Abstract
Background
The cell of origin of sarcoma is still unclear. High grade osteosarcomas frequently demonstrate the potential for multipotent differentiation and along with several other lines of evidence suggest that human mesenchymal stem cells (hMSC) might be the cell of origin.
Methods
The hMSCs were transformed with retrovirus containing human telomerase reverse transcriptase (hTERT), simian virus 40 large t antigen (SV40 TAg) and lentivirus containing oncogenic H-Ras serially. The changes of cellular phenotypes and multilineage differentiation capacity were observed and compared with the standard osteosarcoma cell lines.
Results
Two distinct genotypic and phenotypic sarcoma cell lines resulted from the same genetic events. The gene expression profiles became more complicated and the karyotype became more chaotic during hMSCs' tumorigenesis. The motility of transformed hMSC was promoted. hMSC and its derivatives could be induced to osteogenic, adipogenic and chondrogenic differentiation except that MSC-TSR4 lost osteogenic differentiation capacity.
Conclusion
Multilineage differentiation potential was retained during tumorigenesis of hMSCs and distinct sarcoma cell lines could arise with the same genetic events, providing good models in better understanding the concept of hMSC and in further investigation of the relationship of hMSCs and osteosarcomas.
Keywords: human mesenchymal stem cell, tumorigenesis, osteosarcoma, multilineage differentiation potential
Introduction
The cell of origin of sarcoma is still far from clear. Osteosarcoma, the most common primary malignant bone tumor in children and adolescents,1 could potentially be derived from a cell anywhere on the differentiation pathway between an hMSC and a mature osteoblast. Histologically, great variability exists in osteosarcomas with chondroblastic, fibroblastic and osteoblastic components.2 High grade osteosarcomas frequently demonstrate the potential for multilineage differentiation.1 These tumors could differentiate toward fibrous tissue, cartilage or bone, suggesting the cell of origin of osteosarcoma may be a more pluripotent cell than a mature osteoblast which is commonly considered as the cell of origin. This hypothesis can be supported by the clinical finding that although the most common site of osteosarcoma is the metaphysis of a long bone,1 osteosarcomas can arise primarily in soft tissues such as skeletal muscles3 and even in breasts.4 In these regions hMSCs and not osteoblasts are thought to be present.5
Mesenchymal stem cells are thought to be multipotent cells, which are present in adult bone marrow, that can replicate as undifferentiated cells and that have the potential to differentiate into the full lineage of mesenchymal tissues, including bone, cartilage, fat, tendon, muscle, and marrow stroma.6 Spontaneous malignant transformation has been reported in cultured murine mesenchymal stem cells, which transformed into osteosarcoma or fibrosarcoma.7,8 Malignant transformations have also been reported in long-term cultured hMSCs9 or in hTERT, a catalytic subunit of telomerase, immortalized hMSCs.10 Ewing sarcoma was recently reported to be derived form hMSCs as well.11 All these findings suggest that hMSCs could be the cell of origin of osteosarcoma.
Creation of human tumor cells from normal cells has been performed by introduction of hTERT, SV40 TAg and oncogenic H-RasV12, which maintains the telomere length, functionally inactivates the pRB and p53 tumor suppressor proteins and activates the mitogen-response pathway, respectively. This has been performed in numerous types of human cells including fibroblasts, astrocytes, ovarian surface epithelial cells, and skeletal muscle myoblasts.12-16 In reported literature, the combination of hTERT, SV40 TAg and H-Ras is still the most common way to transform normal cells into malignancy using defined genetic elements. Osteosarcoma has been demonstrated to utilize both telomerase and alternative lengthening telomere (ALT) mechanisms. Approximately 40% of osteosarcomas do express telomerase17,18 suggesting this may be a biologically rational first experimental step. Combined inactivation of pRB and p53 pathways has been reported to be common in osteosarcomas.19,20 Although SV40 does not likely have a role in the pathogenesis of osteosarcoma, this was viewed as a rational means of inactivating these pathways. Mutational activation of Ras was not found in most osteosarcomas,21 but analyses of patient derived osteosarcoma cell cultures have demonstrated constitutive activation of MAP kinase under conditions of serum starvation in most cases suggesting the pathway is activated and supporting the planned incorporation of an oncogenic activated Ras. The aim of this study is to define hMSCs' role as the potential cell of origin of osteosarcoma. The hypothesis that osteosarcoma is derived from hMSCs was tested in the following manner. Cell lines were established by introducing genetic alterations serially to transform hMSC into a malignant phenotype. Changes in cellular phenotype, gene expression profiles, karyotype, and multilineage differentiation capacity were compared to osteosarcoma.
Materials and Methods
Culture of hMSC and its derivatives
hMSCs were purchased from Cambrex. They were reported to be derived from a 7 years old white male. hMSC and its derivatives were cultured in Mesenchymal Stem Cell Medium (Cambrex, East Rutherford, NJ) at 37C° with 5% CO2. Mesenchymal Stem Cell Medium is based on low-glucose Dulbecco's Modified Eagle's Medium (DMEM) with 10% fetal bovine serum (FBS). Proliferation rates were measured every week by counting cell numbers of different cell lines. Standard osteosarcoma cell lines 143B, HOS, SaOS2 and U2OS were purchased from ATCC (Manassas, VA) and cultured in Eagle's Minimum Essential Medium (EMEM) (Cambrex, East Rutherford, NJ) with 10% FBS. Cell morphologies were observed and photos were taken using a Nikon Inverted Microscope ECLIPSE TE200 attached to a CCD (Diagnostic Instruments, Sterling Heights, MI).
Plasmid Constructions, Viral Transfections
Virus induced transfections were carried out serially with drug selection used to purify colonies after each transfection. Retroviral plasmid pLXIN-hTERT-neo was kindly provided by Dr. Izumi Horikawa from the National Institutes of Health (Bethesda, MD). This retroviral plasmid was stably transfected in PT67 packaging cells (Clontech, Mountain View, CA) using Lipofectamin 2000 (Invitrogen, Carlsbad, CA). pBABE-TAg-puro was purchased from DF/HCC DNA Resource Core of Harvard Medical School. To create amphotropic TAg retrovirus, PT67 packaging cells were stably transfected with pBABE-TAg-puro. The lentiviral plasmid pLenti-Ras-blast was constructed by subcloning the H-RasV12 gene from pcDNA3- RasV12 using pLenti6.2/V5-DEST Gateway® Vector Kit and ViraPower™ Lentiviral Expression Systems (Invitrogen, Carlsbad, CA). pLenti-Ras-blast was transiently transfected into 293FT packaging cells (Invitrogen, Carlsbad, CA) with 3 μg of pLenti expression plasmid DNA. Viral stocks of all three plasmids were harvested at 30hr and hMSCs were infected serially with 6μg/ml polybrene when they were at 30-50% confluence. Following 24 hours of co-culture, drug selections of infected hMSCs were performed with 100 μg/ml G418, 0.5 μg/ml puromycin and 2 μg/ml blasticidin. Drug resistant colonies were pooled together after transfection of hTERT and SV40 TAg whereas 16 colonies were picked up and subcultured separately after transfection with H-Ras.
RT-PCR, Immunoblotting, Telomerase and Telomere Analysis
RT-PCR for hTERT was performed as described.17 Expression of SV40 TAg and H-Ras were measured by immunoblotting of 75 μg total cellular protein with TAg antibody PAb 101 (Santa Cruz, Santa Cruz, CA) and H-Ras antibody C-20 (Santa Cruz, Santa Cruz, CA). Telomerase activity was measured by the PCR-based telomeric repeat amplification protocol (TRAP) assay using a TRAPeze® Telomerase Detection Kit (Chemicon, Temecula, CA) according to the manufacturer's protocol. Telomere length was measured by a TeloTAGGG Telomere Length Assay (Roche Applied Science, Indianapolis, IN) according to the manufacturer's protocol. Briefly, 1 μg of purified genomic DNA was digested with restriction enzymes Hinf I and Rsa I, separated on a 0.8% agarose gel and hybridized with a telomere-specific oligonucleotide probe.
Soft Agar Assays
An anchorage-independent growth assay was performed using soft agar as a culture medium. A base layer of 0.5% DMEM agar was placed onto 35-mm plates. Cells were seeded at a density of 5×103 cells/plate in 0.35% top agar containing DMEM and 10% FBS. Two ml of DMEM medium was added the next day when the agar was solid. Medium was changed every 3-4 days. After 3 weeks, the plates were stained with 0.5ml of 0.005% crystal violet and colonies were counted. All cell lines were measured in triplicate.
Subcutaneous and Orthotopic Tumorigenicity Assays
Six to 8 week old CB-17 SCID mice (Taconic, Germantown, NY) were injected with hMSC and its derivatives at a density of 4×106 cells/mouse in 200 μl of 1:1 mixed cell suspension and Matrigel (Becton Dickinson and Co.) subcutaneously in the flank for tumorigenicity studies or with 2×106 cells/mouse in 50 μl of Matrigel in the medullary canal of the left tibia for orthotopic studies. Parallel subcutaneous injections of four standard osteosarcoma cell lines were performed as positive controls. Mice were regularly checked for tumor formation and diameters of any tumors which formed were measured. Developed tumors were removed and parts of them were fixed in 10% formalin overnight and subjected to routine histological examination. The remainder of the transformed tumors were cut to small pieces and digested with 6 mg/ml collagenase overnight. After filtering the undigested residue with a 0.2 mm filter, cells were spun down and resuspended with fresh medium to place cells back in culture. All experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the Albert Einstein College of Medicine.
Microarray Assays
Total RNA of hMSC and its derivatives were extracted and gene expression was analyzed using Affymetrix Human genome 133A expression arrays (Affymetrix, Santa Clara, CA). Data were analyzed with Affymetrix Genechip Operating Software.
Spectral karyotyping (SKY) and Comparative genomic hybridization (CGH) Analysis
Hybridization and detection were carried out according to the manufacturer's protocol with slight modifications. Chromosomes were counterstained with 4', 6-diamidino-2-phenylindole (DAPI). For each case, 5 to 10 metaphase cells were analyzed by SKY. Images were acquired with a SD200 Spectra cube (ASI) mounted on a Zeiss Axioplan II microscope using a custom-designed optical filter (SKY-1) (Chroma Technology, Brattleboro, VT) and analyzed using SKY View 1.5 software (ASI, Carlsbad, CA). The breakpoints on the SKY-painted chromosomes were determined by comparison of the corresponding inverted-DAPI banding of the same chromosome and by comparison with the G-banded karyotype for each case. A breakpoint was considered recurrent if it was identified in 2 or more cases. A breakpoint cluster was defined as the occurrence of 4 or more breakpoints in the same chromosomal band.
High-molecular-weight DNA was extracted from hMSC and its derivatives by standard methods and subjected to CGH according to the previously published method with some modifications.22 The metaphase preparations were captured and processed by use of QUIPS software (Applied Imaging, Santa Clara, CA). Copy number changes were detected based on the variance of the red:green ratio profile from the standard of 1. Ratio values of 1.20 and 0.80 were used as upper and lower thresholds to define gains and losses, respectively. High-level amplification was defined as the occurrence of fluorescein intensity values in excess of 2.0 along with a strong localized FITC (fluorescein isothiocynate) signal at the chromosomal site.
Motility and Migration Assays
Motility (random migration) was measured by a wound healing assay as described previously.23 Cells were cultured in serum-free medium 24 hours before creating wounds. Photos were taken every 6 hours until 48 hours. Migration (haptotaxis) was measured using the QCM™ Quantitative Cell Migration Assay (Chemicon, Temecula, CA). For the migration assays, cells were serum starved 24 hours before being plated into Boyden Chambers. Cells that migrated to the outside of the chamber were stained and extracted in 300 μl of extraction buffer. Absorbance at 570 nm was measured using a microplate spectrophotometer (BIO-RAD, Hercules, CA).
Differentiation Assays
The osteogenic, adipogenic and chondrogenic differentiation capacity were measured according to manufacturer's protocols using a Mesenchymal Stem Cell Osteogenesis Kit (Chemicon, Temecula, CA), Mesenchymal Adipogenesis Kit (Chemicon, Temecula, CA) and Chondrogenic Differentiation Medium plus transforming growth factor (TGF) β3 (Cambrex, East Rutherford, NJ), respectively. Cells were cultured in differentiation induction medium for 3 weeks. Differentiated cells were stained with Alizarin Red, Oil Red O and immunohistochemical staining of type II collagen using antibody Collagen Type II (003-02) (Santa Cruz, Santa Cruz, CA) which can stain calcium, fat and type II collagen respectively to verify formation of osteocytes, adipocytes and chondrocytes. Photos were taken using a Nikon Inverted Microscope ECLIPSE TE200 attached to a CCD (Diagnostic Instruments, Sterling Heights, MI).
Statistics and Analysis
Differences in proliferation rate, colony formation in soft agar, and migration were estimated by comparison of hMSC with its derivatives. Two-sided p values and 95% confidence intervals were calculated using SPSS 10.0. p<0.05 was considered as statistically significant.
Results
Generation of Genetically Modified Human Mesenchymal Stem Cell Lines
hMSCs stably expressing hTERT, SV40 TAg and H-RasV12 were created serially through the use of independent selectable markers (neomycin, puromycin and blasticidin, respectively) after transfection by viral constructs. For each infection, parallel cultures were infected with empty vector specifying only a drug resistance gene as a control except the infection with H-Ras because of the CcdB suicide gene contained in the destination vector. Serially transfected cell lines were termed as MSC-T (T representing hTERT), MSC-S (S representing SV40 TAg), MSC-TS and MSC-TSR (R representing H-Ras), respectively. The over-expression of hTERT was confirmed by RT-PCR (Fig 1A) and functional telomerase activity was present in vitro as assessed by TRAP assays in MSC-T, MSC-TS and MSC-TSR (Fig 1B). Expression of TAg was detected in MSC-S, MSC-TS and MSC-TSR and expression of H-Ras was detected in MSC-TSR through western blots (Fig 1A).
Fig. 1.
Creation of hMSC's derivatives expressing hTERT, SV40 TAg and H-Ras. A, hTERT expression was confirmed in MSC-T, MSC-TS, MSC-TSR6 and MSC-TSR4 by RT-PCR with βactin serving as a control. SV40 TAg protein expression was confirmed in MSC-S, MSC-TS, MSC-TSR6 and MSC-TSR4, H-Ras protein expression was confirmed in MSC-TSR6 and MSC-TSR4 by western blots. B, telomerase activity was observed in MSC-T, MSC-TS, MSC-TSR6 and MSC-TSR4 by TRAP assay. HT, heat treated; IC, internal control. C, telomere lengths were analyzed by hybridization of genomic DNA with a telomerase-specific oligonucleotide probe. The positions of size standards (kb) are indicated at the left.
After transfection of hTERT, telomere length was extended, forming a longer and homogeneous telomere, unlike the heterogeneous pattern of telomere length in hMSCs. Telomere length became even longer with expression of H-Ras in MSC-TSR6 as compared to MSC-TSR4 (Fig 1C).
No distinguishable changes in cellular morphology, growth rate and growth pattern among separate selected colonies were observed after transfection of hTERT and TAg, so the colonies were pooled together as MSC-T and MSC-TS. Two distinct cellular morphologies appeared after transfection with H-Ras. Eleven out of 16 colonies (represented by MSC-TSR4) maintained a spindle-like shape similar to hMSCs. The cells had a stripe-like appearance when reaching 100% confluence (Fig 2A a). The other 5 colonies (represented by MSC-TSR6) developed into smaller and polygon-like cells. These cells appeared disorderly and interlaced when at 100% confluence (Fig 2A b). The fastest growing colony in each group, MSC-TSR4 and MSC-TSR6, were selected and subjected to the subsequent analysis.
Fig. 2.
Growth curve of hMSC and its derivatives and soft agar assay. A, growth curve of transfected cell lines. a, stripe-like pattern represented by MSC-TSR4, b, disorderly pattern represented by MSC-TSR6. B, anchorage independent growth in soft agar of transfected cell lines. a, MSC-T; b, MSC-S; c, MSC-TS; d, MSC-TSR4; e, MSC-TSR6; f, HOS. *, p<0.01 versus MSC-TS.
Changes in Proliferation
Transfection of hTERT and TAg slightly increased the cellular proliferation rate in MSC-T and MSC-TS compared with hMSC. After transfection of H-Ras, cellular growth rate increased markedly by comparison of MSC-TSR4 and MSC-TSR6 with MSC-T and MSC-TS (p<0.05) (Fig 2A). MSC-TSR6 grew even faster than MSC-TSR4 with a 22 hour and 29 hour population doubling time respectively (p<0.05) (Fig 2A).
Immortalization
Expression of hTERT kept hMSC from becoming senescent. MSC-Ts were cultured for more than 80 passages without growth arrest or change in cellular morphology whereas hMSC could not be cultured beyond 20 passages before becoming senescent proved by β-glucosidase staining (data not shown). This suggested MSC-Ts had become immortalized cells. Expression of TAg alone permitted MSC-S to keep growing until passage 50 at which time they detached and did not continue proliferating (Fig 2A). hMSCs transfected with H-Ras alone or combination of hTERT and H-Ras detached and died after a short period of rapid growth, showing H-Ras was not tolerated without TAg even in the immortalized cell line MSC-T (data not shown).
Anchorage independent growth and tumorigenesis
After transfection of H-Ras, cells lost contact inhibition when reaching 100% confluence, forming a multilayer growth pattern. Anchorage-independent growth of hMSC and its derivatives were measured through a colony forming assay in soft agar with the osteosarcoma standard cell line HOS used as a positive control. This is widely accepted as one of the essential characteristic of malignant cells.12 MSC and MSC-T did not form colonies in soft agar whereas MSC-S, MSC-TS, MSC-TSR6 and MSC-TSR4 formed variable numbers of colonies (Fig 2B). Colonies numbers of MSC-TSR4 and MSC-TSR6 were more than that of MSC-TS (p<0.01) but no significant difference was found between these cell lines. MSC-S was capable of forming colonies in soft agar in a considerable numbers although the volume of the colonies was smaller than H-Ras transfected cells and HOS (Fig 2B).
In tumorigenicity assays, tumors formed only with MSC-TSR6 and MSC-TSR4 either in s.c. injected or in orthotopic injected mice 4 weeks after the implantation. No palpable tumor was found with MSC, MSC-T, MSC-S and MSC-TS at the same timepoint (Tab 1). In orthotopic injected mice, tumors formed around the tibia but not inside the tibia (Fig 3A), which may be caused by the leak of cell suspension from needle canals. After animals were sacrificed, the tumor bearing legs were amputated and plain radiographs were taken. No distinguishable cortical destruction or ectopic ossification was found (Fig S1). Xenografts of MSC-TSR6 and MSC-TSR4 grew well in the same medium with all three selective drugs and at equivalent rates (Fig 3B) and growth patterns (Fig 3C) as their parental cells in culture. In pathology, there was no markedly histological difference between MSC-TSR6 and MSC-TSR4 although they were distinct in morphological feature when cultured in dishes. They are high-grade spindle cell sarcomas, anaplastic, with foci of necrosis, mitotic figures are frequent, but without any evidence of osteoid, tumor bone, chondroid, or any other matrix production (Fig 3D). The immunohistochemical markers which might provide some clues as to specific features of cell differentiation, namely cytokeratin (CAM 5.2), S-100 protein, smooth muscle acitn, muscle-specific actin, and even vimentin, are all negative (data not shown). Interestingly, tumors formed with all standard osteosarcoma cell lines when injected into mice, but no osteoid was found in any of them histologically as well (data not shown).
Table 1.
Formation of tumors in CB-17 SCID mice
| Cell lines | Genotype | Injection site | Number of tumors/number of injections |
|---|---|---|---|
| MSC | hTERT-, TAg-, Ras- | s.c. | 0/8 |
| MSC-T | hTERT+, TAg-, Ras- | s.c. | 0/8 |
| MSC-S | hTERT-, TAg+, Ras- | s.c. | 0/8 |
| MSC-TS | hTERT+, TAg+, Ras- | s.c. | 0/8 |
| MSC-TSR6 | hTERT+, TAg+, Ras+ | s.c. | 7/8 |
| hTERT+, TAg+, Ras+ | tibia | 4/4 | |
| MSC-TSR4 | hTERT+, TAg+, Ras+ | s.c. | 6/8 |
| hTERT+, TAg+, Ras+ | tibia | 4/4 |
Abbreviation: s.c. subcutaneous
Fig. 3.
Tumorigenicity assays in C.B-17 SCID mice. A, gross findings of H-Ras transfected cell lines implanted in mouse tibia. B, growth curve of parental H-Ras transfected cell lines and their relevant xenografts in culture. C, Cell morphologies of xenografts of H-Ras transfected cell lines when reaching 100% confluence in culture. D, histopathological findings of H-Ras transfected cell lines. T, tumor; B, tibia cortex; M, bone marrow. a, MSC-TSR6, b, MSC-TSR4 in all panels.
Changes of gene expression profiles, karyotpyes of transformed cell lines
The gene expression profiles of hMSC and its derivatives were compared. Hierarchical clustering analysis revealed that the gene expression profile of MSC-TSR6 was more similar to that of HOS than MSC-TSR4 (Fig 4A). In SKY and CGH assays, the karyotype of MSC-T appeared normal, but it became more and more complex after transfection of TAg and Ras (Fig 4B) (Table S1,S2) which made it more similar to the complex karyotype of osteosarcoma. The karyotype of MSC-TSR6 was more complicated than that of MSC-TSR4, suggesting it may be closer to osteosarcoma as compared to MSC-TSR4 (Fig 4B).
Fig. 4.
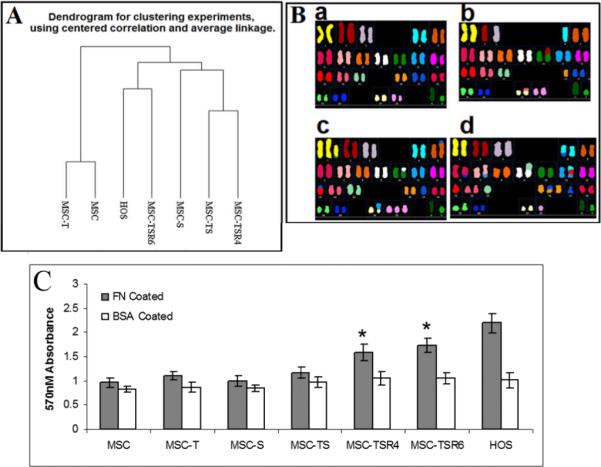
Gene expression profiles, SKY assays and Migration capacity of hMSC and its derivatives. A, clustering analysis of gene expression profile of hMSC and transfected cell lines base on microarray analyses. B, karyotypes of hMSC and its derivatives. a, MSC-T; b, MSC-TS; c, MSC-TSR4; d, MSC-TSR6. C, changes in migration (haptotaxis) of transfected cell lines measured by fibronectin coated Boyden chamber. BAS-coated chambers served as negatives controls, HOS served as a positive control. FN, fibronectin; BAS, bovine serum albumin. *, p<0.05 versus MSC, MSC-T, MSC-S and MSC-TS.
Changes in the Motility of the transformed cell lines
There are at least two different kinds of cellular motility, including random migration which is usually measured by wound healing assays and haptotaxis, a cell movement towards an immobilized extracellular matrix (ECM) protein gradient, which is usually measured by a Boyden chamber system. In our experiments, no significant changes were observed when comparing the random migration capacity of hMSC with its derivatives (Fig S2). But when comparing haptotaxis using a Boyden chamber coated with Fibronectin as ECM, MSC-TSR6 and MSC-TSR4 had greater mobility than MSC, MSC-T and MSC-TS (p<0.05) but no significant difference was observed between MSC-TSR6 and MSC-TSR4 (Fig 4C).
Changes in Multilineage Differentiation Capacity of transformed hMSCs
In osteogenic differentiation assays, the expression of hTERT did not influence the capacity of calcareous material deposit, which can be observed by Alizarin Red staining, in comparison to hMSC (Fig 5a, b). Osteogenesis was accelerated and enhanced markedly in MSC-S as compared to hMSCs (Fig 5c). Expression of H-Ras triggered different changes in the H-Ras transfected cell lines with an inhibition of osteogenic differentiation in MSC-TSR4, whereas osteogenesis was enhanced in MSC-TSR6 (Fig 5d, e). Taken together, hMSC maintained capacity for osteogenic differentiation during tumorigenesis except in MSC-TSR4 (Table S3). An osteogenic differentiation assay was also performed using the four standard osteosarcoma cell lines, 143B, HOS, SaOS-2 and U2OS. Only HOS could be induced to osteogenic differentiation (Fig S3).
Fig. 5.
Changes in multilineage differentiation capacity in transfected hMSC. a-e, osteogenic differentiation staining with Alizarin Red. f-j, adipogenic differentiation staining with Oil Red O. k-o, chondrogenic differentiation immunohistochemical staining with type II collagen. Cell lines are MSC, MSC-T, MSC-S, MSC-TSR6 and MSC-TSR4 from top to bottom in all of three differentiation columns.
In adipogenic differentiation assays, hMSC and all of its derivatives can differentiate into adipocytes with variable efficiency which could be demonstrated by the finding of Oil Red O stained fat vesicle inside cells. Genetic modifications of hTERT, TAg or H-Ras did not change MSCs capacity for adipogenic differentiation. Expression of TAg significantly decreased the efficiency of adipogenic differentiation which was opposite to its effect on osteogenic differentiation (Fig 5f, g). In our experiments, the capacity of adipogenic differentiation was markedly but not completely inhibited in MSC-TSR6 and MSC-TSR4 with a considerable amount of fat vesicles formed although their volume was small and could only be observed with higher magnification microscopy (Fig 5i, j), demonstrating hMSCs maintained capability of adipogenesis during this process of malignant transformation (Table S3).
For the chondrogenic differentiation assays, all cell lines were proved to retain the capacity for chondrogenic differentiation with the findings of positive immunohistochemical staining for type II collagen after three weeks of three-dimensional culture in chondrogenic induction media (Fig 5k-o). The production of type II collagen was enhanced in H-Ras transfected derivatives MSC-TSR6 and MSC-TSR4 when compared with other derivatives and MSC-TSR6 showed stronger staining than MSC-TSR4 (Fig 5n, o). The capacity for chondrogenic differentiation of transformed hMSC's derivatives was retained during tumorigenesis (Table S3).
Discussion
Human mesenchymal stem cells, a multipotent cell, are precursor cells of all the mesenchymal tissue including bone, cartilage, fat, muscle, etc..5 Recently, more and more evidence has shown that hMSCs could be the progenitor cells of sarcomas.8,10,11 In the present study, we successfully transformed hMSC to a sarcoma with defined genetic elements. As far as we know, this is the first reported malignant transformation of hMSCs by introducing the combination of genetic alterations of hTERT, SV40 TAg and H-Ras, which is the most common used genetic combination in tumorigenesis.12 Transformed sarcoma cell lines, MSC-TSR6 and MSC-TSR4 showed malignant features including loss of contact inhibition, capability for anchorage-independent growth and tumorigenicity in SCID mice. hMSC has been previously reported to be transformed into sarcoma with hTERT, Bmi1 and H-Ras which is consistent with our findings.24
After transfection of H-Ras, two distinct types of malignant cells formed, with different telomere length, different cellular morphologies, different growth patterns and proliferation rates, different gene expression profiles, different karyotypes, different capacity for chondrogenic and osteogenic differentiation. Although there was no significant difference in histological appearance in vivo-they were both high-grade spindle cell sarcomas. It was demonstrated that two distinct sarcoma cell lines had resulted from the same genetic alterations.
The formation of two distinct sarcoma cell lines could be explained in multiple ways. Genetic instability can lead to random variation accounting for the difference. The fact that 16 colonies were picked up after transfection of H-Ras and separated into only two distinct morphological groups suggest this is not the case. Another explanation is based upon different gene profiles of precursor cells. The hypothesis has been raised that the pre-existing gene profiles in normal precursor cells may dictate the subtype of tumor rather than epigenetic mutation or introduction as is the case with different breast cancer subtypes derived from the same human breast epithelial cells harvested with different methods.25 The concept of mesenchymal stem cells is still imprecise, which is defined more functionally by the capacity for self-renewal and multilineage differentiation capacity rather than a defined panel of well characterized surface markers in order to identify them unambiguously.5 Hence, hMSCs can be postulated to be not uniform cells but mixtures of heterogeneous cells which might contain different pre-existing gene profiles that could be the basis of subsequent development of different characteristics. Undergoing the same genetic introductions with different pre-existing gene profiles, hMSCs resulted in different sarcomas.
Comparing gene expression profiles, MSC-TSR6 was more similar to the osteogenic inducible standard osteosarcoma cell line HOS than MSC-TSR4, in concordance with the findings that MSC-TSR6 can be induced to osteocytes but MSC-TSR4 can not, MSC-TSR6 was perhaps transformed towards an osteogenic lineage and MSC-TSR4 was transformed towards other lineages.
The haptotaxis of MSC-TSR6 and MSC-TSR4 increased as compared to the parental cell lines. Promotion of cell motility by activation of H-Ras, loss of p53 function or a combination of both of them have been previously reported,26-28 which is consistent with our findings.
Multilineage differentiation capacity of hMSC was retained during tumorigenesis. This is perhaps not surprising because of the multi-lineage potential of hMSCs. However, this at least suggests that hMSC can possibly be the precursor of osteosarcoma, not exclusively being osteoblasts as traditionally believed. Recently animal model of osteosarcoma with gene specific ablation also support this finding.29 In the current study, although differentiation capacities were impacted somewhat, they were not completely inhibited with the sole exception that MSC-TSR4 lost the capacity for osteogenic differentiation. It has been previously reported that osteogenic differentiation was completely inhibited by hTERT, Bmi1 and H-Ras transformation in hMSC24, which is consistent with MSC-TSR4. In the four standard osteosarcoma cell lines tested, only HOS could be induced to differentiate into an osteocyte. This finding may suggest a suppressive action of H-Ras in osteogenesis. This is supported by the fact that the osteogenic differentiation capacity of 143B, a Ki-Ras transformed HOS cell line,30 was completely inhibited as compared with HOS. Osteogenesis was enhanced in the other H-Ras transfected cell line MSC-TSR6, which may indicate that H-Ras plays a complicated role in osteogenic differentiation instead of a simple suppressive action.
Lacking specific genetic alterations and having complex karyotypes in osteosarcoma has impeded efforts to understand the pathogenesis of this lesion. The real genetic alterations of osteosarcoma in vivo are still unclear and can not be simulated in vitro so far. Therefore, we converted hMSCs to sarcomas with the most commonly used genetic alterations hTERT, SV40 TAg and H-Ras to observe changes of cellular phenotype as a first step in studying hMSC's role in the pathogenesis of sarcomas. Although SV40 Tag was not found to be involved in the pathogenesis of osteosarcoma clinically, RB and TP53 were frequent disrupted in osteosarcoma patient specimens.19,20 In animal models, simultaneous ablation of Rb and P53 in osteoblastic lineage cells produced spontaneous osteosarcoma in mice with high penetration rate.29,31 In the current study, we used SV40 TAg to silence RB and TP53 in hMSC. The successful transformation of hMSC in vitro recapitulated the relevance of these two factors in this disease. Even if this combination does not accurately reflect the pathogenesis of osteosarcoma, this approach still revealed the changes in hMSCs that occurred during transformation.
The sarcoma that we created using defined genetic elements in the current study displayed some essential features of a neoplasm. However, we acknowledge that these sarcomas were devoid of bone matrix, and ability of invasion into cortical bone, which are clinically characteristic of osteosarcoma. There are three factors that could account for the failure: the cells, the genetic elements and random factors. This result may be due to any of these, but it is our belief that the selected genetic elements are the most likely explanation. Nevertheless, the multi-lineage potential and chaotic chromosomal alteration observed in this model mirror the extremely diverse clinical features of osteosarcoma. This model therefore serves as a starting point of additional ongoing efforts to further understand the pathogenesis of this fascinating disease. As mutant H-Ras does not seem to play an important role in the pathogenesis of osteosarcoma, other genetic elements such as TGF β, IGF pathway members or WNT pathway members will be transfected into hMSCs as the third element instead of H-Ras as part of future work.
Acknowledgement
We thank members of the AECOM Microarray Core Facility for assistance in the Affymetrix analyses. We thank Dr. Rani S. Sellers, Dr. Radma Mahmood and members of the AECOM Histopathology Facility for assistance in performing histological preparations and interpretations.
Grant support: National Cancer Institute grant R01 CA-83132, Foster Foundation, and Cure Search Foundation.
References
- 1.Meyers PA, Gorlick R. Osteosarcoma. Pediatr Clin North Am. 1997;44:973–89. doi: 10.1016/s0031-3955(05)70540-x. [DOI] [PubMed] [Google Scholar]
- 2.Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- 3.Hulse N, Paul AS. Soft tissue osteosarcoma: a case report. Acta Orthop Belg. 2006;72:783–5. [PubMed] [Google Scholar]
- 4.Bahrami A, Resetkova E, Ro JY, et al. Primary osteosarcoma of the breast: report of 2 cases. Arch Pathol Lab Med. 2007;131:792–5. doi: 10.5858/2007-131-792-POOTBR. [DOI] [PubMed] [Google Scholar]
- 5.Vaananen HK. Mesenchymal stem cells. Ann Med. 2005;37:469–79. doi: 10.1080/07853890500371957. [DOI] [PubMed] [Google Scholar]
- 6.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 7.Miura M, Miura Y, Padilla-Nash HM, et al. Accumulated chromosomal instability in murine bone marrow mesenchymal stem cells leads to malignant transformation. Stem Cells. 2006;24:1095–103. doi: 10.1634/stemcells.2005-0403. [DOI] [PubMed] [Google Scholar]
- 8.Tolar J, Nauta AJ, Osborn MJ, et al. Sarcoma derived from cultured mesenchymal stem cells. Stem Cells. 2007;25:371–9. doi: 10.1634/stemcells.2005-0620. [DOI] [PubMed] [Google Scholar]
- 9.Rubio D, Garcia-Castro J, Martin MC, et al. Spontaneous human adult stem cell transformation. Cancer Res. 2005;65:3035–9. doi: 10.1158/0008-5472.CAN-04-4194. [DOI] [PubMed] [Google Scholar]
- 10.Serakinci N, Guldberg P, Burns JS, et al. Adult human mesenchymal stem cell as a target for neoplastic transformation. Oncogene. 2004;23:5095–8. doi: 10.1038/sj.onc.1207651. [DOI] [PubMed] [Google Scholar]
- 11.Tirode F, Laud-Duval K, Prieur A, et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–9. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 12.Hahn WC, Counter CM, Lundberg AS, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–8. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 13.Rich JN, Guo C, McLendon RE, et al. A genetically tractable model of human glioma formation. Cancer Res. 2001;61:3556–60. [PubMed] [Google Scholar]
- 14.Sonoda Y, Ozawa T, Hirose Y, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001;61:4956–60. [PubMed] [Google Scholar]
- 15.Liu J, Yang G, Thompson-Lanza JA, et al. A genetically defined model for human ovarian cancer. Cancer Res. 2004;64:1655–63. doi: 10.1158/0008-5472.can-03-3380. [DOI] [PubMed] [Google Scholar]
- 16.Linardic CM, Downie DL, Qualman S, et al. Genetic modeling of human rhabdomyosarcoma. Cancer Res. 2005;65:4490–5. doi: 10.1158/0008-5472.CAN-04-3194. [DOI] [PubMed] [Google Scholar]
- 17.Ulaner GA, Huang HY, Otero J, et al. Absence of a telomere maintenance mechanism as a favorable prognostic factor in patients with osteosarcoma. Cancer Res. 2003;63:1759–63. [PubMed] [Google Scholar]
- 18.Terasaki T, Kyo S, Takakura M, et al. Analysis of telomerase activity and telomere length in bone and soft tissue tumors. Oncol Rep. 2004;11:1307–11. [PubMed] [Google Scholar]
- 19.Overholtzer M, Rao PH, Favis R, et al. The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc Natl Acad Sci U S A. 2003;100:11547–52. doi: 10.1073/pnas.1934852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendoza SM, Konishi T, Miller CW. Integration of SV40 in human osteosarcoma DNA. Oncogene. 1998;17:2457–62. doi: 10.1038/sj.onc.1202179. [DOI] [PubMed] [Google Scholar]
- 21.Antillon-Klussmann F, Garcia-Delgado M, Villa-Elizaga I, et al. Mutational activation of ras genes is absent in pediatric osteosarcoma. Cancer Genet Cytogenet. 1995;79:49–53. doi: 10.1016/0165-4608(94)00115-r. [DOI] [PubMed] [Google Scholar]
- 22.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–21. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 23.Yang R, Hoang BH, Kubo T, et al. Over-expression of parathyroid hormone Type 1 receptor confers an aggressive phenotype in osteosarcoma. Int J Cancer. 2007;121:943–54. doi: 10.1002/ijc.22749. [DOI] [PubMed] [Google Scholar]
- 24.Shima Y, Okamoto T, Aoyama T, et al. In vitro transformation of mesenchymal stem cells by oncogenic H-rasVal12. Biochem Biophys Res Commun. 2007;353:60–6. doi: 10.1016/j.bbrc.2006.11.137. [DOI] [PubMed] [Google Scholar]
- 25.Ince TA, Richardson AL, Bell GW, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–70. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Guo F, Gao Y, Wang L, et al. p19Arf-p53 tumor suppressor pathway regulates cell motility by suppression of phosphoinositide 3-kinase and Rac1 GTPase activities. J Biol Chem. 2003;278:14414–9. doi: 10.1074/jbc.M300341200. [DOI] [PubMed] [Google Scholar]
- 27.Kim MS, Lee EJ, Kim HR, et al. p38 kinase is a key signaling molecule for H-Ras-induced cell motility and invasive phenotype in human breast epithelial cells. Cancer Res. 2003;63:5454–61. [PubMed] [Google Scholar]
- 28.Xia M, Land H. Tumor suppressor p53 restricts Ras stimulation of RhoA and cancer cell motility. Nat Struct Mol Biol. 2007;14:215–23. doi: 10.1038/nsmb1208. [DOI] [PubMed] [Google Scholar]
- 29.Berman SD, Calo E, Landman AS, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A. 2008;105:11851–6. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dass CR, Ek ET, Choong PF. Human xenograft osteosarcoma models with spontaneous metastasis in mice: clinical relevance and applicability for drug testing. J Cancer Res Clin Oncol. 2007;133:193–8. doi: 10.1007/s00432-006-0157-x. [DOI] [PubMed] [Google Scholar]
- 31.Walkley CR, Qudsi R, Sankaran VG, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–76. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]






