Abstract
Proteomics has been proposed as one of the key technologies in the postgenomic era. So far, however, the comprehensive analysis of cellular proteomes has been a challenge because of the dynamic nature and complexity of the multitude of proteins in cells and tissues. Various approaches have been established for the analyses of proteins in a cell at a given state, and mass spectrometry (MS) has proven to be an efficient and versatile tool. MS-based proteomics approaches have significantly improved beyond the initial identification of proteins to comprehensive characterization and quantification of proteomes and their posttranslational modifications (PTMs). Despite these advances, there is still ongoing development of new technologies to profile and analyze cellular proteomes more completely and efficiently. In this review, we focus on MS-based techniques, describe basic approaches for MS-based profiling of cellular proteomes and analysis methods to identify proteins in complex mixtures, and discuss the different approaches for quantitative proteome analysis. Finally, we briefly discuss novel developments for the analysis of PTMs. Altered levels of PTM, sometimes in the absence of protein expression changes, are often linked to cellular responses and disease states, and the comprehensive analysis of cellular proteome would not be complete without the identification and quantification of the extent of PTMs of proteins.
Keywords: quantitative proteomics, isotopic labeling, phosphoproteomics
Proteomics is the global analysis of gene expression at the protein level. It has emerged as a postgenomic technology with the promise to unravel the cellular mechanisms of diseases and may lead to the development of reliable markers for disease diagnosis and therapy. Despite its promise, proteomic analyses remain challenging because of various limitations of existing technology. All commonly used approaches to isolate and characterize proteomes are still cumbersome.
Over the past several years, mass spectrometry (MS) has emerged as the most efficient and versatile tool of all the proteomics approaches available so far (1, 24, 26). With the emergence of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) techniques, analyzing large molecules such as proteins by mass spectral analysis has become possible (31, 54, 112). Although significant progress has been made, current instrumentation technology does not allow the comprehensive and complete characterization of cellular proteomes and their posttranslational modifications (PTMs), and there is still a long way to go to reach this goal. Finishing the Human Genome Project was facilitated by the static nature of the genome, which does not change significantly over time within a normal cell. Conversely, the proteome, though encoded by the cellular genome, is not static and changes continuously through various cellular mechanisms. It is believed that the 25,000 genes in the human genome encode up to one million different proteins through various splice forms and posttranslational processing and modifications. Analyzing the genome is simpler because the DNA consists of just 4 building blocks, while proteins are composed of 20 different naturally occurring amino acids. To add to the complexity of the proteome, there is a large dynamic range of concentrations that changes over time from cell to cell and state to state.
Ideally, a comprehensive proteomic analysis needs efficient separation of the complex sample and subsequent identification, quantification, and complete characterization of proteins in a single experiment. Thus each comprehensive proteomic characterization involves a series of analytical techniques. Below, we describe and discuss commonly used approaches and technologies that help in the identification and characterization of proteins with MS.
Comprehensive Proteomic Characterization
A typical standard mass spectral analysis of a complex protein sample involves an initial fragmentation step, a separation methodology to fractionate the complex mixture, and a subsequent mass spectral analysis of the resulting fractions. Commonly, proteins in a complex mixture are fragmented with proteolytic enzymes such as trypsin. Upon treatment, a typical sample from a whole cell lysate contains thousands to millions of peptides. These complex mixtures need to be fractionated before further analysis. Initial proteomic separation techniques were based on gel electrophoresis, either one-dimensional (1-DE) or two-dimensional electrophoresis (2-DE), in which proteins or peptides are separated based on their charge and molecular weight (19, 53, 69, 78). The separated proteins are usually visualized with different dyes. The 1-DE or 2-DE gel bands are excised and in-gel digested, and the resulting peptides are analyzed by either MALDI-MS or ESI-MS. Other methods to fractionate the peptide mixture include liquid chromatography and capillary electrophoresis (CE) (51, 86, 97, 129). While a successful protein and peptide separation and fractionation is essential to a mass spectral analysis, this review focuses on the analytical aspects and the challenges of MS for proteomics. Other reviews discuss the commonly used approaches and challenges of fractionation (see Ref. 69a).
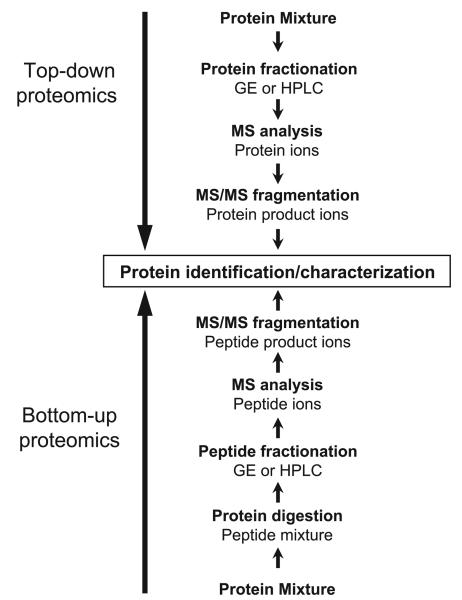
MS is a powerful approach to obtain protein sequence data from unknown samples and to correlate the experimental data with sequence information in public databases (“bottom-up proteomics”; Fig. 1). Such protein or peptide sequence information can be obtained from tandem mass spectral analysis. In a typical tandem MS analysis, the first step consists of the detection of the initial peptide ion. Subsequently, the peptide ions are fragmented by collision-induced dissociation (CID) to break the polypeptide backbone at the amide bond, thereby creating a ladder of fragment ions that reflect the peptides' amino acid sequence (11, 20, 118). The resulting spectra are then compared with publicly available protein sequence information. The observed masses of the proteolytic fragments are compared with theoretical in silico sequences to identify the peptide sequence and thereby the protein (14, 29, 36, 44, 65, 77). With the advancements in the separation techniques, up to 2,000 proteins can now be identified in a standard experiment.
Fig. 1.
Schematic representation of the common top-down and bottom-up approaches for the identification and characterization of proteins using mass spectrometry (MS). GE, gel electrophoresis; HPLC, high performance liquid chromatography.
Alternative approaches to this bottom-up approach, in which the original protein is predicted from sequence information of proteolytic peptides, have utilized “top-down” approaches in which the intact protein is directly dissected and amino acid sequence information is obtained by dissociation (Fig. 1). In this approach, the intact proteins are separated by gel electrophoresis or offline liquid chromatography before MS analysis. The major obstacle in this approach is the determination of product ion masses from multiply charged species of intact proteins. Because of the formation of multiply charged protein precursor ions, it is difficult to interpret top-down fragmentation spectra. This limitation can be evaded by reducing the charge states on the product ions through the introduction of gas-phase anions to strip protons from the product ions through ion-ion proton transfer reaction (106).
Another approach of choice for top-down proteomics is the use of the Fourier transform-ion cyclotron resonance (FT-ICR) mass spectrometer or orbitrap mass spectrometer with high mass accuracy (<2 ppm). The product ion charge state can be determined from the isotope spacing in the multiply charged species that facilitates the identification (27, 66, 85). The dissociation techniques in top-down proteomics are favored toward electron-capture dissociation (ECD) as implemented on FT-ICR-MS or electron-transfer dissociation (ETD) used in orbitrap instruments.
Although the top-down proteomics approach has some limitations because of the formation of very complex spectra, use of expensive instrumentation, and difficulties with proteins of high molecular mass (<50 kDa), it is advantageous over classical bottom-up approaches since it provides access to the complete protein sequence, locates PTMs due to gentle fragmentation methods, and avoids long protein digestion methods.
Comprehensive Protein Quantification
In addition to the identification of proteins, it is essential to quantify proteins in complex biological samples to better understand their role in an organism and in physiological systems (9, 21, 63, 68, 82, 100, 127). Both relative and absolute quantification methods have been established that use mass spectral techniques for the protein identification. Traditionally, protein quantification is carried out with gel-based approaches. In contrast, stable isotope labeling and label-free methods have been established that involve mass spectral analyses and result in quantification at the peptide level. Below, we discuss the different approaches in more detail and highlight the differences between the individual approaches.
Gel-based quantification approaches
Gel-based isolation of proteins was used extensively in molecular biology and biochemistry even before the development of modern MS-based protein identification. Even now it is used as a quantitative proteomic technology that can resolve hundreds to thousands of proteins in a single gel. To overcome the limitations of gel-to-gel reproducibility, differential gel electrophoresis (DIGE) has been established (83, 117, 122). In a DIGE experiment, up to three different samples can be analyzed on a single gel, each labeled with a different fluorescent cyanine dye (Cy2, Cy3, and Cy5). Although the method is successful in quantifying proteins and identifying PTMs on proteins that result in lateral shift in spots on the gel, the approach has several limitations. Like any gel-based method, DIGE has limitations in resolving hydrophobic proteins, interference from high-abundance proteins, and poor resolution of spots. Furthermore, not all individual proteins that are differentially expressed can be subsequently identified with MS. Finally, multiple protein isoforms can often be found in different spots on the gel, complicating the comprehensive analysis.
Stable isotope labeling
With stable isotope labeling, both absolute and relative methods of quantification have been developed, each having its own merits and shortcomings. Most of the absolute methods of quantification are small-scale analyses targeted to specific proteins. In contrast, relative quantification methods are focused on large-scale global proteomic analyses. Any stable isotope labeling method involves labeling of proteins/peptides with one or more stable isotopes and pooling the sample with an unlabeled control sample. The mixed sample is then subjected to a mass spectral analysis. The signal intensities of the labeled and unlabeled peptides are used to measure the relative abundance, while further fragmentation of the peptide provides the sequence information necessary for identification of the quantified protein.
As illustrated in Fig. 2, stable isotope labeling can be classified into metabolic labeling, chemical mass tagging, and enzymatic labeling.
Fig. 2.
Illustration of commonly used labeling strategies for quantitative proteomic analysis using MS (adapted from Ref. 1). Proteins are depicted at top, the resulting labeled peptides from each approach are shown in the middle, and a schematic of the resulting mass spectrum is shown at bottom, with the resulting signal from the light peptides shown in gray and signal from the heavy peptides in red. Peak areas can be compared for relative quantification of peptide abundance.
Metabolic isotope labeling requires in vivo incorporation of isotope-labeled essential amino acids during cell growth. The technology was first developed by Matthias Mann and his laboratory (1, 81, 82) and termed stable isotope labeling by amino acids in cell culture (SILAC). Cells are cultured in a medium supplemented with a labeled amino acid (lysine or arginine containing 13C or 15N). After several passages, cells are pooled with control cells grown in a medium of naturally occurring amino acids. The pooled samples are digested and analyzed by tandem MS. Although the technique is promising, it is limited to studies that involve cell culture so that cells can incorporate the exogenous amino acid into proteins. This normally precludes the use of this methodology for the analysis of tissue samples. However, a recent metabolic labeling study showed that entire model organisms (such as rats) can be labeled by using labeled chow. McClatchy et al. (71) demonstrated this quantification strategy by using 15N-labeled rat brain as an internal standard for large-scale analysis of the mammalian brain. The method is termed stable isotope labeling in mammals (SILAM) (71).
Mass tagging via chemical labeling is another approach using isotopes for quantification. In this method, proteins or peptides are tagged with a stable isotope-containing molecule. Tagged peptides can be efficiently isolated and enriched with affinity groups attached to the tagging moiety that can bind to specific amino acid residues. One such technology is the isotope-coded affinity tag (ICAT) methodology developed by the Aebersold group (104). An affinity group is chemically linked to cysteinyl residues on the protein. The ICAT reagent consists of a sulfhydryl-reactive iodoacetate group, a biotin affinity group, and a linker carrying light or heavy isotopes (101, 115, 123). After derivatization, both light and heavy tagged protein samples are pooled, digested, and enriched on an avidin affinity column to capture peptides containing tagged cysteine residues that can be analyzed by subsequent tandem mass spectral analysis. The method has both advantages and limitations due to the specificity of the affinity tag to specific amino acid residues. On one hand, the complexity of the sample will be reduced because of the enrichment of only tagged peptides. However, as a result, only a fraction of peptides will be analyzed by MS, and no quantitative or sequence information is obtained for proteins or peptides that do not contain cysteinyl residues. While additional affinity tags have been developed in recent years that allow specific binding to other amino acid residues (34, 45, 61), the enrichment step is an essential part of the ICAT protocol and untagged proteins or peptides will not be analyzed and identified.
Recently, isobaric tags for relative and absolute quantification (iTRAQ) have been established as a more comprehensive and efficient method for proteomic quantification (2, 17, 94, 102, 128). The method was developed by Ross et al. (94). It involves tagging of the NH2-terminal and side-chain lysine amino groups with stable isotopically labeled mass tags. The reagent consists of an amine-specific reactive group, a mass balance group, and a reporter group. The reporter and balance groups carry stable isotopes, with different combinations of isotopes in the reporter group but uniform molecular weight in the combined molecule. Because of the chemical composition, several different tags can be generated, allowing the simultaneous labeling of multiple samples. Chemically, the only difference in these reagents is the substitution of 12C, 14N, or 16O with their heavy isotopes 13C, 15N, or 18O, but since their molecular weights remain unchanged, the chromatographic properties of the peptides remain the same and the light and heavy peptides elute at the same time, enabling the quantification of different samples. After coelution, the peptides are further fragmented to release the iTRAQ reagent that allows the distinction of the different samples in MS and provides the necessary quantitative information. With the extensive success with four-plex reagents, eight-plex reagents are now commercially available that permit the quantification of eight different samples in a single run. The method can be used not only to perform relative quantification but also for absolute quantification by using an internal standard peptide labeled with one of the iTRAQ reagents. While the methodology has been used extensively, the iTRAQ system is relatively expensive, and the reporter ion masses are below the low-mass cutoff of ion-trap mass spectrometers, thus requiring more advanced mass spectrometers for the analysis.
Another nonisobaric triplex labeling strategy was introduced by Kellermann and Lottspeich (98, 99), named isotope-coded protein labeling technology (ICPL). In this approach, amino groups of intact proteins were derivatized with three isotopically different nicotinoyl reagents (12C6H4, 12C6D4, and 13C6H4). Since the method is based on labeling proteins even before tryptic digestion followed by fractionation, it significantly reduces the complexity of the sample but retains sequence coverage in the subsequent mass spectral analysis. This results in improved protein identification that is indispensable for the detection of PTMs.
Stable isotope incorporation by enzymatic labeling is another commonly used method to overcome the challenges of other methods discussed above. Enzyme-catalyzed labeling of peptides provides a comprehensive and global quantitation of the cellular proteome. Enzymatic incorporation of 18O atoms during proteolytic cleavage, most commonly by trypsin, results in peptides with either one or two 18O atoms at the carboxy terminus (8, 43, 74, 90, 108). The labeled sample is mixed with known amounts of an unlabeled peptide sample (for absolute quantification) or a reference sample (for relative quantification) and analyzed by tandem MS. In the 18O labeling strategy, the difference in molecular mass between light and heavy peptide samples is either 2 Da (one 18O atom incorporated) or 4 Da (two 18O atoms incorporated). The ratio of the intensities of the labeled and unlabeled peptides allows the quantification of the peptide. Although there are limitations in the method due to the back exchange of 18O with naturally occurring 16O, if sufficient care is taken (109), the method is advantageous over other labeling technologies because of its simplicity and global proteomic quantification both in vivo and in in vitro samples. Our laboratory (41, 43) has developed protocols and analysis tools that allow the efficient quantitative analysis of complex biological samples using 18O labeling. Figure 3 illustrates the common work flow and the analytical software tool ZoomQuant.
Fig. 3.
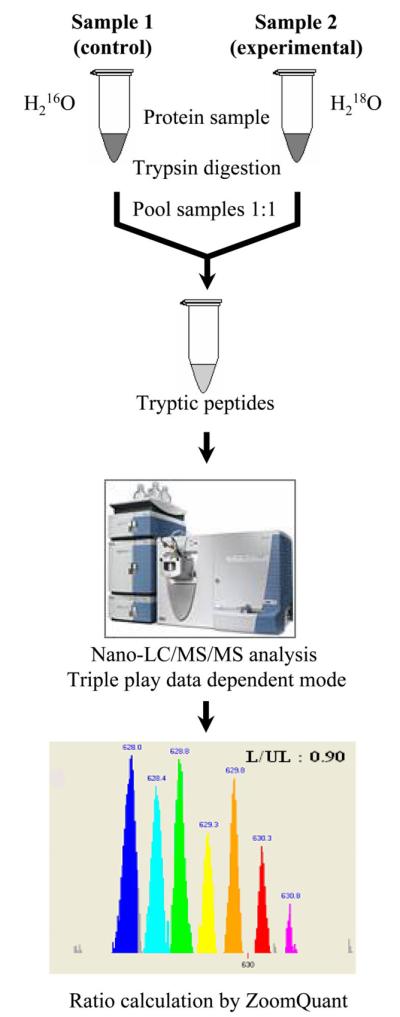
18O labeling strategy for quantitative proteomic analysis using mass spectrometry and ZoomQuant software tool (the MS image is adapted from www.thermo.com). L/UL is the ratio of labeled (18O) to unlabeled (16O) peptide measured by ZoomQuant.
Alternative mass spectral quantification approaches
Recently, label-free methods of quantification have been proposed (80, 116, 124). These methods are based on a correlation between peptide mass spectral peak data and the abundance of the protein in the sample. In one of the approaches, mass spectral peak intensities of peptide ions are used to quantify the protein amount (12, 16, 117). Alternatively, another approach involves spectral counting, where the number of mass spectra assigned to a protein is taken as a measure of protein abundance (33, 64). The label-free methods are advantageous, especially in samples where isotope labeling is either not possible or too cumbersome, and they provide quantitative information. However, the accuracy of the approach needs to be further evaluated, especially for low-abundance proteins in complex samples.
Mass Spectral Analysis of Posttranslational Modifications
The complex and dynamic nature of the cellular proteome has been a major challenge. The comprehensive assessment and analysis of the proteome is further complicated by various PTMs that regulate cellular processes (18, 56, 59, 72, 93, 114). Altered levels of PTMs, sometimes in the absence of protein expression changes, are often linked to cellular responses and disease states. Therefore, the comprehensive analysis of the cellular proteome would not be complete without the identification and quantification of the extent of PTMs of the individual proteins. Proteins are known to undergo more than 300 PTMs that regulate their cellular functions, of which phosphorylation is the most common and best characterized. Several studies have described enrichment methods for these PTM proteins. However, even after enrichment, PTM proteins are not very well ionized under MS conditions because of their negative charge, complicating the MS detection of these proteins.
Classical approaches for phosphoproteomic analysis use polyacrylamide gel electrophoresis, as discussed above. The phosphorylated species can be visualized by radiolabeling, immunodetection, or phosphospecific staining (39, 67, 95, 120). The success in using gel-based methods is due to the isoelectric point (pI) shift of proteins with each phosphorylated site that shifts the protein spot on the gel laterally (28, 39, 49, 62, 119, 120, 125). However, all difficulties discussed above for gel-based quantification approaches apply to phosphoprotein quantification as well, and limit the use of the approach.
Mass spectral approaches to the analysis of phosphoproteins have been complicated by the technical difficulties of detecting phosphoproteins in the presence of nonphosphorylated species. Thus the phosphoproteins are commonly enriched before analysis. Various approaches have been established such as immunoprecipitation, chemical derivatization, affinity purification, and most commonly immobilized metal affinity chromatography (IMAC) methods (52, 91, 107). Below, we describe and discuss the individual approaches.
Immunoprecipitation
Immunoprecipitation of proteins from complex mixtures with antibodies against phosphorylated amino acid residues is a common approach in phosphoproteomics (25, 32, 47, 48). Most analyses have successfully used pTyr antibodies (84, 126), and recently attempts have been made to precipitate proteins with pSer and pThr antibodies (35). The precipitated proteins are further separated on 1-DE or 2-DE gels and analyzed by MS to map the phosphorylation site. While the method does allow the identification of phosphorylated proteins, and permits the characterization of the phosphorylated amino acid residue, the methodology does not allow the quantification of phosphorylated proteins.
Chemical derivatization
Chemical derivatization methods convert the phosphorylated amino acid into more traceable species. β-Elimination of pSer and pThr by alkali treatment is one of the most common derivatization approaches used (73, 92). β-Elimination of phosphoric acid from pSer or pThr followed by the introduction of a biotin moiety aids in the selective enrichment of phosphoproteins. The biotin-tagged phosphopeptides are selectively separated from nonphosphorylated species by affinity chromatography (34, 79). Another approach uses β-elimination followed by reaction with cysteamine to convert pSer or pThr to lysine analogs, which enables site-specific cleavage with trypsin at the phosphoamino acid site (55). However, the method has limitations due to the loss of chromatographic performance and loss of sensitivity during mass spectral analysis. Moreover, the method is only applicable to pSer and pThr residues.
Immobilized metal affinity chromatography
IMAC has been used extensively and efficiently for the enrichment of phosphoproteins and peptides (5, 15, 87). The approach exploits the high affinity of the phospho-moiety to positively charged metal ions like Fe3+ (5), Ga3+ (88), Al3+ (4), and Zr4+ (30). The metal ions are immobilized on a solid support (silica, Sepharose, or agarose) with metal-chelating agents such as iminodiacetic acid (IDA) (6), nitrilotriacetic acid (NTA) (3), tris(carboxymethyl)ethylendiamine (TED) (15), or poly (glycidylmethacrylate/divinylbenzene) (GMD) (6). Although the method has been used extensively, there are limitations to the approach. The acidic groups on the peptides are nonspecifically bound to the metal ions and hence need to be further esterified to block the acidic group, increasing the complexity of the experimental procedure (42). Moreover, the method is biased toward multiply phosphorylated peptides because of their high affinity to the metal ions. Finally, as with all described enrichment approaches, the method does not provide quantitative information.
Affinity purification
Affinity purification with metal oxides is an alternative method of phosphoprotein enrichment. Titanium oxide, aluminum oxide, and zirconium oxide have proven to be more chemically stable materials than silica-based materials. Recently, TiO2 has been demonstrated (57, 96, 121) as an alternative reagent to the IMAC method of enrichment. Lately, Kweon and Hakansson (58) compared zirconium oxide to titanium oxide and concluded that each allows the selective enrichment of phosphopeptides, zirconium being more selective for monophosphorylated peptides while titanium had more affinity toward multiply phosphorylated peptides. Thus mixing both the metal oxides could increase the efficiency in the enrichment of both monophosphorylated and multiply phosphorylated species. Although the method is proven to be efficient in recovering over 90% of phosphopeptides, it has yet to be applied to complex protein mixtures.
An efficient MS-based method that not only identifies the PTM proteins but also measures the degree to which the protein is modified would be very informative for understanding protein function and signaling cascades in various cell functions. With the recently developed relative quantification methods by MS, phosphoproteomic quantification has become possible (22, 37, 113). Recent studies have used SILAC, ICAT, iTRAQ, or 18O methods for the quantification of IMAC-enriched phosphopeptides followed by tandem mass spectral analysis (34, 89). Another approach is to dephosphorylate the peptides for the indirect identification and quantification of phosphopeptides (13, 103). The method is more advantageous over other methods because it only requires the MS analysis of unphosphorylated peptides, circumventing technical difficulties like signal suppression or biased enrichment procedures.
Despite the various enrichment methods for phosphopeptides, mass spectral analysis of these peptides is often not very efficient. The loss of the phosphomoiety from the peptides is very prominent during CID experiments. Thus a very intense peak with loss of 98 Da or to a minor extent at 80 Da dominates over the rest of the fragmentation peaks, thereby providing only limited data for peptide identification (10, 46, 50, 76). To overcome the limitations with CID analysis, new analytical methods like ECD (7, 40, 60, 105, 110) and ETD (23, 38, 70, 75, 111) have been established to identify phosphopeptides. The methods are based on gentle fragmentation of the peptides, preserving the labile amino acid modifications. In this technology, c and z ions are generated, keeping the modification site intact on the fragmented ions. Besides, there is no neutral loss of phosphoric acid from the peptide; even larger and multiply charged species are identified efficiently.
Perspectives
Mass spectral analysis of complex protein mixtures has been improved significantly over the past decade, and technological advances and improved methodologies now allow the comprehensive quantitative analysis of complex biological samples by MS. However, the methodologies are far from optimal, and only a small fraction of the entire proteome of a biological sample is analyzed quantitatively with any of the approaches described above. While further advances are needed to provide the data necessary for the complete dissection of proteomic changes in disease, current technology already allows an initial glimpse of the underlying pathophysiological changes on a cellular level. The information obtained from these initial proteomic analyses of carefully selected biological samples will significantly advance our current understanding of the complex regulation of biological systems, even with this rudimentary information. Analyses of temporal changes of cellular proteomes and the regulation and dynamic alteration of PTMs of the proteome will provide novel information beyond the genomic data currently being obtained. Once both approaches have been merged efficiently, we will be able to dissect the complex underlying pathways that regulate disease processes, and use this information for disease prevention and management.
Acknowledgments
GRANTS
S. P. Mirza and M. Olivier are supported by funding from the National Heart, Lung, and Blood Institute (N01-HV-28182).
REFERENCES
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct Genomic Proteomic. 2006;5:112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- 3.Ahn YH, Park EJ, Cho K, Kim JY, Ha SH, Ryu SH, Yoo JS. Dynamic identification of phosphopeptides using immobilized metal ion affinity chromatography enrichment, subsequent partial beta-elimination/chemical tagging and matrix-assisted laser desorption/ionization mass spectrometric analysis. Rapid Commun Mass Spectrom. 2004;18:2495–2501. doi: 10.1002/rcm.1651. [DOI] [PubMed] [Google Scholar]
- 4.Andersson L. Recognition of phosphate groups by immobilized aluminium(III) ions. J Chromatogr A. 1991;539:327–334. [Google Scholar]
- 5.Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 6.Aprilita NH, Huck CW, Bakry R, Feuerstein I, Stecher G, Morandell S, Huang HL, Stasyk T, Huber LA, Bonn GK. Poly(glycidyl methacrylate/divinylbenzene)-IDA-FeIII in phosphoproteomics. J Proteome Res. 2005;4:2312–2319. doi: 10.1021/pr050224m. [DOI] [PubMed] [Google Scholar]
- 7.Axelsson J, Palmblad M, Hakansson K, Hakansson P. Electron capture dissociation of substance P using a commercially available Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun Mass Spectrom. 1999;13:474–477. doi: 10.1002/(SICI)1097-0231(19990330)13:6<474::AID-RCM505>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 8.Back JW, Notenboom V, de Koning LJ, Muijsers AO, Sixma TK, de Koster CG, de Jong L. Identification of cross-linked peptides for protein interaction studies using mass spectrometry and 18O labeling. Anal Chem. 2002;74:4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 9.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 10.Bennett KL, Stensballe A, Podtelejnikov AV, Moniatte M, Jensen ON. Phosphopeptide detection and sequencing by matrix-assisted laser desorption/ionization quadrupole time-of-flight tandem mass spectrometry. J Mass Spectrom. 2002;37:179–190. doi: 10.1002/jms.271. [DOI] [PubMed] [Google Scholar]
- 11.Biemann K. Sequencing of peptides by tandem mass spectrometry and high-energy collision-induced dissociation. Methods Enzymol. 1990;193:455–479. doi: 10.1016/0076-6879(90)93433-l. [DOI] [PubMed] [Google Scholar]
- 12.Bondarenko PV, Chelius D, Shaler TA. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem. 2002;74:4741–4749. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- 13.Bonenfant D, Schmelzle T, Jacinto E, Crespo JL, Mini T, Hall MN, Jenoe P. Quantitation of changes in protein phosphorylation: a simple method based on stable isotope labeling and mass spectrometry. Proc Natl Acad Sci USA. 2003;100:880–885. doi: 10.1073/pnas.232735599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cagney G, Amiri S, Premawaradena T, Lindo M, Emili A. In silico proteome analysis to facilitate proteomics experiments using mass spec-trometry. Proteome Sci. 2003;1:5. doi: 10.1186/1477-5956-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaga GS. Twenty-five years of immobilized metal ion affinity chromatography: past, present and future. J Biochem Biophys Methods. 2001;49:313–334. doi: 10.1016/s0165-022x(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 16.Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J Proteome Res. 2002;1:317–323. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- 17.Chong PK, Gan CS, Pham TK, Wright PC. Isobaric tags for relative and absolute quantitation (iTRAQ) reproducibility: implication of multiple injections. J Proteome Res. 2006;5:1232–1240. doi: 10.1021/pr060018u. [DOI] [PubMed] [Google Scholar]
- 18.Clark RS, Bayir H, Jenkins LW. Posttranslational protein modifications. Crit Care Med. 2005;33:S407–S409. doi: 10.1097/01.ccm.0000191712.96336.51. [DOI] [PubMed] [Google Scholar]
- 19.Cohen SL, Chait BT. Mass spectrometry of whole proteins eluted from sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Anal Biochem. 1997;247:257–267. doi: 10.1006/abio.1997.2072. [DOI] [PubMed] [Google Scholar]
- 20.Collin OL, Beier M, Jackson GP. Dynamic collision-induced dissociation of peptides in a quadrupole ion trap mass spectrometer. Anal Chem. 2007;79:5468–5473. doi: 10.1021/ac0707683. [DOI] [PubMed] [Google Scholar]
- 21.Conrads TP, Issaq HJ, Hoang VM. Current strategies for quantitative proteomics. Adv Protein Chem. 2003;65:133–159. doi: 10.1016/s0065-3233(03)01018-0. [DOI] [PubMed] [Google Scholar]
- 22.Conrads TP, Issaq HJ, Veenstra TD. New tools for quantitative phosphoproteome analysis. Biochem Biophys Res Commun. 2002;290:885–890. doi: 10.1006/bbrc.2001.6275. [DOI] [PubMed] [Google Scholar]
- 23.Coon JJ, Shabanowitz J, Hunt DF, Syka JE. Electron transfer dissociation of peptide anions. J Am Soc Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 25.De Graan PN, Dekker LV, Oestreicher AB, Van der Voorn L, Gispen WH. Determination of changes in the phosphorylation state of the neuron-specific protein kinase C substrate B-50 (GAP43) by quantitative immunoprecipitation. J Neurochem. 1989;52:17–23. doi: 10.1111/j.1471-4159.1989.tb10892.x. [DOI] [PubMed] [Google Scholar]
- 26.Dreger M. Emerging strategies in mass-spectrometry based proteomics. Eur J Biochem. 2003;270:569. doi: 10.1046/j.1432-1033.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- 27.Du Y, Parks BA, Sohn S, Kwast KE, Kelleher NL. Top-down approaches for measuring expression ratios of intact yeast proteins using Fourier transform mass spectrometry. Anal Chem. 2006;78:686–694. doi: 10.1021/ac050993p. [DOI] [PubMed] [Google Scholar]
- 28.Ducret A, Desponts C, Desmarais S, Gresser MJ, Ramachandran C. A general method for the rapid characterization of tyrosine-phosphorylated proteins by mini two-dimensional gel electrophoresis. Electrophoresis. 2000;21:2196–2208. doi: 10.1002/1522-2683(20000601)21:11<2196::AID-ELPS2196>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 29.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 30.Feng S, Ye M, Zhou H, Jiang X, Jiang X, Zou H, Gong B. Immobilized zirconium ion affinity chromatography for specific enrichment of phosphopeptides in phosphoproteome analysis. Mol Cell Proteomics. 2007;6:1656–1665. doi: 10.1074/mcp.T600071-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 32.Formisano P, Condorelli G, Condorelli G, Beguinot F. Antiphosphotyrosine immunoprecipitation of an insulin-stimulated receptor phosphatase activity from FRTL5 cells. Endocrinology. 1991;128:2949–2957. doi: 10.1210/endo-128-6-2949. [DOI] [PubMed] [Google Scholar]
- 33.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 34.Goshe MB, Conrads TP, Panisko EA, Angell NH, Veenstra TD, Smith RD. Phosphoprotein isotope-coded affinity tag approach for isolating and quantitating phosphopeptides in proteome-wide analyses. Anal Chem. 2001;73:2578–2586. doi: 10.1021/ac010081x. [DOI] [PubMed] [Google Scholar]
- 35.Gronborg M, Kristiansen TZ, Stensballe A, Andersen JS, Ohara O, Mann M, Jensen ON, Pandey A. A mass spectrometry-based proteomic approach for identification of serine/threonine-phosphorylated proteins by enrichment with phospho-specific antibodies: identification of a novel protein, Frigg, as a protein kinase A substrate. Mol Cell Proteomics. 2002;1:517–527. doi: 10.1074/mcp.m200010-mcp200. [DOI] [PubMed] [Google Scholar]
- 36.Grosse-Coosmann F, Boehm AM, Sickmann A. Efficient analysis and extraction of MS/MS result data from Mascot result files. BMC Bioinformatics. 2005;6:290. doi: 10.1186/1471-2105-6-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Gunawardena HP, Emory JF, McLuckey SA. Phosphopeptide anion characterization via sequential charge inversion and electron-transfer dissociation. Anal Chem. 2006;78:3788–3793. doi: 10.1021/ac060164j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guy GR, Philip R, Tan YH. Analysis of cellular phosphoproteins by two-dimensional gel electrophoresis: applications for cell signaling in normal and cancer cells. Electrophoresis. 1994;15:417–440. doi: 10.1002/elps.1150150160. [DOI] [PubMed] [Google Scholar]
- 40.Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptic to yield complementary sequence information. Anal Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 41.Halligan BD, Slyper RY, Twigger SN, Hicks W, Olivier M, Greene AS. ZoomQuant: an application for the quantitation of stable isotope labeled peptides. J Am Soc Mass Spectrom. 2005;16:302–306. doi: 10.1016/j.jasms.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haydon CE, Eyers PA, Aveline-Wolf LD, Resing KA, Maller JL, Ahn NG. Identification of novel phosphorylation sites on Xenopus laevis Aurora A and analysis of phosphopeptide enrichment by immobilized metal-affinity chromatography. Mol Cell Proteomics. 2003;2:1055–1067. doi: 10.1074/mcp.M300054-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Hicks WA, Halligan BD, Slyper RY, Twigger SN, Greene AS, Olivier M. Simultaneous quantification and identification using 18O labeling with an ion trap mass spectrometer and the analysis software application “ZoomQuant.”. J Am Soc Mass Spectrom. 2005;16:916–925. doi: 10.1016/j.jasms.2005.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirosawa M, Hoshida M, Ishikawa M, Toya T. MASCOT: multiple alignment system for protein sequences based on three-way dynamic programming. Comput Appl Biosci. 1993;9:161–167. doi: 10.1093/bioinformatics/9.2.161. [DOI] [PubMed] [Google Scholar]
- 45.Hoang VM, Conrads TP, Veenstra TD, Blonder J, Terunuma A, Vogel JC, Fisher RJ. Quantitative proteomics employing primary amine affinity tags. J Biomol Tech. 2003;14:216–223. [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA. 2006;103:7159–7164. doi: 10.1073/pnas.0600895103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ignatoski KM. Immunoprecipitation and western blotting of phosphotyrosine-containing proteins. Methods Mol Biol. 2001;124:39–48. doi: 10.1385/1-59259-059-4:39. [DOI] [PubMed] [Google Scholar]
- 48.Ikuta K, Luftig RB. Detection of phosphorylated forms of Moloney murine leukemia virus major capsid protein p30 by immunoprecipitation and two-dimensional gel electrophoresis. J Virol. 1988;62:40–46. doi: 10.1128/jvi.62.1.40-46.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang XS, Tang LY, Cao XJ, Zhou H, Xia QC, Wu JR, Zeng R. Two-dimensional gel electrophoresis maps of the proteome and phosphoproteome of primitively cultured rat mesangial cells. Electrophoresis. 2005;26:4540–4562. doi: 10.1002/elps.200500286. [DOI] [PubMed] [Google Scholar]
- 50.Jin M, Bateup H, Padovan JC, Greengard P, Nairn AC, Chait BT. Quantitative analysis of protein phosphorylation in mouse brain by hypothesis-driven multistage mass spectrometry. Anal Chem. 2005;77:7845–7851. doi: 10.1021/ac051519m. [DOI] [PubMed] [Google Scholar]
- 51.Jin X, Chen Y, Lubman DM, Misek D, Hanash SM. Capillary electrophoresis/tandem mass spectrometry for analysis of proteins from two-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis. Rapid Commun Mass Spectrom. 1999;13:2327–2334. doi: 10.1002/(SICI)1097-0231(19991215)13:23<2327::AID-RCM792>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 52.Johnson SA, Hunter T. Phosphoproteomics finds its timing. Nat Biotechnol. 2004;22:1093–1094. doi: 10.1038/nbt0904-1093. [DOI] [PubMed] [Google Scholar]
- 53.Joubert R, Strub JM, Zugmeyer S, Kobi D, Carte N, Van Dorsselaer A, Boucherie H, Jaquet-Guffreund L. Identification by mass spectrometry of two-dimensional gel electrophoresis-separated proteins extracted from lager brewing yeast. Electrophoresis. 2001;22:2969–2982. doi: 10.1002/1522-2683(200108)22:14<2969::AID-ELPS2969>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 54.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 55.Knight ZA, Schilling B, Row RH, Kenski DM, Gibson BW, Shokat KM. Phosphospecific proteolysis for mapping sites of protein phosphor-ylation. Nat Biotechnol. 2003;21:1047–1054. doi: 10.1038/nbt863. [DOI] [PubMed] [Google Scholar]
- 56.Krueger KE, Srivastava S. Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol Cell Proteomics. 2006;5:1799–1810. doi: 10.1074/mcp.R600009-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Kuroda I, Shintani Y, Motokawa M, Abe S, Furuno M. Phosphopep-tide-selective column-switching RP-HPLC with a titania precolumn. Anal Sci. 2004;20:1313–1319. doi: 10.2116/analsci.20.1313. [DOI] [PubMed] [Google Scholar]
- 58.Kweon HK, Hakansson K. Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal Chem. 2006;78:1743–1749. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 59.Larsen MR, Trelle MB, Thingholm TE, Jensen ON. Analysis of posttranslational modifications of proteins by tandem mass spectrometry. Biotechniques. 2006;40:790–798. doi: 10.2144/000112201. [DOI] [PubMed] [Google Scholar]
- 60.Leymarie N, Costello CE, O'Connor PB. Electron capture dissociation initiates a free radical reaction cascade. J Am Chem Soc. 2003;125:8949–8958. doi: 10.1021/ja028831n. [DOI] [PubMed] [Google Scholar]
- 61.Li S, Zeng D. CILAT—a new reagent for quantitative proteomics. Chem Commun (Camb) 2007:2181–2183. doi: 10.1039/b700109f. [DOI] [PubMed] [Google Scholar]
- 62.Li ZB, Flint PW, Boluyt MO. Evaluation of several two-dimensional gel electrophoresis techniques in cardiac proteomics. Electrophoresis. 2005;26:3572–3585. doi: 10.1002/elps.200500104. [DOI] [PubMed] [Google Scholar]
- 63.Linscheid MW. Quantitative proteomics. Anal Bioanal Chem. 2005;381:64–66. doi: 10.1007/s00216-004-2921-6. [DOI] [PubMed] [Google Scholar]
- 64.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 65.MacCoss MJ, Wu CC, Yates JR., 3rd Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 66.Macek B, Waanders LF, Olsen JV, Mann M. Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol Cell Proteomics. 2006;5:949–958. doi: 10.1074/mcp.T500042-MCP200. [DOI] [PubMed] [Google Scholar]
- 67.Mamone G, Caira S, Garro G, Nicolai A, Ferranti P, Picariello G, Malorni A, Chianese L, Addeo F. Casein phosphoproteome: identification of phosphoproteins by combined mass spectrometry and two-dimensional gel electrophoresis. Electrophoresis. 2003;24:2824–2837. doi: 10.1002/elps.200305545. [DOI] [PubMed] [Google Scholar]
- 68.Mann M. Quantitative proteomics? Nat Biotechnol. 1999;17:954–955. doi: 10.1038/13646. [DOI] [PubMed] [Google Scholar]
- 69.Marvin L, Millar A, Saulot V, Machour N, Charlionet R, Tron F, Lange C. Identification of proteins from one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis using electrospray quadrupole-time-of-flight tandem mass spectrometry. Rapid Commun Mass Spectrom. 2000;14:1287–1292. doi: 10.1002/1097-0231(20000730)14:14<1287::AID-RCM27>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 69a.Matt P, Fu Z, Fu Q, Van Eyk J. Biomarker discovery: proteome fractionation and separation in biological samples. Physiol Genomics. 2007 December 27; doi: 10.1152/physiolgenomics.00282.2007. doi:10.1152/physiolgenomics.00282.2007. [DOI] [PubMed] [Google Scholar]
- 70.McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Implementation of electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2007;79:3525–3534. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McClatchy DB, Liao L, Park SK, Venable JD, Yates JR. Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 2007;17:1378–1388. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meri S, Baumann M. Proteomics: posttranslational modifications, immune responses and current analytical tools. Biomol Eng. 2001;18:213–220. doi: 10.1016/s1389-0344(01)00106-x. [DOI] [PubMed] [Google Scholar]
- 73.Meyer HE, Hoffmann-Posorske E, Korte H, Heilmeyer LM., Jr Sequence analysis of phosphoserine-containing peptides. Modification for picomolar sensitivity. FEBS Lett. 1986;204:61–66. doi: 10.1016/0014-5793(86)81388-6. [DOI] [PubMed] [Google Scholar]
- 74.Miyagi M, Rao KC. Proteolytic 18O-labeling strategies for quantitative proteomics. Mass Spectrom Rev. 2007;26:121–136. doi: 10.1002/mas.20116. [DOI] [PubMed] [Google Scholar]
- 75.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moyer SC, Cotter RJ, Woods AS. Fragmentation of phosphopeptides by atmospheric pressure MALDI and ESI/ion trap mass spectrometry. J Am Soc Mass Spectrom. 2002;13:274–283. doi: 10.1016/S1044-0305(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 77.Nicholson IC, Ayhan M, Hoogenraad NJ, Zola H. In silico evaluation of two mass spectrometry-based approaches for the identification of novel human leukocyte cell-surface proteins. J Leukoc Biol. 2005;77:190–198. doi: 10.1189/jlb.0804450. [DOI] [PubMed] [Google Scholar]
- 78.Nordhoff E, Egelhofer V, Giavalisco P, Eickhoff H, Horn M, Przewieslik T, Theiss D, Schneider U, Lehrach H, Gobom J. Large-gel two-dimensional electrophoresis-matrix assisted laser desorption/ionization-time of flight-mass spectrometry: an analytical challenge for studying complex protein mixtures. Electrophoresis. 2001;22:2844–2855. doi: 10.1002/1522-2683(200108)22:14<2844::AID-ELPS2844>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 79.Oda Y, Nagasu T, Chait BT. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 80.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 82.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 83.Orenes-Pinero E, Corton M, Gonzalez-Peramato P, Algaba F, Casal I, Serrano A, Sanchez-Carbayo M. Searching urinary tumor markers for bladder cancer using a two-dimensional differential gel electrophoresis (2D-DIGE) approach. J Proteome Res. 2007;6:4440–4448. doi: 10.1021/pr070368w. [DOI] [PubMed] [Google Scholar]
- 84.Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR, Mann M, Lodish HF. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc Natl Acad Sci USA. 2000;97:179–184. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parks BA, Jiang L, Thomas PM, Wenger CD, Roth MJ, Boyne MT, 2nd, Burke PV, Kwast KE, Kelleher NL. Top-down proteomics on a chromatographic time scale using linear ion trap Fourier transform hybrid mass spectrometers. Anal Chem. 2007;79:7984–7991. doi: 10.1021/ac070553t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pelzing M, Neususs C. Separation techniques hyphenated to electro-spray-tandem mass spectrometry in proteomics: capillary electrophoresis versus nanoliquid chromatography. Electrophoresis. 2005;26:2717–2728. doi: 10.1002/elps.200410424. [DOI] [PubMed] [Google Scholar]
- 87.Porath J, Carlsson J, Olsson I, Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature. 1975;258:598–599. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 88.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem. 1999;71:2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 89.Qian WJ, Goshe MB, Camp DG, 2nd, Yu LR, Tang K, Smith RD. Phosphoprotein isotope-coded solid-phase tag approach for enrichment and quantitative analysis of phosphopeptides from complex mixtures. Anal Chem. 2003;75:5441–5450. doi: 10.1021/ac0342774. [DOI] [PubMed] [Google Scholar]
- 90.Qin J, Herring CJ, Zhang X. De novo peptide sequencing in an ion trap mass spectrometer with 18O labeling. Rapid Commun Mass Spectrom. 1998;12:209–216. doi: 10.1002/(SICI)1097-0231(19980314)12:5<209::AID-RCM141>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 91.Reinders J, Sickmann A. State-of-the-art in phosphoproteomics. Proteomics. 2005;5:4052–4061. doi: 10.1002/pmic.200401289. [DOI] [PubMed] [Google Scholar]
- 92.Resing KA, Johnson RS, Walsh KA. Mass spectrometric analysis of 21 phosphorylation sites in the internal repeat of rat profilaggrin, precursor of an intermediate filament associated protein. Biochemistry. 1995;34:9477–9487. doi: 10.1021/bi00029a024. [DOI] [PubMed] [Google Scholar]
- 93.Robers MB, Horton RA, Bercher MR, Vogel KW, Machleidt T. High-throughput cellular assays for regulated posttranslational modifications. Anal Biochem. 2008;372:189–197. doi: 10.1016/j.ab.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 94.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 95.Salih E. Phosphoproteomics by mass spectrometry and classical protein chemistry approaches. Mass Spectrom Rev. 2005;24:828–846. doi: 10.1002/mas.20042. [DOI] [PubMed] [Google Scholar]
- 96.Sano A, Nakamura H. Titania as a chemo-affinity support for the column-switching HPLC analysis of phosphopeptides: application to the characterization of phosphorylation sites in proteins by combination with protease digestion and electrospray ionization mass spectrometry. Anal Sci. 2004;20:861–864. doi: 10.2116/analsci.20.861. [DOI] [PubMed] [Google Scholar]
- 97.Sanz-Nebot V, Benavente F, Barbosa J. Liquid chromatography-mass spectrometry and capillary electrophoresis combined approach for separation and characterization of multicomponent peptide mixtures. Application to crude products of leuprolide synthesis. J Chromatogr A. 2002;950:99–111. doi: 10.1016/s0021-9673(02)00025-0. [DOI] [PubMed] [Google Scholar]
- 98.Sarioglu H, Brandner S, Jacobsen C, Meindl T, Schmidt A, Kellermann J, Lottspeich F, Andrae U. Quantitative analysis of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced proteome alterations in 5L rat hepatoma cells using isotope-coded protein labels. Proteomics. 2006;6:2407–2421. doi: 10.1002/pmic.200500680. [DOI] [PubMed] [Google Scholar]
- 99.Schmidt A, Kellermann J, Lottspeich F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics. 2005;5:4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- 100.Sechi S, Oda Y. Quantitative proteomics using mass spectrometry. Curr Opin Chem Biol. 2003;7:70–77. doi: 10.1016/s1367-5931(02)00010-8. [DOI] [PubMed] [Google Scholar]
- 101.Sethuraman M, McComb ME, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol Cell Proteomics. 2004;3:273–278. doi: 10.1074/mcp.T300011-MCP200. [DOI] [PubMed] [Google Scholar]
- 102.Shadforth IP, Dunkley TP, Lilley KS, Bessant C. i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics. 2005;6:145. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith JR, Olivier M, Greene AS. Relative quantification of peptide phosphorylation in a complex mixture using 18O labeling. Physiol Genomics. 2007;31:357–363. doi: 10.1152/physiolgenomics.00096.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smolka MB, Zhou H, Purkayastha S, Aebersold R. Optimization of the isotope-coded affinity tag-labeling procedure for quantitative proteome analysis. Anal Biochem. 2001;297:25–31. doi: 10.1006/abio.2001.5318. [DOI] [PubMed] [Google Scholar]
- 105.Stensballe A, Jensen ON, Olsen JV, Haselmann KF, Zubarev RA. Electron capture dissociation of singly and multiply phosphorylated peptides. Rapid Commun Mass Spectrom. 2000;14:1793–1800. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 106.Stephenson JL, Jr, McLuckey SA. Ion/ion proton transfer reactions for protein mixture analysis. Anal Chem. 1996;68:4026–4032. doi: 10.1021/ac9605657. [DOI] [PubMed] [Google Scholar]
- 107.Stern DF. Phosphoproteomics. Exp Mol Pathol. 2001;70:327–331. doi: 10.1006/exmp.2001.2370. [DOI] [PubMed] [Google Scholar]
- 108.Stewart II, Thomson T, Figeys D. 18O labeling: a tool for proteomics. Rapid Commun Mass Spectrom. 2001;15:2456–2465. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]
- 109.Storms HF, van der Heijden R, Tjaden UR, van der Greef J. Considerations for proteolytic labeling-optimization of 18O incorporation and prohibition of back-exchange. Rapid Commun Mass Spectrom. 2006;20:3491–3497. doi: 10.1002/rcm.2738. [DOI] [PubMed] [Google Scholar]
- 110.Sweet SM, Cooper HJ. Electron capture dissociation in the analysis of protein phosphorylation. Expert Rev Proteomics. 2007;4:149–159. doi: 10.1586/14789450.4.2.149. [DOI] [PubMed] [Google Scholar]
- 111.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, Matsuo T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1988;2:151–153. [Google Scholar]
- 113.Tao WA, Wollscheid B, O'Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spec-trometry. Nat Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 114.Veenstra TD. Proteome analysis of posttranslational modifications. Adv Protein Chem. 2003;65:161–194. doi: 10.1016/s0065-3233(03)01019-2. [DOI] [PubMed] [Google Scholar]
- 115.von Haller PD, Yi E, Donohoe S, Vaughn K, Keller A, Nesvizhskii AI, Eng J, Li XJ, Goodlett DR, Aebersold R, Watts JD. The application of new software tools to quantitative protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry. I. Statistically annotated datasets for peptide sequences and proteins identified via the application of ICAT and tandem mass spectrometry to proteins copurifying with T cell lipid rafts. Mol Cell Proteomics. 2003;2:426–427. doi: 10.1074/mcp.D300002-MCP200. [DOI] [PubMed] [Google Scholar]
- 116.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: reproducibility, linearity, and application with complex proteomes. J Proteome Res. 2006;5:1214–1223. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 117.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 118.Wells JM, McLuckey SA. Collision-induced dissociation (CID) of peptides and proteins. Methods Enzymol. 2005;402:148–185. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 119.Wirth PJ, Luo LD, Hoang T, Benjamin T. Two-dimensional polyacrylamide gel electrophoresis of cancer-associated proteins. Recent Results Cancer Res. 1997;143:145–160. doi: 10.1007/978-3-642-60393-8_10. [DOI] [PubMed] [Google Scholar]
- 120.Wu BC, Spohn WH, Busch H. Two-dimensional gel electrophoresis of cytosol phosphoproteins of Novikoff hepatoma and regenerating liver. Cancer Res. 1979;39:116–122. [PubMed] [Google Scholar]
- 121.Wu J, Shakey Q, Liu W, Schuller A, Follettie MT. Global profiling of phosphopeptides by Titania affinity enrichment. J Proteome Res. 2007;6:4684–4689. doi: 10.1021/pr070481m. [DOI] [PubMed] [Google Scholar]
- 122.Yan W, Chen SS. Mass spectrometry-based quantitative proteomic profiling. Brief Funct Genomic Proteomic. 2005;4:27–38. doi: 10.1093/bfgp/4.1.27. [DOI] [PubMed] [Google Scholar]
- 123.Yan W, Lee H, Deutsch EW, Lazaro CA, Tang W, Chen E, Fausto N, Katze MG, Aebersold R. A dataset of human liver proteins identified by protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry. Mol Cell Proteomics. 2004;3:1039–1041. doi: 10.1074/mcp.D400001-MCP200. [DOI] [PubMed] [Google Scholar]
- 124.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 125.Zhang W, Czernik AJ, Yungwirth T, Aebersold R, Chait BT. Matrix-assisted laser desorption mass spectrometric peptide mapping of proteins separated by two-dimensional gel electrophoresis: determination of phosphorylation in synapsin I. Protein Sci. 1994;3:677–686. doi: 10.1002/pro.5560030415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 127.Ziegler Z. Quantitative proteomics goes global. Anal Chem. 2001;73:251A. doi: 10.1021/ac012451n. [DOI] [PubMed] [Google Scholar]
- 128.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57:1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 129.Zolla L, Bianchetti M. High-performance liquid chromatography coupled on-line with electrospray ionization mass spectrometry for the simultaneous separation and identification of the Synechocystis PCC 6803 phycobilisome proteins. J Chromatogr A. 2001;912:269–279. doi: 10.1016/s0021-9673(01)00532-5. [DOI] [PubMed] [Google Scholar]