TIRF; Axelrod et al., 1983), photoactivation localization microscopy (PALM; Betzig et al., 2006), and 4-pi imaging (Schrader et al., 1998), as well as the creation of novel genetically encoded reported molecules and new classes of fluorescent probes (for review, see Tsien [2006] and Giepmans et al. [2006]). These approaches offer researchers unprecedented optical resolution and sensitivity. However, it is still the case that much of the fine cellular machinery operates beyond the resolution of the light microscope in the realm visualized by electron microscopy. Therefore, to maximize and validate observations of protein expression and colocalization as well as characterize subtle alterations to cellular morphology, it is important to use multiple microscopies encompassing a wide range of overlapping scales, making methods that allow for highly correlated light- and electron-microscopic observations very desirable. One such approach for correlated multiscale imaging uses a relatively new class of semiconductor-based fluorescent probes termed quantum dots (Chan and Nie, 1998; Bruchez et al., 1998). These nanomaterials not only possess unique optical properties but are also directly visible by transmission electron microscopy (Liu et al., 2000), opening up a number of unique imaging opportunities (Nisman et al., 2004; Giepmans et al., 2005).
CHARACTERISTICS OF QUANTUM DOTS
Quantum dots are fluorophore nanocrystals whose excitation and emission is fundamentally different than traditional organic fluorophores. Instead of electronic transitions from one valence orbital to another, quantum-dot fluorescence involves exciting an electron from the bulk valence band of the semiconductor material across an energy gap, making it a conduction electron and leaving behind a hole. The electron–hole pair (also known as an exciton) is quantum-confined by the small size of the nanocrystal (smaller than the exciton Bohr radius). When the electron–hole pair eventually recombines, a characteristic photon is emitted. Minute changes to the size of the confining crystal alter the energy bandgap, thus determining the color of the fluorescence photon. In general, the smaller the quantum dot, the larger the bandgap energy for a given material, and thus, the shorter the wavelength of the emitted fluorescence. Of the many types of quantum dots that can be made from various semiconductor materials, CdSe/ZnS quantum dots are presently the most common commercially available as secondary antibody conjugates. They are composed of a core of cadmium selenide ranging from about 10 to 50 atoms in diameter and about 100 to 100,000 atoms in total, and as mentioned, the size of the core determines the fluorescence emission spectra. They have a thin zinc sulfide passivating layer that improves the fluorescence quantum efficiency and stability of the quantum dots and an organic polymer coating to make them water soluble and enabling bioconjugation to targeting molecules such as anti-IgG (immunoglobulin G) secondary antibodies, Fab fragments, peptides, or streptavidin (Figure 1a).
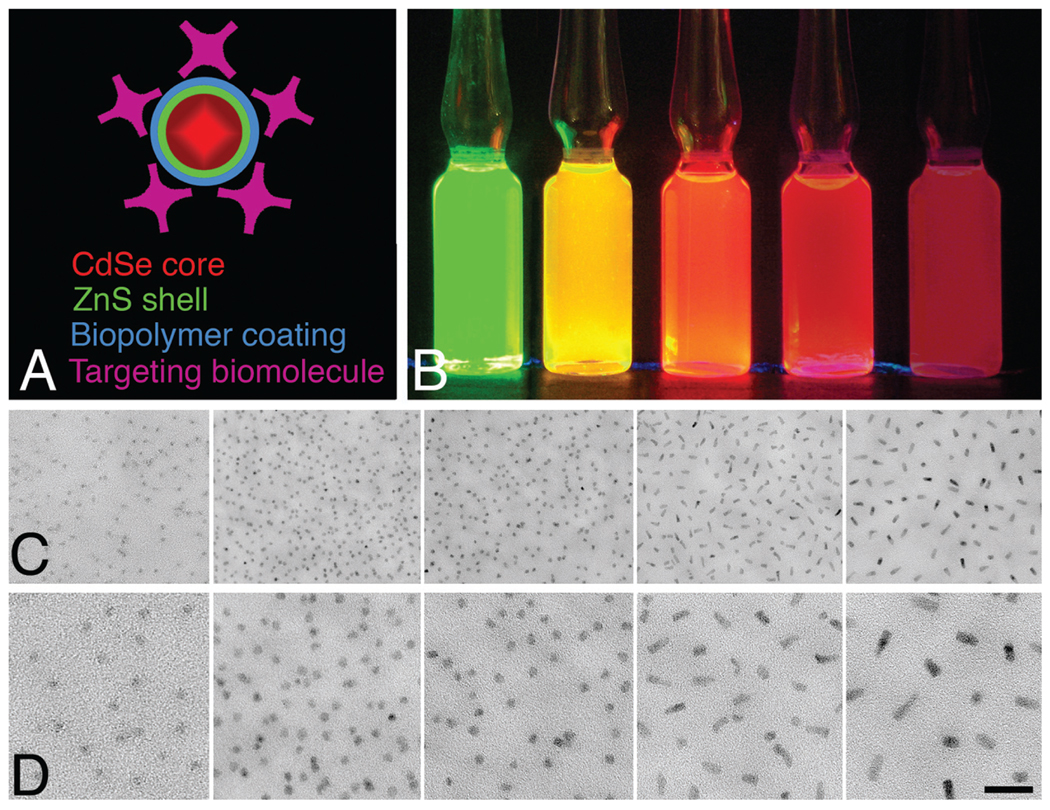
FIGURE 1.
(a) Diagram representing the composition of a CdSe/ZnS quantum dot showing the core, shell, coating, and targeting molecules. The overall size is about 15 to 20 nm. (b) Micromolar aqueous solutions of 525, 565, 585, 605, and 655 quantum dots (left to right) under ultraviolet (UV) illumination. (c) Electron microscopic appearance of the same quantum dots (left to right) spread on a thin carbon substrate and air-dried. (d) Higher magnification image. Bar = 20 nm.
They exhibit high fluorescence quantum yields, and as would be expected from a solid-state material, they are extremely resistant to reactive oxygen-mediated photobleaching. They have large absorption cross-sections and broad absorption spectra with narrow band fluorescence emission that can be tuned over a broad range from blue to near-infrared. Under ambient light, micromolar solutions are nearly colorless, but under UV excitation, they exhibit brilliant and distinct fluorescence (Figure 1b). Very different from traditional fluorophores, they have symmetrical Gaussian-shaped emission spectra, and more importantly, all have exceptionally large Stoke’s shifts and can be excited equally well at a single UV wavelength, making them excellent for multiple labeling experiments (Chan et al., 2002; Klostranec and Chan, 2006). Typically, commercially available quantum dots are named for their peak emission wavelength (i.e., 525, 565, 585, 605, and 655 nm). Additionally, quantum dot cores have sufficient electron density to be directly visible by electron microscopy in the 60-keV to 400-keV accelerating voltage range without any special contrasting treatments (Figure 1c) and can be discriminated by their different sizes and shapes (525-, 565-, and 585-nm Cd/Zn quantum dots are spherical, and 605- and 655-nm quantum dots are oblong).
CONSIDERATIONS FOR IMMUNOLABELING
Antibody conjugates of quantum dots exhibit modest but significant penetration into fixed and mildly permeablized cells and tissues (i.e., several microns), making them suitable for pre-embedding labeling methods (Figure 2a). Whereas quantum-dot secondary antibody conjugates do not penetrate nearly as well as their organic fluorophore counterparts under identical conditions, the ability to distinguish different quantum dots by both light and electron microscopy makes them excellent for multiple labeling-correlated imaging experiments (Giepmans et al., 2005). This feature also allows for efficient trouble-shooting and optimization of various antibody-antigen–dependent labeling parameters by light microscopy before proceeding with the more time-consuming and laborious preparation for electron microscopy.
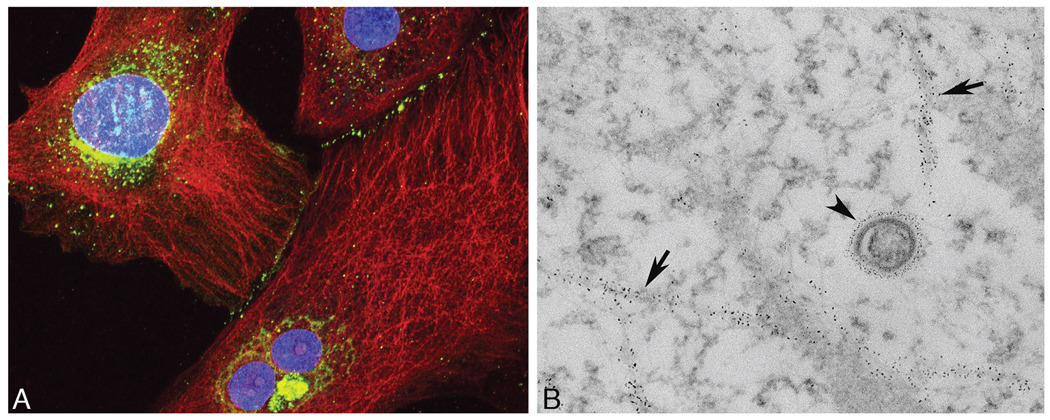
FIGURE 2.
(a) Cultured RFL6 (rat fetal lung) fibroblasts immunolabeled for β-tubulin with quantum dot (QD) 655 (red) and anti connexin 43 with QD 565 (green). Cell nuclei were counterstained with Hoecshst 33342 (blue). (b) Electron microscopic appearance of a similarly labeled sample with QD labeling on microtubules (arrows) and a connexin-43–rich vesicle (arrowhead).
Optimal fixation and permeablization conditions must be determined for each primary antibody with the understanding that four main parameters (chemical fixation, detergent permeablization, temperature, and time of treatments) are a compromise between good labeling and good ultrastructural preservation. Although no universal protocol exists for detecting all proteins with antigen-specific antibodies with consistent, high-quality ultrastructural preservation for electron microscopy, starting-point reference protocols for immunolabeling of cultured cells and for tissue sections have been introduced, along with specific guidelines (Deerinck et al., 2007). These use primary fixation with formaldehyde (from freshly depolymerized paraformaldehyde) containing low concentrations of glutaraldehyde followed by mild detergent permeablization using Triton X-100 or saponins. In general, the strongest fixation and the lowest concentration of detergent that still gives adequate labeling should be used. Because the availability of methods for optical section imaging of relatively thick (60 to 80 micron) sections (i.e., confocal microscopy) and the need to preserve the best possible cellular ultrastructure for subsequent electron microscopy, the use of thick Vibratome sections for pre-embedding immunolabeling is recommended instead of cryostat or paraffin sections. Once free-floating sections are immunolabeled with primary and secondary antibody conjugates, the sections can be temporarily mounted in buffer and viewed by confocal microscopy (Figure 3a). After subsequent postprocessing, epoxy embedding, and cutting of ultrathin sections, the quantum dots can be directly visualized in tissue by electron microscopy (Figure 3b and 3c). It is important that the sections for electron microscopy are taken from the first few microns of the surface of the tissue, and this is easily accomplished using flat embedding of the material between two glass slides.
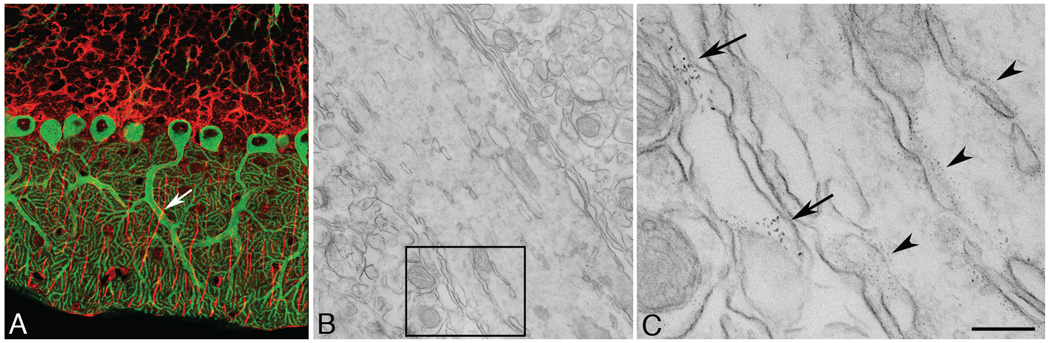
FIGURE 3.
(a) Multiphoton image of rat cerebellum double labeled for the inositol 1,4,5-trisphosphate receptor in Purkinje neurons with 565-nm quantum dots (green) and anti-GFAP (glial fibrillary acidic protein) in glial cells with 655-nm quantum dots (red). (b) A region similar to the area shown in panel A (arrow) of a Purkinje cell dendrite. (c) High-magnification image of the boxed area in panel B showing 565-nm quantum-dot labeling on the endoplasmic reticulum of the dendrite and 655-nm quantum-dot labeling in a neighboring glial cell process. Bar = 200 nm.
In addition to the four parameters mentioned above, one must take into consideration the nature of the cell type being investigated. Generally speaking, cells with relatively dense cytoplasm (i.e., muscle) will allow for far less penetration of quantum-dot conjugates than others (i.e., liver, kidney) and may require harsher permeablization.
FLUORESCENCE IMAGING
Quantum dots can be used for ordinary epifluorescence microscopy with either a xenon or, preferably, a mercury light source. Standard fluorescein or rhodamine filter sets for the 525-nm and 605-nm quantum dots can be used, but they will be 5 to 10 times dimmer than optimized filter sets. The optimal excitation filter for all quantum dots is approximately 425 nm, with a bandwidth of 40 nm (full width half maximum), and the emission filter should be 20 nm (full width half maximum) centered at the emission peak. Because they have such large Stoke’s shifts, the dichroic edge steepness and placement are not critical, and most major manufacturers offer completely optimized filter sets specifically designed for quantum-dot imaging. For confocal microscopy, the most common lasers are the argon and krypton-argon type offering 488-, 514-, 568-, and 647-nm lines, none of which are completely optimal. If possible, it is best to use a blue diode laser with a line at 442 nm, or even better, at 405 nm. Quantum dots are particularly well suited for multiphoton (2-photon) excitation, because their 2-photon absorption cross-sections are two to three orders of magnitude greater than traditional organic flluorophores (Zipfel et al., 2003).
MULTIPHOTON MICROSCOPY AND QUANTUM DOTS
Multiphoton excitation microscopy is a type of fluorescence microscopy in which two photons of low-energy, typically infrared, wavelength are used to mimic the higher energy single-photon excitation of a fluorophore in a quantum event, resulting in the emission of one photon from fluorescence (Denk et al., 1990). The probability of the near simultaneous (<10−15-second) absorption of two photons is extremely low. Therefore, an extremely high flux of photons is required to achieve efficient excitation. To achieve this, a diode-pumped, high-power titanium-sapphire laser is usually used, generally having an infrared output that can be adjusted between 700 and 1,000 nm. The beam is pulsed 80 million times a second with a pulse width of ~100 femtoseconds, so that the actual average power is low and is scanned over the sample point by point in a raster pattern as in confocal microscopy.
Multiphoton microscopy offers a number of interesting advantages (Zipfel et al., 2003). Because the pulse bandwidth is relatively large (~20 nm), one can obtain simultaneous excitation of multiple fluorophores normally excited with UV to blue light using infrared excitation. In addition, infrared light exhibits deeper penetration into biological specimens with less out-of-plane phototoxicity. More importantly, because two photons must be absorbed simultaneously to excite a fluorophore, the probability of emission is a nonlinear process and is related to the intensity squared of the excitation beam. This means that for a laser focused by high numerical aperture (NA) optics, only a small excitation volume results for 2-photon, thereby confining the excitation to the focal plane of the objective lens (Denk et al., 1990). The result is confocal-type imaging without the need for pinhole apertures.
As previously mentioned, quantum dots are particularly well suited for 2-photon excitation because of their enormous 2-photon absorption cross-sections. Typically all CdSe/ZnS quantum dots are efficiently excited at 800 nm by 2-photon excitation. Given their excellent resistance to photobleaching, there is no need to use antifade reagents during imaging.
TOWARD A SPECIMEN COMPATIBLE WITH BOTH FLUORESCENCE AND ELECTRON MICROSCOPY
Quantum-dot fluorescence is extremely durable. It can with-stand many types of chemical treatments, including aldehyde fixation, solvent dehydration, and even epoxy resin embedding and heat (60°C to 70°C) polymerization (Giepmens et al., 2005). This property makes it possible to create specimens in which the fluorescence can be preserved throughout processing for electron microscopy as long as postfixation with osmium tetroxide is omitted (which nearly instantaneously and irreversibly destroys their fluorescence). The specimens can be embedded in either epoxy resin, or better, with LR (London Resin) White acrylic resin, which possesses lower autofluorescence than the former. Specimens in polymerized resin can be imaged by conventional epifluorescence (cells) or by confocal or multiphoton microscopy (cells and tissue sections) using optical sectioning (Figure 4a). Conventional ultrathin sections can be cut by ultramicrotomy, and the sections can be mounted on copper electron microscopy (EM) grids and again imaged by fluorescence microscopy (Figure 4b). The same specimen can then be imaged by electron microscopy for higher resolution examination (Figure 4c and 4d). This approach allows for relatively wide-field surveying of specimens by light microscopy, followed by directly correlated imaging of a smaller region of interest by electron microscopy. Protocols and reagents that could serve as suitable replacements for osmium tetroxide postfixation to better preserve and contrast cell membranes and that do not affect quantum-dot fluorescence are currently being explored. This approach could help create libraries of specimens with room-temperature–stable fluorescence that could be archived indefinitely for later examination by both light and electron microscopy. Such specimens will be important in protein expression and localization studies in healthy versus pathological tissue, and patient-tissue libraries thus created would be of high value in the field of comparative toxicological pathological research. Already, quantum dots are beginning to play a role in molecular pathology by aiding in the assessment of multiple malignant-tumor biomarkers (True et al., 2007). Increasing their multiplexing capability and automating image acquisition will further this and other potential applications to toxicological and experimental pathology.
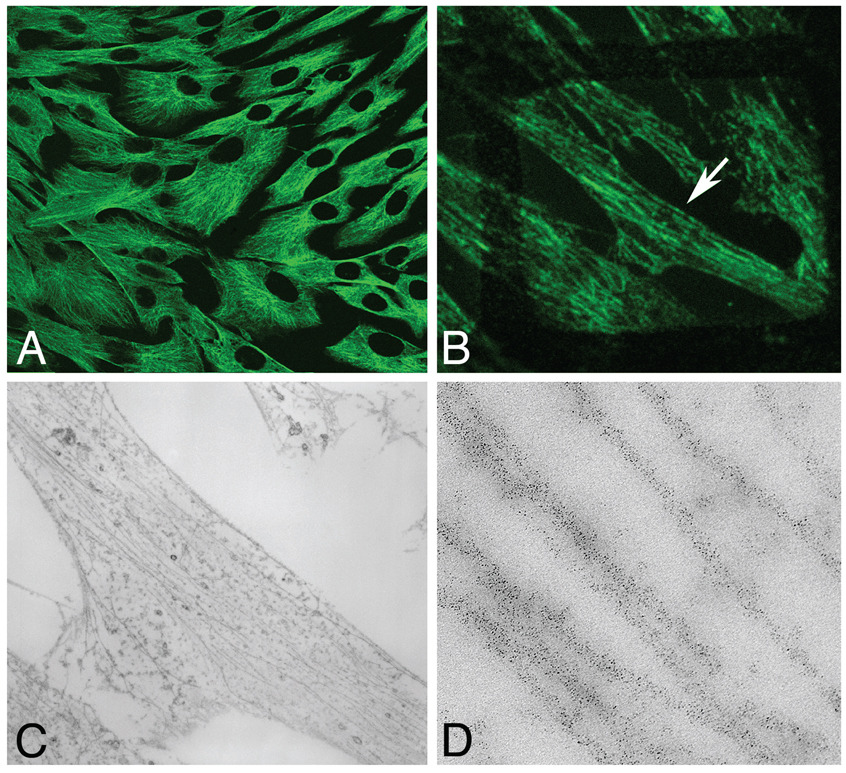
FIGURE 4.
Preservation of quantum-dot fluorescence in fixed and embedded cells. (a) Cultured HeLa (Henrietta Lacks) cells immunolabeled for b-tubulin with 565-nm quantum dots and embedded in epoxy resin. (b) An 80-nm–thick section of the same preparation mounted on a copper electron microscopy (EM) grid (grid bars visible as a boxed shadow) and imaged by confocal microscopy. (c) EM image of the same region shown in panel B (arrow). (d) Higher magnification of the same region showing quantum-dot–labeled microtubules.
FUTURE PROSPECTS
Progress with the development of new nanomaterials continues at a rapid pace, and certainly many new types of commercially available quantum dots and enhanced multifunctional nanoscale probes are to be expected. Aside from an expansion of the repertoire of colors, smaller bioconjugates as well as enhanced surface chemistries should allow for even greater penetration into cells and tissues. More distinct-shaped (pyramid, rod, dot, oblate, square) quantum dots from a wide range of semiconductor materials have already been demonstrated, and their availability as secondary antibody or direct primary antibody conjugates will allow for easier multiprotein discrimination by electron microscopy. Furthermore, as methods for delivering quantum dots into living cells continue to evolve and improve, sophisticated correlated live-cell and electron-microscopic imaging of single molecules will be possible.
ACKNOWLEDGEMENTS
Thanks to the excellent assistance of B. N. G. Giepmans, Benjamin Smarr, and Ying Jones. This work was supported by National Institutes of Health Grant 2P41RR004050-16 to Mark H. Ellisman.
Abbreviations
- EM
electron microscopy
- HeLa
Henrietta Lacks
- IgG
immunoglobulin G
- LR
London Resin
- NA
numerical aperture
- QD
quantum dot
- RFL6
rat fetal lung fibroblasts
- TIRF
total internal reflection fluorescence microscopy
- UV
ultraviolet
REFERENCES
- Axelrod D, Thompson NL, Burghardt TP. Total internal inflection fluorescent microscopy. J Microsc. 1983;129:19–28. doi: 10.1111/j.1365-2818.1983.tb04158.x. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos P. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- Chen H, Swedlow JR, Grote M, Sedat JW, Agard DA. The collection, processing, and display of digital three-dimensional images of biological specimens. In: Pawley JM, editor. Handbook of Biological Confocal Microscopy. 2nd ed. New York: Plenum Press; 1995. pp. 197–210. [Google Scholar]
- Deerinck TJ, Giepmans BN, Smarr BL, Martone ME, Ellisman MH. Light and electron microscopic localization of multiple proteins using quantum dots. Methods Mol Biol. 2007;374:43–54. doi: 10.1385/1-59745-369-2:43. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler J, Webb W. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. review. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH. Correlated light and electron microscopic imaging of multiple endogenous proteins using quantum dots. Nat Methods. 2005;10:743–749. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- Klostranec J, Chan W. Quantum dots in biological and biomedical research: recent progress and present challenges. Adv Materials. 2006;18:1953–1964. [Google Scholar]
- Liu C, Miller PD, Henstrom WL, Gibson JM. Transmission electron microscopy of semiconductor quantum dots. J Microsc. 2000;199:130–140. doi: 10.1046/j.1365-2818.2000.00729.x. [DOI] [PubMed] [Google Scholar]
- Nisman R, Dellaire G, Ren Y, Li R, Bazett-Jones DP. Application of quantum dots as probes for correlative fluorescence, conventional, and energy-filtered transmission electron microscopy. J Histochem Cytochem. 2004;52:13–18. doi: 10.1177/002215540405200102. [DOI] [PubMed] [Google Scholar]
- Schrader M, Bahlmann K, Giese G, Hell SW. 4Pi-confocal imaging in fixed biological specimens. Biophys J. 1998;75:1659–1668. doi: 10.1016/S0006-3495(98)77608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True LD, Gao X. Quantum dots for molecular pathology: their time has arrived. J Mol Diagn. 2007;9:7–11. doi: 10.2353/jmoldx.2007.060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RY. Breeding and building molecules to spy on cells and tumors. Keio J Med. 2006;55:127–140. doi: 10.2302/kjm.55.127. [DOI] [PubMed] [Google Scholar]
- Zipfel W, Williams R, Webb W. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]