Abstract
Double-stranded DNA cleavage of light-activated lysine conjugates is strongly enhanced at the slightly acidic pH (<7) suitable for selective targeting of cancer cells. This enhancement stems from the presence of two amino groups of different basicities. The first amino group plays an auxiliary role by enhancing solubility and affinity to DNA whereas the second amino group which is positioned next to the light-activated DNA-cleaver undergoes protonation at the desired pH threshold.
This protonation results in two synergetic effects which account for the increased DNA-cleaving ability at the lower pH. First, lysine conjugates show tighter binding to DNA at the lower pH, which is consistent with the anticipated higher degree of interaction between two positively charged ammonium groups with the negatively charged phosphate backbone of DNA. Second, the unproductive pathway which quenches the excited state of the photocleaver through intramolecular electron transfer is eliminated once the donor amino group next to the chromophore is protonated.
Experiments in the presence of traps for diffusing radicals show that reactive oxygen species do not contribute significantly to the mechanism of DNA cleavage at the lower pH, which is indicative of tighter binding to DNA under these conditions. This feature is valuable not only because many solid tumors are hypoxic but also because cleavage which does not depend on diffusing species is more localized and efficient. Sequence-selectivity experiments suggest combination of PET and base alkylation as the chemical basis for the observed DNA-damage. The utility of these molecules for phototherapy of cancer is confirmed by the drastic increase in toxicity of five conjugates against cancer cell lines upon photoactivation.
INTRODUCTION
Control of chemical reactions becomes especially challenging when chemical processes have to in the complexity of biological environments. This is one of the reasons why the ability to design molecules with structure, reactivity and biological activity “switchable” via an externally controlled factor continues to draw significant attention, both from the practical and fundamental points of view. Possible applications of such molecules include design of molecular machines and switches, logic gate mimics, optical sensors, drug delivery systems, etc. Since pH-dependent “switchable” molecules are of particular for processes that occur in biochemical systems and in the environment, interesting pH-sensitive systems were developed to control such diverse phenomena as strand orientation and exchange in peptide assemblies,1 charge densities in self-assembled monolayers,2 encapsulation of guests in supramolecular polymers, 3 phase transitions in stimuli-sensitive polymers for drug delivery, 4 rotaxane switching, 5 properties of luminescent devices based on organic fluorophores6 and metal complexes,7 permeability of mesoporous materials,8 growth of nanomaterials,9 control of recognition-mediated reactions,10 and the design of bioreactors.11 Life itself is a pH sensitive phenomenon, as many biochemical processes work within a very narrow pH window.
In this study, we provide the first example of a “switchable” molecular system for pH-controlled double stranded DNA-cleavage designed for selective targeting of cancer cells. The more acidic extracellular environment of solid tumors, relative to that of the normal cells, leads to a pH gradient that dramatic effect on drug uptake in tumor cells12 and can be explored in the design of tumor-specific cleaving agents.13 It is known that hyperglycemia (e.g., glucose infusion) and/or certain drugs, e.g., amiloride, nigericin, and hydralyzine, are also able to lower the intracellular pH of cancer cells. For example, administration of amiloride and nigericin at dosages that do not affect the normal cells drops the intracellular pH in a number of tumor cell types from 7.2 to 6.2−6.6.14, 15, 16, 17 Moreover, hyperglycemia well as hypoxia lead to an even further acidification to pH as low as 5.5.18, 19
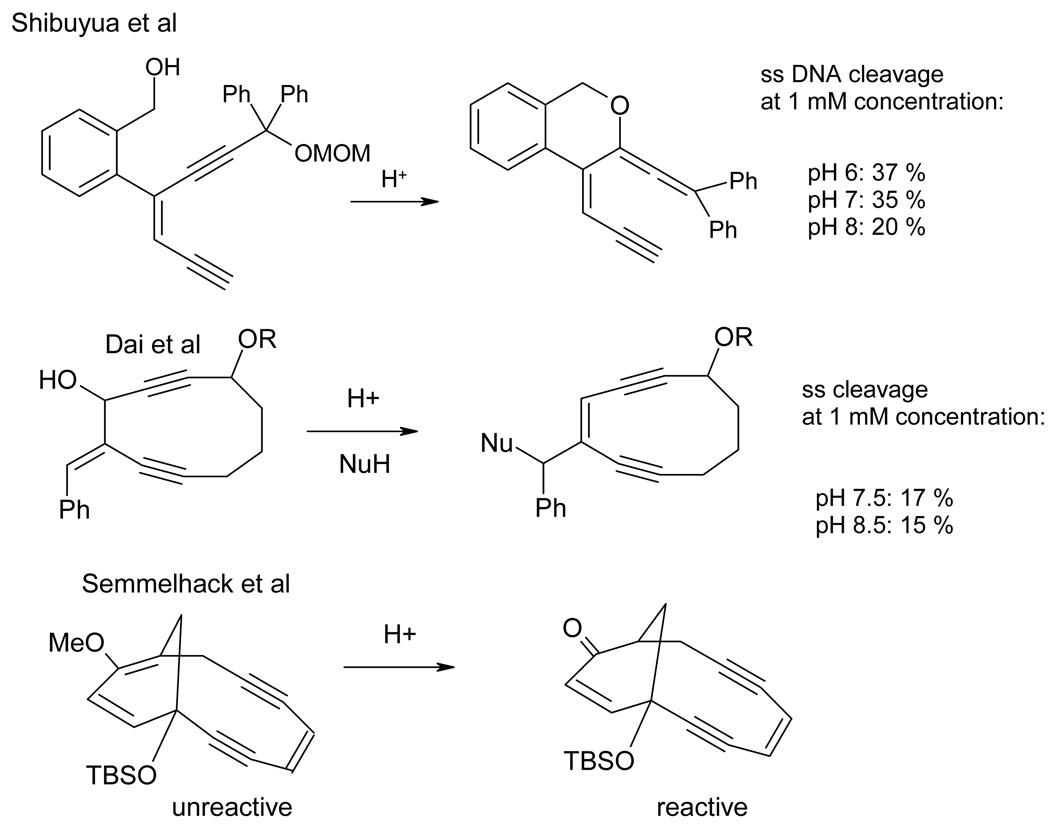
Although research in this direction has been hampered by the scarcity of suitable pH-dependent cytotoxic agents, a number of approaches can be used for the rational design of such molecules. 20 An illustration of how these ideas can be applied to highly potent class of natural enediyne antibiotics either through isomerization into more reactive functional groups or through unlocking of structural constraints is given in Figure 1.21 In addition, there have been promising reports of acid-labile drugs that either hydrolyze at lower pH to give toxic products22 or produce free radicals due to accelerated Co-R bond homolysis.23 An interesting recent finding involves acid-promoted DNA cleavage by natural antibiotic Varacin C.24 The authors found a two-fold increase in the extent of single stranded (ss) DNA cleavage at 5.5 (47% at 5 µM antibiotic loading) compared with that at pH 7 (23%). Unfortunately, this increase only applies to the ss damage which is, unlike the double stranded (ds) damage, usually repairable by the cell chemical machinery.
Figure 1.
Approaches to pH-activated enediynes based on acid-catalyzed transformations of unreactive prodrugs.
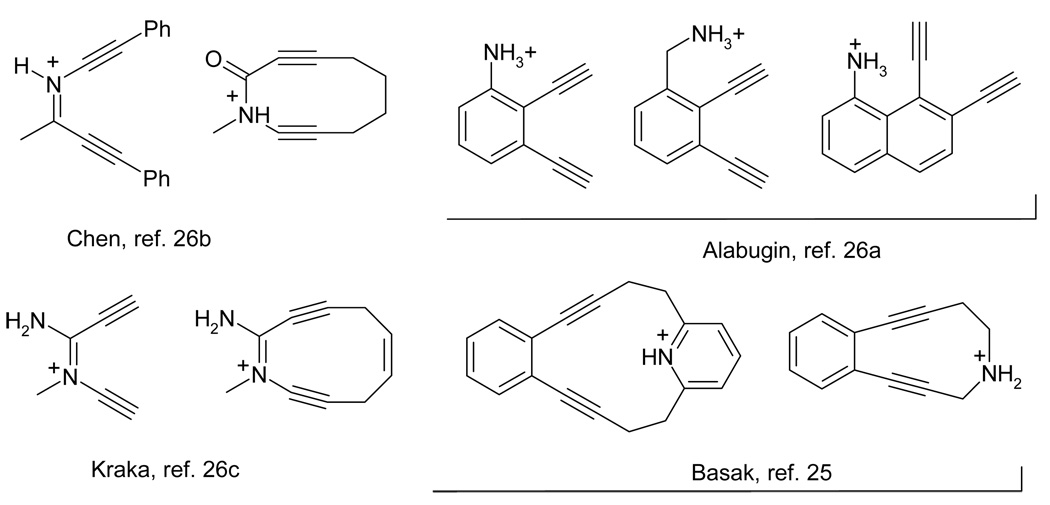
Another approach to pH-regulated DNA-cleaving agents involves protonation of basic functional groups. Amine functionality is one of the obvious choices and several elegant experimental designs based on the protonation of this functional group appeared in the literature (Figure 2). In particular, the groups of Kerwin, Chen, and Kraka & Cremer reported systems where protonation increased the efficiency of radical damage through simultaneous acceleration of the H-abstraction step and deceleration of p-benzyne diradical deactivation through the retro-Bergman ring opening.25 After detailed computational studies indicated that properly positioned cationic groups decrease activation barrier for the Bergman cyclization,26 Basak and coworkers27 pursued an alternative approach based on significant acceleration of cycloaromatization step imposed by a spatially close ammonium moiety.
Figure 2.
Literature examples of pH-controlled amino enediynes.
Unfortunately, simple aliphatic amines are too basic for the change in the protonation state to occur at the pH window necessary for targeting hypoxic cancer cells. As the result, even when protonation has been shown to accelerate the Bergman cyclization of enediynes, the change in reactivity did not occur in the optimal pH-range. Nevertheless, the basicity of nitrogen bases can be controlled in a number of ways and, thus, the above is not insurmountable. For example, anilines are significantly less basic than aliphatic amines and can be fine-tuned through substitution28 to accept the proton only at the desired pH-range. Amides, 25b suggested by Chen, and aldimines25c designed by Kraka and Cremer may offer an excellent solution for fine-tuning the nitrogen basicity.
The present work describes the development of the first pH-controlled system capable of inducing the much more therapeutically important double-stranded (ds) DNA cleavage. Several challenges presented themselves at the beginning of this study. First, the change in reactivity has to occur at a relatively narrow and predefined pH point (ideally ~ pH 7). Second, an efficient DNA-cleaver was needed which could operate within the physiological pH range and be attached to a pH-sensitive functionality without sacrificing its potency.
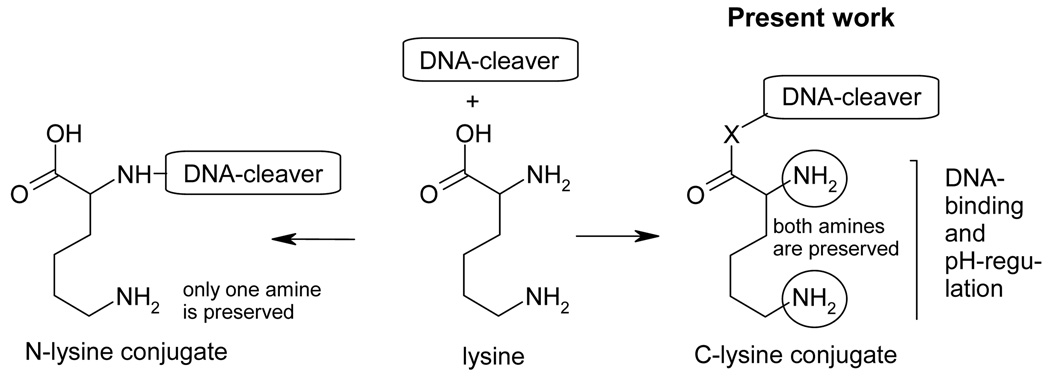
In order to solve the first problem, we utilized a simple but, to the best of our knowledge, new strategy for the design of pH-regulated molecules which is based on the presence of two amino moieties (or related functional groups) with different basicities. The first amino group should be sufficiently basic to be protonated at a wider range of physiological conditions. This auxiliary group plays three roles through a) enhancing solubility of the conjugates in water, b) increasing their affinity to the negatively charged backbone of DNA, and c) modulating the basicity of the pH-trigger. The pH-trigger itself corresponds to the second amino group which should enable pH-switchable behavior at the desired pH range.
The bifunctional pH-regulated part in our design is derived from a diamino carboxylic acid (lysine) which is connected to the DNA-cleaving moiety through the carboxyl group of lysine (Figure 3). This mode of attachment is different from the most common way of lysine incorporation into conjugates or proteins through the formation of a peptide bond at the expense of the α-amino group.29 Importantly, the use of the carboxyl group for the assembly preserves the two amino groups, both of which are essential for solubility, binding to DNA and pH-regulation of DNA-cleavage.
Figure 3.
Two types of lysine conjugates
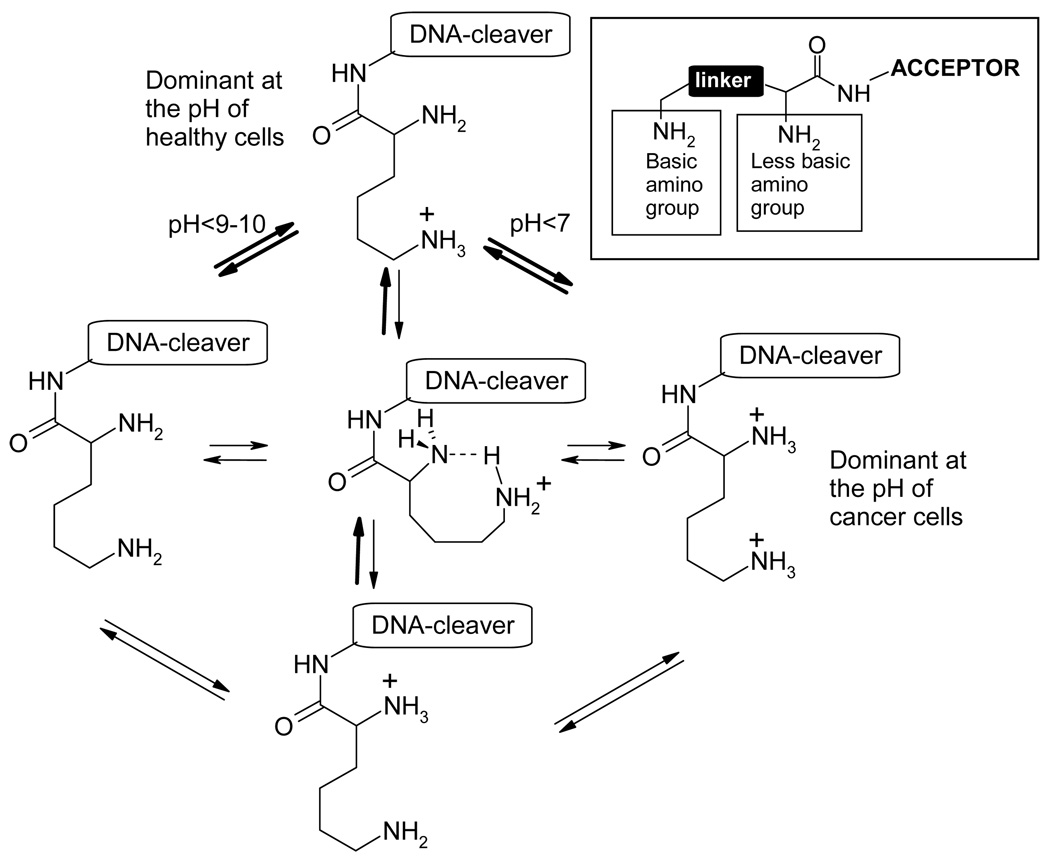
The auxiliary amino group is at the remote terminus whereas the pH-switchable amine is next to the carbonyl group.30 We were interested in determining whether the presence of a highly polarized acceptor moiety can differentiate the basicities of the two amino groups to the extent where distinct protonation states exist at the biologically relevant pH regions (Figure 4). If this approach is successful, sequential protonation of these groups at different pH can be used to control the photophysical properties of such molecules, their binding to DNA and, ultimately, the efficiency of ds DNA cleavage.
Figure 4.
Design of pH-dependent DNA-cleavers based on different stages of protonation of the lysine side chain. The dominant protonation pathway is indicated by the bold arrows.
The pH-activated part was combined with light-activated DNA-cleavers in order to take advantage of the high degree of spatial and temporal controls over reactivity inherent to the photochemical activation.31 We based the choice of the photochemically activated DNA part on our earlier discovery of the C1–C5 cyclization of enediynes which results in the net transfer of four hydrogen atoms to the enediyne moiety.32, 33 In particular, the tetrafluropyridine (TFP) substituted enediynes were found to be both photochemically and thermally stable, unless photoexcitation occurs in the proximity of a suitable electron donor, such as DNA. We have recently shown that these molecules are efficient reagents for the double-strand (ds) DNA photocleavage34 and that they are amenable to two-photon activation.35 We have also shown that the lysine conjugates of enediynes, as well as related fulvenes and acetylenes, perform guanosine-specific (oxidative) cleavage at the ends of AT-rich sequences of DNA. 36 After a serendipitous discovery of the photochemical removal of the terminal radioactive label from 32P-labelled oligonucleotides suggested that the lysine conjugates are capable of recognition of terminal phosphate groups, we utilized this phenomenon for the design of reagents which can recognize the DNA-damage sites (nicked or gapped ss cleavage). These reagents allowed for the first time to achieve a controlled photochemical transformation of the easily repairable ss DNA cleavage to the much more therapeutically important ds cleavage. 37
This considerations guided our design and prompted us to investigate DNA-cleavage activity andbinding to DNA of five lysine conjugates 1–5 in the range of pH 5.5–8 which can be used to differentiate between cancer and healthy cells (Figure 5). After testing whether the pH-dependent DNA cleavage based on this molecular design is feasible, this paper will concentrate on mechanistic basis of this phenomenon through a combination of photophysical techniques, NMR titrations, analysis of DNA binding trend and effect of chemical additives at the cleavage efficiency.
Figure 5.
Structures of lysine conjugates 1– 5 and related control compounds 6 – 8.
RESULTS AND DISCUSSION
pH-Dependent DNA cleavage
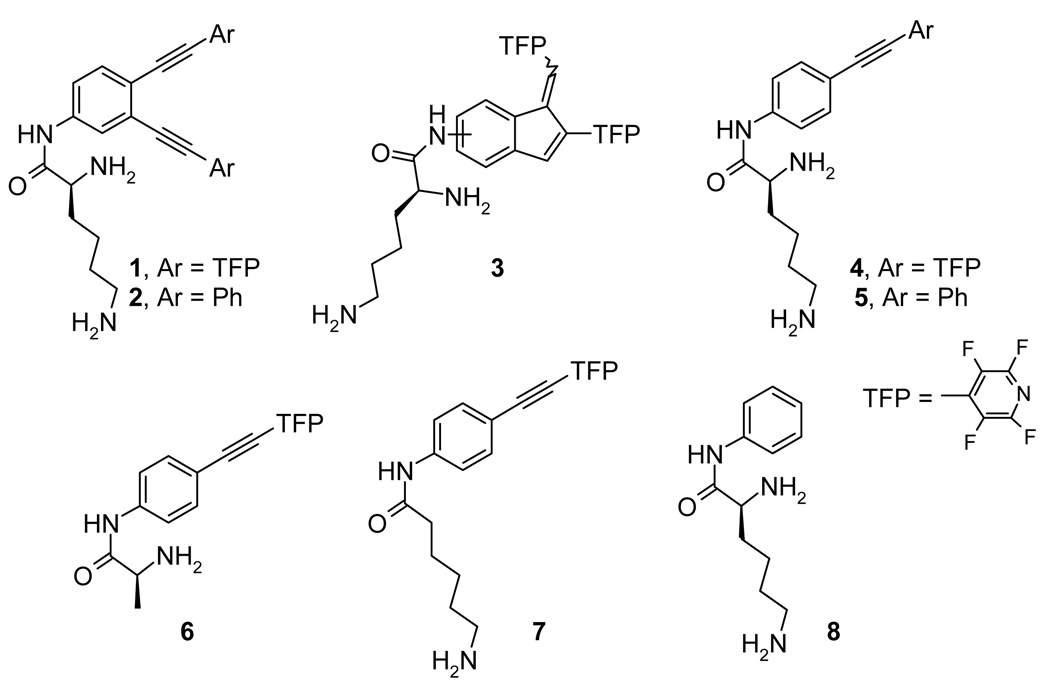
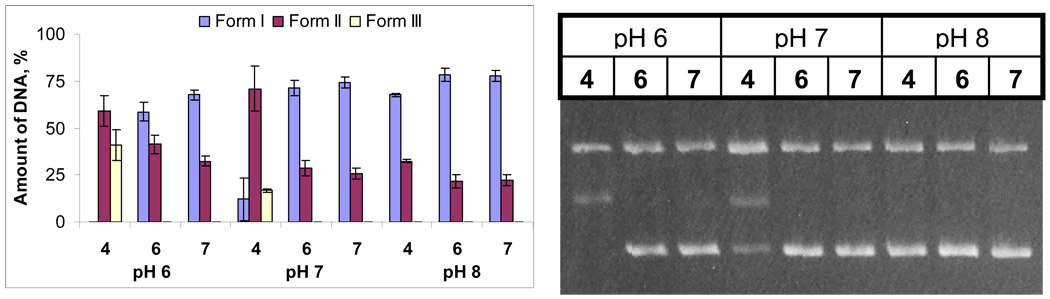
The ability of lysine conjugates to cleave DNA upon irradiation was investigated using conversion of supercoiled plasmid DNA into the respective relaxed circular and linear forms (Forms II and III). The relative amounts of the three DNA forms were determined by densitometric analysis 38 of the gel electrophoresis bands. Although enhancement of DNA-cleaving ability has been observed for all lysine conjugates (see the Figure 8), we will focus our discussion below on compound 4.
Figure 8.
Quantified plasmid relaxation assays with 15 µM of compounds 1, 2, 5 and 30 µM/bp of pBR 322 plasmid DNA in 20 mM sodium phosphate buffer at pH 6, 7 and 8 after 3 min. irradiation.
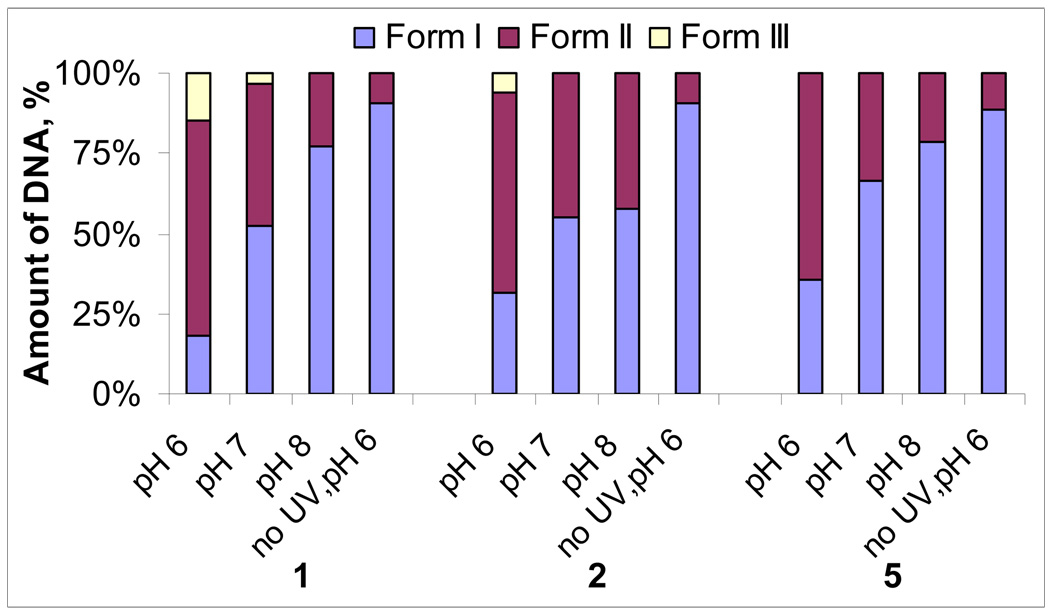
The most remarkable observation is the dramatic increase in efficiency of ds DNA cleavage caused by compound 4 when pH changes from neutral to slightly acidic (Figure 6). While control experiments clearly indicate that no additional DNA cleavage is caused at this pH by the conjugate alone in the dark or by UV itself, the damage is amplified strongly when both light and compound 4 interact with DNA at the lower pH. At pH 6, only 16 % of intact DNA (Form I) remained after 3 minutes of irradiation in contrast to 60% and 86% of unreacted DNA at pH 7 and 8, respectively. Even more remarkable is that at the concentrations where hardly any ds cleavage is observed at pH 7, the amount of double stranded cleavage at the lower pH reaches the 2:1 ss:ds ratio, which rivals DNA cleavage mediated by the natural antibiotics calicheamicin γ39 and bleomycin40! Higher concentrations of 4 lead to as much as 50% of ds cleavage which, to the best of our knowledge, is unprecedented for a small molecule-based DNA-cleaver (natural or designed).
Figure 6.
Plasmid relaxation assays with conjugate 4 (15 µM) and pBR 322 plasmid DNA (30 µM/bp) in 20 mM sodium phosphate buffer after 3 minutes of irradiation. Form I = intact supercoiled DNA, Form II = relaxed form (ss cleavage), Form III = linear form (ds cleavage). Lanes 1–8: from pH 5.5 to pH 9, lane 9: no compound + UV, lane 10: compound + no UV. Cleavage data were quantified through densitometry. Reported values represent the average of four experiments.
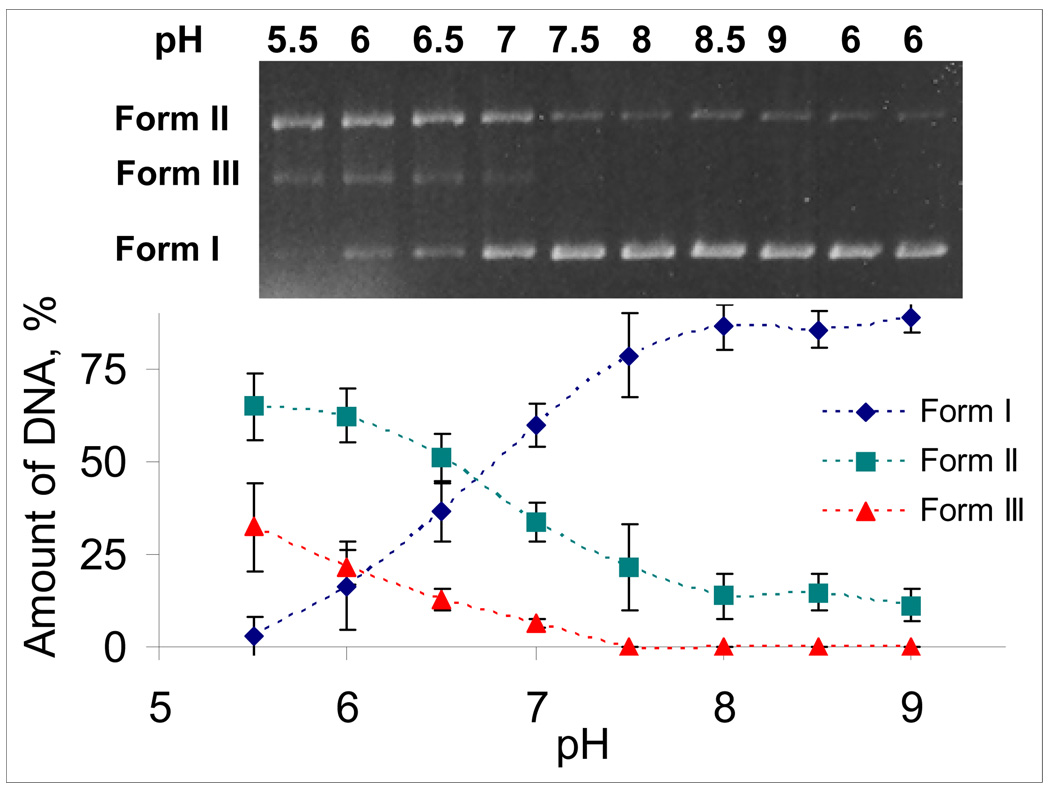
Interestingly, the dependences of all three forms of DNA cleavage from pH follow the classic titration curves, which afford the apparent pKa values of 6.6, 6.8 and 6.2 respectively for the Forms I, II and III (Figure 7).41 All of these values agree that a significant change in the efficiency of DNA cleavage occurs at the pH 6–7 window ideal for targeting cancer cells.
Figure 7.
Percentage of unreacted DNA (Form I), single-stranded cleavage (Form II) and double-stranded cleavage (Form III) as a function of pH, and the apparent pKa values corresponding to these plots.
Although all lysine-conjugates have similar pH-dependent trends (Figure 8), conjugates 1 and 4 possess the greatest DNA-cleaving ability. As the result, we focused our mechanistic discussion on acetylene 4 which is not only less toxic than enediyne 1 in the dark (vide infra), but also does not aggregate and precipitate at the higher pH values (~8). When necessary, we incorporated data for other conjugates as well.
In order to investigate the relative role of the two amino groups in these conjugates, we extended this study to include DNA cleavage activities of alanine and caproic acid conjugates 6 and 7. These compounds have only one amino group (at the α- and ε-carbon, respectively) and, as expected, show different behavior (Figure 9). Even though the monoamines 6, 7 are comparable in reactivity to diamine 4 at pH 8, the two conjugates lacking one of the two amino groups rapidly fall far behind in their efficiency at the lower pH values42 and do not produce any ds-cleavage even at pH 6.
Figure 9.
Plasmid relaxation assays for DNA photocleavage with 15 µM of compounds 4, 6 and 7 and pBR322 plasmid DNA (30 µM/bp) in 20 mM sodium phosphate buffer (pH 6, 7, 8) after 10 min of UV irradiation. Quantified cleavage data are presented on the left, the original gels are on the right.
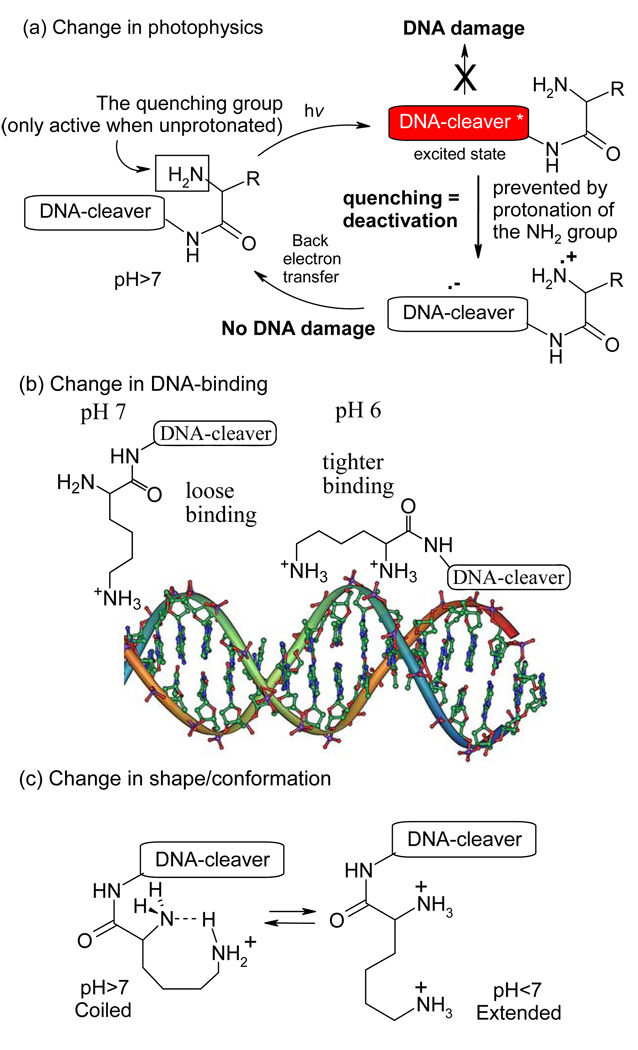
The fact that only the lysine conjugates produces clear ds cleavage under these conditions suggests that both amino groups are required for this process to occur. Several explanations are possible. For example, the remarkable ability of lysine to recognize a nick in DNA and facilitate a break of the complementary strand (the ss→ds cleavage conversion) discovered in our previous work37 may play some role in the high efficiency of the ds cleavage. However, the enhancement of this type of cleavage at the lower pH is an unprecedented phenomenon. This effect is likely to stem from the presence of a protonated α-amino group in compound 4 which enhances the efficiency of DNA cleavage through one of the three mechanisms outlined in Figure 11 (vide infra).
Figure 11.
Three possible mechanisms for pH-regulated DNA modification by lysine conjugates.
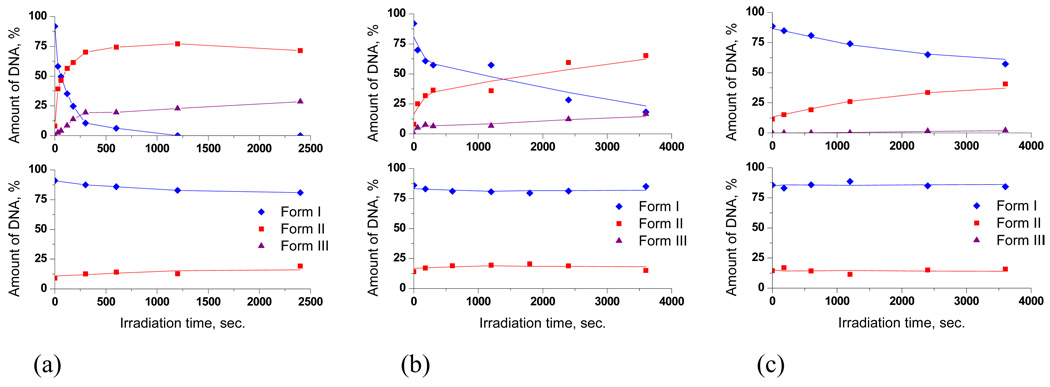
The difference in kinetics of DNA photocleavage by conjugate 4 at different pH values provides another illustration of the remarkable degree of pH-control (Figure 10). DNA cleavage at pH 6 is remarkably fast – plasmid DNA is completely consumed within 5 minutes of irradiation, even though control experiments clearly show that there is very little of DNA damage by UV itself under these conditions. In a sharp contrast, DNA cleavage at pH 8 is slow – even after an hour of irradiation ~60% of DNA remains unreacted (note, however, that this reaction is still considerably faster than the damage by UV itself shown at the bottom part of Figure 10). These observations indicate that, depending on the pH, compound 4 can induce DNA damage in two kinetically different ways. The DNA-damaging activity at pH 7 can be described as a superposition of the two different cleavage patterns described above. These experiments also provide the control data to confirm that change in the pH per se does not induce acceleration of DNA photocleavage (lower part of Figure 10). Instead, the observed pH-dependence stems from the properties of lysine conjugates.
Figure 10.
Quantified plots of pBR 322 plasmid (30 µM/bp) relaxation assays for 10 µM of compound 4 in 20 mM sodium phosphate buffer at pH 6 (a), 7 (b) and 8 (c) as a function of the irradiation time. Plots on the bottom show effect of UV on the DNA without the conjugate at different pH. Plots on the top illustrate reactivity in the presence of compound 4.
Mechanistic alternatives for the observed pH-dependence of DNA-cleavage
Several possible mechanisms which could lead to the observed pH-dependency for the interaction of lysine conjugates with DNA are outlined in Figure 11. Mechanism (a) involves the control of deactivating intramolecular electron transfer from the donor α-amino group to the excited chromophore through protonation. Electron transfer intercepts the excited state of the DNA-cleaver, making it less active at higher pH. In contrast, at lower pH, both amino groups are protonated and the nitrogen lone pair is not available for deactivating the reactive excited state.
Another possibility is illustrated by a model for stronger binding of the diprotonated lysine moiety to DNA at lower pH43, as shown in Figure 11b. At neutral pH, only the more basic, remote amino group of the lysine moiety is protonated, leading to a relatively loose binding to DNA. At lower pH, the electrostatic interaction of positively charged ammonium groups and negatively charged phosphate groups of DNA brings the DNA-cleaving warhead close to the ribose backbone of DNA, providing a possible mechanism for the increase in the extent of DNA damage by this warhead.
Finally, we envisioned that a conformational change may occur upon the second protonation (Figure 11c). Although it was not clear a priori whether the coiled conformation may gain sufficient importance in the strongly hydrogen bonding aqueous environment, it is interesting to consider such possibility as a protonation-induced shape change may lead to a change in the DNA binding mode.
Photophysical properties of lysine conjugates with acceptor substituents will help us to determine whether the choice of photochemical DNA-cleavers activated through photoinduced electron transfer (PET) introduces a new component in the pH-controlled behavior (Figure 11a). NMR analysis of different protonated species will elucidate the possibility of conformation change in Figure 11c whereas pH-dependence in DNA binding will help to determine whether tighter binding is achieved for dications as illustrated in Figure 11b.
However, before comparing the relative importance in the three possible pathways for the observed pH-dependence of DNA cleavage for the lysine conjugates, it is important to obtain reliable answers regarding the basicities of the two amino groups in the lysine conjugates. Is the presence of an acceptor moiety next to the α-amino group and another already protonated amino group sufficient to decrease the basicity of the α-amino group to the extent where the change in its protonation will occur in the vicinity of pH 6–7? If yes, how are these changes in the protonation state transferred in the changes in the efficiency of DNA binding? In order to address these questions, we proceeded to determine pKas for the two amino groups of lysine moiety as well as binding affinities of the different protonated states to DNA.
Determination of pKa values
Direct potentiometric pH determination of both pKa values is inaccurate for most of the conjugates because, at a pH where both ammonium groups are deprotonated and these molecules lose all their charge, the solubility in water becomes very low (< 1 mM). The decrease in solubility leads to the apparent aggregation and distortion of potentiometric curves at the higher pH. This distortion prevents accurate curve fitting (see the Figure S1). Thus, we had to resort to alternative spectral methods. Fortunately, there is a selection of convenient procedures which provide complementary information which can be used for cross-checking the accuracy of the alternative approaches.
NMR titrations
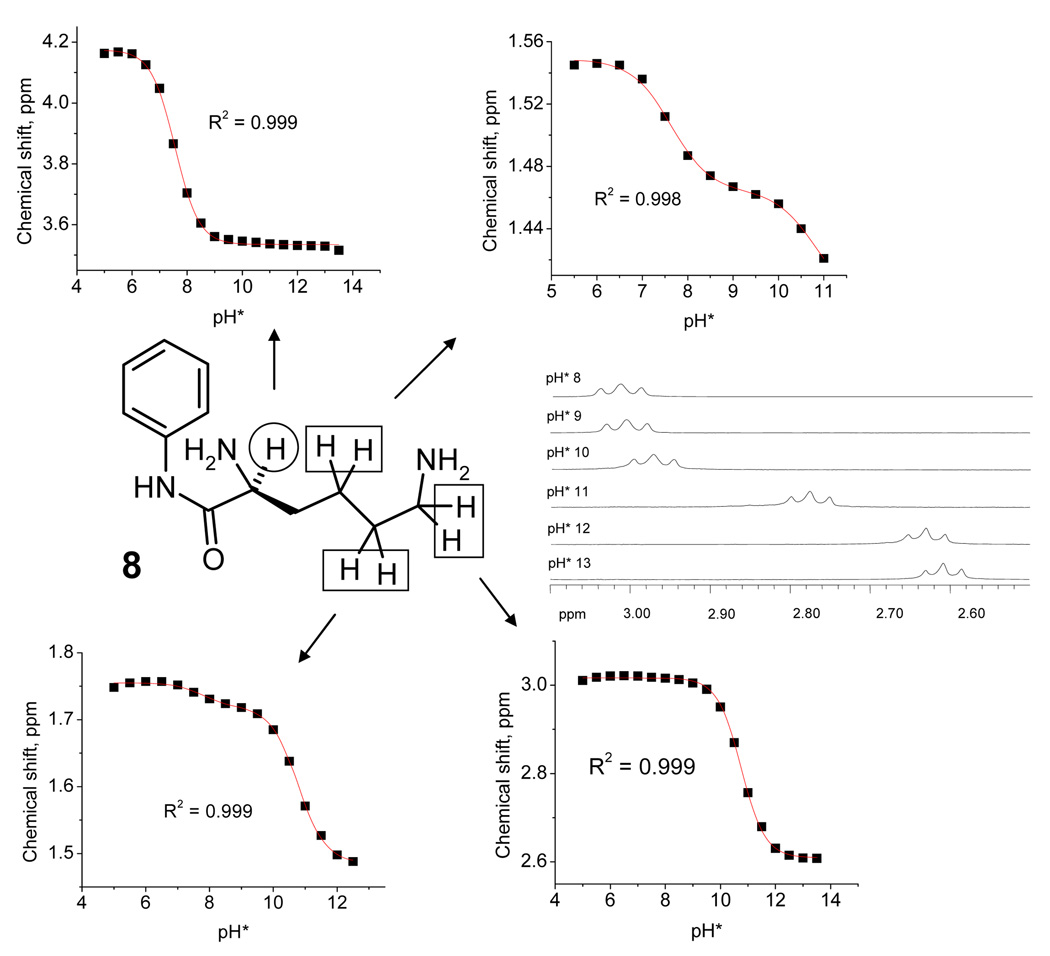
The advantage of NMR titrations44 is that they provide direct structural information allowing differentiation of the two protonation events through observations of chemical shifts for hydrogens spatially close to one of the two amino groups. 45 Such differentiation can confirm unambiguously whether the presence of a carbonyl group next to the amino group leads to decrease in the pKa of the respective ammonium ion.
Favorable solubitity of N-phenyl-lysine amide 8 in D2O rendered this molecule a particularly convenient substrate for the 1H-NMR studies. These studies followed changes in the chemical shifts of the lysine chain protons upon the protonation/deprotonation of the two amino groups during the pH (pD) titration. The titrations curves clearly showed that α-amino group is less basic (pKa=7.56) than ε-amino group (pKa=10.75). To allow comparison of these pKas with pKas obtained by other methods, one has to take into account the difference between pKa H*, which is obtained by a direct reading of the H2O-calibrated pH-meter in a D2O solution, and pKa (pKa=0.929pKa H*+0.42)46
Interestingly, monitoring of protons on the β-, γ- and δ-carbons produced two protonation curves with their relative sensitivity being directly proportional to the proximity to the respective amino groups. The observation that α-hydrogens are not sensitive to the change in protonation of the ε-amino group suggests a lack of communication between the two amino groups, thus eliminating the cyclic H-bonded structure in the monoprotonated species where the ammonium group is hydrogen bonded to the other NH2 group of the same molecule (the central structure in Figure 4 which corresponds to the mechanism (c) of pH-regulation in Figure 11).47
Fluorometric determination of pKa of the α-amino group
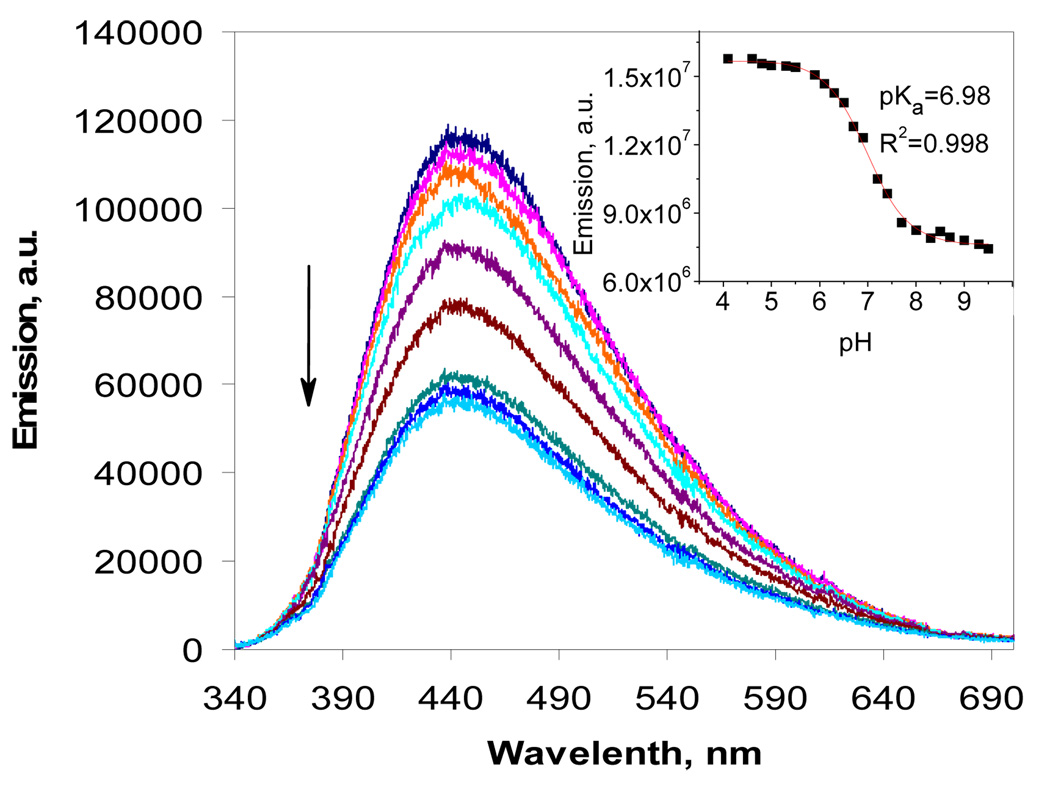
Highly sensitive fluorescence methods are especially convenient for compounds of limited solubility. We determined the basicity of the α-amino group using intramolecular quenching of fluorescence through photoinduced electron transfer (PET) from the nitrogen lone pair to the excited chromophore. At the lower pH, amino groups are protonated and the lone pair of electrons at nitrogen is unavailable for inter- or intramolecular electron transfer. However, as the ammonium group adjacent to the chromophore loses the proton at the higher pH, electron transfer becomes possible and quenches the fluorescence (Figure 13). Although the validity of this approach is based on the assumption that changes in intermolecular PET and other fluorescence quenching pathways with the pH are insignificant relative to intramolecular PET at the 10−5 M concentration, the advantage of fluorescence is that it is more sensitive to changes in the immediate vicinity of the chromophore and, thus, directly reports protonation of the α-amino group.
Figure 13.
Fluorescence spectra of compound 4 (10 µM) at pH 4.1, 5.0, 6.1, 6.5, 6.9, 7.2, 8.0, 9.0, and 9.5. Insert: Quantified changes in fluorescence intensity as a function of pH and their fit to the Henderson-Hasselbalch equation for an acid-base equilibrium involving one amino group.
Lifetimes
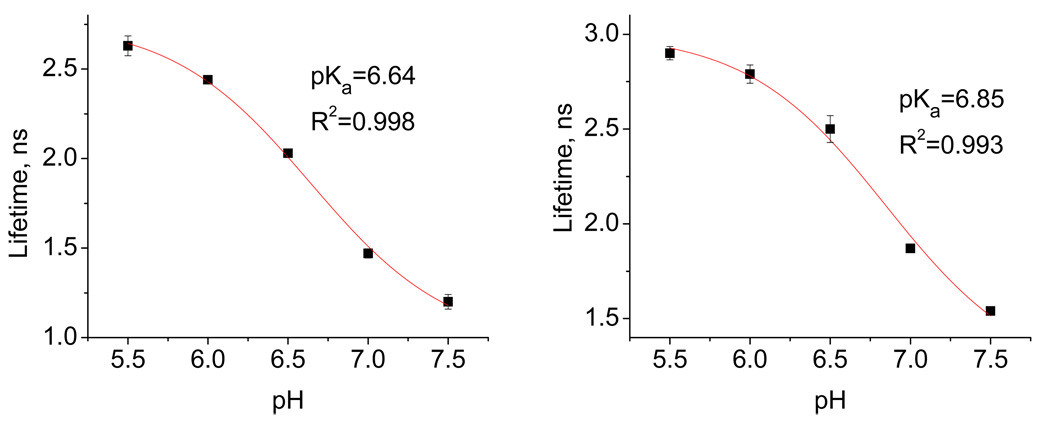
To gain further insight into the effect of protonation on the excited state properties, we measured the singlet excited state lifetimes for compounds 4 and 6 as a function of pH. Within the error of measurement, all TFP-substituted compounds show a clear decrease in the lifetime of the singlet excited state at the higher pH. An illustration of the pH-dependence for the singlet excited state lifetimes of compound 4 and 6 is provided in Figure 14.
Figure 14.
The pH-dependent lifetimes of the singlet excited state for conjugates 4 and 6 (10 µM), measured in 20 mM sodium phosphate buffer.
The observed pH-dependency in the lifetimes means that the S1 state is less available for triggering DNA-cleavage at the higher pH, a factor which should contribute to the observed decrease in activity under these conditions. At the lower pH, both amino groups are protonated, and hence electron transfer from the nitrogen lone pair does not deactivate the excited state, exactly as it is described in Figure 11a.
UV-absorption titrations
Similar pKa values can be obtained using pH-dependent changes in the UV— Vis absorption spectra of the conjugates. This method is less sensitive as the changes observed are much smaller (about 10% increase in absorption upon transition from pH 11 to 5.5) than the analogous changes in the fluorescence spectra discussed in the previous section. In contrast to fluorescence, there are two titration curves for compound 4 which correspond to absorption changes of almost equal magnitude (Figure S4). The pKa for the first of these curves corresponds exactly to the pKa obtained from the fluorescence titrations. Interestingly, pKa for the ε- ammonium group of compound 7 is >1 pKa units lower than that derived from the fluorescence changes. Although the reasons for this interesting behavior of the terminal amino group extend beyond the scope of this paper,48 this subtle difference is irrelevant at the biologically relevant pH region where we observe the enhancement of ds-DNA cleavage. It is clear that, independent on the method, the pKa of the auxiliary ammonium group is sufficiently high to allow more than 99% of the respective amine to be protonated even at neutral pH. The above experiments do confirm unequivocally that the two amino groups of lysine conjugates possess different basicities and that the less basic α-group has the right properties to serve as a pH trigger for selective targeting of cancer cells.
Summary of experimental pKa trends
The results for conjugates 4, 6, 7 and 8 are summarized in Table 1. The α-pKa for the lysine conjugates 4 is ~0.5 units lower than the pKa for the analogous alanine conjugate 6. This difference indicates that, as expected, the presence of a second cationic moiety does play some role in the decrease of α-amino moiety basicity. However, the relatively close pKa values for the α-ammonium groups in compounds 4 and 6 together with their large deviation from the pKa values for the ε-ammonium groups in compounds 4 and 7 illustrate that the lowered basicity of the α-amino group in lysine is mostly due to its proximity to the amide moiety. It is also interesting that the presence of the acceptor tetrafluoropyridinyl moiety in 4 leads to a noticeable (~0.5–0.6 pKa units) decrease in the basicity of the α-amino group relative to that in a simple phenyl conjugate 8.
Table 1.
The pKa values of amino groups in lysine conjugates 4, 8 and related monoamines 6, 7 according to emission / absorption and NMR spectra.
Computational analysis of pKa trends
In order to provide a theoretical rationale to the observed differences in basicity, we investigated electronic properties of the two amino groups in diamines 4 and 8. Natural Bond Orbital (NBO)50 analysis of electronic structure of these compounds at the B3LYP/6-31G(d, p) level clearly shows that the α-nitrogen lone pair is lower in energy and has a significantly lower population than the ε-nitrogen lone pair (see Table 2 and the SI part). Both of these factors render the α-nitrogen a less efficient donor28, 51 and a weaker base than the ε-nitrogen.
Table 2.
NBO analysis of electronic properties of α-and ε-nitrogen lone pairs of diamines 4 and 8 at the B3LYP/6-31G(d,p) level of theory with the Polarizable Continuum Model (PCM)52 for solvation in H2O.
| Neutral | Protonated at the ε-nitrogen atom | |||||
|---|---|---|---|---|---|---|
| Compound 4 |
Occupancy, e | Energy, a.u. | Hybrid character, n in spn orbital |
Occupancy, e | Energy, a.u. | Hybrid character, n in spn orbital |
| α | 1.92791 | −0.30618 | 4.42 | 1.92765 | −0.30724 | 4.49 |
| ε | 1.96564 | −0.29939 | 3.50 | - | - | - |
| Compound 8 |
Occupancy, e | Energy, a.u. | Hybrid character, n in spn orbital |
Occupancy, e | Energy, a.u. | Hybrid character, n in spn orbital |
| α | 1.92941 | −0.30457 | 4.39 | 1.92906 | −0.30557 | 4.46 |
| ε | 1.96555 | −0.29893 | 3.52 | - | - | - |
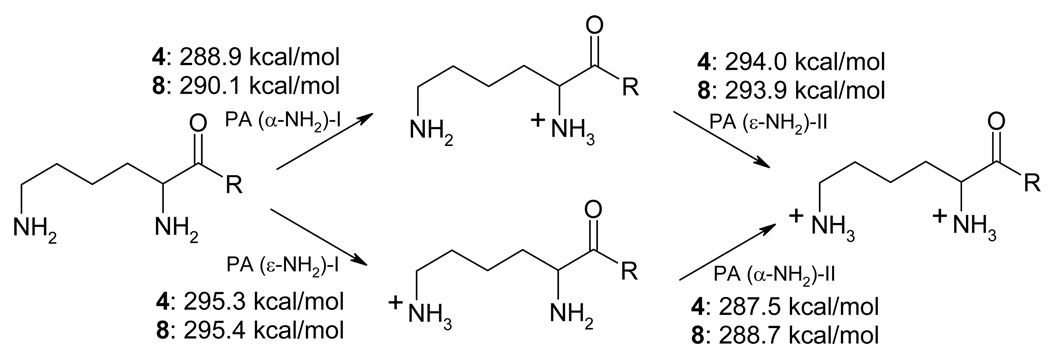
Proton affinities calculated at the B3LYP/6-31G(d, p) level in water (Figure 15) support the above experimental findings very well. For both compounds 4 and 8, proton affinity of the ε-nitrogen is significantly higher than that of the α-nitrogen. In agreement with the experimental pKa values for monoamines 6 and 7 relative to the two pKa values of the diamines 4 and 8, the first protonation has some effect on the basicity of the second amino group but the effect is not dramatic (1.2–1.5 kcal/mol difference in the proton affinity values). In addition, and again in a full agreement with the experiment, the α-amino group in compound 8 is noticeably more basic than the α-amino group in compound 4 (1.2 kcal/mol difference)
Figure 15.
Proton affinities (in kcal/mol) of α- and ε-nitrogen lone pairs of diamines 4 and 8 at the B3LYP/6-31G(d, p) level of theory with PCM solvation in H2O.
pH-Dependent DNA binding
From a practical perspective, the pKa value of 7.0 for the TFP-acetylene 4 suggests that this compound exists mostly as the monocation at the slightly basic pH and it should be converted into >90% of dication at the same pH region (pH < 6) where we observe the amplification of the ds DNA cleavage. The following section concentrates on interaction (binding) of DNA and lysine conjugates in the different protonation states. We approached this question through a combination of several methods outlined below. Note that qualitative treatments of binding affinities in these systems are difficult due to the presence of several species with different spectroscopic properties (the three protonation states of diamines) and inherent complexity of calf thymus (CT) DNA which offers numerous types of binding environments to the DNA-cleaver. As the result, we will limit ourselves in providing sufficient information for illustrating the differences in binding at the semi-quantitative basis.
Absorbance and fluorescence titration
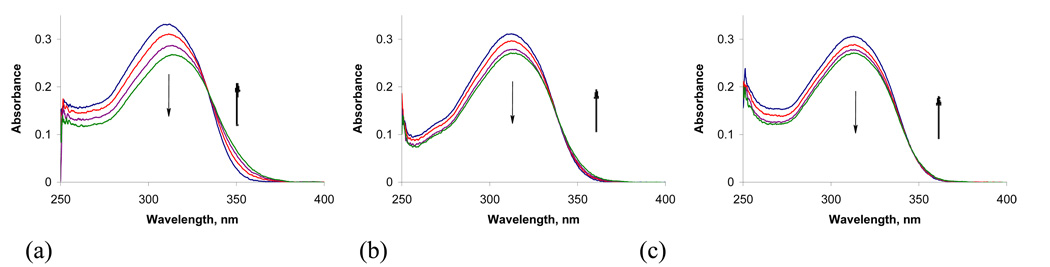
Titrations of 10 µM solutions of compounds 1 – 5 with calf thymus (CT) DNA revealed significant difference in absorbance and emission changes at different pH values. Results for compound 4 are discussed in more detail in this section. While little or no shift of λmax and small hypochromicity (14 %, 12%, respectively) are observed for the titrations of conjugate chromophore with DNA at pH 7 and 8, the λmax changes from 310 nm to 315 nm during the titrations with the same amounts of DNA at pH 5.5 and 20 % hypochromicity is observed. The greater change of absorbance at pH 5.5 indicates the larger interaction between the chromophore and DNA.
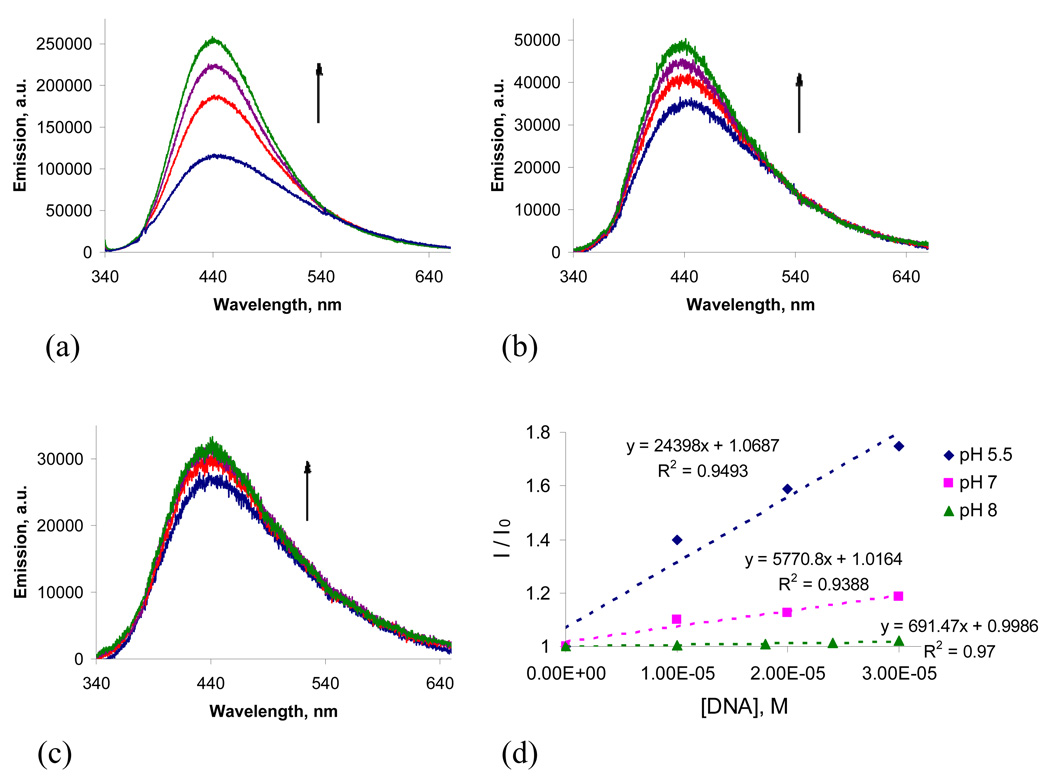
Changes in fluorescence upon the DNA titrations followed a similar pattern where greater changes were observed at the lower pH, indicating a stronger binding affinity to DNA. Interestingly, while emission of TFP-enediyne-lysine conjugate 1 was quenched with DNA (Figure S6), most likely through electron transfer pathway, 53 we observed an increase in fluorescence for compound 4. This finding is likely to indicate that the DNA-binding modes for these two compounds are different. In particular, the fluorescence increase displayed by compound 4 suggests that it may be a DNA groove binder.54 Although fluorescence increase is often observed for intercalated chromophores as well,55 intercalation is unlikely in this system because close proximity to the nucleobases should quench fluorescence of 4 through electron transfer.
Further evidence for a significant difference in binding modes of enediyne 1 and acetylene 4 is provided by the ethidium bromide displacement assays (Figure 18). Only compound 1 displaces intercalated ethidium bromide from its complex with DNA, whereas addition of compound 4 does not lead to the displacement. This observation indicates again that compound 4 is not an intercalator. Nevertheless, despite the above differences in the DNA-binding mode, both lysine conjugates display similar pH-dependence in their DNA-cleavage.
Figure 18.
Changes in fluorescence of ethidium bromide (10 µM, the excitation wavelength = 535 nm) in CT DNA (10 µM) as a function of displacement by compound 1
Overall, both compounds show a much greater spectral response at the lower pH. As pH increases, the changes become less pronounced, indicating a lower degree of interaction between the compounds and DNA at the higher pH. This trend was also shown in measurement of binding affinity of compound 4 to CT DNA by isothermal titration calorimetry (ITC, see the SI part for details). These observations are consistent with the protonation behavior of these diamines discussed in the previous sections. At the lower pH, both amino groups are protonated to provide a higher degree of attraction between the two positive charges of the ammonium groups and the negative charges of the DNA phosphate backbone.
Mechanistic considerations and pH-dependence of the involvement of diffusing radicals in the DNA cleavage
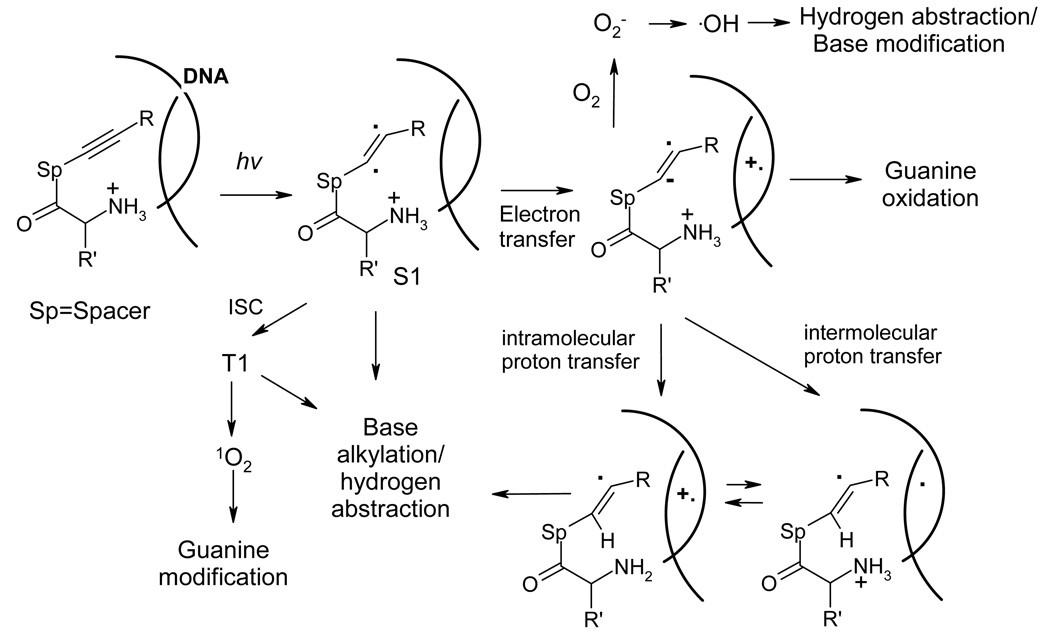
The potential complexity of chemical mechanisms which can be involved into DNA photocleavage by lysine acetylene conjugates is illustrated in Figure 19. A priori, a number of mechanisms may be considered as an explanation for the observed photochemical DNA cleavage by compound 4.56 In the first step, photochemical excitation leads to the formation of acetylene excited singlet state. After this necessary step, the list of diverging possibilities includes a number of choices such as generation of reactive oxygen species like singlet oxygen, superoxide and OH radical;57 direct hydrogen abstraction from DNA sugar backbone by photogenerated radicals, or singlet and triplet excited states;58 oxidative damage due to electron transfer from DNA nucleobases;59 and photoinduced DNA alkylation.60
Figure 19.
Summary of possible mechanistic alternatives for the observed DNA cleavage by amino acid / acetylene conjugates.
We have shown previously that cleavage by the enediyne and acetylene derivatives discussed in the work proceeds with selectivity for guanine-rich parts of DNA which flank AT-tracts due to a compromise between AT-selectivity for binding and G-selectivity for the formation of reactive species through PET.36 Both the selectivity and the fact that the lesions in DNA-oligomers required piperidine treatment clearly indicated that PET is involved in the damage. However, purely oxidative damage could not be the only mechanism because several compounds including 4 caused noticeable damage at a single G inside of a 12-bp AT-tract. This observation suggested that these photocleavers may damage DNA through base alkylation as well. Subsequent work confirmed that compound 4 in capable of localized DNA cleavage and the damage does not dissipate through hole hopping when the same single G site is chosen as a target using phosphate-lysine recognition.37 Furthermore, we also observed frank cleavage in 5’-(AATT)nGG(AT)mGG(AAATTT)l-3’ oligomers. The cleavage is amplified by heat which is indicative of either base pair alkylation or abstraction of a C1’-hydrogen by reactive radical species.40 We have confirmed that triplet excited states of TFP-acetylenes behave as strongly electrophilic radicals which can alkylate π-systems. 61 Taken together these observations suggest base pair alkylation is likely to complement direct PET damage as an important mechanism for the DNA damage, especially at the lower pH and tighter binding.
Further mechanistic details are unclear at the moment and a number of possible scenarios can be suggested. In particular, intersystem crossing (ISC) may convert this excited state into a triplet state of the TFP-acetylene moiety, shown to behave as a highly electrophilic diradical. 61 In addition, the triplet state of the photocleaver may sensitize the formation of singlet oxygen. Alternatively, PET from DNA can lead to the formation of acetylene radical-anion and electron hole at one of the DNA bases. Electron transfer from acetylene radical-anion to dissolved oxygen would form superoxide anion which can be transformed into other reactive oxygen species. Depending on the relative rates of hole hopping and hole trapping, the DNA electron hole can either be trapped at a suitable location to produce an oxidized DNA base (likely, a guanine) or transfer a proton to the DNA-cleaver radical-anion. This proton transfer (which can be classified as Proton-Assisted Electron Transfer) 62 would transform the charge separated ion-pair into two new radicals, one at DNA and another at the partially reduced triple bond. The latter vinyl radical can be also formed when the intermediate radical anion of the DNA-cleaver receives a proton (possibly through a solvent-mediated mechanism) from the adjacent α-ammonium group (Figure 19). Independent on the formation pathway, this vinyl radical can either abstract hydrogen from the deoxyribose backbone or alkylate an adjacent base.
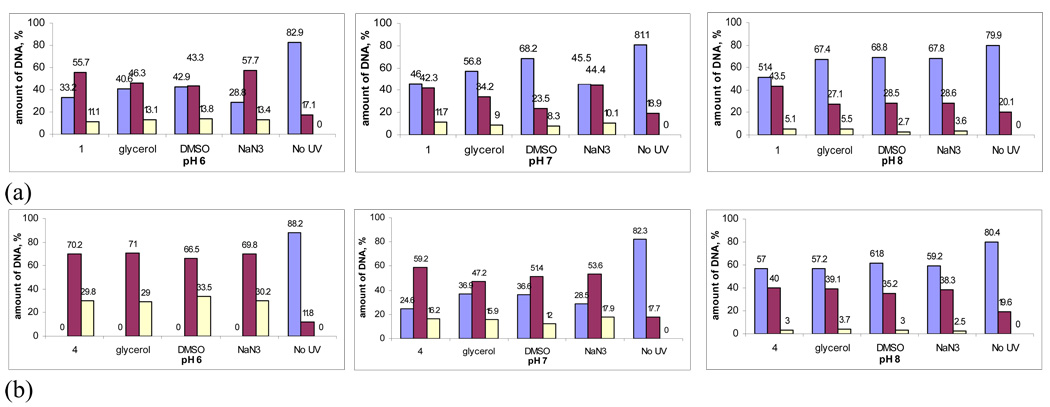
In order to get further insight in the mechanism of the DNA cleavage by the conjugates, we used the plasmid relaxation assays for the cleavage with enediyne conjugate 1 and TFP-acetylene conjugate 4 in the presence of hydroxyl radical (glycerol, DMSO) and singlet oxygen (NaN3) scavengers. The results are summarized in Figure 20.
Figure 20.
Effect of hydroxyl radical/singlet oxygen scavengers (10 mM) upon the efficiency of DNA cleavage at pH 6, 7 and 8 by conjugates 1 (a) and 4 (b) after 5 min. irradiation. Color coding: light blue – Form I, dark purple – Form II, light yellow – Form III.
For compound 1, a small protecting effect of the hydroxyl radical scavengers is observed at all pH values. The protecting effect seems to be slightly lower at pH 6 then at the higher pH values. It is noteworthy, however, that the ds cleavage at pH 6 is not affected by any of the additives. Singlet oxygen scavengers show no effect upon the efficiency of DNA damage by compound 1 at pH 6 and 7.
Remarkably, neither of the additives is able to protect DNA from damage by acetylene-lysine conjugate 4 at pH 6. Some protecting effect is observed only at the higher pH. These results clearly indicate that reactive oxygen species are not involved in the mechanism of DNA cleavage at the lower pH, perhaps due to the tighter binding under these conditions. This observation provides additional support for direct mechanism of DNA damage such as base pair alkylation. From a practical perspective, this is valuable not only because many solid tumors are hypoxic but also because cleavage which does not depend on diffusing species is more localized and efficient.
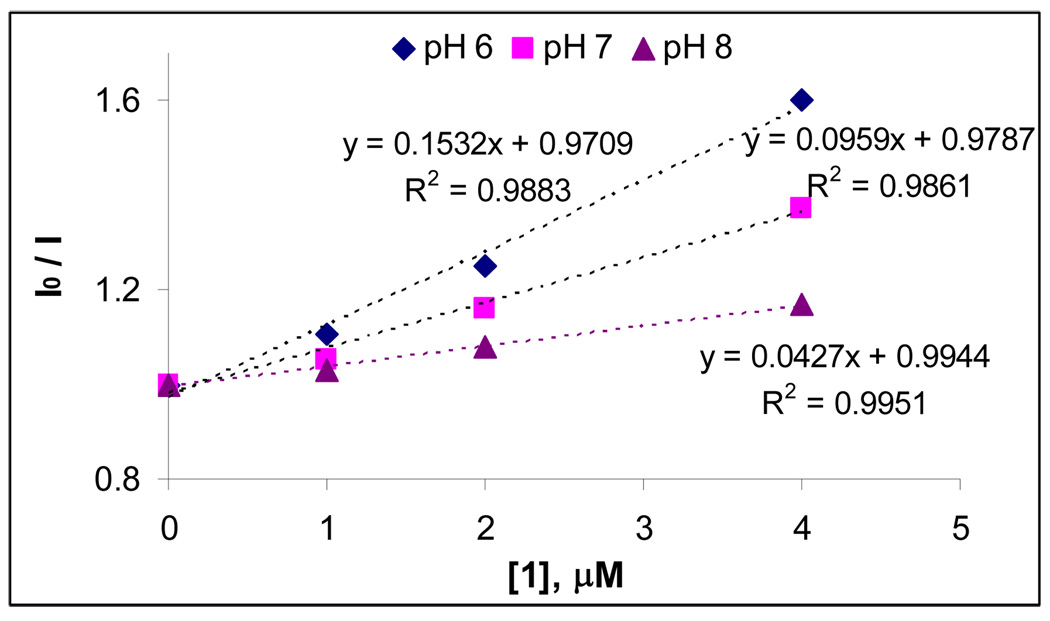
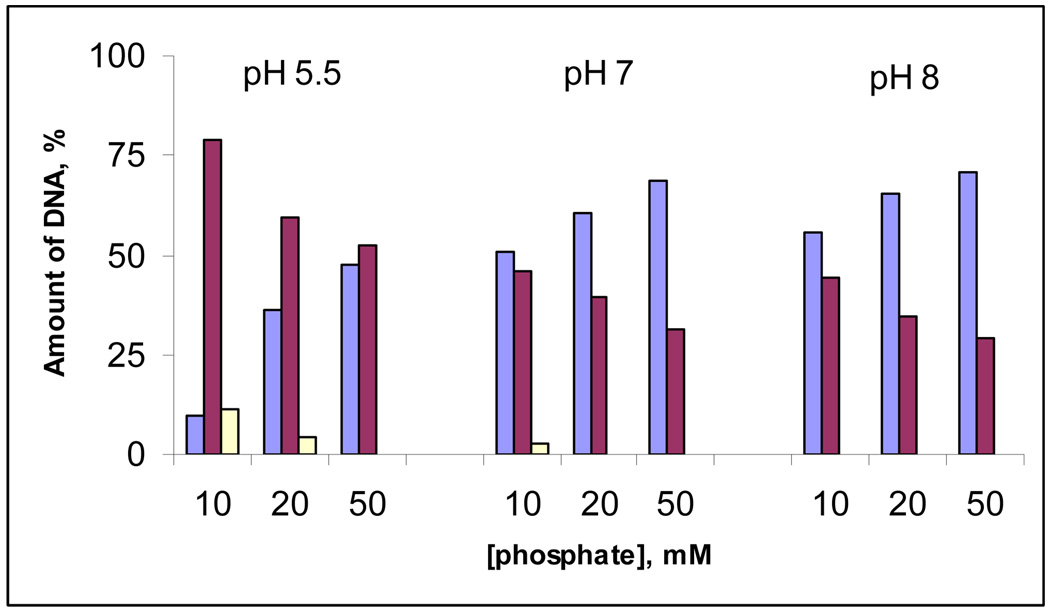
The final insight on the nature of lysine conjugates/DNA interaction came from the effect of ionic strength on the efficiency of DNA cleavage (Figure 21). Increasing concentration of phosphate buffer from 10 mM to 50 mM decreases the total DNA cleavage activity of compound 4 by 48 %, 40 %, 34 % at pH 5.5, 7 and 8, respectively.63 This result suggests that the electrostatic interaction between the positive charges of ammonium group of compound 4 and the negative charges of DNA backbone is an important factor for the cleavage. Again, this observation indicates relatively close binding between the DNA-cleaver and its target which should be particularly important for the DNA-cleavage mechanisms based on alkylation and hydrogen abstraction. In contrast, the cleavage at pH 8 has greater contributions from processes not affected strongly by the ionic strength, such as the generation of diffusing reactive oxygen species.
Figure 21.
Quantified Cleavage data of pBR 322 plasmid (30 µM/bp) relaxation assays of compound 4 (10 µM) at different concentrations of sodium phosphate buffer at pH 5.5, 7 and 8. Color coding: light blue – Form I, dark purple – Form II, light yellow – Form III.
Cancer cell proliferation assays
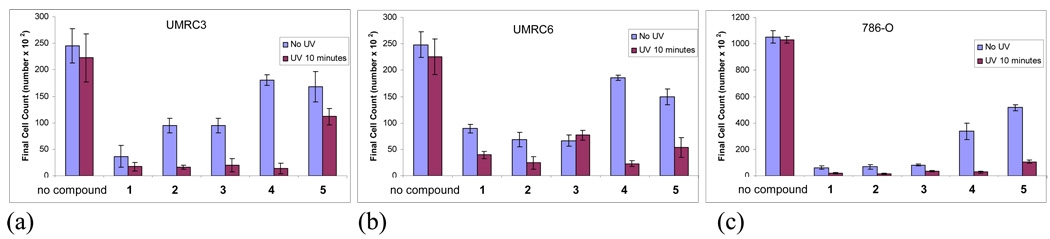
The ability of compounds 1 – 5 to inhibit cell proliferation in three human clear cell renal cell carcinoma (RCC) cell lines was tested in the dark and under photoactivation (Figure 22). No difference was detected in the control experiments between cells not exposed to UV versus those that were exposed to UV at 350 nm for 10 minutes in any of the three RCC cell lines tested (Figure 22 A, B & C; first two columns). On the other hand, all five compounds inhibited growth in each of the three cell lines but to varying degrees both in the dark and after UV radiation. Although UV treatment always enhanced the ability of conjugates 1–3 to inhibit cell proliferation, these compounds had strong growth inhibitory activity even without photoactivation in all three cell lines (Figure 22 A–C) with a range of 62 – 94 % inhibition.64 On the other hand, compounds 4 and 5 were considerably less cytotoxic in the dark. Compound 4 seems to be an especially promising lead for the future development of anticancer agents because the decrease of cell numbers in control UV-irradiated cells as compared to control without UV-irradiation in UMRC3 and UMRC6 cells was not statistically significant (Figure 22 A, B) but, when exposed to UV radiation and the conjugate, these cells displayed mortality of 94% in UMRC3 and 90% in UMRC6 cells.
Figure 22.
Cell proliferation assays using three different RCC cell lines: (a) UMRC3, (b) UMRC6 and (c) 786-O, and compounds 1–5. RCC cells were resuspended in media and treated with UV radiation (350 nm) for 10 minutes with and without indicated compounds. Control cells were exposed to the indicated compound 1–5 at final concentration of 10 µM. Cell numbers determined after 72 hours as described in the SI Section.
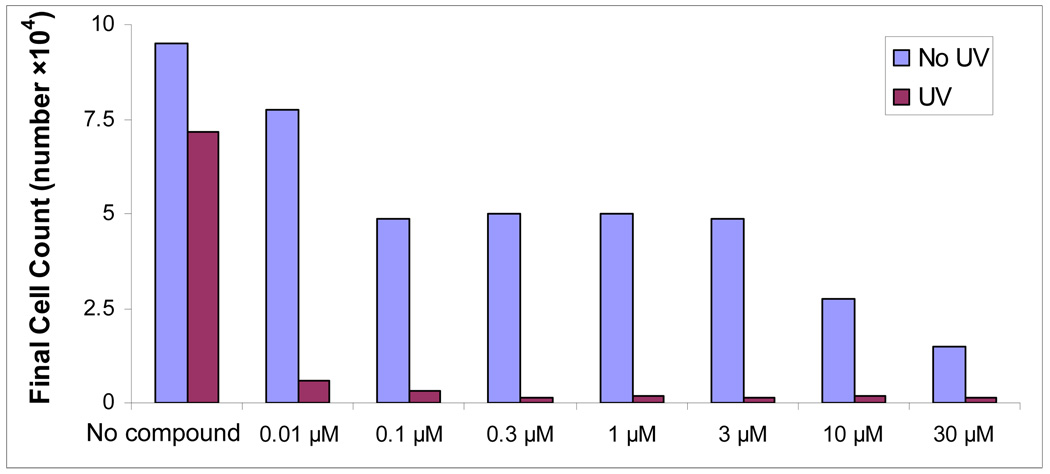
Encouraged by these results, we studied the concentration effects of compound 4 at the proliferation of in LNCap human prostate adenocarcinoma cells in the dark and under photoactivation conditions in more detail. Remarkably, even at the low 10 nM concentration, this system destroys more than 90% of LNCap cells in one treatment whereas almost no toxicity is observed without light.
CONCLUSION
Hybrid molecules which combine pH-regulated lysine moiety with a powerful DNA-photocleaver display dramatically increased DNA-cleaving reactivity at the lower pH. Synergistic effects associated with the protonation of the α-amino group, such as pH-dependent changes in photophysical properties as well as in pH-dependent DNA-binding (Figure 11), account for the increase in reactivity. Deactivation of the internal quenching pathway (Figure 11a) increases the lifetime of reactive singlet state while transformation of the conjugates into their dicationic form (Figure 11b) changes their binding mode with the polyanionic backbone of DNA.
Remarkably, the observed switch from negligible to record-breaking efficiency of the most therapeutically useful form of DNA cleavage (the ds DNA cleavage) occurs upon a relatively small change in the pH which can be used to differentiate healthy cells from hypoxic cancer tissues. The amplification of DNA-cleaving activity seems to be quite general and manifests itself for DNA-cleaving agents which bind DNA in different way and attack DNA through different chemical mechanisms. Cleavage at the lower pH for the conjugate 4 is not affected by the reactive oxygen species scavengers, suggesting possible value of this DNA-cleaver for targeting hypoxic cancer tissues. Cancer cell proliferation assays further confirm remarkable potential of these molecules for the development of light-activated anticancer agents.
Supplementary Material
Figure 12.
1H-NMR spectra of α- and ε- hydrogens in compound 8 (5 mM) in D2O at pD 5, 6, 7, 8, 9, 10, 11, 12, 13, and chemical shift-pD titration data for α- and ε- hydrogens in compound 8. These pKa values change to 7.44 and 10.40 when correction for going from pKa H* to pKa is applied
Figure 16.
UV-Vis absorbance titrations of 4 (10 µM) in 20 mM sodium phosphate buffer with 0, 1, 2, 3 equivalents of 30 μM/bp CT DNA at pH 5.5 (a), pH 7 (b) and pH 8 (c).
Figure 17.
Emission titrations of 4 (10µM) in 20 mM sodium phosphate buffer with 0, 10, 20, 30 μM of CT DNA at pH 5.5 (a), pH 7 (b) and pH 8 (c). The initial slopes of plots illustrating changes in fluorescence of 4 as a function of DNA concentration (d).
Figure 23.
LNCap cell proliferation assays with varying concentration of compound 4. The cells were exposed to UV for 10 minutes. Cells were counted 48 hours after the exposure. Full experimental details are described in the SI Section.
ACKNOWLEDGEMENTS
I. A. is grateful to the National Science Foundation (CHE-0848686) for partial support of this research. J.A.C. is funded in part by NIH/National Cancer Institute grant R01CA104505 and Dr. and Mrs. Ellis Brunton Rare Cancer Research Fund. I. A. and W.-Y. Y. acknowledge partial support from NIH grant RO1EB006158 and Professor J. Schlenoff for helpful discussions.
Footnotes
SUPPORTING INFORMATION. Details for experimental procedures, synthesis of DNA-photocleavers and photophysical experiments, extended survey of DNA binding studies, cartesian coordinates and energies of different protonation states of lysine conjugates, 1H and 13C NMR spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Wang-Yong Yang, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306-4390, USA.
Boris Breiner, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306-4390, USA.
Serguei V. Kovalenko, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306-4390, USA
Chi Ben, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306-4390, USA.
Mani Singh, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306-4390, USA.
Shauna N. LeGrand, Department of Cancer Biology, Mayo Clinic Comprehensive Cancer Center, Jacksonville, Florida 32224
Qing-Xiang Amy Sang, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306-4390, USA.
Geoffrey F. Strouse, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306-4390, USA
John A. Copland, Department of Cancer Biology, Mayo Clinic Comprehensive Cancer Center, Jacksonville, Florida 32224
Igor V. Alabugin, Email: alabugin@chem.fsu.edu, Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306-4390, USA.
REFERENCES AND NOTES
- 1.Schnarr NA, Kennan A. Org. Lett. 2005;7:395. doi: 10.1021/ol047720l. [DOI] [PubMed] [Google Scholar]; Schnarr NA, Kennan AJ. J. Am. Chem. Soc. 2003;125:6364. doi: 10.1021/ja029344a. [DOI] [PubMed] [Google Scholar]
- 2.Rooth M, Shaw AM. J.Phys. Chem. C. 2007;111:15363. [Google Scholar]
- 3.Xu H, Stampp SP, Rudkevich DM. M. Org. Lett. 2003;5:4583. doi: 10.1021/ol0357301. [DOI] [PubMed] [Google Scholar]
- 4.Choi HS, Huh KM, Ooya T, Yui N. J. Am. Chem. Soc. 2003;125:6350. doi: 10.1021/ja034149x. [DOI] [PubMed] [Google Scholar]
- 5.(a) Martinez-Diaz M-V, Spencer N, Stoddart JF. Angew. Chem., Int. Ed. Engl. 1997;36:1904. [Google Scholar]; Saha S, Stoddart JF. Chem. Soc. Rev. 2007;36:77. doi: 10.1039/b607187b. [DOI] [PubMed] [Google Scholar]; (b) Ashton PR, Ballardini R, Balzani V, Baxter I, Credi A, Fyfe MCT, Gandolfi MT, Gomez-Lopez M, Martinez-Diaz M-V, Piersanti A, Spencer N, Stoddart JF, Venturi M, White AJP, Williams DJ. J. Am. Chem. Soc. 1998;120:11932. [Google Scholar]; (c) Elizarov AM, Chang T, Chiu H-S, Stoddart JF. J. Org. Chem. 2002;67:9175. doi: 10.1021/jo020373d. [DOI] [PubMed] [Google Scholar]; (d) Silvi S, Arduini A, Pochini A, Secchi A, Tomasulo M, Raymo FM, Baroncini M, Credi A. J. Am. Chem. Soc. 2007;129:13378. doi: 10.1021/ja0753851. [DOI] [PubMed] [Google Scholar]; (e) Leigh DA, Thomson AR. Org. Lett. 2006;8:5377. doi: 10.1021/ol062284j. [DOI] [PubMed] [Google Scholar]
- 6.Fabbrizzi L, Gatti F, Pallavicini P, Parodi L. New J. Chem. 1998;1403 [Google Scholar]; (f) de Silva AP, Gunaratne HQN, McCoy CP. Chem. Commun. 1996:2399. [Google Scholar]; (g) Becuwe M, Cazier F, Bria M, Woisel P, Delattre F. Tetrahedron Lett. 2007;48:6186. [Google Scholar]
- 7.(a) Pallavicini P, Amendola V, Massera C, Mundum E, Taglietti A. Chem. Commun. 2002:2452. doi: 10.1039/b205951g. [DOI] [PubMed] [Google Scholar]; (b) Gunnlaugsson T, Leonard JP, Senechal K, Harte AJ. J. Am. Chem. Soc. 2003;125:12062. doi: 10.1021/ja035425a. [DOI] [PubMed] [Google Scholar]; (c) Tajc SG, Miller BL. J. Am. Chem. Soc. 2006;128:2532. doi: 10.1021/ja058126p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fattakhova-Rohlfing D, Wark M, Rathousky J. Chem. Mater. 2007;19:1640. [Google Scholar]
- 9.Xue C, Mirkin CA. Angew. Chem. Int. Ed. 2007;46:2036. doi: 10.1002/anie.200604637. [DOI] [PubMed] [Google Scholar]
- 10.Turega SM, Philp D. Chem. Commun. 2006;35:3684. doi: 10.1039/b607551g. [DOI] [PubMed] [Google Scholar]
- 11.Broz P, Driamov S, Ziegler J, Ben-Haim N, Marsch S, Meier W, Hunziker P. Nano Lett. 2006;6:2349. doi: 10.1021/nl0619305. [DOI] [PubMed] [Google Scholar]
- 12.Adams DJ, Dewhirst MW, Flowers JL, Gamcsik MP, Colvin OM, Manikumar G, Wani MC, Wall ME. Cancer Chemother. Pharmacol. 2000;46:263. doi: 10.1007/s002800000157. [DOI] [PubMed] [Google Scholar]; Gabr A, Kuin A, Aalders M, El-Gawly H, Smets LA. Cancer Res. 1997;57:4811. [PubMed] [Google Scholar]; Teicher BA, Holden SA, Khandakar V, Herman TS. J. Cancer Res. Clinical Oncol. 1993;119:645. doi: 10.1007/BF01215982. [DOI] [PubMed] [Google Scholar]; Wood PJ, Sansom JM, Newell K, Tannock IF, Stratford IJ. Int. J. Cancer. 1995;60:264. doi: 10.1002/ijc.2910600222. [DOI] [PubMed] [Google Scholar]; Vukovic V, Tannock IF. Brit. J. Cancer. 1997;75:1167. doi: 10.1038/bjc.1997.201. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wachsberger PR, Burd R, Wahl ML, Leeper DB. Int. J. Hyperthermia. 2002;18:153. doi: 10.1080/02656730110091333. [DOI] [PubMed] [Google Scholar]
- 13.Hoffner J, Schottelius J, Feichtinger D, Chen P. J. Am. Chem. Soc. 1998;120:376. [Google Scholar]
- 14.Adams GE, Stratford IJ. Int. J. Radiat. Oncol. Biol. Phys. 1994;29:231. doi: 10.1016/0360-3016(94)90267-4. [DOI] [PubMed] [Google Scholar]; Priyadarsini KI, Dennis MF, Naylor MA, Stratford MRL, Wardman P. J. Am. Chem. Soc. 1996;118:5648. and references therein. [Google Scholar]
- 15.Stubbs M, Rodrigues L, Howe FA, Wang J, Jeong KS, Veech RL, Griffiths JR. Cancer Res. 1994;54:4011. [PubMed] [Google Scholar]
- 16.Lyons JC, Ross BD, Song CW. Int. J. Radiat. Oncol. Biol. Phys. 1993;25:95. doi: 10.1016/0360-3016(93)90150-t. [DOI] [PubMed] [Google Scholar]; Song CW, Lyons JC, Griffin RJ, Makepeace CM. Radiother. Oncol. 1993;27:252. doi: 10.1016/0167-8140(93)90082-j. [DOI] [PubMed] [Google Scholar]; Song CW, Lyons JC, Griffin RJ, Makepeace CM, Cragoe EJ., Jr Cancer Res. 1993;53:1599. [PubMed] [Google Scholar]; Song CW, Lyons JC, Griffin RJ, Makepeace CM, Cragoe EJ., Jr Int. J. Radiat. Oncol. Biol. Phys. 1994;30:133. doi: 10.1016/0360-3016(94)90528-2. [DOI] [PubMed] [Google Scholar]; Song CW, Kim GE, Lyons JC, Makepeace CM, Griffin RJ, Rao GH, Cragoe EJ., Jr Int. J. Radiat. Oncol. Biol. Phys. 1994;30:1161. doi: 10.1016/0360-3016(94)90324-7. [DOI] [PubMed] [Google Scholar]; Lyons JC, Song C. Radiat. Res. 1995;141:216. [PubMed] [Google Scholar]
- 17.Newell K, Wood P, Stratford I, Tannock I. Br. J. Cancer. 1992;66:311. doi: 10.1038/bjc.1992.262. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hasuda K, Lee C, Tannock IF. Oncol. Res. 1994;6:259. [PubMed] [Google Scholar]
- 18.Osinsky SP, Levitin IY, Bubnovskaya LN, Ganusevich, Sigan AL, Tsykalova MV, Zagorujko LI. Exp. Oncol. 1999;21:216. [Google Scholar]; Tannock IF, Rotin D. Cancer Res. 1989;49:4373. [PubMed] [Google Scholar]; Wike-Hooley JL, Haveman J, Reinhold HS. Radiother Oncol. 1984;2:343. doi: 10.1016/s0167-8140(84)80077-8. [DOI] [PubMed] [Google Scholar]
- 19.von Andrenne M. Adv. Pharmacol. 1972;10:339. [Google Scholar]; Osinsky S, Bubnovskaya L. Arch. Geschwulstforsch. 1984;54:463. [PubMed] [Google Scholar]
- 20.Kar M, Basak A. Chem. Rev. 2007;107:2861–2890. doi: 10.1021/cr068399i. [DOI] [PubMed] [Google Scholar]
- 21.Selected examples given below are limited to acid-catalyzed transformations: Enediyne – eneynecumulene conversion: Toshima K, Ohishi K, Tomishima M, Matsumura S. Heterocycles. 1997;45:8. Naoe Y, Kikuishi J, Ishigaki K, Iitsuka H, Nemoto H, Shibuya M. Tetrahedron Lett. 1995;36:9165. Acid-catalyzed ring opening: Unno R, Michishita H, Inagaki H, Suzuki Y, Baba Y, Jomori T, Moku M, Nishikawa T, Isobe M. Biorg. Med. Chem. Lett. 1997;5:903. doi: 10.1016/s0968-0896(97)00027-8. Allylic rearrangement: Dai W-M, Fong KC, Lau CW, Zhau L, Hamaguchi W, Nishimoto S-I. J. Org. Chem. 1999;64:682. doi: 10.1021/jo9820263. Enol-keto tautomerization: Semmelhack MF, Gallagher JJ, Minami T, Date T. J. Am. Chem. Soc. 1993;115:11618. A number of other intellectually interesting examples involve activation at the basic pH: Toshima K, Ohta K, Ohashi A, Nakamura T, Nakata M, Tatsuta K, Matsumura S. J. Am. Chem. Soc. 1995;117:4822. David WM, Kumar D, Kerwin SM. Bioorg. Med. Chem. Lett. 2000;10:2509. doi: 10.1016/s0960-894x(00)00516-3. Swakyama M, Shigenaga A, Nemoto H, Shibuya M. Tetrahedron Lett. 2000;41:10019. Nicolaou KC, Hong YP, Dai WM, Zeng ZJ, Wrasidlo W. Chem. Commun. 1992:1542. Nicolaou KC, Dai WM. J. Am. Chem. Soc. 1992;114:8908. Kerwin SM. Tetrahedron Lett. 1994;35:1023. Pal R, Basak A. Chem. Commun. 2006:2992. doi: 10.1039/b605743h.
- 22. Tietze LF, Fischer-Beller A. Carbohydr. Res. 1994;254:169. and references therein. For an application of this idea to enediynes, see: Nace Y, Kikuishi J, Ishigaki K, Iitsuka H, Nemoto H, Shibuya M. Tetrahedron Lett. 1995;36:9165.
- 23.Volpin ME, Levitin I, Osinsky S. Angew. Chem. Int. Ed. Engl. 1996;35:2395. [Google Scholar]; Angew. Chem. 1996;108:2516. [Google Scholar]; Levitin IY, Belkov VM, Novodarova GN, Shabarova ZA, Volpin ME. Mendeleev Commun. 1996:153. [Google Scholar]
- 24.Lee Alex HF, Chen J, Liu D, Leung TYC, Chan ASC, Li T. J. Am. Chem. Soc. 2002;124:13972. doi: 10.1021/ja020531i. [DOI] [PubMed] [Google Scholar]
- 25.a) David WM, Kerwin SM. J. Am. Chem. Soc. 1997;119:1464. [Google Scholar]; b) Hoffner J, Schottelius MJ, Feichtinger D, Chen P. J. Am. Chem. Soc. 1998;120:376. [Google Scholar]; (c) Kraka E, Cremer D. J. Am. Chem. Soc. 2000;122:8245–8264. [Google Scholar]
- 26. Alabugin IV, Manoharan M. J. Phys. Chem. A. 2003;107:3363–3371. Alabugin IV, Manoharan M, Kovalenko SV. Org. Lett. 2002;4:1119–1122. doi: 10.1021/ol0255054. For a general development of these ideas to other substituents, see recent work on the ortho-effect in the Bergman cyclization: Zeidan T, Kovalenko SV, Manoharan M, Alabugin IV. J. Org. Chem. 2006;71:962–975. doi: 10.1021/jo0520801. Zeidan T, Manoharan M, Alabugin IV. J. Org. Chem. 2006;71:954–961. doi: 10.1021/jo051857n.
- 27. Basak A, Kar M. Bioorg. Med. Chem. 2008;16:4532. doi: 10.1016/j.bmc.2008.02.044. This system shows promising pH-dependency (more cleavage at pH 7.5 than pH 8). However the change occurs at the slightly basic conditions, which is insufficient for differentiating of normal and cancer cells.
- 28.Alabugin IV, Manoharan M, Buck M, Clark R. J. THEOCHEM. 2007;813:21. [Google Scholar]
- 29.Gengrinovitch S, Izakovich E. PCT Int. Appl. 2005:73. [Google Scholar]; Saito I, Takayama M, Sugiyama H, Nakatani H. J. Am. Chem. Soc. 1995;117:6406. [Google Scholar]; Saito I, Takayama M, Kawanishi S. J. Am. Chem. Soc. 1995;117:5590. [Google Scholar]; Plourde G, II, El-Shafey A, Fouad FS, Purohit AS, Jones GB. Bioorg. Med. Chem. Lett. 2002;12:2985. doi: 10.1016/s0960-894x(02)00618-2. [DOI] [PubMed] [Google Scholar]
- 30.Basicity of these groups is known to be sensitive to the environment. See, for example: Highbarger LA, Gerlt JA, Kenyon GL. Biochemistry. 1996;35:41. doi: 10.1021/bi9518306.
- 31.Armitage B. Chem. Rev. 1998;98:1171. doi: 10.1021/cr960428+. [DOI] [PubMed] [Google Scholar]
- 32.Alabugin IV, Kovalenko SV. J. Am. Chem. Soc. 2002;124:9052. doi: 10.1021/ja026630d. [DOI] [PubMed] [Google Scholar]
- 33.For other photochemically activated enediynes, see: Tachi Y, Dai W-M, Tanabe K, Nishimoto S-I. Bioorg. Med. Chem. 2006;14:3199. doi: 10.1016/j.bmc.2005.12.040. Bhattacharyya S, Zaleski JM. Curr. Top. Med. Chem. 2004;4:1637. doi: 10.2174/1568026043387403. Schmittel M, Viola G, Dall'Acqua F, Morbach G. Chem. Comm. 2003;5:646. doi: 10.1039/b211783e. Karpov G, Kuzmin A, Popik VV. J. Am. Chem. Soc. 2008;130:11771. doi: 10.1021/ja802688c. Polukhtine A, Karpov G, Popik VV. Curr. Top. Med. Chem. 2008;8:460. doi: 10.2174/156802608783955700. Yoshimura N, Momotake A, Shinohara Y, Nishimura Y, Arai T. Chem. Lett. 2008;37:174. Sud D, Wigglesworth TJ, Branda NR. Angew. Chem. Int. Ed. 2007;46:8017. doi: 10.1002/anie.200703034. Karpov GV, Popik VV. J. Am. Chem. Soc. 2007;129:3792. doi: 10.1021/ja064470q. Poloukhtine A, Popik VV. J. Org. Chem. 2006;71:7417. doi: 10.1021/jo061285m. O'Connor JM, Friese SJ, Rodgers BL. J. Am. Chem. Soc. 2005;127:16342. doi: 10.1021/ja050060a. Kar M, Basak A, Bhattacharjee M. Bioorg. Med. Chem. Lett. 2005;15:5392. doi: 10.1016/j.bmcl.2005.09.005. Zhao Z, Peacock JG, Gubler DA, Peterson MA. Tetrahedron Lett. 2005;46:1373. Poloukhtine A, Popik VV. J. Org. Chem. 2005;70:1297. doi: 10.1021/jo048065y. Bhattacharyya S, Zaleski JM. Metalloenediynes. Curr. Top. Med. Chem. 2004;4:1637. doi: 10.2174/1568026043387403. Fouad FS, Crasto CF, Lin Y, Jones GB. Tetrahedron Lett. 2004;45:7753. Jones GB, Russell KC. CRC Handbook of Organic Photochemistry and Photobiology. 2nd. New York: CRC-Press; 2004. p. 21/1.p. 29/21. Poloukhtine A, Popik VV. J. Org. Chem. 2003;68:7833. doi: 10.1021/jo034869m. Chandra T, Kraft BJ, Huffman JC, Zaleski JM. Inorg. Chem. 2003;42:5158. doi: 10.1021/ic030035a. Benites PJ, Holmberg RC, Rawat DS, Kraft BJ, Klein LJ, Peters DG, Thorp HH, Zaleski JM. J. Am. Chem. Soc. 2003;125:6434. doi: 10.1021/ja020939f. Banfi L, Guanti G, Rasparini M. Eur. J. Org. Chem. 2003;7:1319. Choy N, Blanco B, Wen J, Krishan A, Russell KC. Org. Lett. 2000;2:3761. doi: 10.1021/ol006061j. Jones GB, Wright JM, Plourde G, II, Purohit AD, Wyatt JK, Hynd G, Fouad F. J. Am. Chem. Soc. 2000;122:9872. Kaneko T, Takahashi M, Hirama M. Angew. Chem. Int. Ed. 1999;38:1267. doi: 10.1002/(SICI)1521-3773(19990503)38:9<1267::AID-ANIE1267>3.0.CO;2-F. Evenzahav A, Turro NJ. J. Am. Chem. Soc. 1998;120:1835. Ramkumar D, Kalpana M, Varghese B, Sankararaman S, Jagadeesh MN, Chandrasekhar J. J. Org. Chem. 1996;61:2247. Kagan J, Wang X, Chen X, Lau KY, Batac IV, Tuveson RW, Hudson JB. J. Photochem. Photobiol. B. 1993;21:135. doi: 10.1016/1011-1344(93)80175-9. Nicolaou VKC, Dai WM, Wendeborn SV, Smith AL, Torisawa Y, Maligres P, Hwang CK. Angew. Chem. 1991;103:1034. Shiraki T, Sugiura Y. Biochemistry. 1990;29:9795. doi: 10.1021/bi00494a006. Falcone D, Li J, Kale A, Jones GB. Bioorg. Med. Chem. Lett. 2008;18:934. doi: 10.1016/j.bmcl.2007.12.045. Fouad FS, Wright JM, Plourde G, 2nd, Purohit AD, Wyatt JK, El-Shafey A, Hynd G, Crasto CF, Lin Y, Jones GB. J. Org. Chem. 2005;70:9789. doi: 10.1021/jo051403q. Poloukhtine A, Popik VV. Chem. Comm. 2005;5:617. doi: 10.1039/b414951c. Kraft BJ, Coalter NL, Nath M, Clark AE, Siedle AR, Huffman JC, Zaleski JM. Inorg. Chem. 2003;42:1663. doi: 10.1021/ic0207045. Plourde G, El-Shafey A, Fouad FS, Purohit AS, Jones GB. Bioorg. Med. Chem. Lett. 2002;12:2985. doi: 10.1016/s0960-894x(02)00618-2. Basak A, Bdour HM, Shain JC, Mandal S, Rudra KR, Nag S. Bioorg. Med. Chem. Lett. 2000;10:1321. doi: 10.1016/s0960-894x(00)00237-7.
- 34.Kovalenko SV, Alabugin IV. Chem. Comm. 2005:1444. doi: 10.1039/b417012a. [DOI] [PubMed] [Google Scholar]
- 35.Kauffman JF, Turner JM, Alabugin IV, Breiner B, Kovalenko SV, Badaeva EA, Masunov A, Tretiak S. J. Phys. Chem. A. 2006;110:241. doi: 10.1021/jp056127y. [DOI] [PubMed] [Google Scholar]
- 36.Interestingly, this selectivity is observed not only for the TFP-substituted molecules, but also for the Ph-substituted enediynes expected to undergo the photochemical Bergman cyclization. Breiner B, Schlatterer JC, Kovalenko SV, Greenbaum NL, Alabugin IV. Angew. Chem. 2006;45:3666. doi: 10.1002/anie.200504479.
- 37.Breiner B, Schlatterer JC, Kovalenko SV, Greenbaum NL, Alabugin IV. Proc. Natl. Acad. Sci. 2007;104:13016. doi: 10.1073/pnas.0705701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The cleavage was quantified by using SAFA software: Laederach RDA, Pearlman S, Herschlag D, Altman R. RNA. 2005;11:344. doi: 10.1261/rna.7214405.
- 39.The ds/ss ratio is 1:2 for plasmid DNA and 1:3 for cellular DNA: Elmroth K, Nygren J, Martensson S, Ismail IH, Hammarsten O. DNA Repair. 2003;2:363. doi: 10.1016/s1568-7864(02)00235-5.
- 40.About 10% of the DNA strand breaks induced by bleomycin are double-strand breaks: Charles K, Povirk LF. Chem. Res. Toxicol. 1998;11:1580. doi: 10.1021/tx980154g. Povirk LF, Wubker W, Kohnlein W, Hutchinson F. Nucleic Acids Res. 1977;5:3573. doi: 10.1093/nar/4.10.3573. See also: Lloyd RS, Haidle CW, Hewitt RR. Cancer Res. 1978;38:3191. Harsch A, Marzilli LA, Bunt RC, Stubbe J, Vouros P. Nucleic Acids Res. 2000;28:1978. doi: 10.1093/nar/28.9.1978. Stubbe J, Kozarich JW. Chem. Rev. 1987;87:1107. Burger RM. Chem. Rev. 1998;98:1153. doi: 10.1021/cr960438a. Quada JC, Jr, Zuber GF, Hecht SM. Pure Appl. Chem. 1998;70:307. Hecht SM. J. Nat. Prod. 2000;63:158. doi: 10.1021/np990549f.
- 41.Value for the ds cleavage (Form III) is less accurate because of the greater experimental error bars.
- 42.Interestingly, monoamines possess a certain (but lesser) degree of pH-dependency and presence of a α-amine in 6 has a slightly larger effect than the presence of a ε-amine in 7.
- 43.Standke KHC, Brunnert H. Nucleic Acids Res. 1975;2:1839. doi: 10.1093/nar/2.10.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ahmad J, Ashok BT, Ali R. Immun. Lett. 1998;62:87. doi: 10.1016/s0165-2478(98)00024-8. [DOI] [PubMed] [Google Scholar]; Tyler JK, Allen KE, Everett RD. Nucl. Acids Res. 1994;22:270. doi: 10.1093/nar/22.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao G, DeRose EF, Kirby TW, London RE. Biochemistry. 2006;45:1785. doi: 10.1021/bi051856p. [DOI] [PubMed] [Google Scholar]; Kakehashi R, Shizuma M, Yamamura S, Maeda H. J. Colloid Interface Sci. 2005;289:498. doi: 10.1016/j.jcis.2005.03.090. [DOI] [PubMed] [Google Scholar]; Rush JR, Sandstrom SL, Yang J, Davis R, Prakash O, Baures PW. Org. Lett. 2005;7:135. doi: 10.1021/ol047812a. [DOI] [PubMed] [Google Scholar]
- 45.As the control experiment, we tested two pKa values of ethylene diamine with NMR titration. After correcting measured pKa values with the equation (ref. 42), the values of 7.03 and 9.99 are obtained experimentally (R2 =0.999). These data are in a good agreement with the corresponding potentiometric literature values of 7.19 and 9.98. Avdeef A, Kearney DL, Brown JA, Chemotti AR. Anal. Chem. 1982;54:2322.
- 46.pKa values measured with pH* are assumed to be similar to the corresponding values in H2O: Primrose WU. NMR of Macromolecules. In: Poberts GCK, editor. Pxford: A Practical Approach; Oxford University Press; 1993. pp. 22–23. Scheller KH, Scheller-Krattiger V, Martin RB. J. Am. Chem. Soc. 1981;103:6833. Alternatively, one can use a formula for correlating pKa values determined in D2O and H2O: Krezel A, Bal Wojciech A. J. Inorg. Biochem. 2004;98:161. doi: 10.1016/j.jinorgbio.2003.10.001.
- 47.Another interesting observation is that chemical shifts of aromatic hydrogens show small, but measurable response to the changes in pH in the vicinity of pH 10 region (See the SI section) suggesting that there may some contribution from cation-π interaction. Gallivan JP, Dougherty DA. Proc. Natl. Acad. Sci. 1999;96:9459. doi: 10.1073/pnas.96.17.9459. Ma JC, Dougherty DA. Chem. Rev. 1997;97:1303. doi: 10.1021/cr9603744. Mercozzi S, West AP, Jr, Dougherty DA. J. Am. Chem. Soc. 1996;118:2307. Dougherty DA. Science. 1996;271:163. doi: 10.1126/science.271.5246.163.
- 48.A possible explanation is that fluorescence reports properties of an excited molecule, whereas absorption does if for a ground state counterpart. Acidities of ground and excited states are often different: Tolbert LM, Solntsev KM. Acc. Chem. Res. 2002;35:19. doi: 10.1021/ar990109f. The validity of this explanation depends on the relative rates of protonation/deprotonation and the key photophysical events. This complicated question is outside of the scope of this paper.
- 49.Carey FA. Organic Chemistry. 6th Ed. New York: Mc Graw Hill; 2006. [Google Scholar]
- 50.Reed AE, Curtiss LA, Weinhold F. Chem. Rev. 1988;88:899. [Google Scholar]
- 51.Alabugin IV, Manoharan M, Zeidan TA. J. Am. Chem. Soc. 2003;125:14014. doi: 10.1021/ja037304g. [DOI] [PubMed] [Google Scholar]
- 52.(a) Miertus S, Tomasi J. Chem. Phys. 1982;65:239. [Google Scholar]; (b) Miertus S, Scrocco E, Tomasi J. Chem. Phys. 1981;55:117. [Google Scholar]
- 53.Both singlet and triplet excited states of DNA are too high in energy to allow for efficient quenching by energy transfer.
- 54.For major groove binders: Kashanian S, Gholivand MB, Ahmadi F, Taravati A, Colagar A. Hosseinzadeh. Spectro. Acta Part A. 2007;67A:472. doi: 10.1016/j.saa.2006.08.017. Vijayalakshmi R, Kanthimathi M, Subramanian V, Nair BU. Biochim. Biophys. Acta, Gen. Subj. 2000;1475:157. doi: 10.1016/s0304-4165(00)00063-5. Tysoe SA, Morgan RJ, Baker AD, Strekas TC. J. Phys. Chem. 1993;97:1707. For minor groove binders: Karlsson HJ, Eriksson M, Perzon E, Akerman B, Lincoln P, Westman G. Nucleic Acids Res. 2003;31:6227. doi: 10.1093/nar/gkg821. Tawar U, Jain AK, Chandra R, Singh Y, Dwarakanath BS, Chaudhury NK, Good L, Tandon V. Biochemistry. 2003;42:13339. doi: 10.1021/bi034425k. Adhikary A, Buschmann V, Mueller C, Sauer M. Nucleic Acids Res. 2003;31:2178. doi: 10.1093/nar/gkg308. Rucker VC, Foister S, Melander C, Dervan PB. J. Am. Chem. Soc. 2003;125:1195. doi: 10.1021/ja021011q. Baliga R, Crothers DM. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7814. doi: 10.1073/pnas.97.14.7814. Kumar CV, Turner RS, Asuncion EH. J. Photochem. Photobiol. A. 1993;74:231.
- 55.Li J, Wei Y-X, Wei Y-L, Dong C. J. Lumi. 2007;124:143. [Google Scholar]; Melvin MS, Ferguson DC, Lindquist N, Manderville RA. J. Org. Chem. 1999;64:6861. doi: 10.1021/jo990944a. [DOI] [PubMed] [Google Scholar]; Spillane CB, Dabo MNV, Fletcher NC, Morgan JL, Keene FR, Haq I, Buurma NJ. J. Inorg. Biochem. 2008;102:673. doi: 10.1016/j.jinorgbio.2007.10.011. [DOI] [PubMed] [Google Scholar]; Fuerstenberg A, Deligeorgiev TG, Gadjev NI, Vasilev AA, Vauthey E. Chem. Eur. J. 2007;13:8600. doi: 10.1002/chem.200700665. [DOI] [PubMed] [Google Scholar]; Bhadra K, Maiti M, Kumar GS. Biochim. Biophys. Acta, Gen. Subj. 2007;1770:1071. doi: 10.1016/j.bbagen.2007.03.001. [DOI] [PubMed] [Google Scholar]; Erve A, Saoudi Y, Thirot S, Guetta-Landras C, Florent J-C, Nguyen C-H, Grierson DS, Popov AV. Nucleic Acids Res. 2006;34:e43/1. doi: 10.1093/nar/gkl011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Marinic M, Piantanida I, Rusak G, Zinic M. J. Inorg. Biochem. 2006;100:288. doi: 10.1016/j.jinorgbio.2005.11.013. [DOI] [PubMed] [Google Scholar]; Zipper H, Brunner H, Bernhagen J, Vitzthum F. Nucleic Acids Res. 2004;32:e103/1. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]; Piantanida I, Palm BS, Zinic M, Schneider H-J. J. Chem. Soc. Perkin Trans. 2. 2001;9:1808. [Google Scholar]
- 56.Saito I, Nakatani K. Bull. Chem. Soc. Jpn. 1996;69:3007. [Google Scholar]; Hiraku Y, Ito K, Hirakawa K, Kawanishi S. Photochem. Photobiol. 2007;83:205. doi: 10.1562/2006-03-09-IR-840. [DOI] [PubMed] [Google Scholar]
- 57.Devasagayam TPA, Steenken S, Obendorf MSW, Schulz WA, Sies H. Biochemistry. 1991;30:6283. doi: 10.1021/bi00239a029. [DOI] [PubMed] [Google Scholar]
- 58.Saito I. Pure Appl. Chem. 1992;64:1305. [Google Scholar]; Quadda JC, Levy MJ, Hecht SM. J. Am. Chem. Soc. 1993;115:12171. [Google Scholar]
- 59.Steenken S. Chem. Rev. 1989;89:503. [Google Scholar]; Dunn DA, Lin VH, Kochevar IE. Biochemistry. 1992;31:11620. doi: 10.1021/bi00161a048. [DOI] [PubMed] [Google Scholar]; Sugiyama H, Tsutsumi K, Fujimoto K, Saito I. J. Am. Chem. Soc. 1993;115:4443. [Google Scholar]; Cadet J, Berger M, Bunchko GW, Joshi RC, Raoul S, Ravanet J-L. J. Am. Chem. Soc. 1994;116:7403. [Google Scholar]; Armitage B, Changjun Y, Devadoss C, Schuster GB. J. Am. Chem. Soc. 1994;116:9847. [Google Scholar]
- 60.Nielsen PE, Jeepsen C, Egholm M, Buchardt O. Nucleic Acids Res. 1988;16:3877. doi: 10.1093/nar/16.9.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chatterjee M, Rokita SE. J. Am. Chem. Soc. 1990;112:6397. [Google Scholar]; Henriksen U, Larsen C, Karup G, Jeepsen C, Nielsen PE, Buchardt O. Photochem. Photobiol. 1991;53:299. doi: 10.1111/j.1751-1097.1991.tb03632.x. [DOI] [PubMed] [Google Scholar]; Chatterjee M, Rokita SE. J. Am. Chem. Soc. 1994;116:1690. [Google Scholar]; Saito I, Takayama M, Sakurai T. J. Am. Chem. Soc. 1994;116:2653. [Google Scholar]; Nakatani K, Shirai J, Tamaki R, Saito I. Tetrahedron Lett. 1995;36:5363. [Google Scholar]
- 61.Zeidan T, Kovalenko SV, Manoharan M, Clark RJ, Ghiviriga I, Alabugin IV. J. Am. Chem.Soc. 2005;127:4270. doi: 10.1021/ja043803l. [DOI] [PubMed] [Google Scholar]; Zeidan T, Clark RJ, Kovalenko SV, Ghiviriga I, Alabugin IV. Chemistry Eur. Journal. 2005;11:4953. doi: 10.1002/chem.200500180. [DOI] [PubMed] [Google Scholar]
- 62.Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Phil. Trans. Royal Soc. B. 2006;361:1351. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.The total percentage of cleavage for each pH was calculated as (2x[Form III]+[Form II])/([Form III]+[Form II] +[Form I]).
- 64.We currently have no clear explanation for activity of the conjugates in the dark but, for compounds 1 and 2, it is unlikely to be associated with thermal Bergman reaction which is too slow under these conditions.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.