Abstract
Stable isotope labeling with 18O is a promising technique for obtaining both qualitative and quantitative information from a single differential protein expression experiment. The small 4 Da mass shift produced by incorporation of two molecules of 18O, and the lack of available methods for automated quantification of large data sets has limited the use of this approach with electrospray ionization-ion trap (ESI-IT) mass spectrometers. In this paper, we describe a method of acquiring ESI-IT mass spectrometric data that provides accurate calculation of relative ratios of peptides that have been differentially labeled using18O. The method utilizes zoom scans to provide high resolution data. This allows for accurate calculation of 18O/16O ratios for peptides even when as much as 50% of a 18O labeled peptide is present as the singly labeled species. The use of zoom scan data also provides sufficient resolution for calculating accurate ratios for peptides of +3 and lower charge states. Sequence coverage is comparable to that obtained with data acquisition modes that use only MS and MS/MS scans. We have employed a newly developed analysis software tool, ZoomQuant, which allows for the automated analysis of large data sets. We show that the combination of zoom scan data acquisition and analysis using ZoomQuant provides calculation of isotopic ratios accurate to ~21%. This compares well with data produced from 18O labeling experiments using time of flight (TOF) and Fourier transform-ion cyclotron resonance (FT-ICR) MS instruments.
There is a growing impetus to develop methods for relative quantification of proteins and peptides that are compatible with shotgun methods and high throughput experiments. Two general approaches have thus far been used. Both approaches depend on the use of “light” and “heavy” isotopes to differentially label proteins from two different populations of cells grown under variant conditions. The resultant mass shift provides a mass based separation for otherwise identical molecules from both cell populations that can be used for relative quantification. In-vivo labeling methods, such as metabolic labeling relying on the incorporation of isotopically labeled specific amino acids [1], or complete substitution of 14N with 15N [2], cannot be used for many mammalian systems. Yates et al. have reported success with in-vivo metabolic labeling of rat proteins by providing them with a diet enriched in 15N labeled amino acids [3]. Post-translational methods of labeling are applicable to eukaryotic systems. These methods introduce mass shifts through the modification of the side chains of specific amino acids, such lysine [4] and cysteine [5], labeling of the carboxy, or amino terminus of peptides. Isotope-coded affinity tag (ICAT), which introduces “heavy” and “light” variants of an affinity tag that modifies cysteines, is the most well developed technology for post translational modification of peptides for differential protein expression analysis b y mass spectrometry [5]. It selectively enriches for cysteine containing peptides, and thereby produces mixtures of reduced complexity, but also a sample that is less representative of the original protein mixture. Its labeling target, cysteine, is a rare amino acid that occurs only at a frequency of 1.7 % [6]. We used the IPI human database to calculate that just under 7% of proteins have no cysteine containing tryptic peptides between 400 and 3000 Da and approximately 11% contain only one such peptide [7].
Isotopic labeling with 18O provides an alternative method for obtaining quantitative information that offers many advantages to the methods described above. Every peptide in a peptide mixture is labeled through the enzyme mediated oxygen substitution (EMOS) [8] of serine proteases, with the exception of carboxy-terminal peptides [9]. This produces a more representative mixture of the biological sample, and an enhanced possibility for protein detection and identification in a protein profiling experiment. Stable isotope labeling with 18O is very simple, inexpensive, and requires few handling steps. Isotopic labeling can be done simultaneously with proteolysis, or decoupled from digestion by post-proteolytic labeling [8, 10 –12]. Isotopically labeled peptide pairs have been shown to co-chromatograph, providing accurate quantitation from a single chromatographic peak [13]. This list of characteristics makes 18O labeling compatible with small samples [11, 12]. A recent paper by Zang et al. demonstrated that 18O labeling was effective with as little as 10,000 cells [11].
Thus far, the ability to routinely utilize 18O for differential expression analyses using ESI-ion trap mass spectrometry has been resolution limited by the small mass shift of two incorporated molecules of 18O. In the case of ESI instruments, the magnitude of this small 4 Da mass shift is further reduced by the presence of multiply charged species. Isotopic labeling with 18O also often produces peptides that are present as a mixture of singly labeled peptides (18O1/16O1) that result in a 2 Da mass shift, and fully labeled peptides (18O2) that result in a 4 Da mass shift [8, 11, 12, 16]. Accurate calculation of isotopic enrichment, therefore, requires quantification of both the singly labeled species and the doubly labeled species.
There have been recent efforts to use 18O stable isotope labeling with ion trap mass spectrometers, however medium to high resolution instruments that use time of flight (TOF) or Fourier transform ion cyclotron resonance (FT-ICR) have been the primary instruments used for quantification with 18O labeling [8, 9, 11, 14].
Quantification methods for 18O labeling data from ion trap instruments have thus far utilized extracted ion chromatograms (EIC) [11, 13] or peak intensity data from full scan MS and MS/MS spectra [13]. EICs are resolution limited and are subject to overlap from other coeluting peaks.
We describe a method that makes use of the zoom scan function of the Thermo Finnigan (West Palm Beach, FL) Deca Xp Plus LCQ mass spectrometer to obtain high resolution spectra. Using this method, peptides that contain only a single 18O atom can now be included in 18O/16O calculations. It is also possible to sufficiently resolve isotopic clusters so that +1, +2, and +3 charged tryptic peptides can be included in the analysis and yield accurate isotopic ratios. We tested this approach using individual proteins and a protein mixture that were digested with trypsin, isotopically labeled with 18O, and mixed in different ratios with parallel samples digested in . The resulting mixtures were then analyzed using an LCQ mass spectrometer and individual peptides were identified with SEQUEST. Isotopic ratio calculations were performed with newly developed software, “ZoomQuant” [15] that allows for automated calculation of the isotopic ratios. Prior to the development of this software; isotopic ratio calculation for 18O/16O isotope labeled data sets that included information from the 18O1 16O1 mixed species could only be done manually. The development of this method and software now makes it possible to obtain qualitative and quantitative information from differentially labeled protein profiling experiments using ESI-IT instruments.
Experimental
Chemicals
Equine skeletal muscle myoglobin (product number M 0630), and rabbit muscle-glyceraldehyde-3-phosphate dehydrogenase were purchased from Sigma Chemical (St. Louis, MO). Human recombinant protein tyrosine phosphatase-1β was purchased from BIOMOL (catalog number SE-332, Plymouth Meeting, PA). Recombinant rat vascular endothelial cell growth factor was purchased from Biosource International (catalog number PRG0115, Camarillo, CA). Dithiothreitol was purchased from Pierce (Rockford, IL), and iodoacetamide was purchased from Sigma Chemical. Sequencing grade endoproteinase Lys-C was obtained from Roche Biochemicals (Indianapolis, IN). POROS immobilized trypsin (product code 2-3127-00) was purchased from Applied Biosystems (Foster City, CA), and ≥95% atom percent excess (APE) H218O (catalog number OLM-240) was purchased from Cambridge Isotope Laboratories (Andover, MA). Mobile phase solvents were purchased from Burdick and Jackson (Muskegon, MI).
Digestion and Labeling of Myoglobin
One hundred twenty pmol of myoglobin was denatured in a 60 μL solution of 50 mM Tris (pH 8.0) containing 6 M urea and 2 m M DTT at 55 °C for 1.5 h. Alkylation was performed with 20 mM iodoacetamide for 0.5 h w ith agitation in the dark at room temperature. The denatured and alkylated protein was divided into two equal fractions. Both fractions were lyophilized to complete dryness. One fraction was reconstituted with 50 mM NH4HCO3 (pH 8.0) to six times the original volume. The other fraction was treated in a like manner except that the 50 mM NH4HCO3 (pH 8.0) solution was made with ≥95% H218O (APE). Both solutions were mixed with acetonitrile (ACN) (20% of protein d igest volume) and POROS immobilized trypsin (30% of digest volume). The reaction mixtures were incubated overnight (18 h) at 37 °C on a 2-dimensional platform rocker. The solutions were centrifuged at 16,100 × g in a microcentrifuge for 5 m in to pellet the immobilized trypsin. The supernatant was removed and concentrated in a speed vac to ≤80% volume to remove ACN. The digest solution was adjusted to pH 3–4 with 10% formic acid (formic acid was diluted using H218O for the 18O labeled sample.)
Digestion and Labeling of Standard Protein Mixture
Each of the following four proteins were included in a protein mixture: 340 pmol myoglobin, 145 pmol glyceral-dehyde-3-phosphate dehydrogenase (GAPDH), 120 pmol protein tyrosine phophatase-1β (PTP-1β), and 120 pmol rat recombinant vascular endothelial growth factor (VEGF). Proteins were added to a microcentrifuge tube and lyophilized to dryness. The mixture was denatured in a solution of 50 mM Tris (pH 8.0) containing 6 M urea and 2 m M DTT at 55 °C for 1.5 h. Alkylation was done with 20 mM iodoacetamide for 0.5 h with agitation in the dark and at room temperature. The mixture was then digested with endoproteinase Lys-C at an enzyme substrate ratio of 1:100 (wt/wt) overnight at 37 °C. Tryptic digestion and 18O/16O isotopic labeling was carried out as described above for myoglobin.
Desalting and Mixing Samples
The myoglobin samples were taken to near dryness with a speed vac concentrator, and diluted to the original volume (prior to speed vac concentration) with either 50 mM NH4HCO3 or 50 mM NH4HCO3 in H218O as appropriate. Aliquots from the tryptic digest were mixed in the desired 18O/16O ratios, and desalted using “Omix” C18, 100 μL pipette tips from Varian (Walnut Creek, CA). The desalted samples were then dried in a speed vac concentrator, and suspended in 5% ACN in 0.1% formic acid. The samples of the peptide mixtures were mixed in the desired ratios, and desalted without prior volume adjustment.
Chromatography
Approximately 3 pmol (1.5 pmol of each isotopically labeled digest) in 10 μL o f the myoglobin tryptic digest was loaded onto a 100 × 0.18 mm BioBasic C18 (ThermoElectron Corp., Bellefonte, PA) column using a Surveyor 2 autosampler (ThermoFinnigan). A two component mobile phase system was used, with two quaternary Surveyor pumps. The sample pump was used to load the column. Sample was loaded with 100% A with a flow rate of 4 μL/min onto the column. Solvent A was 5% ACN in 0.1% formic acid, Solvent B was 95% formic acid. The analytical pump was programmed with the following elution gradient: 0 –1 min 0% B–10% B, 1–35 min 10% B– 45% B, 35– 43 min 45% B– 65% B, 43– 47 min 65% B–100% B, hold for 2 min at 100% B, 49 min 100% B–50 min 100% A, hold for 10 min.
The tryptic digest of the protein mixture was loaded at approximately 6 pmol for VEGF and PTP-1β, 17 pmol for myoglobin, and 7.25 pmol for GAPDH in 10 μL total volume. A 2 h elution gradient was used. All other conditions were the same as described above. The elution gradient was as follows: 0 –1 min 0% B–5% B, 1–75 min 5% B–30 % B, 75– 90 min 30% B– 65% B, 90 –100 min 65% B–100% B, hold for 5 min at 100% B, 105–110 min 100% B– 0% B, hold for 10 min.
Mass Spectrometry
Mass spectrometric data was acquired with a ThermoFinnigan Deca Xp Plus operated in the triple play data-dependent mode. Column eluent was sprayed at a flow rate of 2 μL/min using a 34 gauge metal needle with a tip i.d. of 24 μm. The triple play data-dependent mode of the instrument provides a duty cycle in which a full scan is followed by a high resolution zoom scan and an MS/MS scan. We used the following instrument settings: number of scan events 3, full scan mass window 400 –2000, dependent zoom scan of the most intense ion from full scan, dependent MS/MS of the most intense ion from full scan, precursor isolation width 2, normalized collision energy 35%, minimum signal required 3 × 107. Global data dependent settings: exclusion mass width: 3, reject mass width: 3, dynamic exclusion enabled. Dynamic exclusion parameters were as follows: repeat count: 2, repeat duration: 0.5 min, exclusion list size: 50, exclusion duration: 5 min, exclusion mass width: 3 Da. Tuning parameters were as follows: capillary temperature 160 °C, no sheath gas flow, no sweep gas flow, automatic gain control (AGC) was on, source voltage 3.1, zoom micro scans 5, zoom AGC target 7 × 107, MSn micro scans 5, MSn AGC target 2 × 108, full micro scans 3, full AGC target 5 × 108. The mass spectra data for the protein mixtures was collected during a total run time of 120 min, and a total run time of 60 min was used for the myoglobin data.
Data Analysis
Raw files were searched using SEQUEST. Peptide identifications were filtered using an xcorr versus charge state filter with the following settings: charge state 1 and xcorr of 1, charge state 2 and xcorr of 1.8, charge state 3 and xcorr of 2.6. We used a low stringency filter and a group scan setting of 2 in order to maximize the number of peptides available to test with the ZoomQuant [15] software. The SEQUEST results file and corresponding RAW file was submitted to the ZoomQuant suite of programs. ZoomQuant reads in the SEQUEST results file and the data from zoom scans extracted from the RAW file and matches the SEQUEST identified peptides with the corresponding zoom scan. ZoomQuant also separates adjacent zoom scans for quantification from the grouped scans used by SEQUEST for a single peptide identification. The 18O/16O isotopic ratios are calculated using three different algorithms. The first method uses the peptides mass, charge, and atomic composition, based on the SEQUEST identification, to calculate a theoretical isotope distribution for each of the labeled species [14]. The second method uses the averagine approach described by Johnson and Muddiman [8]. The third method is a modification of the algorithm of Yao et al. [17] in which peak areas are used for the calculation instead of intensities to determine the 18O/16O ratios from the M and M + 2 and M + 4 peaks.
Results and Discussion
Myoglobin
Tryptic digests of myoglobin were prepared, isotopically labeled, and mixed in two different 18O/16O ratios of 1 and 0.3. Three separate tryptic digests were used to generate LCMS data for the samples with an 18O/16O ratio of one. This was done to test the inherent variability of the digest procedure itself and the sample handling involved in the digestion and labeling procedure. Different digests might give different ratios because of sample handling steps, but the calculated isotopic ratio from zoom scan to zoom scan for a given peptide within an individual LCMS run should not vary significantly. Table 1 shows the averaged isotopic ratios for each peptide. Data from the individual peptides were averaged together to give an overall ratio of 1.15 for the 1:1 18O/16O mixture of myoglobin. The coefficient of variation of 35% highlights the frequent observation that the level of EMOS is peptide-dependent, as well as the variability between individual sample preparations [11, 16]. When multiple peptides and data sets are available, however, the calculated ratio closely approximates the expected value. This would suggest that more peptides produce results with higher confidence levels. The data generated for myoglobin digest that were mixed in a 0.3 18O/16O ratio was from a single digest from which we performed three replicate LCMS analyses. We used only two peptides in the isotopic ratio calculations for the samples mixed in a 0.3 ratio (18O/16O) since only these two peptides were observed, yet for these two peptides the coefficient of variation was only 14%.
Table 1.
The averaged 18O/16O ratio from three individual LCMS runs for individual myoglobin peptides, organized by charge state
| Peptide | Number | Charge | Ratioa | Efficiency |
|---|---|---|---|---|
| Expected ratio of 1 | ||||
| LFTGHPETLEK | 4 | 2 | 1.24 | 0.76 |
| VEADIAGHGQEVLIR | 2 | 2 | 1.17 | 0.79 |
| VEADIAGHGQEVLIR | 1 | 3 | 0.74 | 0.90 |
| HGTVVLTALGGILK | 8 | 2 | 1.39 | 0.80 |
| ALELFR | 1 | 1 | 1.74 | 0.87 |
| YLEFISDAIIHVLHSK | 1 | 3 | 0.65 | 0.68 |
| protein mean | 1.15 | 0.80 | ||
| standard deviation | 0.41 | 0.08 | ||
| coefficient of variation | 0.35 | |||
| Expected ratio of 0.3 | ||||
| LFTGHPETLEK | 3 | 2 | 0.49 | 0.80 |
| HGTVVLTALGGILK | 3 | 2 | 0.40 | 0.72 |
| protein mean | 0.44 | 0.76 | ||
| standard deviation | 0.06 | 0.06 | ||
| coefficient of variation | 0.14 | |||
Ratios were generated using method 1 o f ZoomQuant. The data for the expected ratio of 1 were generated from three separate digests of myoglobin. The data for the 0.3 ratio is the average of 3 replicates from the same digest. “Number” indicates the number of times each peptide was observed within the data set, and “efficiency” indicates the percent of the peptide that was fully labeled relative to the sum of the fully labeled and singly labeled species.
Overall, the more frequently observed peptides cluster more tightly. If one considers only those peptides that appear more than once from the myoglobin data with an expected 18O/16O ratio of one, then the standard deviation decreases to 0.1122, and the coefficient of variation decreases to 8.8%.
Protein Mixture
Table 2 shows the average isotopic ratios for each of the four standard proteins from three replicate LCMS experiments. The averaged isotopic ratio for each individual protein is very close to the expected value of one for the 1:1 mixture. Three of the four proteins have a coefficient of variation of 18% or less. VEGF had the highest standard deviation and the least number of peptides. It is not clear whether this is due to the nature of the individual peptides, or possibly the digestion efficiency for VEGF. VEGF is a very stable cytoplasmic protein. It forms homodimers that contain three intra-chain disulfide bonds and two interchain disulfide bonds. We have performed numerous digests and LCMS analyses of this protein and always see the same three peptides, although we do not always see all three in every experiment. Our data suggests that the nature of the peptide is an inherent factor in the reproducibility of isotopic ratios.
Table 2.
The 18O/16O ratios calculated from zoom scans for the tryptic digest of the four protein mixture with an expected 18O/16O ratio of 1. Peptides that occur in multiple charge states are listed separately for each charge state. Ratios were generated using method 1 o f ZoomQuant
| Peptide | Number | Charge | Ratio | Efficiency |
|---|---|---|---|---|
| Myoglobin | ||||
| LFTGHPETELK | 4 | 2 | 0.89 | 0.75 |
| LFTGHPETELK | 1 | 3 | 0.82 | 0.76 |
| GLSDGEWQQVLNVWGK | 2 | 2 | 1.00 | 0.49 |
| VEADIAGHGQEVLIR | 7 | 2 | 1.01 | 0.84 |
| VEADIAGHGQEVLIR | 3 | 3 | 1.06 | 0.66 |
| HGTVVLTALGGILK | 6 | 2 | 0.84 | 0.82 |
| HGTVVLTALGGILK | 1 | 3 | 0.82 | 0.85 |
| YLEFISDAIIHVLHSK | 2 | 2 | 0.51 | 0.81 |
| YLEFISDAIIHVLHSK | 1 | 3 | 0.61 | 0.20 |
| protein mean | 0.84 | 0.69 | ||
| standard deviation | 0.18 | 0.21 | ||
| coefficient of variation | 0.22 | |||
| Protein Tyrosine Phosphatase - 1 β | ||||
| LTISEDIK | 4 | 2 | 0.57 | 0.47 |
| FSYLAVIEGAK | 6 | 2 | 0.90 | 0.84 |
| FSYLAVIEGAK | 1 | 1 | 0.84 | 0.82 |
| ESGSLSPEHGPVVVHCSAGIGR | 3 | 3 | 1.11 | 0.81 |
| DVSPFDHSR | 3 | 2 | 1.02 | 0.78 |
| LHQEDNDYINASLIK | 4 | 2 | 0.94 | 0.71 |
| HEASDFPCRVAK | 1 | 2 | 0.74 | 0.57 |
| MGLIQTADQLR | 1 | 2 | 0.97 | 0.90 |
| protein mean | 0.88 | 0.74 | ||
| standard deviation | 0.17 | 0.15 | ||
| coefficient of variation | 0.19 | |||
| Glyceraldehyde-3-Phosphate Dehydrogenase | ||||
| GAAQNIIPASTGAAK | 4 | 2 | 1.06 | 0.83 |
| VPTPNVSVVDLTCR | 4 | 2 | 0.98 | 0.78 |
| protein mean | 1.02 | 0.80 | ||
| standard deviation | 0.05 | 0.03 | ||
| coefficient of variation | 0.05 | |||
| Vascular Endothelial Growth Factor | ||||
| HFLVQDPQCTCK | 1 | 2 | 1.55 | 0.55 |
| QLELNER | 2 | 2 | 0.73 | 0.78 |
| FMDVYQR | 1 | 2 | 1.02 | 0.76 |
| protein mean | 1.10 | 0.70 | ||
| standard deviation | 0.58 | 0.17 | ||
| coefficient of variation | 0.52 | |||
We used the same standard protein mixture in a 3:1 18O/16O mixture. The results for the averaged data are given in Table 3. The calculated isotopic ratios are all below the expected value of three, however the coefficient of variation for all four proteins is 20% or less. There are two major factors that contribute to uncertainty in the ratio calculations. The first is that the abundance of the 18O labeled species depends on the efficient incorporation of 18O into individual peptides and the level of enrichment of the H218O used. The abundance of the labeled species is likely to always be somewhat less than the unlabeled species which is independent of EMOS. Also, additional variation is reportedly introduced by sample handling and in mixing steps [11–13]. Differentially labeled samples were maintained separately up to the point of desalting. Equivalent aliquots were mixed, then applied to the desalting column. Steps prior to this included the initial splitting of the sample and the subsequent lyophilization. All of these steps are possible sources of small errors [16] and the difference between the actual ratio and the targeted ratio for mixing of the differentially labeled samples.
Table 3.
The 18O/16O ratios calculated from zoom scans for the tryptic digest of the four protein mixture with an expected 18O/16O ratio of 3. Peptides that occur in multiple charge states are listed separately for each charge state
| Peptide | Number | Charge | Ratio | Efficiency |
|---|---|---|---|---|
| Myoglobin | ||||
| ALELFR | 6 | 1 | 2.02 | 0.84 |
| LFTGHPETLEK | 5 | 2 | 2.50 | 0.79 |
| LFTGHPETLEKFDK | 1 | 3 | 1.50 | 0.87 |
| VEADIAGHGQEVLIR | 6 | 2 | 2.85 | 0.86 |
| VEADIAGHGQEVLIR | 4 | 3 | 2.16 | 0.77 |
| HGTVVLTALGGILK | 10 | 2 | 2.46 | 0.87 |
| protein mean | 2.25 | 0.83 | ||
| standard deviation | 0.47 | 0.04 | ||
| coefficient of variation | 0.21 | |||
| Protein Tyrosine Phosphatase - 1β | ||||
| FSYLAVIEGAK | 11 | 2 | 2.29 | 0.83 |
| FSYLAVIEGAK | 2 | 1 | 2.48 | 0.77 |
| LHQEDNDYINASLIK | 6 | 2 | 2.87 | 0.72 |
| LHQEDNDYINASLIK | 1 | 3 | 2.91 | 0.75 |
| DVSPFDHSR | 7 | 2 | 2.41 | 0.76 |
| ESGSLSPEHGPVVVHCSAGIGR | 4 | 3 | 2.12 | 0.87 |
| MGLIQTADQLR | 2 | 2 | 1.93 | 0.86 |
| protein mean | 2.43 | 0.80 | ||
| standard deviation | 0.36 | 0.06 | ||
| coefficient of variation | 0.15 | |||
| Glyceraldehyde-3-Phosphate Dehydrogenase | ||||
| GAAQNIIPASTGAAK | 9 | 2 | 2.61 | 0.83 |
| VPRPNVSVVDLTCR | 4 | 2 | 2.65 | 0.73 |
| protein mean | 2.63 | 0.78 | ||
| standard deviation | 0.03 | 0.07 | ||
| coefficient of variation | 0.01 | |||
| Vascular Endothelial Growth Factor | ||||
| FMDVYQR | 4 | 2 | 2.36 | 0.68 |
| HLFVQDPQTCK | 2 | 2 | 3.26 | 0.69 |
| QLELNER | 1 | 2 | 1.10 | 0.91 |
| protein mean | 2.24 | 0.76 | ||
| standard deviation | 1.08 | 0.13 | ||
| coefficient of variation | 0.48 | |||
Labeling Efficiency and Ratio Calculations
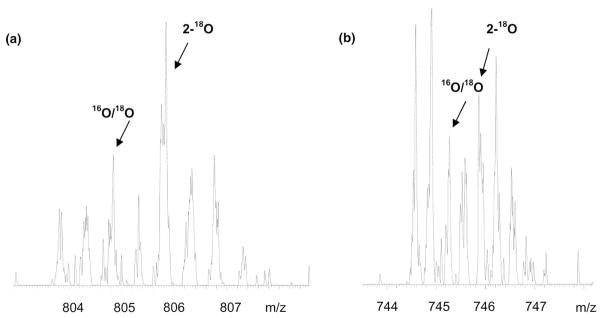
ZoomQuant automatically calculates labeling efficiencies for individual peptides as part of the final report. We use this term in the same manner as Stewart et al. [16]. The efficiency is defined as the percentage of 18O labeled peptides that are doubly labeled relative to the sum of all peptides that contain one 18O or two 18O atoms: 2-18O/(2-18O + 18O16O). The average efficiency for all of the peptides in our experiments with a one to one ratio mixture is 73%. The calculated efficiency for the myoglobin tryptic peptide GLSDEWQQVLNVWGK is 49%. The peptide dependence of EMOS has been reported to be linked to its efficiency as a pseudo-substrate for trypsin [8, 9, 12]. This underscores the necessity of using mass spectral information of sufficiently high resolution to include the singly 18O labeled species in the isotopic ratio calculation. The high resolution of the zoom scan even provides mass spectral information to effectively resolve the isotopic envelope of triply charged peptide pairs. The information given in Tables 2 and 3 show several peptides that are present in +2 and +3 charge states. Figure 1 shows a tryptic peptide from PTP-1β, ESGSLSPEHGPVVHCSAGIR, that is only present in the +3 charge state. The high resolution provided from zoom scans allows the accurate calculation of ion ratios from these triply charged peptides and for peptides for which as much as 51% of the 18O is present in the singly labeled species.
Figure 1.
(a) Zoom scan of the +2 charged peptide, VEADIAGHQEVLIR, from a myoglobin tryptic digest with an expected 18O/16O ratio of three. The unlabeled peptide (16O2) appears at m/z 803.8. The peaks from the doubly 18O labeled peptide and the singly labeled peptide are indicated by the arrows. The height of the M+2 peak shows clear enrichment from the singly labeled species. (b) Zoom scan of the +3 charged peptide, ESGSLSPEHGPVVVHCSAIGIGR, of PTP-1β from a tryptic digest of four standard proteins mixed with an expected 18O/16O ratio of one. The unlabeled peptide (16O2) appears at 744.6. This spectrum clearly illustrates the well resolved isotopic cluster provided by zoom scans for a +3 charged peptide. The peaks from the doubly 18O labeled peptide and the singly labeled peptide are indicated by arrows.
Reproducibility and Accuracy
The essential aspect of any quantification method is that it should be reproducible and that the limits of its accuracy should be known. We selected one tryptic peptide from myoglobin, HGTVVLTALGGILK, and grouped the isotopic ratios and efficiency for all zoom scans within a single LCMS run and compared them across all runs in which the peptide was observed. This particular peptide was selected because it is the most abundant peptide in our data set present in a single charge state. Table 4 shows the collected data for this peptide from the single protein digests and for the protein mixture. The calculated ratios cluster well for individual zoom scans within an LCMS experiment. The data for the differentially labeled set with an expected ratio of 0.3 shows the largest variance and the highest coefficient of variation is 48%. The higher coefficient of variation suggests that greater confidence can be gained by doing a reverse labeling experiment, or by replicates, an experimental approach commonly used for other methods that yield relative ratios. In contrast, the ratios for the mixture with an expected ratio of 1 have a much smaller coefficient of variation of 16% or less.
Table 4.
The zoom scan to zoom scan variance of the myoglobin tryptic peptide HGTVVLTALGGILK. Sample numbers indicate individually performed tryptic digests of myoglobin. Ratios from individual zoom scans of the above peptide within a single LCMS run are listed. Replicates indicate repeated LCMS analysis for a single tryptic digest. The mean and standard deviation was calculated for each individual sample. Ratios were generated using method 1 of ZoomQuant
| Expected ratio of 1 |
Expected ratio of 0.3 |
Expected ratio of 3.0 |
|||||
|---|---|---|---|---|---|---|---|
| Myoglobin |
4 Protein mix |
Myoglobin |
4 Protein mix |
||||
| Obs. ratio | Obs. ratio | Obs. ratio | Obs. ratio | ||||
| Sample 1 | 1.69 | replicate 1 | 0.83 | replicate 1 | 0.40 | replicate 1 | 2.43 |
| 1.57 | 0.75 | replicate 2 | 0.48 | 3.36 | |||
| 1.68 | 0.96 | 0.32 | 2.60 | ||||
| Mean | 1.65 | 0.82 | mean | 0.41 | mean | 2.80 | |
| s.d. | 0.07 | mean | 0.84 | s.d. | 0.20 | s.d. | 0.50 |
| c.v. | 0.04 | s.d. | 0.09 | c.v. | 0.48 | c.v. | 0.18 |
| Sample 2 | 1.03 | c.v. | 0.10 | replicate 3 | 0.36 | replicate 2 | 2.24 |
| 1.30 | replicate 2 | 0.75 | 1.81 | ||||
| 1.00 | 0.75 | 2.66 | |||||
| 0.91 | mean | 0.75 | 1.70 | ||||
| Mean | 1.06 | s.d. | 0.01 | mean | 2.10 | ||
| s.d. | 0.17 | c.v. | 0.01 | s.d. | 0.44 | ||
| c.v. | 0.16 | replicate 3 | 0.99 | c.v. | 0.21 | ||
| Sample 3 | 1.98 | replicate 3 | 2.80 | ||||
| 2.74 | |||||||
| 2.25 | |||||||
| mean | 2.60 | ||||||
| s.d. | 0.30 | ||||||
| c.v. | 0.12 | ||||||
Table 5 shows the collected data from every peptide we observed more than once from our mixed protein data sets in this paper. Those data with an expected ratio of 3 were adjusted to a value of 1 by multiplying by 0.3. This allowed us to group all of our observed peptides from data sets with an expected 18O/16O ratio of 1 and an expected ratio of 3 and examine the peptide-dependent variance for ratio measurements with respect to length, labeling efficiency, and sequence across all experiments. No clear trends were apparent for efficiency or identity. Peptide length did appear to have an effect on variance of ratio measurements. Charge state does not appear to be a factor, as only one peptide was present as the triply charged species only, and the CV for the peptide was 25%. Seven of the 20 peptides have coefficients of variation greater than 40%, and two of these peptides are seven residues or less in length. Fifty percent of the peptides of 12 amino acid residues in length or less have coefficients of variation of 50%. Two of three peptides that are seven residues or less in length have coefficients of variation of 80%. Two of the ten peptides with 12 or more residues have coefficients of variation between 40 and 50%. Once a peptide reaches a length of 12 residues, it tends to provide ratio measurements with much less variance. Only two of the ten peptides with 12 or more residues have coefficients of variation that exceed 40%.
Table 5.
The calculated 18O/16O ratios from ten separate LCMS runs of the four standard protein tryptic digest were pooled. Samples that were mixed in 18O/16O ratios of 3 were adjusted to a value of one by multiplying by 0.3, and the standard deviation was calculated for each peptide
| Peptide | Number | Ratio | Std. dev. |
|---|---|---|---|
| ALELFR | 9 | 1.25 | 0.91 |
| DVSPFDHSR | 10 | 0.87 | 0.22 |
| ESGSLSPEHGPVVHCSAGIGR | 7 | 0.88 | 0.25 |
| FMDVYQR | 5 | 0.83 | 0.13 |
| FSYLAVIEGAK | 22 | 0.84 | 0.19 |
| GAAQNIIPASTGAAK | 15 | 1.04 | 0.41 |
| GLSDEWQQVLNVVWGK | 3 | 0.90 | 0.18 |
| HGTVVLTALGGILK | 23 | 0.84 | 0.13 |
| HLFVQDPQTCK | 8 | 1.91 | 0.79 |
| IVSNASCTTNCLAPLAK | 4 | 1.29 | 0.28 |
| LFTGHPETLEK | 13 | 1.20 | 0.56 |
| LFTGHPETLEKFDK | 2 | 0.66 | 0.23 |
| LHQEDNDYINASLIK | 11 | 0.95 | 0.23 |
| LTLISEDIK | 8 | 0.71 | 0.53 |
| MGLIQTADQLR | 3 | 0.75 | 0.19 |
| QLELNER | 5 | 1.21 | 0.84 |
| VEADIAGHGQEVLIR | 26 | 0.90 | 0.20 |
| VGVNGFGR | 2 | 0.81 | 0.20 |
| VPTPNSVSVVDLTCR | 9 | 1.08 | 0.49 |
| YLEFISDAIIHVLHSK | 5 | 0.48 | 0.12 |
Mass Spectrometric Analysis and Sequence Coverage
We used the “triple play” method with dynamic exclusion to acquire data on the Deca XP Plus mass spectrometer. Dynamic exclusion parameters were programmed with a repeat count of 2 and an exclusion time of 5 min. This gave us the ability to collect as many as six zoom scans for each individual peptide in a single charge state. This would only be possible if all of the isotopically labeled variants (16O2, 16O1/18O1, and 18O2) were present in sufficient abundance to be isolated for zoom scan and MS/MS scans. Our instrument parameters were set so that we only selected the completely unlabeled (16O2) and the completely labeled (18O2) species for zoom scans and MS/MS. The zoom scan window of 10 Da allowed quantitation with either of these variants isolated as the center of mass. Our standard triple play method produced an average of 12 cycles/min. This gives us the ability to identify and quantify six peptides per 60 s chromatographic peak, assuming each peptide is isolated twice. We compared this with the standard method of using full MS scans followed by MS/MS that is used for qualitative LCMS where identification of a large set of peptides is the priority. Table 6 shows that there was no significant increase in sequence coverage when using the standard double play method in which a full scan MS is followed by MS/MS. This is likely reflective of chromatographic conditions, and that our mass spectrometer collection times provided ample separation space for the inherent complexity of the protein mixture.
Table 6.
Sequence coverage by protein was calculated for data acquired using the triple play MS method, and compared to sequence coverage when zoom scans were excluded from the MS method
| Myoglobin |
PTP-1β |
GAPDH |
VEGF |
|||||
|---|---|---|---|---|---|---|---|---|
| Sample | % a.a. | Peptides | % a.a. | Peptides | % a.a. | Peptides | % a.a. | Peptides |
| Expected 18O/16O ratio of 1 | ||||||||
| 6M urea replicate 1 | 39.87 | 6 | 6.21 | 1 | 18.67 | 5 | 8.41 | 2 |
| Replicate 2 | 35.29 | 5 | 2.5 | 1 | 9.34 | 2 | 5.14 | 1 |
| Replicate 3 | 50.33 | 7 | 4.6 | 2 | 15.96 | 4 | 9.47 | 2 |
| Mean | 41.83 | 6 | 4.43 | 1.33 | 14.65 | 3.66 | 7.67 | 1.66 |
| 8M urea replicate 1 | 38.56 | 4 | 13.1 | 4 | 4.22 | 1 | 3.27 | 1 |
| Replicate 2 | 49 | 6 | 25 | 8 | 8.73 | 2 | 8.41 | 2 |
| Replicate 3 | 36.7 | 4 | 16.78 | 5 | 8.73 | 2 | 6.54 | 2 |
| mean | 41.42 | 4.66 | 18.29 | 5.66 | 7.23 | 1.66 | 6.07 | 1.66 |
| Double play | 36.6 | 5 | 18.85 | 7 | 8.73 | 2 | 3.27 | 1 |
| Expected 18O/16O ratio of 3 | ||||||||
| 8 M urea replicate 1 | 32 | 5 | 22.76 | 5 | 8.73 | 3 | 3.27 | 3 |
| Replicate 2 | 30.07 | 5 | 20.69 | 7 | 8.73 | 2 | 5.14 | 1 |
| Replicate 3 | 32.03 | 4 | 22.76 | 6 | 8.73 | 2 | 3.27 | 1 |
Conclusions
We have developed a mass spectrometry method and compatible analysis software that allows for the simultaneous collection of qualitative and quantitative information from complex peptide mixtures. The method uses zoom scans to extend the resolution capabilities of the Deca XP Plus mass spectrometer. The methods presented in this paper, furthermore, provide the ability for relative quantification of 18O labeled peptides of + 3 and lower charge states. This has not been done previously for 18O differential expression profiling using ion trap mass spectrometers. We acquired data using the AGC mode; this limits the number of ions entering the trap and thereby reduces problems with space charging. Finally, peptides that label slowly or fail to exchange completely require the ability to include the singly 18O carboxy-terminal labeled peptides in ratio calculations to obtain consistent ratios. The methods presented here make use of this information for more precise ratio calculations.
Our experiments demonstrate that this method produces ratios that have good sample to sample reproducibility. The reproducibility of ratio calculations from zoom scan to zoom scan is also very good within a single LCMS run. Coefficients of variation are less than 21% for a given peptide within a single LCMS run.
The reproducibility and accuracy of quantification from zoom scan data on the LCQ instrument compares well with variance of ratio measurements obtained on Q-TOF [11], FT-ICR instruments [8], and with other methods for ion trap instruments [13]. Coefficients of variation are 21% for the expected ratios of one or three. Variance increases for ratios less than one, however, ratios can be verified by reverse labeling, or replicate experiments. MALDI-TOF data for 18O-labeling based quantification gives a comparable coefficient of variation of ~20% [10, 13]. Other methods that have been developed for relative quantification using 18O and ion trap mass spectrometers typically rely on extracted ion chromatograms and produce relative standard deviations of 25– 45% [11, 13]. However +3 charged peptides cannot be quantified using this approach. Our method can reliably detect 1.5- to 2-fold changes in protein expression. Furthermore, it expands the utility of using 18O labeling with ESI-IT technology to +3 charged peptides and those with a low efficiency of labeling, to improve the overall statistics for individual proteins in a differential expression study.
The experiments described in this paper did not address the issue of sensitivity of the method, which is instrument-dependent. Further experiments should be carried out to address the dynamic range of the method. The experiments described here show that we can reliably measure a 1.5-fold increase in protein expression. Peptides without an isotopic partner suggest that the change in protein expression exceeds the limits of the method and should be verified by a reverse labeling experiment.
The development of the ZoomQuant software permits automatic quantification of 18O data and makes it convenient to use 18O labeling for the analysis and quantification of complex mixtures. This is an inherently better method than using extracted ion chromatograms from MS data, which are subject to carryover from nearby peaks. The use of zoom scans does potentially decrease the number of peptides that can be identified from a single chromatographic peak. However, extended gradients and gas phase fractionation [18, 19] with replicate runs can be used to offset this limitation. Newly developed linear trap instruments offer much higher scan speeds and sensitivity. When the method of using zoom scans for 18O quantification is combined with these new instruments, the ability to collect quantitative and qualitative information of complex mixtures in a single LCMS experiment should greatly improve. The methods presented in this paper do offer a cost-efficient and effective means to perform qualitative and quantitative protein profiling experiments with the LCQ system that is already present in so many proteomics laboratories.
Acknowledgments
The authors gratefully acknowledge technical assistance and support from Brian Gau, Linda Smith, Regina Cole, and Victor Ruotti. This work was supported by the NHLBI Proteomics Center grant NIH-BAA-HL-02-04 (ASG, MO) and by a NIH minority postdoctoral supplement (WAH).
References
- 1.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandy A, Mann M. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Mol Cell Proteomics. 2002;10:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR. Analysis of Quantitative Proteomic Data Generated via Multidimensional Protein Identification Technology. Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 3.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR. Metabolic Labeling of Organisms with Stable Isotopes for Quantitative Proteomic Analysis. Anal Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 4.Berger SJ, Lee SW, Anderson GA, Pasa-Tolic L, Tolic N, Shen Y, Zhao R, Smith RD. High-Throughput Global Peptide Proteomic Analysis by Combining Stable Isotope Amino Acid Labeling and Data-Dependent Multiplexed-MS/MS. Anal Chem. 2002;74:4994–5000. doi: 10.1021/ac020105f. [DOI] [PubMed] [Google Scholar]
- 5.Gygi SP, Rist B, Gerber SA, Turicek F, Gelb MH, Aebersold R. Qualitative Analysis of Complex Protein Mixtures Using Isotope-Coded Affinity Tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 6.Creighton TE. Proteins. 2. W. H. Freeman and Company; New York: 1993. p. 4. [Google Scholar]
- 7.Available: http:www.ebi.ac.uk/IPI/
- 8.Johnson KL, Muddiman DC. A Method for Calculating 16O/18O Peptide Ion Ratios for the Relative Quantification of Proteomes. J Am Soc Mass Spectrom. 2004;15:437–445. doi: 10.1016/j.jasms.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Yao X, Alfonso C, Fenselau C. Dissection of Proteolytic 18O Labeling: Endoprotease-Catalyzed 16O-to-18O Exchange of Truncated Peptide Substrates. J Prot Res. 2002;2:147–152. doi: 10.1021/pr025572s. [DOI] [PubMed] [Google Scholar]
- 10.Wang KY, Ma Z, Quinn DF, Fu EW. Inverse 18O Labeling Mass Spectrometry for the Rapid Identification of Marker/Target Proteins. Anal Chem. 2001;73:3742–3750. doi: 10.1021/ac010043d. [DOI] [PubMed] [Google Scholar]
- 11.Zang L, Toy DP, Hancock WS, Sgroi DS, Karger BL. Proteomic Analysis of the Breast Using Laser Capture Micro-dissection, LC-MS and 16O/18O Isotopic Labeling. J Proteome Res. 2003;3:604–612. doi: 10.1021/pr034131l. [DOI] [PubMed] [Google Scholar]
- 12.Bantscheff M, Dumpelfeld B, Kuster B. Femtomol Senstivity Post-Digest 18O Labeling for Relative Quantification of Differential Protein Complex Composition. Rapid Commun Mass Spectrom. 2004;18:869–876. doi: 10.1002/rcm.1418. [DOI] [PubMed] [Google Scholar]
- 13.Heller M, Mattou H, Menzel C, Yao X. Trypsin Catalyzed 16O-to-18O Exchange for Comparative Proteomics: Tandem Mass Spectrometry Comparison Using MALDI-TOF, ESI-QTOF, and ESI-ion trap mass spectrometers. J Am Soc Mass Spectrom. 2003;14:704–718. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 14.Yao X, Freas A, Demirev AP, Fenselau CF. Proteolytic 18O Labeling for Comparative Proteomics: Model Studies with Two Serotypes of Adenovirus. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 15.Halligan BD, Slyper RY, Twigger SN, Hicks WA, Olivier M, Greene AS. ZoomQuant: An Application for the Quantitation of Stable Isotope Labeled Peptides. J Am Soc Mass Spectrom. doi: 10.1016/j.jasms.2004.11.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart II, Thomson T, Figeys D. 18O Labeling: A Tool for Proteomics. Rapid Commun Mass Spectrom. 2001;15:2456–2465. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]
- 17.Yao X, Freas A, Ramirez J, Demirev AP, Fenselau C. Proteolytic 18O Labeling for Comparative Proteomics: Model Studies with Two Serotypes of Adenovirus. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 18.Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Atchinson JD, Goodlet DR. Approaching Complete Peroxisome Characterization by Gas Phase Fractionation. Electrophoresis. 2002;23:3205–3216. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Blonder J, Rodriguez-Galan MC, Lucas DA, Young HA, Issaq HJ, Veenstra TD, Conrads TP. Proteomic Investigation of Natural Killer Cell Microsomes Using Gas-Phase Fractionation by Mass Spectrometry. Biochem Biophys Acta. 2004;1698:87–95. doi: 10.1016/j.bbapap.2003.10.009. [DOI] [PubMed] [Google Scholar]