Abstract
We have developed a method to determine the degree of phosphorylation of a peptide in a complex mixture without enrichment or operation of the mass spectrometer in negative ion mode. Yeast lysate containing known amounts of synthetic peptides (VPQLEIVPNSAEERLHSMK and VPQLEIVPN[pS]AEERLHSMK) was labeled with 16O and 18O during hydrolysis. After treatment of one sample with a cocktail of phosphatases, the two samples were pooled. The intensity of the dephosphorylated peptide peaks was used to infer the degree of phosphorylation present before treatment. The linear dynamic range of this method is >10-fold before either of the peptide envelopes becomes indistinguishable from the surrounding noise. Since both the site of posttranslational modification and the proportion of the protein population that is modified are vital in protein function, the employment of this technique will provide a valuable tool for the analysis of the functional implications of protein phosphorylation.
Keywords: mass spectrometry, peptide ratio, isotopic labeling
Posttranslational modification (PTM) of proteins confers control over function, activity, location, and structural stability. The elucidation of PTMs has become increasingly important to understand interactions between cellular components. Commonly studied modifications include acetylation, glycosylation, and phosphorylation; however, the ability to study these modifications in depth is often limited by the technology available and the nature of the PTM of interest (11).
Phosphorylation is one of the most common modifications, affecting up to 30% of the proteome (3). This reversible modification can occur at multiple sites along a single polypeptide at serine, threonine, and tyrosine residues and modulates protein function and activity. The phosphorylation event can cause a conformational change in the protein that regulates its activity, provide a binding site for another protein or cation, determine the subcellular localization of the target protein, or affect the rate of degradation of the protein (3, 15). Since phosphorylation events are often transient, the actual amount of phosphorylated protein at any one moment can be relatively low, exist at different sites along the same protein, or be lost because of phosphatase activity during the sample preparation (3, 11, 12, 15). Present methods of analysis of phosphorylation have focused on the identification of the position of the phosphate group and the elucidation of its importance in relation to biological activity.
Identification of phosphorylated proteins from two-dimensional gel-based systems has been successfully achieved using phosphospecific stains, 32P, and antibodies against phosphorylated residues (14). The ability to specifically stain phospho-proteins separated by gel electrophoresis helps to simplify the complexity of the sample by identifying an initial target. Increasingly, methods are being developed to study phosphorylation indirectly. Chemical modification of the phosphoserine (pS) or phosphothreonine (pT) residues using β-elimination to generate aminoethyl cysteine and β-methylaminoethyl cysteine, respectively, introduces a tryptic site at the position of the phosphorylated residue. Mapping of these sites is then achieved by mass spectrometry without many of the problems usually associated with phosphopeptides (2, 8).
The use of mass spectrometry to study phosphorylated peptides can be problematic, since phosphorylated peptides do not ionize efficiently because of the presence of the anionic phosphate group and also as a result of suppression effects from other peptides. Often the least stable modifications are lost in the ionization process, so only a population of unmodified peptides is detected. To overcome this problem, a method has recently been proposed that utilizes alkaline phosphatase to remove the phosphate group from digested protein, and the peptide mass fingerprint collected by matrix-assisted laser desorption/ionization time of flight (MALDI-ToF) is compared with an untreated sample, thus identifying peptides containing phosphorylated residues. Neither the position of the modification nor quantification of the modification can be determined by this method (10). Differential labeling of peptides using isotopic labels, 15N, 2H, and 18O, has been used to determine ratios of labeled to unlabeled peptide to demonstrate up- or downregulation of the parent protein (4, 6, 9, 16).
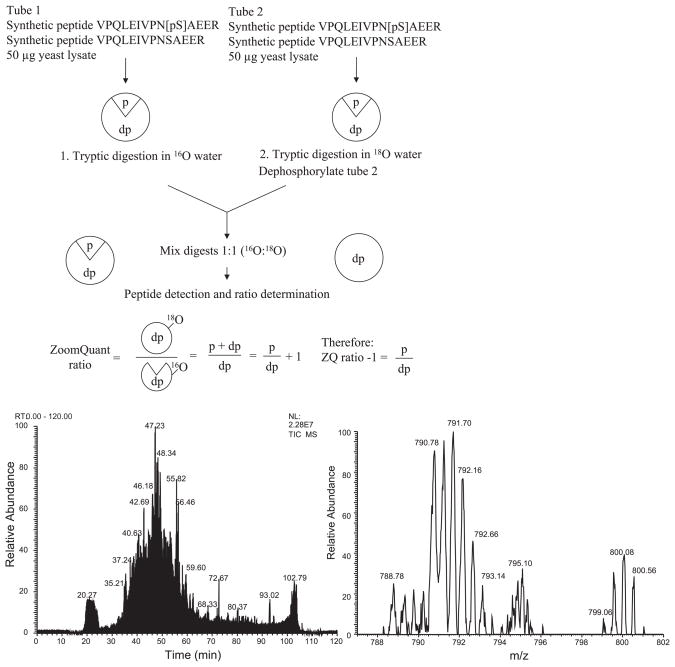
We present here a new method to indirectly determine not only the position but also the degree of phosphorylation of a peptide using a combination of isotopic labeling strategies (18O) and liquid chromatography separation, followed by mass spectrometry (LC MS/MS) as summarized in Fig. 1. The advantage of this method is that, since only the unphosphorylated peptide version of the peptide is measured, any ion suppression due to the phosphate group has been eliminated. The amount of phosphorylation present can be quantified by comparison of the treated with the untreated sample.
Fig. 1.
Schematic of methodology. Two tubes containing yeast lysate (to provide a complex mixture of proteins) and synthetic peptide are prepared. Each tube contains the same amount of yeast protein and both versions of the synthetic peptides. The amount of one of the peptides is varied to generate the different ratios. Tryptic digestion is performed in the presence of 16O or 18O; one sample is treated with phosphorylase before the samples are mixed in a 1:1 ratio and introduced to the mass spectrometer. Bottom left: total ion chromatogram. Bottom right: zoom scan of peptide VPQLEIVPNSAEER (2+ ion) where the ratio = 0.6; peak 790.78 contains 2× 16O molecules, 791.70 contains 1× 16O and 1× 18O molecule, and 792.66 contains 2× 18O molecules. This peptide eluted at 42.33 min in the chromatogram.
MATERIALS AND METHODS
Reagents
Yeast lysate was a gift from Dr. Robert J. Deschenes (Medical College of Wisconsin, Milwaukee, WI). Dithiothreitol, iodoacetamide, ethanol, and methanol were purchased from Sigma-Aldrich (St. Louis, MO); sequencing-grade trypsin was obtained from Fisher (Hanover Park, IL). Ammonium hydrogen carbonate was obtained from Fluka BioChemica (Sigma-Aldrich). The synthetic peptides VPQLEIVPNSAEERLHSMK and VPQLEIVPN[pS]AEERLHSMK were generated by the Biotechnology Center at the University of Wisconsin (Madison, WI). Calf intestinal alkaline phosphatase (CIAP) (product no. EF0341) was purchased from Fermentas Life Sciences (Hanover, MD), lambda phosphatase (λPP) and associated buffers were obtained from Upstate Cell Signaling Solutions (Lake Placid, NY), and potato acid phosphatase type II (P3752) was purchased from Sigma-Aldrich. Water containing ≥95% atom excess H218O was purchased from Cambridge Isotope Laboratories (Andover, MA). POROS material was obtained from PerSeptive Biosystems (Framingham, MA). BCA microprotein assay kit was purchased from Pierce (Rockford, IL). C18 desalting tips were purchased from Varian (Lake Forest, CA). Formic acid was obtained from EMD Chemicals, Fisher (Hanover Park, IL), and all other MS solvents were purchased from Burdick and Jackson (Muskegon, MI).
Digestion and O18 labeling of proteins and peptides
Yeast lysate (100 μg) spiked with known ratios of the synthetic peptides VPQLEIVPNSAEER and VPQLEIVPN[pS]AEER (2.5–50 μg) was reduced using a 10 mM solution of dithiothreitol incubated at 37°C for 30 min; the thiol groups were then blocked by the addition of 20 mM iodoacetamide, and incubation was continued for an additional 30 min at 37°C in the dark. The protein was precipitated by 9 vol of ice-cold acetone for 1 h on ice; the precipitate was pelleted by centrifugation at 12,000 g for 10 min at 4°C. The supernatant was removed and discarded, and the pellet was dried under vacuum. The dried protein pellet was suspended in 5 μl of 200 mM ammonium bicarbonate and again reduced to dryness. The proteins were suspended in ≥95% H218O or H216O to a final concentration of 50 mM ammonium bicarbonate and sonicated for 2 min to aid solubilization. Trypsin was added to this at a ratio of 1:50 (enzyme/target protein), and the digestion was allowed to continue for 18 h at 37°C.
Dephosphorylation of the peptide mixture
After overnight incubation and digestion, a cocktail of phosphatases was added to the sample to be dephosphorylated (0.1 U of CIAP: stock provided as 1 U/μl, 0.1 μg of λPP I, and 1 μg of potato acid phosphatase type II) in 1× λPP buffer and left at 37°C. After 1 h, 0.1% (vol/vol) formic acid was added, and the 16O and 18O samples were pooled before desalting.
Desalting and mixing samples for LC-MS analysis
The 16O and 18O pooled samples were desalted on a C18 tip. The tips were prepared for use per the manufacturer’s instructions. Peptides were loaded onto the column and washed in 10× column volume of 0.1% (vol/vol) formic acid. Peptides were eluted by 10× column volume of solvent B [95% (vol/vol) acetonitrile, 0.1% (vol/vol) formic acid]. The eluent was reduced to dryness in a speedvac, and the peptides were resuspended in 27 μl of solvent A [5% (vol/vol) acetonitrile, 0.1% (vol/vol) formic acid].
MS
MALDI-ToF spectra were collected on an Applied Biosystems Voyager-DE PRO instrument. Samples were mixed 1:1 with a saturated solution of α-cyano-4-hydroxycinnamic acid matrix, 50% (vol/vol) acetonitrile, and 0.1% (vol/vol) formic acid; spotted onto a 100-well MALDI target; and allowed to air dry before analysis.
LC MS/MS mass spectrometric data were collected on either a ThermoFinnigan Deca Xp Plus (LCQ) instrument with a method utilizing the triple-play data-dependant mode (6) or an LTQ instrument. Peptides were loaded onto a 10-cm C18 column packed with a Magic C18AQ 200-Å, 5-μm (Michrom BioResources, Auburn, CA) stationary phase using a Surveyor 2 autosampler and a Surveyor pump delivering two mobile phase solvents, solvents A and B, at 200 μl/min through a 1:200 split to a 10-μl injection loop. A 10-μl “no waste” injection was made in solvent A and the sample eluted from the column by introduction of the following solvent B gradient: 0–25 min, 0% solvent B; 25–30 min, 5% solvent B; 30–140 min, 35% solvent B; 140–210 min, 65% solvent B; 210–220 min, 100% solvent B; 220–225 min, 100% solvent B; 225–230 min, 0% solvent B; 230–240 min, 0% solvent B. The column was left to equilibrate in 100% solvent A before the data collection was terminated. With the use of the LTQ instrument, the sample was eluted from the column using the following gradient: 0–40 min, 30% solvent B; 40–60 min, 65% solvent B; 60–70 min, 100% solvent B; 70–80 min, 100% solvent B; 80–85 min, 0% solvent B; 85–120 min, 0% solvent B. The LCQ instrument settings were as follows: default charge state 2, default isolation width 2, normalization collision energy 38%, minimum MSn signal required 1.5 × 107, exclusion mass width 1.5 (low and high), reject mass width 0.5 (low and high), exclusion duration 3 s, and zoom scan width by mass low 5.0, high 10.00. Tuning parameters were as follows: no sheath or sweep gas and automatic gain control was “on.” For the LTQ, the only parameters that differed were normalization collision energy 35%, minimum MSn signal required 500, and exclusion duration 180 s.
Synthetic peptides for quantification
The synthetic peptide used in this experiment was selected with the following criteria: the peptide was previously reported in an LC-MS experiment using a ThermoFinnigan Deca Xp Plus instrument, contained a naturally phosphorylated residue, and did not match to a rat, human, or yeast sequence.
RESULTS
Dephosphorylation of synthetic peptide “VPQLEIVPN[pS] AEER”
The success of this method depends on the ability of phosphatase to remove 100% of the phosphate groups from the modified peptide. We evaluated the reliability of enzymatic dephosphorylation using a phosphatase cocktail and 18O labeling (Fig. 2). Samples containing equal amounts of the synthetic phosphopeptide were digested in the presence of 16O or 18O; these were mixed in a ratio of 1:1 and detected using a MALDI-ToF instrument. In Fig. 2A, peptide VPQLEIVPN[pS] AEER is evident at 1,660.9226 Da and the 18O-labeled peptide at 1,664.9281 Da; the 4-Da mass shift corresponds to the incorporation of two 18O molecules. The 16O sample was treated with a cocktail containing λ-protein phosphatase, calf alkaline phosphatase, and potato acid phosphatase type II and incubated at 37°C for 1 h. The dephosphorylated 16O sample was mixed 1:1 with an equal amount of the 18O sample for analysis. In Fig. 2B, the removal of the phosphate is demonstrated, as no peptide is evident at 1,660.9226 Da, while the 18O peptide remains present at 1,665.0159 Da. Figure 3 demonstrates the data that can be collected by this method and shows peptides pairs that represent three different ratios. Figure 3A shows a peptide for which the measured ratio was 0.97; equal amounts of the peptide being present in the labeled and unlabeled sample can be compared with Fig. 3, B and C. In Fig. 3B, the measured ratio is 0.55; there is more of the peptide present in the labeled sample, whereas the converse is true in Fig. 3C, where the measured ratio is 2.3.
Fig. 2.
Dephosphorylation of synthetic peptide “VPQLEIVPN[pS]AEER.” Matrix-assisted laser desorption/ionization (MALDI) spectra, as follows. A: showing peptide VPQLEIVPN[pS]AEER evident at mass 1,660.717 Da and no peak at mass 1,580.776 Da, corresponding to peptide VPQLEIVPNSAEER. B: after treatment with the phosphatase cocktail, the appearance of the dephosphorylated peptide VPQLEIVPNSAEER is evident at 1,580.77 Da. C: peptide VPQLEIVPN[pS]AEER generated by hydrolysis in the presence or absence of 18O generates peptides of mass 1,664.92 and 1,660.92 Da, respectively, mixed in a 1:1 ratio, resulting in a “peptide pair.” D: dephosphorylation of the 16O sample before mixing results in the complete removal of the phosphate group, leaving only the 18O peptide at 1,665.01 Da and no corresponding 16O peak.
Fig. 3.
Zoom scans of dephosphorylated synthetic peptide VPQLEIVPN[pS]AEER. A: 2+ ion of VPQLEIVPNSAEER mixed at a ratio of 1.0; ratio was measured as 0.97 (18Ointensity/16Ointensity or untreated/treated). B: 2+ ion of VPQLEIVPNSAEER after treatment with the phosphatase cocktail, expected ratio 0.5; observed ratio was 0.55. C: 2+ ion of VPQLEIVPNSAEER after treatment with the phosphatase cocktail, expected ratio 2.0; observed ratio was 2.3.
Dynamic range of methodology
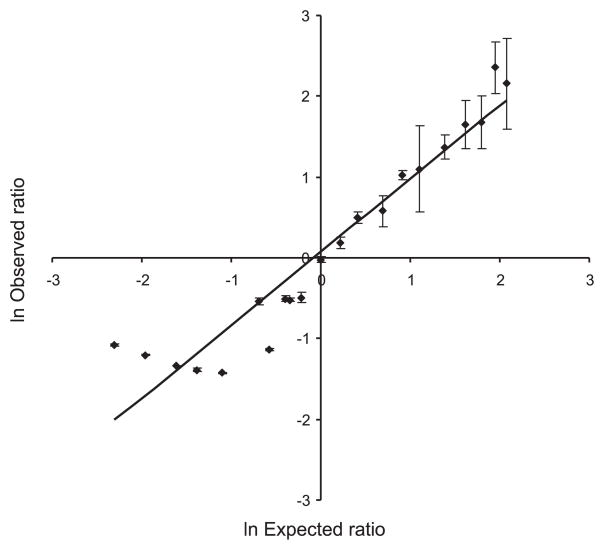
We evaluated the sensitivity of this method to ensure that we could detect the peptide at physiologically relevant levels. We tested a range from 2 μg (1.27 nmol) to 2 fg (1.27 amol), and we were able to detect the labeled peptide pair. The expected ratio was 0.5, and our measured ratio was 0.6 ± 0.1. To determine the dynamic range of this method, we spiked known ratios of the synthetic peptide ± phosphate into a complex mixture of proteins (soluble fraction of yeast lysate) and treated either the 16O or the 18O sample with the phosphatase cocktail before desalting and analysis by LC MS/MS. Zoom scans of the precursor ion were collected before fragmentation to quantify the peptide present in each sample using the ZoomQuant software (5, 6). The dynamic range of this technique is summarized in Fig. 4. The addition of the 16O dephosphorylation cocktail to the 18O sample was initially of concern because of potential back exchange of water, however, over the 1-h incubation and LC separation before MS detection. We tested peptide ratios as low as 0.3 and up to 8.0 to determine the applicable range using forward and reverse labeling. As the intensity of the 18O peak increased and the 16O envelope decreased, the errors encountered within the calculation became large, and the measured ratio remained constant despite the experimental ratio. As shown in Fig. 4, a dynamic range of >10-fold was achievable before the 16O peptide envelope became engulfed in the surrounding noise. The inability to distinguish the 18O peaks from the 16O envelope became a problem at the other end of the tested range of ratios, evident in Fig. 4, as many of the ratios measured were ~0.3. We have found that, in the absence of any 18O label, a peptide will generate a ratio of 0.2–0.3. The linear relationship between the predicted and observed ratios summarized in Fig. 4 confirms that the dephosphorylation of the peptide results from phosphatase activity and not from an effect incurred in the mass spectrometer, such as the loss of a phosphate group from the peptide because of the ionization effect within the mass spectrometer; this would affect both samples and be evident in the calculated ratio.
Fig. 4.
Synthetic peptides VPQLEIVPN[pS]AEER and VPQLEIVPNSAEER spiked into yeast lysate in known ratios. The 16O or the 18O sample was treated with a phosphatase cocktail, and the intensity of peptide VPQLEIVPNSAEER (both 2+ and 1+ ions included in the calculation) in the treated sample was compared with the intensity of the same peptide in the untreated sample; the increase in intensity results from the amount of phosphopeptide that was dephosphorylated. The dynamic range for the measured ratio using this technique is from 0.5 to 5, calculated from the intensity of the dephosphorylated peptide (18Ointensity/16Ointensity). Standard errors are shown for each point. The linear correlation coefficient is 0.92.
To validate our method in a real cell lysate, we interrogated the yeast lysate used as a background medium for our synthetic peptide. On examination of the data, we quickly found two peptides from superoxide dismutase (SODC_YEAST) that are reported to be phosphorylated. The ratios measured for the peptides FEQASESEPTTVSYEIAGNSPNER and LIGPTSVVGR were 0.76 ± 0.09 and 0.72 ± 0.02, respectively. A nonphosphorylated peptide from this protein (SVVIHAGQDDLGK) was measured at a ratio of 0.96 ± 0.01.
DISCUSSION
We have demonstrated that it is possible to determine the degree of phosphorylation of a peptide present in a complex mixture using a combination of 18O labeling, dephosphorylation, and detection by MS. In a population of peptides generated from intracellular proteins, the abundance of phosphopeptides will be comparatively low, since only 30% of proteins may be phosphorylated and not all of these are at multiple sites.
Although there are many methods presently available for the study of phosphopeptides, they suffer from a variety of problems. Isolation of the modified peptide from a complex mixture of peptides is commonly achieved using immobilized metal affinity chromatography (IMAC) (2, 4, 13). A simplified population enriched in phosphopeptides is generated, and the identity of each peptide and location of the phosphate group are then determined by MS. Methods relying on complex chemical reactions aimed at modifying the phosphoserine or phosphothreonine residues may be expensive and time consuming and involve precise chemical reactions. None of these methods allows quantification of the overall degree of phosphorylation. There have been several attempts to improve methods by methylation of the peptides before their introduction to the IMAC column and alternative elution approaches using a phosphatase (1, 2). Both of these methods decrease the proportion of nonphosphorylated peptides in the final population, either by increasing the selectivity of binding to the IMAC column or by elution due to the presence of a phosphate group. However, such approaches introduce a bias toward phosphopeptides that bind efficiently to the column and eliminate those that do not.
The method we present in this paper allows us to determine the degree of phosphorylation of a peptide present in a complex mixture without enrichment for phosphopeptides; therefore, each phosphopeptide present may be measured. The procedure is cheap, labeling performed during tryptic digestion of the protein does not add time to the preparation, and dephosphorylation of the peptides is simple, adding only a short incubation postdigestion. The dephosphorylation generates nonphosphorylated ions that ionize in positive ion mode on a simple mass spectrometer with inline chromatographic separation of the peptides to simplify the mixture. The separation of the peptides by LC limits any ion suppression effects associated with competing peptides by decreasing the complexity of the sample entering the mass spectrometer.
To allow normalization of the data and to determine the dynamic range of this technique, we used a synthetic peptide with and without a phosphate group attached to the serine residue; this was added in known ratios to two protein mixtures. The ratio of the 16O/18O unmodified peptides was determined using the ZoomQuant software. This deconvolution technique developed in-house determines the ratio of the 16O to 18O peptides using peak intensity and separates the singly and doubly labeled peptide envelopes (6). We determined that the dynamic range for accurately measuring ratios is from 0.5 to 5.0 based on multiple measurements of the synthetic peptide. Quantification of IMAC-isolated phosphopeptides has demonstrated that errors for pairs of dimethylated peptides range from 0.2 to 7.5%. These peptides are derived from the same protein and are for ratios ranging from 1:1 to 1:10. The errors for ratios <5 were reported to be <10%, while the standard errors using our method were recorded as ±0.11 to ±0.02, which is equally reliable (7). When using 18O labeling, the two peptide envelopes are not completely resolved, as the mass difference is only 2 Da between the fully unlabeled and the labeled peptides for a 2+ ion; in addition, there is a subpopulation of peptides that contain a single 18O and a single 16O molecule, as seen in Fig. 1, bottom right. The ZoomQuant software resolves the spectra and computes quantitative ratios over a broad dynamic range.
This method allows determination of the degree of phosphorylation of a peptide present in a complex mixture without enrichment for phosphopeptides or the need to operate the mass spectrometer in negative ion mode. We demonstrated this by identifying two peptides from the superoxide dismutase that were present in the yeast lysate used as a background for our synthetic peptide experiments. In the case of these peptides (FEQASESEPTTVSYEIAGNSPNER and LIGPTSVVGR), we can conclude that one-third of the population of the protein was phosphorylated at the sites within these tryptic peptides. By measurement of baseline levels of phosphorylation, this method could be used to investigate the effects of different stimuli on phosphorylation events. The labeling and quantification of a synthetic peptide without prior isolation of the phosphopeptide are reproducible, and the ZoomQuant software allows accurate measurement of the ratio with a working dynamic range of measured ratios from 0.5 to 5.0. We believe this to be a powerful analysis tool for assessing the proportion of each protein that is phosphorylated in a complex mixture.
Acknowledgments
We wish to thank Brian Halligan for help with the ZoomQuant software and Daniela Didier for assistance with the statistical analyses.
GRANTS
This work was funded by National Institutes of Health (NIH/NHLBI) Grant N01-HV-28182.
References
- 1.Bonenfant D, Schmelzle T, Jacinto E, Crespo JL, Mini T, Hall MN, Jenoe P. Quantitation of changes in protein phosphorylation: a simple method based on stable isotope labeling and mass spectrometry. Proc Natl Acad Sci USA. 2003;100:880–885. doi: 10.1073/pnas.232735599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal Chem. 2004;76:2763–2772. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- 3.Cohen P. The regulation of protein function by multisite phosphorylation–a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 4.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 5.Halligan BD, Slyper RY, Twigger SN, Hicks W, Olivier M, Greene AS. ZoomQuant: an application for the quantitation of stable isotope labeled peptides. J Am Soc Mass Spectrom. 2005;16:302–306. doi: 10.1016/j.jasms.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hicks WA, Halligan BD, Slyper RY, Twigger SN, Greene AS, Olivier M. Simultaneous quantification and identification using 18O labeling with an ion trap mass spectrometer and the analysis software application “ZoomQuant. J Am Soc Mass Spectrom. 2005;16:916–925. doi: 10.1016/j.jasms.2005.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang SY, Tsai ML, Wu CJ, Hsu JL, Ho SH, Chen SH. Quantitation of protein phosphorylation in pregnant rat uteri using stable isotope dimethyl labeling coupled with IMAC. Proteomics. 2006;6:1722–1734. doi: 10.1002/pmic.200500507. [DOI] [PubMed] [Google Scholar]
- 8.Knight ZA, Schilling B, Row RH, Kenski DM, Gibson BW, Shokat KM. Phosphospecific proteolysis for mapping sites of protein phosphorylation. Nat Biotechnol. 2003;21:1047–1054. doi: 10.1038/nbt863. [DOI] [PubMed] [Google Scholar]
- 9.Krijgsveld J, Ketting RF, Mahmoudi T, Johansen J, Artal-Sanz M, Verrijzer CP, Plasterk RH, Heck AJ. Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat Biotechnol. 2003;21:927–931. doi: 10.1038/nbt848. [DOI] [PubMed] [Google Scholar]
- 10.Larsen MR, Sorensen GL, Fey SJ, Larsen PM, Roepstorff P. Phospho-proteomics: evaluation of the use of enzymatic de-phosphorylation and differential mass spectrometric peptide mass mapping for site specific phosphorylation assignment in proteins separated by gel electrophoresis. Proteomics. 2001;1:223–238. doi: 10.1002/1615-9861(200102)1:2<223::AID-PROT223>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 11.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 12.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 13.Nuhse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Patton WF. Detection technologies in proteome analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771:3–31. doi: 10.1016/s1570-0232(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 15.Pawson T, Scott JD. Protein phosphorylation in signaling–50 years and counting. Trends Biochem Sci. 2005;30:286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Stewart II, Thomson T, Figeys D. 18O labeling: a tool for proteomics. Rapid Commun Mass Spectrom. 2001;15:2456–2465. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]