Abstract
Laboratory mice serve as important models in biomedical research. Monitoring these animals for infections and infestations and excluding causative agents requires extensive resources. Despite advancements in detection and exclusion over the last several years, these activities remain challenging for many institutions. The infections and infestations present in laboratory mouse colonies are well documented, but their mode of introduction is not always known. One possibility is that wild rodents living near vivaria somehow transmit infections to and between the colonies. This study was undertaken to determine what infectious agents the wild mice on the University of Pennsylvania (Philadelphia) campus were carrying. Wild mice were trapped and evaluated for parasites, viruses, and selected bacteria by using histopathology, serology, and PCR-based assays. Results were compared with known infectious agents historically circulating in the vivaria housing mice on campus and were generally different. Although the ectoparasitic burdens found on the 2 populations were similar, the wild mice had a much lower incidence of endoparasites (most notably pinworms). The seroprevalence of some viral infections was also different, with a low prevalence of mouse hepatitis virus among wild mice. Wild mice had a high prevalence of murine cytomegalovirus, an agent now thought to be confined to wild mouse populations. Helicobacter DNA was amplified from more than 90% of the wild mice (59% positive for H. hepaticus). Given the results of this study, we conclude that wild mice likely are not a source of infection for many of the agents that are detected in laboratory mouse colonies at the University of Pennsylvania.
Abbreviations: EDIM, epizootic diarrhea of infant mice; MAV, mouse adenovirus; MCMV, murine cytomegalovirus; MFIA, multiplex fluorescent immunoassay; MHV, mouse hepatitis virus; MNV, murine norovirus; MPV, mouse parvovirus; MVM, minute virus of mice; TMEV, Theiler mouse encephalomyelitis virus
Laboratory mice constitute the most popular animal models used in biomedical research today. Like all animals, even mice housed in so-called ‘barrier’ facilities are subject to infection. The infectious agents and organisms present in laboratory mouse colonies on the University of Pennsylvania campus are known and documented by the University Laboratory Animal Resources Diagnostic Services Unit. Sentinel mice that are housed on soiled bedding from resident mouse cages are screened onsite at 3 quarterly intervals for fur mites and pinworms and for a panel of viral infections: mouse hepatitis virus (MHV); epizootic diarrhea of infant mice (EDIM) virus; minute virus of mice (MVM); mouse parvovirus (MPV); Theiler mouse encephalomyelitis virus (TMEV); and Sendai virus. Comprehensive bacteriology and parasitology assessments are performed on all sentinels once yearly during the fourth quarter. In addition, these sentinels are screened serologically for 18 viral infections, Mycoplasma pulmonis, cilia-associated respiratory bacillus, and Encephalitozoon cuniculi and by PCR for Helicobacter spp. and M. pulmonis. Mesenteric lymph nodes from sentinels monitoring barrier-maintained colonies are also screened once yearly by PCR for MPV. In addition, University Laboratory Animal Resources maintains a quarantine facility for rodents received from nonapproved sources (sources other than selected commercial breeding facilities). Mice entering the quarantine facility are housed in semirigid isolators, and contact sentinels are tested for all of the agents included in the fourth quarter comprehensive health assessment described, including PCR for MPV.
Wild mice (Mus musculus) could serve as a source of infection or infestation in laboratory mouse colonies, although little is known about the prevalence of infectious diseases in wild mouse populations in Philadelphia. However, we have surveyed wild mouse populations in other geographic areas.1,9 Significant seroprevalence of MHV, EDIM, murine cytomegalovirus (MCMV), parvovirus, and thymic virus (murid herpesvirus 3), in addition to the presence of many types of parasites and bacteria including Myocoptes spp., Myobia spp., Radfordia spp., Spironucleus spp., Giardia spp., Pasteurella pneumotropica, Pseudomonas spp., and Leptospira spp. were found in wild populations of mice from farms in southeastern Connecticut.1 Studies of wild mouse (Mus domesticus) populations in the cereal-growing region of southeastern Australia revealed a high serologic prevalence of MHV, EDIM, and MCMV, as well as significant seroprevalence of mouse adenovirus (MAV), MPV, and reovirus type 3.9
The goal of the current study was to expand preliminary data obtained from wild mice trapped in the University City district of Philadelphia in 2005 (which are included with the current results from a 2007 survey). These data document the prevalence of various infectious agents and parasites commonly found in populations of wild mice on the University of Pennsylvania campus in Philadelphia and are discussed in the context of infectious disease outbreaks in campus vivaria over the past 5 y.
Materials and Methods
Animal trapping.
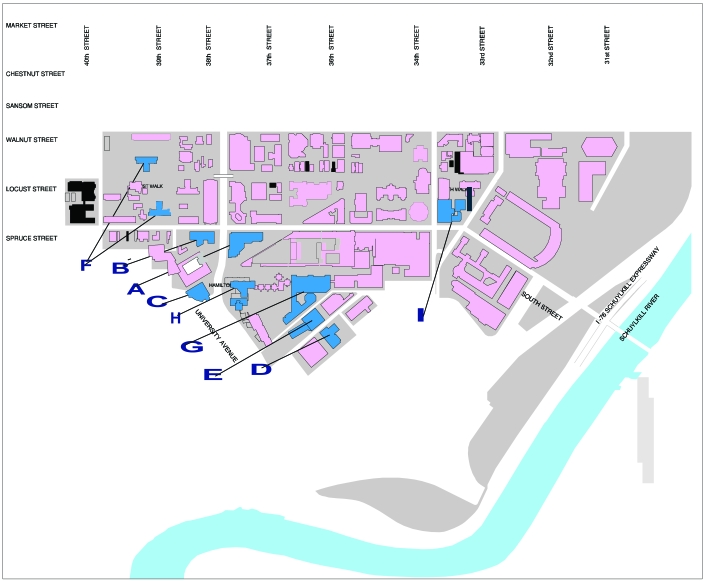
Mice were trapped with glue traps (Aardvark Pest Control, Aston, PA) with or without bait (bacon bits). These traps were set in the evening and checked first thing in the morning. Live traps (Victor 313 traps, Aardvark Pest Control) were used for 1 location (a convenience store loading dock) and checked twice daily when in place. Traps were placed in strategic locations around the University of Pennsylvania campus, where mice were known or expected to be living or traveling. Examples of such sites included locations where substantial quantities of mouse fecal pellets were found, along walls, in dark corners, beside dumpsters, and near holes containing pipes or electric wiring. Trap placement sites (Figure 1) included: the loading dock of the Veterinary Hospital of the University of Pennsylvania (containing laboratories, offices, and conventional rodent housing facility), a convenience store loading dock (close to the Biology conventional mouse housing facility), the High Rise Towers East and South (undergraduate dormitories), the loading dock for the Biomedical Research Building (containing laboratories, offices, and a School of Medicine barrier-maintained mouse vivarium), Stellar–Chance Laboratories (containing laboratories and offices), an auditorium (Class of 1925) in the John Morgan Building (containing laboratories, classrooms, offices and a School of Medicine barrier-maintained mouse vivarium), Hayden Hall (containing laboratories, offices, and a small conventional rodent housing facility), the Leidy Laboratories of Biology (containing laboratories, offices, and conventional mouse housing facility in 2005, decommissioned in 2007), and the Hill Pavilion (containing laboratories, offices, veterinary school library, and a barrier-maintained mouse vivarium shared by the Schools of Medicine and Veterinary Medicine). All animal trapping was done in accordance with an approved institutional animal care and use protocol.
Figure 1.
Map of the University of Pennsylvania (Philadelphia) campus showing locations of traps used to collect wild mice. A, convenience store; B, Rosenthal/Veterinary Hospital of the University of Pennsylvania; C, Hill Pavilion vivarium; D, Biomedical Research Building; E, Stellar–Chance Laboratories; F, High Rise Towers; G, John Morgan Building (Class of 1925 Auditorium); H, Leidy Laboratories of Biology; I, Hayden Hall.
Additional mice were donated to this project from traps placed by University of Pennsylvania Facilities (maintenance) staff, as well as other members of the university community, who reported mice in their laboratories or offices. These mice were collected between 2005 and 2007.
All University of Pennsylvania vivaria participate in an integrated pest management program that is contracted to an outside vendor. However, typically no formal pest control program exists for other areas of the buildings that contain vivaria, and investigators and other university staff regularly mention sighting mice in their laboratories and offices.
Sample collection.
Live mice were transported in cardboard containers to the University Laboratory Animal Resources necropsy area, where they were given subcutaneous fluids (a balanced electrolyte solution) if dehydration was noted. They then were euthanized in a CO2 chamber. All animals were handled according to approved protocols.
Samples were collected at necropsy immediately after euthanasia. Blood was collected through cardiocentesis and refrigerated, and serum was collected after centrifugation. The resulting serum was diluted 1:10 in sterile PBS and frozen at –80 °C for shipping to the University of Missouri Research Animal Diagnostic Laboratory (Columbia, MO) where serologic testing was performed. Fresh feces (or colonic fecal pellets) were collected and frozen at –80 °C until processing for PCR which was also performed by the University of Missouri Research Animal Diagnostic Laboratory. In addition, fur samples, ‘plucks’ from the dorsal neck, and perianal tapes were taken for evaluation for external parasites and pinworm eggs, respectively. Cecal and colonic contents were collected and examined for pinworm larvae and adults. After sample collection, complete necropsies were performed. The following major organs were trimmed for histopathology, sectioned at 5 µm, and stained with hematoxylin and eosin: salivary glands (sublingual, submandibular, parotid, and exorbital lacrimal), heart, thymus, lungs, pancreas, spleen, liver, kidneys, adrenal glands, stomach, cecum, and the entire remaining gastrointestinal tract (which was processed as a ‘Swiss roll’).6 Slides were evaluated microscopically, and digital photographs of lesions and parasites were taken.
Sample analysis.
Serum samples were tested for antibody to the following agents using a multiplex fluorometric immunoassay (MFIA): M. pulmonis, cilia-associated respiratory bacillus, E. cuniculi, Clostridium piliforme (causative agent of Tyzzer disease), ectromelia virus (causative agent of mousepox), EDIM virus, TMEV, lymphocytic choriomeningitis virus, MAV1, MAV2, MCMV, MHV, MVM, murine norovirus (MNV; test performed only on samples collected in 2007), MPV (NS1 protein as antigen), polyoma virus, pneumonia virus of mice, reovirus type 3, and Sendai virus. Samples yielding positive or indeterminate results were retested by the indirect immunofluorescent assay.
Fecal and colonic samples were tested by PCR or RT-PCR to detect C. piliforme, Helicobacter spp., MPV, MVM, EDIM virus, and MHV.
Results
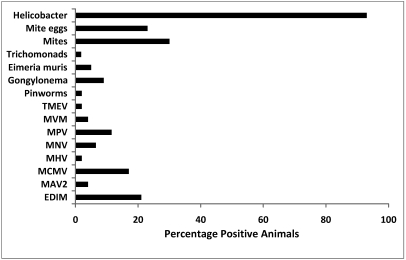
No serologic evidence of M. pulmonis, cilia-associated respiratory bacillus, E. cuniculi, C. piliforme, ectromelia virus, LCMV, MAV1, polyoma virus, pneumonia virus of mice, reovirus type 3, or Sendai virus was detected in any serum sample, and all PCR tests for C. piliforme, EDIM virus, and MVM were negative. Positive results are summarized in Tables 1 to 4 and Figure 2. Of the 52 sera for which serology results were obtained, 21% were positive for EDIM, 17% for MCMV, and 12% for MPV, and 2% to 5% were positive for MAV 2, MHV, MVM, or TMEV (Table 1; Figure 2). Serology for MNV was not performed on samples collected prior to 2007; of the 31 mice tested, 2 (6.5%) were positive for MNV. According to serologic results, 15 mice had been infected with only 1 virus, of which EDIM virus, MCMV, and MPV accounted for 12 (Table 2). Another 7 mice had been infected with 2 or more viral agents, and all of these mice were seropositive for EDIM virus (Table 2). The feces of 2 mice were RT-PCR-positive for MHV (1 of these mice was also seropositive), and feces from 2 mice were PCR-positive for MPV (both animals were MPV-seropositive; Table 1). One or more species of Helicobacter were detected in feces of 93% of the mice by PCR (Table 3). H. hepaticus was the most commonly detected species, present in 59% of the mice. Only 1 of the 56 mice examined was found to have pinworms, whereas 2%, 5%, and 9% were found to have trichomonads, Eimeria spp., and Gongylonema, respectively (Table 4). Mites were found on 30% of the mice sampled, while mite eggs were found on 23% (Table 4).
Table 1.
Viruses for which positive serologic and PCR results were obtained from wild mice
| MHV |
MPV |
|||||||||
| Location | EDIM | MAV2 | MCMV | MFIA | RT-PCR | MNV | MFIA | PCR | MVM | TMEV |
| Biomedical Research Building loading dock | 0/2 | 0/2 | 0/2 | 0/2 | 0/4a | 0/1 | 1/2 | 0/4a | 0/2 | 0/2 |
| Class of 1925 | 0/1 | 0/1 | 1/1 | 0/1 | 1/1 | ND | 0/1 | 0/1 | 0/1 | 0/1 |
| Hill Pavilion | 1/4 | 0/4 | 1/4 | 0/4 | 0/4 | 1/4 | 0/4 | 0/4 | 0/4 | 0/4 |
| High Rise Tower East | 5/11 | 0/11 | 7/11 | 0/11 | 0/10 | 0/6 | 0/11 | 0/10 | 0/11 | 1/11 |
| High Rise Tower South | 1/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/5 | 0/6 | 0/6 | 0/6 | 0/6 |
| Leidy basement | 0/10 | 0/10 | 0/10 | 0/10 | 0/11 | ND | 0/10 | 0/11 | 0/10 | 0/10 |
| Stellar–Chance | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | ND | 0/1 | 0/1 | 0/1 | 0/1 |
| Rosenthal/Veterinary Hospital | 2/11 | 0/11 | 0/11 | 1/11 | 1/11 | 1/11 | 1/11 | 0/11 | 1/11 | 0/11 |
| Convenience store loading dock | 2/6 | 2/6 | 0/6 | 0/6 | 0/7 | 0/4 | 4/6 | 2/7 | 1/6 | 0/6 |
| TOTAL | 11/52 | 2/52 | 9/52 | 1/52 | 2/55 | 2/31 | 6/52 | 2/55 | 2/52 | 1/52 |
EDIM, epizootic diarrhea of infant mice virus; MAV2, mouse adenovirus type 2 (strain K87); MCMV, murine cytomegalovirus; MFIA, multiplex fluorescent immunoassay; MHV, mouse hepatitis virus; MNV, murine norovirus; MPV, mouse parvovirus; MVM, minute virus of mice; TMEV, Theiler mouse encephalomyelitis virus
Building locations are defined in the legend for Figure 1.
Data are given as number of virus-positive samples/total number of samples.
No serologic evidence of M. pulmonis, cilia-associated respiratory bacillus, E. cuniculi, C. piliforme, ectromelia virus, lymphocytic choriomeningitis virus, polyoma virus, pneumonia virus of mice, reovirus type 3, or Sendai virus was detected. PCR or RT-PCR for viral nucleic acid in fecal samples was uniformly negative for EDIM virus, MVM, reovirus type 3, and TMEV.
Fecal pellets from 4 additional mice trapped in laboratories and offices in the BRB were RT-PCR-negative for MHV and PCR-negative for MPV.
Table 2.
Occurrence of single versus mixed viral infections based on serologic testing of wild mice.
| No. of mice | EDIM | MAV2 | MCMV | MHV | MNV | MPV | MVM | TMEV |
| 4 | X | |||||||
| 5 | X | |||||||
| 1 | X | |||||||
| 1 | X | |||||||
| 3 | X | |||||||
| 1 | X | |||||||
| 4 | X | X | ||||||
| 1 | X | X | X | |||||
| 1 | X | X | X | X | ||||
| 1 | X | X | X | X |
Table 3.
Helicobacter PCR results for fecal samples from wild mice
| Location | H. hepaticus | H. typhlonius | H. rodentium | Helicobacter spp. | All Helicobacter |
| Biomedical Research Building | 1/4 | 0/4 | 0/4 | 3/4 | 4/4 |
| Biomedical Research Building loading dock | 0/4 | 0/4 | 3/4 | 0/4 | 3/4 |
| Class of 1925 | 0/1 | 1/1 | 0/1 | 0/1 | 1/1 |
| Hill Pavilion | 1/4 | 0/4 | 3/4 | 1/4 | 4/4 |
| High Rise Tower East | 5/10 | 7/10 | 0/10 | 0/10 | 9/10 |
| High Rise Tower South | 3/6 | 5/6 | 0/6 | 0/6 | 6/6 |
| Leidy basement | 10/11 | 0/11 | 0/11 | 0/11 | 10/11 |
| Stellar–Chance | 1/1 | 0/1 | 0/1 | 0/1 | 1/1 |
| Rosenthal/Veterinary Hospital | 9/11 | 0/11 | 0/11 | 1/11 | 10/11 |
| Convenience store loading dock | 5/7 | 4/7 | 0/7 | 0/7 | 7/7 |
| TOTAL | 35/59 | 17/59 | 6/59 | 5/59 | 55/59 |
Data are given as number of Helicobacter-positive samples/total number of samples.
Fecal samples also were evaluated by PCR for C. piliforme and Salmonella spp. and were negative.
Table 4.
Parasites found microscopically in and on wild mice
| Location | Pinworms | Gongylonema spp. | Eimeria spp. | Trichomonads | Myobia | Myocoptes | Radfordia | All mites | Mite eggs |
| Biomedical Research Building | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
| Biomedical Research Building loading dock | 0/3 | 0/3 | 0/3 | 0/3 | 2/3 | 0/3 | 0/3 | 2/3 | 1/3 |
| Class of 1925 | 0/1 | 1/1 | 0/1 | 0/1 | 1/1 | 0/1 | 0/1 | 1/1 | 1/1 |
| Hayden basement | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 1/1 | 1/1 | 1/1 |
| Hill Pavilion | 0/4 | 0/4 | 0/4 | 0/4 | 2/4 | 0/4 | 0/4 | 2/4 | 2/4 |
| High Rise Tower East | 0/11 | 3/11 | 3/11 | 1/11 | 2/11 | 0/11 | 2/11 | 4/11 | 4/11 |
| High Rise Tower South | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| Leidy basement | 0/11 | 0/11 | 0/11 | 0/11 | 0/11 | 3/11 | 0/11 | 3/11 | 1/11 |
| Stellar–Chance | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Rosenthal/Veterinary Hospital | 0/6 | 0/6 | 0/6 | 0/6 | 1/6 | 0/6 | 0/6 | 1/6 | 1/6 |
| Convenience store loading dock | 1/7 | 0/7 | 0/7 | 0/7 | 2/7 | 1/7 | 0/7 | 3/7 | 2/7 |
| TOTAL | 1/55 | 5/55 | 3/55 | 1/55 | 10/55 | 4/55 | 3/55 | 17/55 | 13/55 |
Data are given as number of parasite-positive samples/total number of samples.
Figure 2.
Overall summary of infectious diseases detected in wild mice trapped on the University of Pennsylvania (Philadelphia) campus, 2005 through 2007. TMEV, Theiler mouse encephalomyelitis virus; MVM, minute virus of mice; MPV, mouse parvovirus; MNV, murine norovirus; MHV, mouse hepatitis virus; MCMV, murine cytomegalovirus; MAV2, mouse adenovirus type 2 (strain K87); EDIM, epizootic diarrhea of infant mice virus.
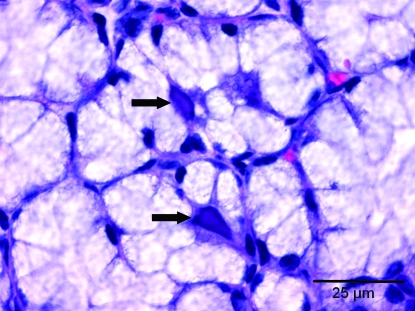
Of the 9 mice that were seropositive for MCMV, salivary gland tissue was available from 5 mice. Microscopically, characteristic intranuclear inclusion bodies were present in 4 of 5 submandibular salivary glands, 1 of 5 sublingual glands, 1 of 5 parotid glands, and none of the exorbital lacrimal glands. The inclusions were found in secretory mucous and serous cells and possibly demilunar cells (Figure 3) but not in ductal cells in any of the glands. When observed in the submandibular and sublingual glands, inclusions were plentiful, but inclusion bodies were only rarely seen in the parotid gland. Occasionally, a few small intracytoplasmic inclusion bodies seemed to accompany the intranuclear inclusions in some submandibular glands. No histopathologic changes characteristic of infection were seen in tissues examined from the 11 animals seropositive for EDIM virus, the 2 mice seropositive for MAV2, or the single MHV-seropositive animal. None of the mice had histopathologic changes (mucosal thickening and accompanying inflammation) characteristic of Helicobacter spp.
Figure 3.
Section of sublingual salivary gland. Two large intranuclear inclusion bodies characteristic of murine cytomegalovirus infection are present in acinar secretory cells (arrows).
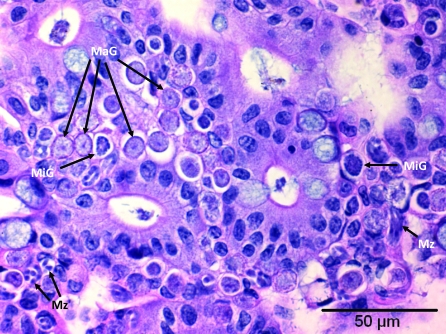
Three mice trapped at 1 location had moderate infections in their large intestines with a protozoan morphologically compatible histologically with Eimeria spp. (Figure 4).
Figure 4.
Section of large intestine. Epithelial cells and underlying lamina propria contain developmental stages of Eimeria spp.: multiple macrogamonts (MaG), several microgamonts (MiG), and merozoites (Mz).
Nematode parasites morphologically compatible with Gongylonema spp. were found in the gastrointestinal tracts of 5 of 56 mice trapped in 3 locations (Figure 5). In the stomach, the parasites produced marked hyperplasia of the stratified squamous epithelium of the nonglandular stomach with accompanying transmural acute to chronic inflammation.
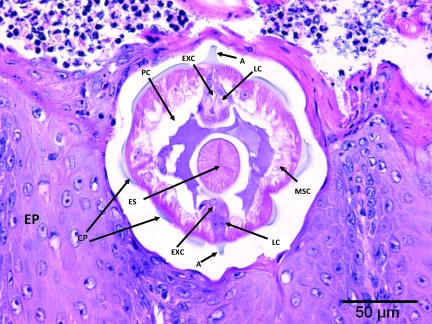
Figure 5.
Section of nonglandular stomach. Within the stratified squamous epithelium (EP) is a cross section of Gongylonema spp. Present are: cuticular plaques (CP), lateral alae (A), hypodermal lateral chords (LC), lateral excretory canals (EXC), coelomyarian musculature (MSC), muscular esophagus (ES), and pseudocoelom (PC).
Discussion
Included in this study are results from a pilot done during the summer of 2005, in which 28 wild mice trapped on or near the University of Pennsylvania (Philadelphia) campus were examined for 19 infectious agents, including the viruses and bacteria listed earlier, as well as many parasites and other bacteria. Results from the pilot study and the current follow-up study were consistent so it seemed appropriate to pool the data.
In 1994, Helicobacter hepaticus was first discovered and shown to be a complicating factor in laboratory mice, after unusual findings in control mice from a hepatic tumor study.10 This species of Helicobacter was detected by PCR in fecal pellets from 59% of the mice in the current survey of University of Pennsylvania campus wild mice. Other species of Helicobacter, many as yet uncharacterized in laboratory mice,2 were found also, with a total of 55 of 59 (93%) tested mice yielding positive PCR test results. Helicobacter spp. were reported by approximately 80% of respondents to a recent survey on the occurrence of infectious diseases in contemporary rodent colonies.3 Historically, no effort has been made to exclude Helicobacter spp. from University of Pennsylvania vivaria. The most recent testing of sentinel mice in barrier-maintained colonies at the University of Pennsylvania revealed that 45% to 82% of sentinels were PCR-positive for the Helicobacter genus, with the majority of those being positive for H. hepaticus. Despite the high proportion of wild mice that had PCR evidence of Helicobacter infection, lesions compatible with Helicobacter were not observed in this study.
Some of the wild mice trapped in this study were found in close proximity to rodent vivaria (in the same buildings, but not within the colonies), so there was some concern that wild mice might have contributed to disease outbreaks in these colonies. In particular, mice in the University of Pennsylvania vivaria, particularly those in the School of Medicine, have sustained multiple pinworm outbreaks over the past 5 y. Likewise, in a recent questionnaire-based nationwide survey of infectious diseases in laboratory rodent colonies in the United States, nearly 80% of respondents reported infections with pinworms.3 In contrast, only 1 of the 56 mice that we examined in the University City district of Philadelphia had detectable pinworms. In addition, nearly 100% of respondents reported colonies that were seropositive to MPV, and between 30% and 40% reported EDIM virus and MHV in their laboratory mouse colonies.3 In the current study, 12% of the wild mice were MPV-seropositive, 21% were seropositive for EDIM virus, and 2% were MHV-seropositive. In addition, one-third of the mice that had serologic evidence of prior viral infection had sustained 2 or more infections, and all of those mice were seropositive for EDIM virus. The University of Pennsylvania has not detected MPV in sentinels from barrier-maintained colonies during the 6 y for which reliable data have been available. This status has been confirmed during fourth quarter testing of mesenteric lymph nodes by PCR in 2007 and 2008. MPV has been detected sporadically in sentinels monitoring conventionally housed mice in vivaria with high population turnover rates. The 2 viral infections that have caused epizootics on campus in the last 5 y, twice in combination, are EDIM virus and MHV. The sources of the EDIM virus outbreaks were not definitively established. Three MHV outbreaks could be attributed to (1) infected animals received from an approved vendor, (2) mice from a nonapproved source that bypassed quarantine and were introduced directly into a colony, and (3) special diet stored improperly in a laboratory. In the last case, the plastic bag containing the diet was chewed, and rodent feces were clearly visible on the floor near the bag; MHV RNA was amplified from fecal pellets collected near the bag. Therefore, it appeared that wild or feral mice that had access to improperly stored food that was subsequently dispensed in the animal facility may have, in fact, been responsible for one of the larger viral disease outbreaks we have sustained.
The seroprevalence of MCMV was 17% in the wild mice; to our knowledge, this agent is no longer found in laboratory mice in most areas of the world8 and is confined to wild mice, sometimes at extremely high prevalence.1,8,9 In addition, we noted MCMV-associated intranuclear inclusions in sublingual salivary glands, a finding not published in standard texts and previously reported only once.7 Unlike the other viral agents detected serologically in the current study, MCMV is transmitted in salivary secretions.8 This mechanism presumably requires close contact, a requirement supported by the fact that in Australia, where mouse ‘plagues’ occur, MCMV seroprevalence is very high (approaching 100%) during plagues and considerably lower (20% to 30%) during interplague years.8,9 Interestingly, 7 of the 9 MCMV-seropositive mice were trapped at a single site, 1 of the undergraduate dormitories. The other point that this finding reinforces, which also is supported by genetic studies,5 is that populations of mice have limited ranges of movement and that they are subdivided into fairly isolated groups; therefore, some murine infectious diseases may be isolated within groups. Other agents seen in the wild mice but not frequently in contemporary laboratory mouse colonies were Gongylonema spp. and Eimeria spp.
The presence of ectoparasites (mites) seemed to be roughly equivalent in both the wild mice that we surveyed (30%) and the colony mice reported through the previous questionnaire (20% to 40%).3 Given the greater incidence of endoparasites in laboratory mouse colonies on the University of Pennsylvania campus, our results suggest that wild mice are not a significant contributing factor to disease outbreaks, particularly pinworm outbreaks, in the rodent vivaria on campus. Their insignificant role is bolstered by the fact that MPV is present only in a small proportion of the conventionally maintained facilities on campus (and in none of the barrier-maintained facilities). The role of wild mice in EDIM virus outbreaks has not been determined. A single MHV outbreak may have been caused indirectly by the presence of wild or feral mice in the laboratory setting. The literature supports introduction of both viruses into research facilities by similar means.4
Acknowledgments
We thank Ms Raina Rigoli-Magyar for preparing and submitting the serum and fecal samples for laboratory analysis, Dr Thomas Nolan for his aid in identifying endoparasites, and Dr James Lok for his help with identifying ectoparasites.
References
- 1.Barthold SW, Smith AL.1988. Unpublished data.
- 2.Besselsen DG, Franklin CL, Livingston RS, Riley LK. 2008. Lurking in the shadows: emerging rodent infectious diseases. ILAR J 49:277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carty AJ. 2008. Opportunistic infections of mice and rats: Jacoby and Lindsey revisited. ILAR J 49:272–276 [DOI] [PubMed] [Google Scholar]
- 4.Clifford CB, Watson J. 2008. Old enemies: still with us after all these years. ILAR J 49:291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein J, Bailey DW. 1971. Histocompatibility differences in wild mice: further evidence for the existence of deme structure in natural populations of the house mouse. Genetics 68:287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moolenbeek C, Ruitenberg EJ. 1981. The ‘Swiss roll’: a simple technique for histological studies of the rodent intestine. Lab Anim 15:57–59 [DOI] [PubMed] [Google Scholar]
- 7.Ruebner BH, Hirano T, Slusser R, Osborn J, Medearis DN. 1966. Cytomegalovirus infection. Viral ultrastructure with particular reference to the relationship of lysosomes to cytoplasmic inclusions. Am J Pathol 48:971–989 [PMC free article] [PubMed] [Google Scholar]
- 8.Shellam GR, Redwood AJ, Smith LM, Gorman S. 2007. Murine cytomegalovirus and other herpesviruses, p 1–48. : Fox JG, Barthold SW, Davisson MT, Newcomer CE, Quimby FW, Smith AL. The mouse in biomedical research, 2nd ed. Volume II. Diseases. New York (NY): Elsevier [Google Scholar]
- 9.Smith AL, Singleton GR, Hansen GM, Shellam G. 1993. A serologic survey for viruses and Mycoplasma pulmonis among wild house mice (Mus domesticus) in southeastern Australia. J Wildl Dis 29:219–229 [DOI] [PubMed] [Google Scholar]
- 10.Ward JM, Fox JG, Anver MR, Haines DC, George CV, Collins MJ, Jr, Gorelick PL, Nagashima K, Gonda MA, Gilden RV, Tully JG, Russell RJ, Benveniste RE, Paster BJ, Dewhirst FE, Donovan JC, Anderson LM, Rice JM. 1994. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst 86:1222–1227 [DOI] [PubMed] [Google Scholar]