Abstract
Aims
To identify the genes regulated by RR11, the regulator of the Streptococcus mutans HK/RR11 two-component system.
Methods and Results
The S. mutans RR11-encoding gene was inactivated, and the effects of gene disruption on the cell's ability to form biofilms under stresses and acquire extracellular DNA were tested. Biofilm was reduced in cells lacking RR11 following exposure to oxidative stress. RR11-defective cells showed approx. 20-fold reduction in transformation efficiency. Microarray used to decipher the RR11-regulated genes in biofilm showed that approx. 5% of the UA159 genome underwent a significant change in expression. RR11 was found to regulate 174 genes, including genes involved in competence, stress-response and cell division.
Conclusions
Target genes controlled by RR11during biofilm growth have been identified by a comparison of transcriptional profiles between an RR11 defective mutant and the parental strain. The results demonstrated that RR11 is involved in the control of diverse cellular processes, including the formation of biofilm under oxidative stress and development of genetic competence.
Significance and Impact of the Study
The regulator of HK/RR11 system controls a large regulon and is an important regulator involved in stress response during S. mutans biofilm growth enabling the survival and persistence of its progeny in the microbial community.
Keywords: biofilm, competence, DNA microarray, stress response, two-component system
Introduction
Dental plaque is the diverse biofilm community found on the tooth surface and comprises over 700 different bacterial species (Aas et al. 2005). Streptococcus mutans, a member of the oral biofilm community, causes dissolution of tooth enamel by producing acid from dietary carbohydrates and is considered one of the principal etiological agents of dental caries (Mitchell 2003). It has evolved a biofilm lifestyle to survive and persist in the oral cavity, where it employs two-component systems to sense and respond to environmental stimuli. Changing environmental conditions are sensed by the membrane-bound histidine kinase (HK) receptor, resulting in its autophosphorylation. The resulting high-energy phosphoryl group is transferred to the cognate cytosolic response regulator (RR), which then acts as a transcriptional regulator (Stock et al. 2000). One of the 13 putative two-component systems found in the S. mutans UA159 genome consists of the membrane-associated sensor HK11 and its cognate regulator RR11 (Ajdic et al. 2002; Lévesque et al. 2007). This system has been demonstrated to be involved in cell segregation and in the response to thermal, oxidative and acid stresses (Li et al. 2002a; Biswas et al. 2008). Although HK/RR11 system has been implicated in S. mutans biofilm development (Bhagwat et al. 2001; Li et al. 2002a), the genes controlled by RR11 during biofilm growth are still unknown. In the present study, DNA microarray technology was used to decipher the genes controlled by RR11 during S. mutans biofilm growth.
Materials and methods
Bacterial strains, growth media and conditions
Streptococcus mutans UA159 was used as wild-type strain. Mutants were constructed by a PCR-based gene replacement strategy (Lau et al. 2002). All strains were grown in Todd-Hewitt yeast extract (THYE) broth and incubated statically at 37°C in air with 5% CO2.
In vitro model for growing biofilms
Static biofilms were developed on polystyrene microtitre plates in semi-defined minimal medium (SDM) supplemented with glucose (Li et al. 2002b). Plates were incubated statically at 37°C with 5% CO2 for 16 h (standard assay), or for 8 h before the addition of SDM-glucose supplemented with either 0·5 mmol l–1 H2O2, 2·5% NaCl or SDM-glucose adjusted to pH 5·0 with HCl for a further 8 h of growth (stress assay). Biofilms were stained with safranin and quantified as described previously (Lévesque et al. 2005). To closely examine the morphology of biofilm-grown cells, scanning electron microscopy (SEM) was performed on 16-h biofilms grown on glass discs according to the standard assay. Biofilms were washed with sterile phosphate-buffered saline (PBS, pH 7·2), dehydrated through ethanol rinses, critical point dried with liquid CO2, mounted, and sputter coated with gold. Samples were examined using a scanning electron microscope (model S-2500; Hitachi Instruments, San Jose, CA, USA).
DNA microarrays
Biofilms of UA159 and its Δrr11 mutant were grown in SDM-glucose as described in the standard assay. Planktonic cells were carefully removed and washed once in sterile PBS. Biofilm cells were then washed once with sterile PBS to remove loosely bound cells and dislodged by gentle scraping using sterile pipette tips. The biofilm and planktonic cell pools were harvested separately, resuspended in Trizol reagent (Invitrogen), and processed with the Bio101 Fast Prep system (Qbiogene). The cDNA labelling for microarray analysis was performed according to the Pathogen Functional Genomics Resource Center (PFGRC) protocol (http://pfgrc.tigr.org/protocols/M007.pdf). Following hybridization, the microarray chips were scanned using a Gene Pix 4000B (Axon, Sunnyvale, CA). The software package TM4 Microarray Software Suite (http://www.tm4.org/) was used to calculate the relative expression levels of each gene and to identify differentially expressed genes. A postnormalization cutoff of twofold up- or down regulation was used to define differential gene expression. Statistical significance of expression profiles was determined using a one-class t-test. Microarray assays of three independent RNA isolations were conducted, and the differential expression was defined based on their consensus. Results were validated by real-time RT-PCR using the QuantiTech SYBR Green PCR kit in a Mx3000P QPCR system (Stratagene).
Transformation experiments
One microgram of plasmid pDL289 conferring kanamycin resistance (Buckley et al. 1995) was added to growing cultures at an OD600 of 0·1 both in the presence and absence of synthetic competence signalling peptide (CSP; 1 μg ml–1), and incubated at 37°C for 2·5 h. Cultures were then gently sonicated and spread on THYE agar plates.
Results
Phenotypic characterization of Δrr11 mutant
The RR component-encoding gene (SMU.487) of the S. mutans HK/RR11 system was successfully inactivated by deletion–insertion mutagenesis. No significant difference in doubling time or growth yield was noted between the wild-type strain and its Δrr11 derivative (SMRR11 mutant) after 16 h of planktonic growth at pH 7·5. To determine if biofilm formation was affected in the SMRR11 mutant, a static in vitro biofilm assay was performed. Our results demonstrated that SMRR11 mutant cells formed stable and reproducible biofilms with a biomass of 12·2 ± 4·9% less than the wild type. A closer examination by SEM revealed a difference in cell morphology (Fig. 1). The SMRR11 mutant formed smaller, more spherical cells than wild-type, suggesting a role for RR11 in the regulation of cell growth and/or cell division in the biofilm.
Figure 1.
Scanning electron micrographs of Streptococcus mutans UA159 and SMRR11 biofilms accumulated on the surface of glass discs. Magnifications, ×1K (top panels), and ×60K (bottom panels).
Microarray identification of RR11-regulated genes involved in stress response
To decipher the genes controlled by RR11 during biofilm growth, global gene expression profiles of UA159 and SMRR11 mutant strains were analysed using microarray. Expression data comparing biofilm and planktonic growth phases revealed that the expression of approx. 5% of the genome was significantly altered in UA159 biofilms. Comparison of the gene expression profile between UA159 and SMRR11 strains suggested that RR11 directly and/or indirectly regulated 174 genes (approx. 9% of the genome) during biofilm growth (Table S1). Of these, several genes encoding proteins involved in stress response were found differentially regulated (Table 1).
Table 1.
Stress-response genes regulated by RR11 in Streptococcus mutans cells growing in biofilms
| Relative fold change* |
|||
|---|---|---|---|
| Gene | Description/putative function | UA159 | SMRR11 |
| SMU.15 | Cell division protein FtsH | n.c. | +2·2 |
| SMU.91 | Peptidyl-prolyl isomerase RopA (trigger factor) | n.c. | +2·2 |
| SMU.151 | Non-lantibiotic mutacin IV, NlmB | –2·4 | n.c. |
| SMU.228 | Alkaline-shock protein homolog | n.c. | +3·8 |
| SMU.403 | DNA-damage-inducible protein P | n.c. | +2·5 |
| SMU.602 | Sodium-dependent transporter | n.c. | –2·1 |
| SMU.728 | Oxidoreductase | n.c. | –2·0 |
| SMU.745 | Drug-export protein multidrug resistance protein | n.c. | –2·6 |
| SMU.949 | ATP-dependent protease Clp, ATPase subunit Clpx | –2·2 | n.c. |
| SMU.1063 | ABC transporter, proline/glycine betaine | n.c. | +2·5 |
| SMU.1129 | Response regulator CiaR | +2·6 | n.c. |
| SMU.1672 | ATP-dependent Clp protease, proteolytic subunit | n.c. | +2·0 |
| SMU.1916 | Histidine kinase ComD of the competence regulon | –2·1 | n.c. |
| SMU.1964 | Response regulator RR9 | n.c. | –2·0 |
| SMU.1993 | ABC transporter, permease protein | n.c. | +3·5 |
| SMU.1994 | ABC transporter, ATP-binding protein | n.c. | +3·0 |
| SMU.2030 | Transcriptional regulator CtsR | n.c. | +2·0 |
| SMU.2116 | Osmoprotectant amino acid ABC transporter | n.c. | +5·1 |
n.c., no change in expression observed.
Fold change calculated as the log2 of gene expression biofilm vs. planktonic phase. Transcript levels were measured by cDNA hybridized to a fourfold redundant UA159 microarray and averaged for three replicate hybridization. Positive and negative values indicate up- and downregulation, respectively.
The microarray results also identified other two-component systems whose expression was altered in S. mutans biofilms, and which have been implicated in stress-response cascades in streptococci. Among these genes was SMU.1916, encoding the membrane-associated ComD receptor for the CSP pheromone. ComDE system was originally characterized for its role in the development of genetic competence in Streptococcus pneumoniae (Havarstein et al. 1995, 1996) and S. mutans (Li et al. 2002b), but has also been implicated in the control of the stress-responsive autolysis pathway in pneumococci (Guiral et al. 2005). SMU.1129 encoding the regulator of the CiaHR system also showed altered expression in our microarray. CiaHR is involved in stress tolerance and competence development in S. mutans (Ahn et al. 2006). Finally, SMU.1964 encoding the regulator of the HK/RR9 system was also found potentially regulated by RR11. This system has recently been shown to be involved in S. mutans acid stress survival, but not required for biofilm development (Lévesque et al. 2007).
SMRR11 mutant biofilms under oxidative, osmotic and acid stresses
As changes in the expression of the aforementioned RR11-regulated stress response genes could impact the formation of S. mutans biofilms through a reduced ability to respond to stress in the localized biofilm environment, we examined the ability of SMRR11 biofilms to grow in the presence of oxidative, osmotic and acid stress conditions. A significant reduction in biomass (37·8 ± 17·1%, P < 0·05) was observed when SMRR11 biofilms were grown in the presence of H2O2 compared with UA159 grown under the same conditions. No significant difference between UA159 and SMRR11 biofilm biomass was observed following acid stress and osmotic stress. Under all three stress conditions assayed, planktonic cultures of UA159 and SMRR11 grew with identical kinetics (data not shown), indicating that the observed defects in biofilm formation following oxidative stress was the result of biofilm-specific growth impairments.
Regulatory role for RR11 in competence development
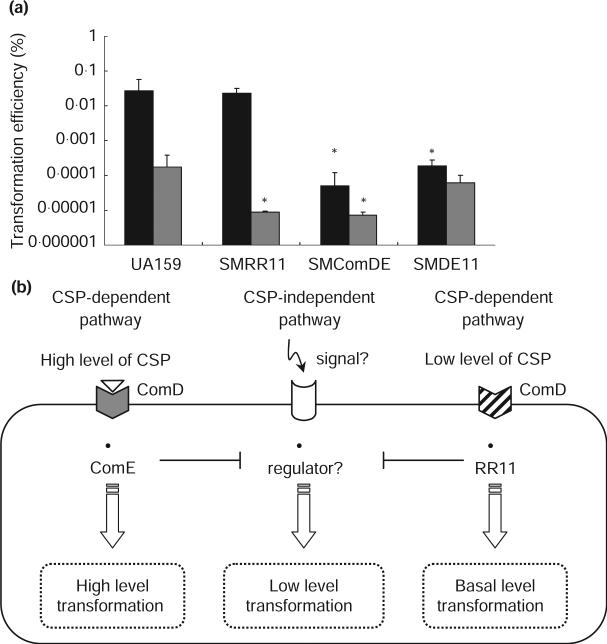
Our microarray analysis demonstrated that SMU.1916 encoding the ComD receptor for the CSP pheromone was likely regulated by RR11. Because of the interconnection of competence, stress response and ComDE system in S. pneumonie, we also examined the competence phenotype of SMRR11 mutant. Ahn et al. (2006) suggested that more than one two-component system might be involved in triggering competence induction in S. mutans. The authors proposed that ComDE is the primary system responding to CSP, while CiaHR, and possibly some other unidentified regulators, integrate secondary signals. We hypothesized that RR11 may be one such additional regulator that could be responsible for integrating stress signals to trigger competence. We thus investigated the role of RR11 in the development of competence by testing the ability of SMRR11, SMComDE (ΔcomDE), and SMDE11 (ΔcomDE/rr11) mutants to be transformed with DNA (Fig. 2a). Our results demonstrated that SMRR11 had an approx. 20-fold reduction in transformation efficiency vs. UA159 in the absence of CSP. As expected, inactivation of comDE diminished the transformation efficiency several-fold. Surprisingly, the SMDE11 double mutant behaved like UA159 in the absence of CSP, regardless of whether CSP was added. This finding led us to hypothesize that both ComE and RR11 may negatively regulate a third regulator, which integrates environmental signals other than CSP (CSP-independent pathway) (Fig. 2b). To test whether CiaR could be involved in CSP-independent competence induction, transformation efficiency was measured in the CiaHR-deficient mutant. However, inactivation of ciaHR had no impact on competence as transformation levels were similar to UA159 (results not shown).
Figure 2.
Development of genetic competence in Streptococcus mutans. (a) Transformation efficiency of UA159 and its mutants with pDL289 is plotted with (black bars) and without (grey bars) the addition of synthetic competence signalling peptide (CSP). The results are expressed as the mean + SD of at least two independent experiments. Asterisks denote statistical significance compared with UA159 under the same conditions. (b) Model for S. mutans competence development. This model integrates the CSP-dependent and CSP-independent pathways (see text for details).
Discussion
Environmental conditions in the oral biofilms are highly variable with respect to pH, oxygen and osmotic balance. Shifts from neutral pH to as low as 3·0 occur during host ingestion of dietary carbohydrates, oxygen gradients ebb and flow in the dental biofilm, and salts may accumulate from tooth demineralization. Streptococcus mutans uses two-component systems to detect environmental signals and transmits the information to cytosolic effector proteins, which trigger appropriate cellular responses. The involvement of two-component systems in environmental stress response has been characterized during S. mutans planktonic growth, but few studies have examined the role of these systems during biofilm growth. In S. mutans, the HK/RR11 system governs important biological parameters enabling its survival and persistence in the oral cavity. Although this regulatory system has been implicated in S. mutans biofilm development, the genes controlled by RR11 during biofilm growth were still unknown. Our aim was to decipher the genes controlled by the regulator component of the HK/RR11 system during biofilm development using DNA microarray technology.
Several stress responsive genes were identified in our microarray analysis, including ropA and clpP. RopA is a molecular chaperone with peptidyl-prolyl cis-trans isomerase activity that functions in protein biogenesis and stress survival (Hesterkamp and Bukau 1996). ClpP is the proteolytic subunit of the ATP-dependent Clp protease, which performs protein reactivation and targeting of specific proteins for degradation (Porankiewicz et al. 1999). In S. mutans, both ropA and clpP mutants are defective in competence and biofilm development, and are more susceptible to stress (Lemos and Burne 2002; Wen et al. 2005). Interestingly, Wen et al. (2005) found that a RopA mutant also showed longer chain length in broth and altered biofilm architecture, which they attributed to alterations in protein trafficking. These results suggested that the changes in ropA and/or clp expression through RR11 might be responsible for the altered cell morphology seen in the SMRR11 mutant biofilm. Moreover, the increased susceptibility to oxidative stress during SMRR11 mutant biofilm growth may occur owing to RR11's role in regulating the turnover of damaged proteins via RopA and Clp.
Specific stress response proteins like the osmoprotectant ABC transporters encoded by SMU.2116 and SMU.1063 were also upregulated in SMRR11 mutant biofilms. Both SMU.2116 and SMU.1063 have homology to the quaternary amine uptake transporter family (Biemans-Oldehinkel et al. 2006). SMU.2116 is highly homologous to OpuCA, a solute transporter that serves a protective role to Streptococcus agalactiae cells growing in a hyperosmolar environment. SMU.1063 shares high identity with a Lactococcus lactis proline/glycine betaine transporter. In L. lactis, the homologous ATPase subunit of the transporter has a cytoplasmic K+ sensor for osmotic stress. Both ABC transporters have been found upregulated under osmotic stress conditions in planktonically grown S. mutans cells, and implicated in the survival of the organism under those stress conditions (Abranches et al. 2006).
Exciting evidence has recently emerged linking the competence regulon to the general stress response cascade in pneumococci. Claverys et al. (2006) have shown that a subpopulation of cells under antibiotic stress triggers the expression of high levels of the CSP signalling pheromone. High levels of CSP are sensed by the ComDE system, resulting in the death of damaged cells and the uptake of potentially fitness-enhancing DNA from the environment by competent surviving cells. A similar link between competence and cell death exists in S. mutans (Perry and Lévesque, unpublished data). Our microarray data combined with the transformation experiments have led us to propose a new model of regulation of the S. mutans competence regulatory network which integrates the CSP-dependent and CSP-independent systems (Fig. 2b). In this model, the ComDE system is the primary circuit sensing CSP, and induces a high level of transformation at high levels of CSP. At low levels of CSP, the major competence system remains inactive (the unphosphorylated to phosphorylated ratio of ComD is altered), and unphosphorylated ComD may cross-regulate RR11 to induce a basal level of genetic competence. Investigations are ongoing in our lab to elucidate the effects of environmental stresses in the development of genetic competence, including the role of RR11 in this pathway.
Supplementary Material
Acknowledgements
This research was supported by NIDCR grant DE013230. DNA microarray resources were supported through NIDCR via NIAID contract number N01-AI15447 to J. Craig Venter Institute. D.G. Cvitkovitch is supported by a Canada Research Chair. J.A. Perry and P. Suntharaligam are supported by a CIHR strategic program, ‘Cell Signaling in Mucosal Inflammation and Pain’.
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article:
Table S1 Genes potentially regulated by RR11 in Streptococcus mutans cells growing in biofilms.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J, Lemos JA, Burne RA. Osmotic stress response of Streptococcus mutans UA159. FEMS Microbiol Lett. 2006;255:240–246. doi: 10.1111/j.1574-6968.2005.00076.x. [DOI] [PubMed] [Google Scholar]
- Ahn S-J, Wen ZT, Burne RA. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun. 2006;74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat SP, Nary J, Burne RA. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans. FEMS Microbiol Lett. 2001;205:225–230. doi: 10.1111/j.1574-6968.2001.tb10952.x. [DOI] [PubMed] [Google Scholar]
- Biemans-Oldehinkel E, Doeven MK, Poolman B. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 2006;580:1023–1035. doi: 10.1016/j.febslet.2005.11.079. [DOI] [PubMed] [Google Scholar]
- Biswas I, Drake L, Erkina D, Biswas S. Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J Bacteriol. 2008;190:68–77. doi: 10.1128/JB.00990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley ND, Lee LN, Leblanc DJ. Use of a novel mobilizable vector to inactivate scrA gene of Streptococcus sobrinus by allelic replacement. J Bacteriol. 1995;177:5028–5034. doi: 10.1128/jb.177.17.5028-5034.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J-P, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu Rev Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- Guiral S, Mitchell TJ, Martin B, Claverys J-P. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci USA. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havarstein LS, Gaustad P, Nes IF, Morrison DA. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol. 1996;21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- Hesterkamp T, Bukau B. The Escherichia coli trigger factor. FEBS Lett. 1996;389:32–34. doi: 10.1016/0014-5793(96)00582-0. [DOI] [PubMed] [Google Scholar]
- Lau PCY, Sung CK, Lee JH, Morrison DA, Cvitkovitch DA. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods. 2002;49:193–205. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- Lemos JA, Burne RA. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J Bacteriol. 2002;184:6357–6366. doi: 10.1128/JB.184.22.6357-6366.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque CM, Voronejskaia E, Huang Y-CC, Mair RW, Ellen RP, Cvitkovitch DG. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect Immun. 2005;73:3773–3777. doi: 10.1128/IAI.73.6.3773-3777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque CM, Mair RW, Perry JA, Lau PCY, Li Y-H, Cvitkovitch DG. Systemic inactivation and phenotypic characterization of two-component systems in expression of Streptococcus mutans virulence properties. Lett Appl Microbiol. 2007;45:398–404. doi: 10.1111/j.1472-765X.2007.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-H, Lau PCY, Tang N, Svensater G, Ellen RP, Cvitkovitch DG. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol. 2002a;184:6333–6342. doi: 10.1128/JB.184.22.6333-6342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-H, Tang N, Aspiras MB, Lau PCY, Lee JH, Ellen RP, Cvitkovitch DG. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol. 2002b;184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TJ. The pathogenesis of streptococcal infections: from tooth decay to meningitidis. Nat Rev Microbiol. 2003;1:219–230. doi: 10.1038/nrmicro771. [DOI] [PubMed] [Google Scholar]
- Porankiewicz J, Wang J, Clarke AK. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol Microbiol. 1999;32:449–458. doi: 10.1046/j.1365-2958.1999.01357.x. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Ann Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Wen ZT, Suntharaligham P, Cvitkovitch DG, Burne RA. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect Immun. 2005;73:219–225. doi: 10.1128/IAI.73.1.219-225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.