Abstract
Streptococcal competence-stimulating peptides (CSPs) were once thought to passively communicate population density in a process known classically as quorum sensing. However, recent evidence has shown that these peptides may also be inducible ‘alarmones,’ capable of conveying sophisticated messages in a population including the induction of altruistic cellular suicide under stressful conditions. We have previously characterized the alarmone response in Streptococcus mutans, a cariogenic resident of the oral flora, in which a novel bacteriocin-like peptide causes cell death in a subset of the population. Our objective in this work was to characterize the mechanism of immunity to cell death in S. mutans. Toward this goal, we have identified the conditions under which immunity is induced, and identified the regulatory system responsible for differential (and protective) expression of immunity. We also showed that CSP-induced death contributes to S. mutans biofilm formation through the release of chromosomal DNA into the extracellular matrix, providing a long sought-after mechanistic explanation for the role of CSP in S. mutans biofilm formation.
Keywords: Streptococcus mutans, biofilm, extracellular DNA, peptide pheromone, autolysis
Introduction
Bacteria have long been studied as single-celled, primitive organisms, free-floating in laboratory culture. However, bacterial species in nature have a strong tendency to colonize surfaces and form complex, multispecies communities referred to as biofilms (Costerton et al., 1994). In nature, biofilms are found on rocks in streams, in industrial bioreactors and in animal host environments such as the oropharyngeal, gastrointestinal and vaginal tracts, and on medical devices. A biofilm in its simplest form is composed of a surface (or ‘substratum’), surface-attached cells and a surrounding extracellular matrix of biopolymers (Dunne, 2002). By microscopic analysis, a biofilm appears to be a highly hydrated and open structure, composed mainly of noncellular material including water channels and exopolymeric substances (EPS) that form the extracellular matrix (Lawrence et al., 1991). The EPS forms the outermost layer of the biofilm, and is composed of a hydrated, anionic mesh of bacterial exopolymers and trapped environmental molecules (Branda et al., 2005). EPS comprise a wide variety of polysaccharides, proteins, glycoproteins, glycolipids and, in some cases, large amounts of extracellular DNA (eDNA). DNA was first shown to be present in the extracellular matrix of biofilms formed by Pseudomonas aeruginosa (Whitchurch et al., 2002), and is now widely recognized as a major constituent of the matrix (Flemming et al., 2007). The matrix functions as a permeability barrier to limit both the diffusion of beneficial nutrients away from the biofilm and prevent or slow the diffusion of harmful substances such as antibiotics and predatory cells of the immune system from accessing matrix-embedded cells (Costerton et al., 1999).
The spatial separation of sessile cells, combined with nutrient/waste and oxygen gradients within the biofilm, results in a heterogeneous population of cells, distinct from their planktonic counterparts in gene expression patterns and ‘behaviors’ (Stoodley et al., 2002; Beloin & Ghigo, 2005). The metabolic task of sharing, communication and phenotypic heterogeneity within a biofilm have led to their being likened to multicellular-type organisms, because co-operation results in the success of the group (Shapiro, 1998; Parsek & Greenberg, 2005). The coordinated behavior of single-celled bacteria is accomplished using different classes of small diffusible signaling molecules in a process called quorum sensing (reviewed in Waters & Bassler, 2005).
Streptococcus mutans is a well-characterized resident of the oral biofilm, and is thought to be the main causative agent of the most common human infectious disease, dental caries (Loesche, 1986). The S. mutans quorum-sensing system is composed of the competence-stimulating peptide (CSP) pheromone and the ComDE two-component signal transduction system (TCS). The S. mutans CSP-ComDE system regulates several phenotypes including genetic competence (Li et al., 2001b), biofilm formation (Li et al., 2002b), acid tolerance (Li et al., 2001a) and bacteriocin production (Kreth et al., 2006).We have recently shown that the CSP pheromone is also stress-inducible and triggers autolysis in a fraction of the S. mutans population at high concentrations (Perry et al., 2009). Autolysis in S. mutans occurs through the intracellular accumulation of a self-acting bacteriocin, CipB, and is prevented by the action of the bacteriocin immunity protein CipI. Previously, we showed that CipI was differentially regulated from CipB at a low cell density (Perry et al., 2009). Here, we report the characterization of the CipI immunity protein, detailing its expression and regulation, and propose a role for CSP-induced autolysis in the release of eDNA in the S. mutans biofilm.
Materials and methods
Bacterial strains and culture conditions
Streptococcus mutans UA159 wild-type strain and its mutants were grown in Todd-Hewitt–yeast extract (THYE) broth at 37 °C with 5% CO2 without agitation. Growth kinetics were assessed using a Bioscreen microbiology work-station (Bioscreen C Labsystems, Finland) as described previously (Hasona et al., 2005). Results represent an average of five technical replicates and two to four independent experiments.
CipI expression studies
The expression of the CipI-encoding gene in UA159 wild-type, ΔLiaS and ΔLiaR strains was quantified by real-time reverse-transcription (RT)-PCR. Briefly, cells were harvested and total RNA was extracted using the Bio101 Fast Prep System (Qbiogen) and Trizol reagent (Invitrogen). DNA-free RNA samples were then reverse-transcribed using the First-Strand cDNA Synthesis Kit (MBI Fermentas) in preparation for real-time RT-PCR using the QuantiTech SYBR Green PCR kit in an Mx3000P QPCR system (Stratagene). Gene expression was determined using the following formula:
where E = (10−1/slope) represents the efficiency of gene amplification. The 16S rRNA gene was used as an internal reference as we found the expression of this gene to be stable under the test conditions. All assays were performed in triplicate with RNA isolated from three independent experiments and using a P ≤ 0.01.
Biofilm assays
Static biofilms were developed in polystyrene microtiter plates at 37 °C with 5% CO2 using semi-defined minimal medium (SDM) containing 1% sucrose as described previously (Perry et al., 2008). After 16 h of growth, the planktonic phase was carefully removed, and fresh SDM-sucrose alone (control) or containing 2 µM synthetic CSP (sCSP) pheromone was overlayed onto established biofilms and the plates were incubated for a further 5 h. The same experiment was also repeated with SDM-sucrose or SDM-sucrose + sCSP supplemented with 50UmL−1 DNAse I (MBI Fermentas). The upper phase was then removed, and biofilms were allowed to dry overnight before staining with crystal violet for biomass quantification. Purification and quantification of eDNA in 16-h biofilms were performed according to Rice et al. (2007). The amount of eDNA was determined using real-time RT-PCR, using four sets of primers designed to amplify genes randomly distributed across the S. mutans genome. CT expression values were averaged, and normalized to the expression in the wild-type strain. All assays were performed in triplicate from three independent experiments.
Results and discussion
Low cell density-dependent expression of cipI is regulated by LiaR
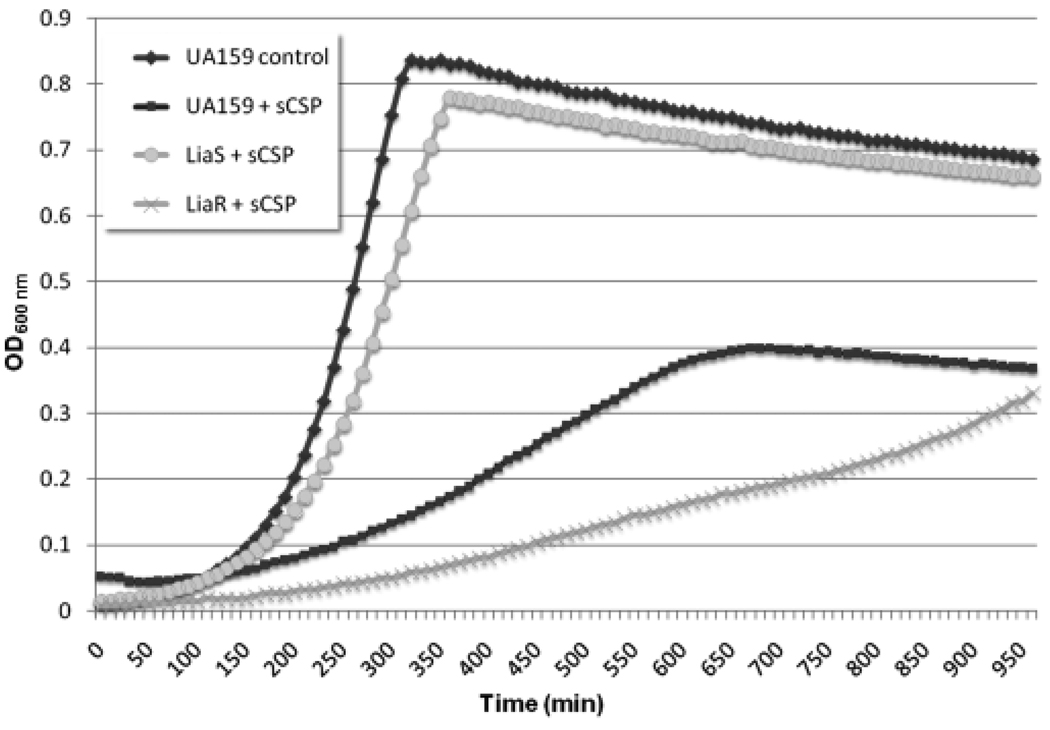
In this work, we sought to characterize the regulation of the cell death immunity protein CipI. Because previous findings indicated that CipI was differentially regulated from the death effector protein CipB at low cell density, we first attempted to identify the regulator responsible for its differential expression. We hypothesized that the regulator responsible for cipI expression at a low cell density would be one of the 13 known TCSs in the S. mutans UA159 genome. TCSs are typically composed of a membrane-bound histidine kinase sensor, which phosphorylates a cytoplasmic response regulator when triggered by environmental stimuli (Stock et al., 2000). We screened mutants defective in each TCS (Lévesque et al., 2007) for differential growth kinetics under high concentrations of sCSP pheromone, conditions known to increase gene expression of both the CipB bacteriocin and the CipI immunity protein (Perry et al., 2009). Inactivation of the response regulator LiaR (originally referred to as RR11 (Li et al., 2002a; Perry et al., 2008; Suntharalingam et al., 2009) resulted in an increased sensitivity to sCSP, while inactivation of its cognate histidine kinase sensor LiaS (formerly known as HK11) showed an increased resistance to sCSP compared with the wild type (Fig. 1). These results implicate the LiaSR TCS in the regulation of cipI expression. To prove the direct regulation of cipI by LiaSR at a low cell density, we compared the expression of cipI at high and low cell density in the wild-type strain, a ΔLiaS mutant and a ΔLiaR mutant. While the wild-type strain showed a highly significant increase in cipI gene expression upon dilution from high to low cell density (120.6 ± 24.3-fold), little to no change in cipI expression was found in ΔLiaS and ΔLiaR mutant strains (7.3 ± 7.6-fold and 17.3 ± 3.8-fold, respectively). Moreover, we found a 64.6 ± 4.4-fold increase in liaR itself upon dilution of the wild-type strain from an overnight culture. Together, our results suggest that the LiaSR TCS is responsible for regulation of cipI gene expression at a low cell density.
Fig. 1.
Growth of Streptococcus mutans TCS mutants in the presence of 2 µM sCSP. All 13 TCSs in the UA159 genome were inactivated (Lévesque et al., 2007) and assayed for their growth kinetics in the presence of sCSP. The TCS composed of LiaS and LiaR showed a differential sensitivity to sCSP compared with the wild-type strain.
CipI is protective at low cell density, while CipB is lethal at high cell density
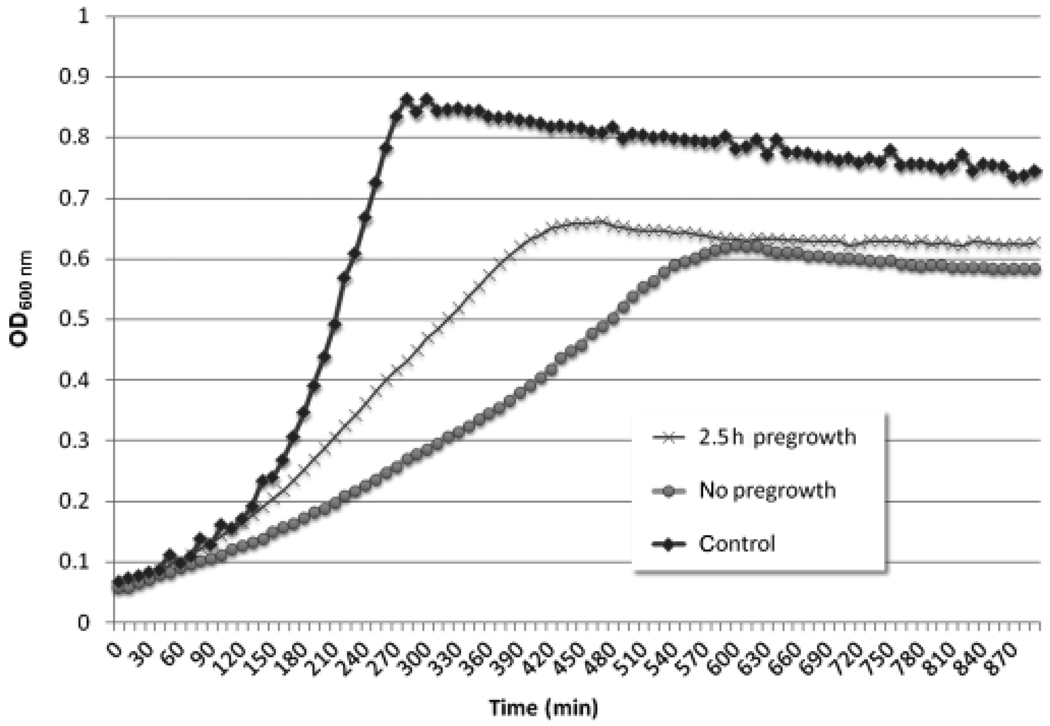
Having now shown that cipI is differentially regulated at a low cell density through the LiaSR TCS, we next sought to determine whether the upregulation of cipI expression at a low cell density protected S. mutans cells from CSP-induced autolysis via the action of CipB bacteriocin. To test this hypothesis, we pregrew aliquots of a UA159 overnight culture in THYE broth without sCSP for up to 2.5 h to induce expression of CipI at a low cell density before exposing them to 2 µM sCSP. We found that inducing CipI expression before sCSP exposure had a protective effect on the culture (Fig. 2).
Fig. 2.
Growth kinetics of Streptococcus mutans UA159 wild-type strain in the presence of 2 µM sCSP. Overnight cultures were pregrown for 2.5 h before sCSP addition, to induce expression of the CipI immunity protein-encoding gene at a low cell density. Control cultures were exposed to sCSP directly upon dilution from an overnight culture (no pregrowth) or were grown in the absence of sCSP (control).
Although CipI upregulation at a low cell density has a protective effect, cultures begin to undergo significant levels of autolysis as growth progresses in the presence of sCSP, culminating at the stationary phase (Perry et al., 2009). How is the transition to autolysis accomplished in these cultures? We hypothesized that death due to the CipB/CipI system would occur at the stationary phase if the CipB bacteriocin was expressed at higher levels than its CipI immunity factor at a high cell density. Indeed, the gene expression of CipB is induced 60.5 ± 1.2-fold at the stationary phase vs. the early log phase, while the gene expression of CipI remains unchanged (−0.6 ± 2.3-fold change in expression) as determined by real-time RT-PCR analysis.
It is tempting to speculate on the implications of these results in the physiological context of cell death. Autolysis is often found to be linked to signals that convey a high population density, because the purpose of cell death in a unicellular organism is to provide benefits (e.g. provide nutrients, space or transforming DNA; eliminate competition) to sibling cells (Claverys & Havarstein, 2007). Lysis of a sparse population serves no purpose because sibling cells are unlikely to benefit, and is therefore counter-selected by evolution. The unlinked transcriptional units encoding CipB and CipI may therefore serve to protect cells from lysis at a low cell density to allow expansion of the clonal population. Conversely, when nutrients become scarce at a high cell density, autolysis is triggered through CipB upregulation to confer a competitive growth advantage to the surviving siblings. Nowhere is the above high cell density-mediated altruistic process more likely to occur than in a biofilm. In a previous report, we found that the expression of cipI is downregulated 2.1-fold through LiaR in biofilm cells compared with their planktonic counterparts (Perry et al., 2008). In addition, others have reported the upregulation of cipB expression in the biofilm phase (Shemesh et al.,2007). We therefore hypothesized that the CipB/CipI autolysis cascade is inhibited during rapid growth in planktonic cultures, but is induced during the high cell density biofilm growth mode of S. mutans.
Cell death participates in biofilm formation through eDNA release
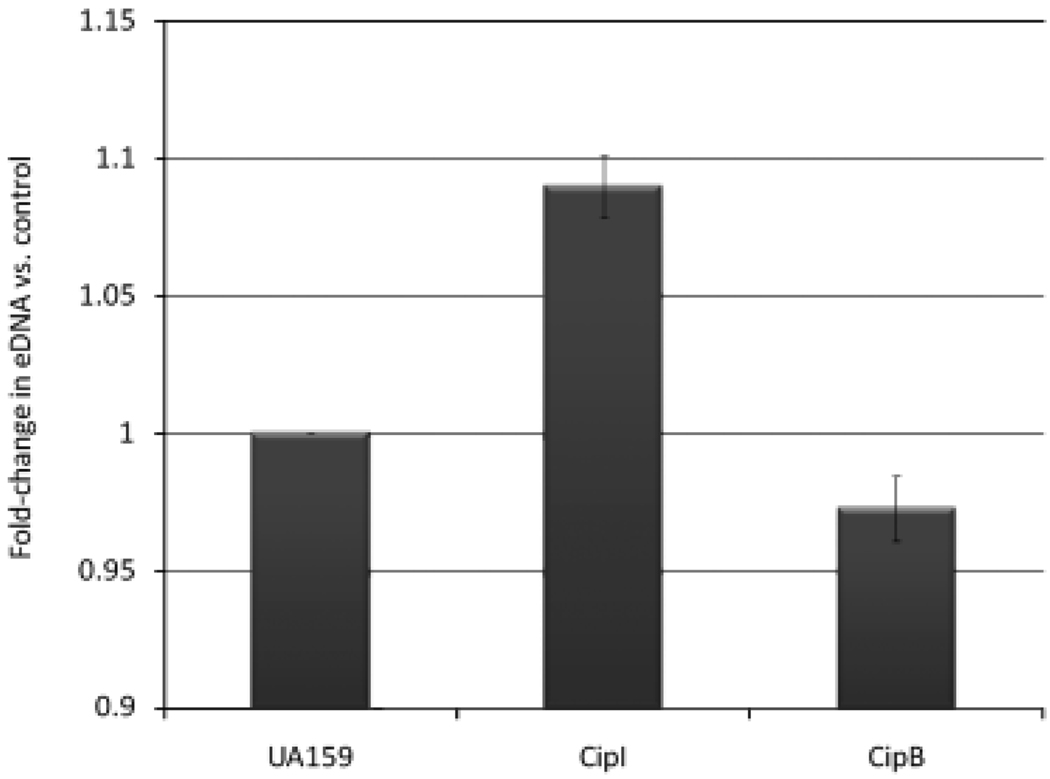
From the outset, both the CSP-ComDE and the LiaSR systems have been implicated in S. mutans biofilm formation (Li et al., 2002a, b). Recently, Zhang et al. (2009) reported that exogenous CSP addition to S. mutans resulted in an increase in both cell death and biofilm biomass. These authors also reported that scanning electron microscopy of a mutant unable to synthesize the CSP signal molecule produced biofilms composed of loosely attached, single cells, but that complementing the mutant with exogenous sCSP resulted in the formation of large aggregates with abundant extracellular matrix. Reports have also suggested that autolysis is necessary for S. mutans biofilm formation (Wen & Burne, 2002), and that CSP induces eDNA release (Petersen et al., 2005). However, none of these studies has provided a mechanistic explanation for CSP-induced release of eDNA. We first sought to link cell lysis in the biofilm to the CipB/CipI system by measuring the amount of eDNA in 16-h biofilms formed by the wild-type strain, ΔCipB and ΔCipI mutants. Using real-time RT-PCR, we found a significant decrease in eDNA in the ΔCipB mutant biofilm and a significant increase in eDNA in the ΔCipI mutant biofilm compared with the wild type (Fig. 3). We conclude from this result that death due to the CipB/CipI system can influence the amount of eDNA in the biofilm matrix via autolysis.
Fig. 3.
Fold-change in the quantity of eDNA in ΔCipI and ΔCipB mutant biofilms compared with the UA159 wild-type control. Quantitative real-time RT-PCR was used to amplify four randomly selected chromosomal genes from DNA extracted from the extracellular matrix of UA159, ΔCipI and ΔCipB biofilms. Amplification values for UA159 were arbitrarily set at one, and results are expressed as a fold-change relative to the UA159 wild type. SD represents the variation in amplification across the four chromosomal genes selected. Both CipI and CipB results represent significant differences (P-value ≤ 0.01) from the UA159 wild-type control.
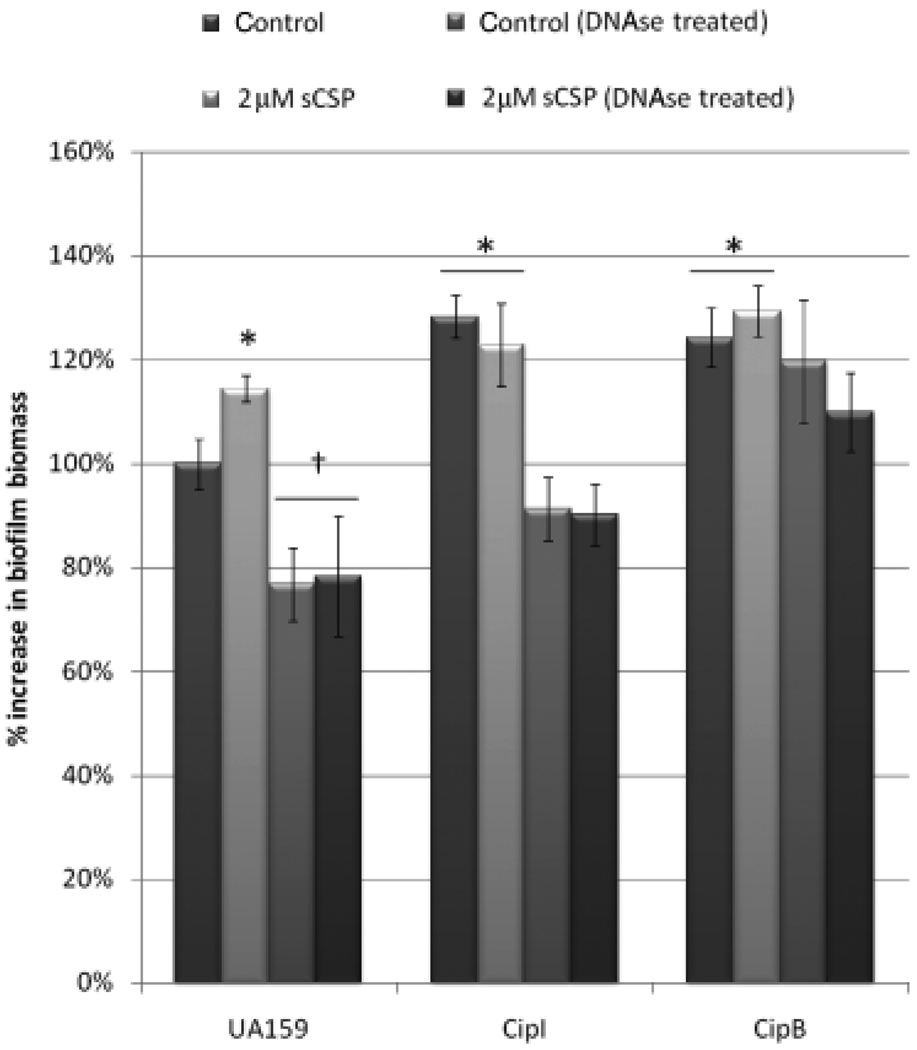
To test whether the modulation of eDNA release via the CipB/CipI autolysis cascade affects S. mutans biofilm biomass, 16-h biofilms were treated with DNAse I and the biomass was quantified by crystal violet staining. DNAse treatment of biofilms formed by the wild-type strain decreased their biomass by > 20% (Fig. 4). In either case, this result confirms previous reports that eDNA plays an important role in S. mutans biofilm formation (Petersen et al.,2005).We next assayed the ΔCipB and ΔCipI mutant strains for their ability to form biofilms. Importantly, ΔCipI formed biofilms with greater biomass than the wild-type strain even in the absence of sCSP, as observed by an increased biomass of ~30% (Fig. 4). The ΔcipI mutant cells possess an intact CSP-encoding gene, and are able to produce endogenous CSP pheromones during biofilm formation. When grown planktonically, the ΔCipI mutant is approximately 10 times more sensitive to sCSP than the wild type (unpublished data), and is likely succumbing to the accumulation of endogenous CSP in the biofilm. The increase in biomass is solely due to eDNA release, because treatment with DNAse I restored the biofilm biomass to wild-type DNAse-treated levels (Fig. 4). Interestingly, the ΔCipB mutant also formed more biofilm biomass than the wild type. However, when the ΔCipB mutant biofilms were treated with DNAse I, the biofilm biomass remained unchanged (Fig. 4). This result indicates that ΔCipB has a growth advantage in the biofilm due to its resistance to autolysis, and the increase in biofilm biomass appears to be mostly due to increasing cell number.
Fig. 4.
Biofilm biomass of Streptococcus mutans UA159 wild-type strain, ΔCipB and ΔCipI mutants. Biofilms were allowed to develop for 16 h in SDM-sucrose. The planktonic phase was then removed, and fresh SDM-sucrose alone (control) or supplemented with 2 µM sCSP was overlayed onto the biofilms and incubated for a further 5 h. The same experiment was repeated with SDM-sucrose supplemented with 50 U mL−1 DNAse in the overlay. Biofilms were washed once with sterile dH2O before quantification. Quantification of biomass was performed by OD of crystal violet-stained UA159, ΔCipB and ΔCipI biofilms. Results are expressed as a % increase in biofilm biomass compared with the biofilm biomass of the UA159 wild-type control, which was set at 100%. *Significant (P < 0.02) increase in biomass compared with the UA159 wild-type control condition; †significant decrease in biomass compared with the UA159 wild-type control.
Finally, we added sCSP to established biofilms to induce CipB-mediated autolysis. As expected, adding sCSP to UA159 and ΔCipI mutant biofilms increased the biofilm biomass beyond CSP-uninduced conditions (Fig. 4). The increase in biomass was again due to eDNA in the extracellular matrix, because DNAse I restored the biomass to wild-type levels. These results suggest that cell death has a positive impact on biofilm biomass through the release of eDNA into the extracellular matrix, through the endogenous CSP-induced CipB/CipI-mediated cell death pathway.
Conclusions
The lifecycle of a biofilm includes periods of both exponential growth and of nutrient limitation (Stoodley et al., 2002). The cooperative nature of biofilm growth has been likened to the task-sharing behavior common in higher-order multicellular organisms. As such, the biofilm lifestyle may allow altruistic behaviors such as autolysis in unicellular prokaryotic organisms, which can contribute nutrients for the continued survival of siblings in a stressed population. Our recent findings led us to propose a mechanistic explanation implicating CSP pheromone during the development of S. mutans biofilm. At a low cell density, S. mutans upregulates expression of the CipI immunity protein through the LiaSR TCS. Upregulation of CipI has a protective effect on the cell, and allows the culture to proliferate under favorable environmental conditions. Conversely, in the high cell density biofilm environment, the high concentrations of CSP pheromone signal upregulation of the CipB autolysis effector through the ComDE TCS. Altruistic autolysis in the S. mutans biofilm contributes nutrients for the continued survival of the population as a whole, as well as eDNA to the extracellular matrix.
Acknowledgements
This work was supported by the Canadian Institutes of Health Research (CIHR) Grant MOP-93555 to C.M.L. and by the National Institutes of Health (NIH) Grant R01 DE013230-08 to D.G.C. J.A.P. is the recipient of a CIHR Strategic Training Fellowship in Cell Signaling in Mucosal Inflammation and Pain.
References
- Beloin C, Ghigo JM. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005;13:16–19. doi: 10.1016/j.tim.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Branda SS, Vik A, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d’etre. Nat Rev Microbiol. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, DeBeer D, Caldwell D, Korber D, James G. Biofilms, the customized microniche. J Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Dunne WM., Jr Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Neu TR, Wozniak D. The EPS matrix: the ‘house of biofilm cells’. J Bacteriol. 2007;189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasona A, Crowley PJ, Lévesque CM, Mair RW, Cvitkovitch DG, Bleiweis AS, Brady LJ. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. P Natl Acad Sci USA. 2005;102:17466–17471. doi: 10.1073/pnas.0508778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Zhu L, Shi W, Qi F. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol Lett. 2006;265:11–17. doi: 10.1111/j.1574-6968.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- Lawrence JR, Korber DR, Hoyle BD, Costerton JW, Caldwell DE. Optical sectioning of microbial biofilms. J Bacteriol. 1991;173:6558–6567. doi: 10.1128/jb.173.20.6558-6567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque CM, Mair RW, Perry JA, Lau PCY, Li Y-H, Cvitkovitch DG. Systemic inactivation and phenotypic characterization of two-component systems in expression of Streptococcus mutans virulence properties. Lett Appl Microbiol. 2007;45:398–404. doi: 10.1111/j.1472-765X.2007.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Hanna MN, Svensater G, Ellen RP, Cvitkovitch DG. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J Bacteriol. 2001a;183:6875–6884. doi: 10.1128/JB.183.23.6875-6884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001b;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lau PC, Tang N, Svensater G, Ellen RP, Cvitkovitch DG. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol. 2002a;184:6333–6342. doi: 10.1128/JB.184.22.6333-6342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol. 2002b;184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Perry JA, Lévesque CM, Suntharaligam P, Mair RW, Bu M, Cline RT, Peterson SN, Cvitkovitch DG. Involvement of Streptococcus mutans regulator RR11 in oxidative stress response during biofilm growth and in the development of genetic competence. Lett Appl Microbiol. 2008;47:439–444. doi: 10.1111/j.1472-765X.2008.02455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Lévesque CM. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol. 2009;72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen FC, Tao L, Scheie AA. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol. 2005;187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. P Natl Acad Sci USA. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- Shemesh M, Tam A, Steinberg D. Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiology. 2007;153:1307–1317. doi: 10.1099/mic.0.2006/002030-0. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Suntharalingam P, Senadheera MD, Mair RW, Lévesque CM, Cvitkovitch DG. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol. 2009;191:2973–2984. doi: 10.1128/JB.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Bi. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Wen ZT, Burne RA. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl Environ Microb. 2002;68:1196–1203. doi: 10.1128/AEM.68.3.1196-1203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Zhang K, Ou M, Wang W, Ling J. Effects of quorum sensing on cell viability in Streptococcus mutans biofilm formation. Biochem Bioph Res Co. 2009;379:933–938. doi: 10.1016/j.bbrc.2008.12.175. [DOI] [PubMed] [Google Scholar]