Abstract
Phosphorylation and SUMO conjugation contribute to the spatial and temporal regulation of substrates containing phosphorylation-dependent SUMO consensus motifs (PDSM). MEF2 is a transcription factor and PDSM substrate whose modification by SUMO drives postsynaptic dendritic differentiation. NMR analysis revealed that the human SUMO E2 interacted with model substrates for phosphorylated and non-phosphorylated MEF2 in similar extended conformations. Mutational and biochemical analysis identified a basic E2 surface that enhanced SUMO conjugation to phosphorylated PDSM substrates MEF2 and HSF1, but not to non-phosphorylated MEF2 or HSF1 or the non-PDSM substrate p53. Mutant Ubc9 isoforms defective in promoting SUMO conjugation to phosphorylated MEF2 in vitro and in vivo also impair postsynaptic differentiation in organotypic cerebellar slices. These data support an E2-dependent mechanism that underlies phosphorylation-dependent SUMO conjugation in pathways that range from heat shock response to nuclear hormone signaling to brain development.
Keywords: SUMO, signal transduction, phosphorylation, MEF2, HSF1, PDSM, ubiquitin, Ubc9
Many signal transduction pathways rely on reversible chemical modifications to relay information within and across cells. Signal transduction can be mediated via covalent modification of protein substrates by the ubiquitin-like protein SUMO1 in processes that contribute to diverse cellular functions such as nuclear transport, cytokinesis, chromosome segregation, and transcriptional regulation among many others2,3. While SUMO conjugation contributes to the regulation of many pathways, the mechanisms that control when and where SUMO modification occurs on any particular substrate remain poorly understood.
SUMO conjugation shares mechanistic similarities with the ubiquitin conjugation system and both utilize a cascade of factors that include E1 activating enzymes, E2 conjugating enzymes, and E3 ligases4-6. While the ubiquitin pathway includes dozens of E2s, only one E2 is utilized in the SUMO pathway, Ubc9 (Ref. 7). As such, Ubc9 represents a critical node in the SUMO pathway because it interacts with and catalyzes conjugation to all SUMO targets identified to date. Some SUMO substrates interact with Ubc9 through a consensus sequence ψ-K-x-E8-12 (where ψ is a hydrophobic amino acid, K is the lysine to which SUMO is attached, X is any amino acid, and E is an acidic residue) while other SUMO substrates require a SUMO E3 ligase or SUMO interacting motif (SIMs) to enhance conjugation to SUMO consensus sites or redirect conjugation to non-consensus sites13-17.
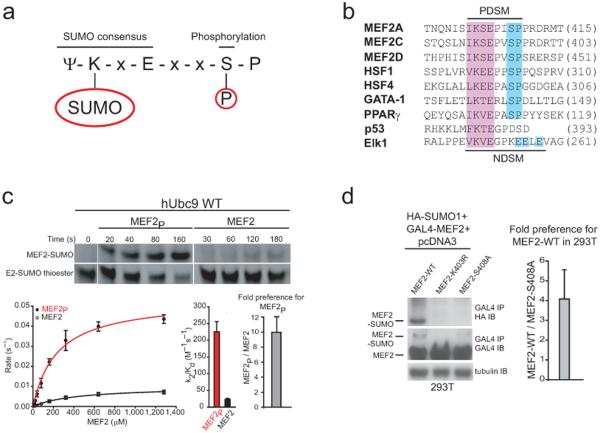
An extended consensus sequence, ψ-K-x-E-x-x-S-P, was recently identified in several proteins that are substrates for both SUMO conjugation and proline directed kinases18-19 (Fig. 1a). Because phosphorylation of the serine residue leads to increased levels of the SUMO conjugation20, this extended SUMO consensus site has been coined PDSM for phosphorylation-dependent SUMO motif21. The PDSM was initially identified in mammalian heat-shock factors (HSFs) and subsequently identified in several other proteins including the transcription factors myocyte enhancement factor 2 (MEF2), GATA-1, and peroxisome proliferator-activated receptor γ (PPARγ)21-23 (Fig. 1b). In these cases, kinase signaling regulates SUMO modification of substrates that contain these extended PDSM sequences.
Figure 1.
Phosphorylation-dependent SUMO conjugation is mediated by the SUMO E2. (a) Extended SUMO consensus motif highlighting the substrate lysine and the phosphorylated serine residues. (b) Sequence alignment for 20 amino acids encompassing the Phosphorylation-dependent SUMO conjugation motif (PDSM) from human MEF2 family members, HSF1, HSF4, GATA-1, PPARγ, the Negatively charged amino acid dependent SUMO conjugation motif (NDSM) from human Elk1 and 15 amino acids encompassing the p53 SUMO consensus motif. The amino acid number for each C-terminal residue is indicated in parentheses. (c) Upper panel; insets showing examples of SDS-PAGE analysis and fluorescent detection for SUMO conjugation to MEF2P and MEF2 at substrate concentrations of 80 μM. Additional examples for a subset of raw data used in constructing plots in the kinetic analysis provided in Supplementary Figures 1 and 2. Lower panel; plot shown on the left for initial rates of reaction versus substrate concentration. The bar chart to the right of the plot indicates individual specificity constants (k2/Kd) for wild-type Ubc9 mediated SUMO conjugation to MEF2P (red) and MEF2 (black). The bar chart (right) indicates the fold preference for MEF2P (gray) expressed as MEF2P(k2/Kd)/MEF2(k2/Kd). (d) HA-SUMO1 conjugation to wild-type GAL4-MEF2A and GAL4-MEF2A-S408A in 293T cells. Left panel; GAL4 immunoprecipitates of 293T cells transfected with HA-SUMO1 and wild-type, K403R or S408A mutant GAL4-MEF2A were immunoblotted using the indicated antibodies. Immunoblot of total lysate with anti-tubulin antibody (as loading control). Right panel; bar chart indicating 4.08 ± 1.48 enhancement for HA-SUMO1 modification to wild-type GAL4-MEF2A compared to GAL4-MEF2A-S408A calculated as described in Methods. Biochemical assays were conducted in triplicate. Error bars are ±1 standard deviation.
MEF2 proteins are transcription factors that are abundantly expressed in the mammalian brain during synaptogenesis24. SUMO modification converts MEF2A from a transcriptional activator to a transcriptional repressor form which promotes synapse maturation in neurons23,25. SUMO modified MEF2A drives postsynaptic dendritic differentiation in the cerebellar cortex in a process characterized by morphogenesis of claw-like structures at the termini of granule neuron dendrites. Granule neuron dendritic claws house sites of synaptic contact with mossy fiber terminals and Golgi neuron axons23. SUMO modification of MEF2A primarily occurs on a lysine residue that is part of a PDSM (Fig. 1b), a site highly conserved from C. elegans to humans23. These data suggest an important functional link between phosphorylation, SUMO conjugation, and biological functions for MEF2.
Although SUMO conjugation is enhanced in response to PDSM phosphorylation, the molecular basis for this effect remains unclear21,26-28. In the present study, we examined SUMO conjugation to the human PDSM substrates, MEF2 and HSF1 in their phosphorylated and non-phosphorylated forms. Biochemical studies indicated that phosphorylation-dependent SUMO conjugation is E2-dependent and NMR titration experiments suggested that phosphorylated and non-phosphorylated MEF2 substrates interacted with Ubc9 in an extended conformation similar to that observed for other Ubc9-substrate complexes. Inspection of the Ubc9 structure suggested that Lys65, Lys74, and Lys76 side chains composed a positively charged or basic patch that could interact with the negatively charged phosphorylated serine side chain. Site-directed mutagenesis coupled with biochemical and kinetic analysis revealed that this E2 surface was important for enhanced SUMO conjugation to phosphorylated MEF2 and HSF1 substrates but not for conjugation to non-phosphorylated MEF2 or HSF1 or to the non-PDSM substrate p53. Mutations in Ubc9 that disrupted PDSM discrimination in vitro also disrupted SUMO modification of phosphorylated MEF2 substrates in transient transfection assays in vivo. Functional analysis of the Ubc9 mutations that disrupted PDSM modification of MEF2 in assays of dendritic claw development in organotypic cerebellar slices revealed a strong correlation between Ubc9 activities in vitro and those observed in vivo.
Results
PDSM phosphorylation enhances E2-dependent conjugation
To identify components of the SUMO conjugation pathway that were responsive to phosphorylation within PDSM sequences, we began our analysis with two well defined and homogenous twenty amino acid MEF2 peptide substrates. One contained the non-phosphorylated extended SUMO consensus motif (MEF2) and, the other contained the MEF2A PDSM in which Ser408 was phosphorylated (MEF2P) (Fig. 1a,b). Biochemical analysis of E2-mediated conjugation under conditions of multiple turnover suggested that the phosphorylated MEF2 was a better substrate for SUMO conjugation (Supplementary Fig. 1 online). SUMO conjugation to the consensus lysine residues in both substrates was confirmed by mass spectrometry (data not shown).
Reactions conducted under conditions of single turnover enabled calculation of the apparent binding and rate constants. In these assays, Kd represents the apparent binding constant and k2 the apparent rate. For wild-type Ubc9, it was determined that MEF2P was a better substrate than MEF2 (Fig. 1c and Supplementrary Fig. 1 online) and at room temperature the apparent Kd and k2 for MEF2P were 240 ± 30 μM and 0.054 ± 0.002 s-1, respectively while the Kd and k2 for MEF2 were 440 ± 60 μM and 0.010 ± 0.001 s-1, respectively. Wild-type Ubc9 exhibited a 10-fold higher specificity constant (k2/Kd) for MEF2P (230 ± 30 M-1s-1) compared to MEF2 (23 ± 4 M-1s-1) (Fig. 1c and Table 1). In this case, binding was affected by 2-fold while rates differed by 5-fold. Because binding was affected less than the rate constants, these data suggest that the phosphorylated substrate may interact with Ubc9 in a slightly different but more favorable conformation to facilitate catalysis within the E2 active site. Precise positioning of the lysine residue within the SUMO E2 active site was previously shown to play an important role during conjugation primarily through altering the catalytic rate12. We envision a similar mechanism underlying the observed effects in the present case.
Table 1. Catalytic constants for SUMO conjugation to MEF2, HSF1 or p53.
| Ubc9 variant | Substrate | Kd (μM) | k2(s-1) | k2/Kd(M-1s-1) |
|---|---|---|---|---|
| Ubc9 wild-type | MEF2P | 240 ± 30 | 0.054 ± 0.002 | 230 ± 30 |
| MEF2 | 440 ± 60 | 0.010 ± 0.001 | 23 ± 4 | |
| Ubc9-K59A | MEF2P | 240 ± 40 | 0.046 ± 0.002 | 190 ± 30 |
| MEF2 | 650 ± 90 | 0.0139 ± 0.0009 | 21 ± 3 | |
| Ubc9-R61A | MEF2P | 300 ± 50 | 0.020 ± 0.001 | 70 ± 10 |
| MEF2 | 600 ± 100 | 0.0073 ± 0.0008 | 12 ± 3 | |
| Ubc9-K65A | MEF2P | 430 ± 60 | 0.0125 ± 0.0007 | 29 ± 4 |
| MEF2 | 400 ± 40 | 0.0126 ± 0.0006 | 32 ± 4 | |
| Ubc9-K74A | MEF2P | 900 ± 200 | 0.0034 ± 0.0004 | 4 ± 1 |
| MEF2 | 800 ± 100 | 0.0050 ± 0.0004 | 6 ± 1 | |
| Ubc9-K76A | MEF2P | 270 ± 40 | 0.0067 ± 0.0004 | 25 ± 4 |
| MEF2 | 400 ± 60 | 0.0052 ± 0.0003 | 13 ± 2 | |
| Ubc9-S89A | MEF2P | 440 ± 60 | 0.0068 ± 0.0004 | 15 ± 2 |
| MEF2 | 1000 ± 300 | 0.0011 ± 0.0002 | 1.1 ± 0.4 | |
| Ubc9-T91A | MEF2P | 500 ± 100 | 0.0096 ± 0.0008 | 19 ± 4 |
| MEF2 | 400 ± 60 | 0.0052 ± 0.0003 | 13 ± 2 | |
| Ubc9 wild-type | HSF1P | 263 ± 33 | 0.078 ± 0.004 | 296 ± 37 |
| HSF1 | 457 ± 87 | 0.018 ± 0.002 | 23 ± 4 | |
| Ubc9-K65A | HSF1P | 532 ± 94 | 0.028 ± 0.002 | 52 ± 4 |
| HSF1 | 546 ± 10 | 0.022 ± 0.002 | 41 ± 8 | |
| Ubc9 wild-type | p53 | 68 ± 10 | 0.018 ± 0.001 | 265 ± 40 |
| Ubc9-K65A | p53 | 42 ± 3 | 0.012 ± 0.003 | 280 ± 20 |
The preference observed for conjugation to the phosphorylated substrate in vitro was similar to that observed in vivo without over-expression of Ubc9 (ref. 23 and Fig. 1d). As such, there is presently no reason to invoke an E3 in the process of discriminating PDSM phosphorylation status. Data consistent with this hypothesis includes the observation that the SUMO E3 PIASx interacts with MEF2A independent of Ser408 phosphorylation25. While SUMO-conjugated wild-type MEF2A increased in the presence of exogenous PIASx, SUMO conjugation was still dependent on phosphorylation, suggesting that PDSM discrimination occurred in a manner dependent on the specificity of the E2. In other words, phosphorylation was important for SUMO conjugation independent of whether PIASx was absent or present in cells25. As further evidence that the process is E2-dependent, the SUMO E3 IR1* domain of Nup358/RanBP2 increased conjugation to both substrates in vitro, and the preference for phosphorylated MEF2 was maintained at a similar ratio to that observed in E2-medatied reactions (Supplementary Fig. 2 online).
Ubc9 binds MEF2P and MEF2 in similar conformations
Human Ubc9 was uniformly 15N-labeled and used in isolation to obtain nearly complete assignments for non-proline residues based on previously deposited assignments10 (Methods and Supplementary Fig. 3 online). NMR chemical shift perturbations were then assessed for Ubc9 in the presence of increasing concentrations of the non-phosphorylated and phosphorylated MEF2 substrates (Supplementary Fig. 3 online). Chemical shift perturbations for both MEF2 and MEF2P map to Ubc9 surfaces known to be important for recognition of substrates containing SUMO consensus sites (Supplementary Figs. 3, 4 and 5 online), including complexes between Ubc9 and RanGAP1 obtained from crystallographic studies9,11,12 as well as complexes between Ubc9 and peptide models for p53 and c-Jun as determined by NMR solution studies and biochemical analysis9,10,12. These data suggest that non-phosphorylated and phosphorylated MEF2 substrates adopt similar extended conformations as observed for other Ubc9 substrates.
Comparing Ubc9 chemical shift perturbations in the presence of MEF2 or MEF2P revealed few differences within and adjacent to previously characterized E2 binding surfaces for substrates that contain SUMO consensus sites (Supplementary Figs. 4, 5). When Ubc9:MEF2 and Ubc9:MEF2P chemical shift differences were compared, some of the largest differences were observed for Ala5, Asp66, Cys75, Ser89, Val92, Leu94, and Tyr134. Val92 and Leu94 are located immediately adjacent to the Ubc9 E2 active site cysteine (Cys93) while Tyr134 is proximal to the binding site for the hydrophobic side chain in the SUMO ψ-K-x-E consensus motif. Ser89 is part of a hydrophilic and positively charged pocket that includes Thr91 and Lys74 (Fig. 2a,b). These amino acid side chains participate in hydrogen bonding interactions with the SUMO consensus motif glutamate side chain as observed in structures of Ubc9 in complex with RanGAP1. While the magnitude of chemical shifts differs somewhat between Ubc9:MEF2 and Ubc9:MEF2P, no changes in direction were observed for chemical shift perturbations of residues involved in substrate binding. These data suggest that the Ψ-K-x-E SUMO motif in MEF2 PDSM substrates occupy similar binding pockets on the surface of Ubc9 independent of their phosphorylation status.
Figure 2.
Model for PDSM recognition by the E2 Ubc9. (a) Surface representation and electrostatic potential of Ubc9 proximal to the RanGAP SUMO consensus motif (ψ-K-x-E SUMO consensus motif obtained from a previously characterized complex between RanGAP and Ubc9; PDB ID 2GRN). The positions for three C-terminal amino acids including the phosphorylated serine residue were obtained from a previously characterized complex between Pin1 and a phosphorylated peptide (PDB ID 1F8A) and combined with the RanGAP SUMO consensus sequence to model the PDSM. The relevant amino acids discussed in the text are labeled using the three-letter code on or adjacent to their position on the Ubc9 surface. Blue represents basic surfaces while red represents acidic surfaces. (b) Model for the Ubc9:PDSM complex. The structure of Ubc9 is shown in ribbon representation and the PDSM residues in stick representation. The SUMO consensus motif and the three C-terminal residues were obtained as in a. All structures graphically depicted using PyMol30.
The glutamate side chain in SUMO ψ-K-x-E consensus motifs is coordinated by Ser89 and Thr91 (ref. 9,12). Because the phosphorylated serine side chain and glutamate are both negatively charged, we reasoned that the glutamate side chain binding pocket might coordinate the phosphorylated PDSM serine side chain if it displaced the glutamate side chain upon binding. To test this hypothesis, we substituted Ser89 or Thr91 to alanine and assayed these mutant isoforms for their ability to discriminate PDSM phosphorylation status. Ubc9-S89A and Ubc9-T91A exhibited reduced SUMO conjugation to both MEF2 substrates although Ubc9-T91A maintained a ~3-fold preference for MEF2P while Ubc9-S89A maintained a ~14-fold preference for MEF2P (Supplementary Fig. 2 online).
PDSM discrimination requires Ubc9 basic patch in vitro
NMR analysis did not reveal the precise identity of Ubc9 residues responsible for PDSM discrimination, so we examined the Ubc9 structure for residues that flank the active site and SUMO consensus motif binding pocket. Side chains of Lys65, Lys74 and Lys76 form a positively charged patch on the Ubc9 surface 7-12 Å from the glutamate within the extended ψ-K-x-E consensus motif, a suitable distance from the E2 active site to mediate complementary interactions with a phosphorylated serine side chain if the PDSM adopted a somewhat but not fully extended conformation (Model presented in Fig. 2).
Individual alanine substitutions were generated for Lys65, Lys74, and Lys76 and the resulting proteins were assayed for activity. Ubc9-K74A and Ubc9-K76A exhibited a 2-4 fold decrease in activity for modification of the non-phosphorylated MEF2 substrate while Ubc9-K65A maintained activities similar to wild-type Ubc9. When Ubc9-K65A, Ubc9-K74A and Ubc9-K76A were assayed for SUMO conjugation to the phosphorylated MEF2 substrate, the specificity constants remained low and near to those obtained with the non-phosphorylated substrate (Fig. 3a-c and Supplementary Fig. 1 online). These data suggest that Ubc9-K65A and Ubc9-K74A lost the ability to discriminate between MEF2P and MEF2 such that the preference for MEF2P was reduced from 10-fold for wild-type Ubc9 to 0.9-fold for Ubc9-K65A and 0.6-fold for Ubc9-K74A. Ubc9-K76A also lost most of its ability to discriminate between substrates, although it maintained a 2.0-fold preference for MEF2P (Table 1). We determined that the Nup358/RanBP2 IR1* SUMO E3 could not rescue defects observed for Ubc9-K65A with the MEF2P substrate (Table 2 and Supplementary Figs. 1 and 2 online).
Figure 3.
Kinetic and mutational analyses for Ubc9 amino acid residues involved in PDSM discrimination. (a) Upper panel; insets showing examples of SDS-PAGE analysis and fluorescent detection for Ubc9-K65A mediated SUMO conjugation to MEF2P and MEF2 at substrate concentrations of 80 μM. Lower panel; the plot on the left indicates initial rates versus substrate concentration and the bar chart shown on the right indicates the specificity constants (k2/Kd) for Ubc9-K65A mediated SUMO conjugation to MEF2P (red) and MEF2 (black). (b), (c) and (d) Similar data as in a, but for Ubc9-K74A, Ubc9-K76A and Ubc9-K59A respectively. (e) The bar chart indicates the fold preference for MEF2P (gray) expressed as MEF2P(k2/Kd)/MEF2(k2/Kd) for wild-type Ubc9, Ubc9-K65A, Ubc9-K74A, Ubc9-K76A and Ubc9-K59A. The dashed red line indicates an equal preference for Ubc9-mediated SUMO conjugation to MEF2P and MEF2. Biochemical assays were conducted in triplicate. Error bars are ±1 standard deviation.
Table 2. Catalytic constants for SUMO conjugation to MEF2 or Elk1 in the presence of the general SUMO E3 Nup358/RanBP2 IR1*.
| Ubc9 variant | Substrate | Kd (μM ) | k2 (s-1) | k2/Kd(×104)(M-1s-1) |
|---|---|---|---|---|
| Ubc9 wild-type | MEF2P | 1.1 ± 0.1 | 0.103 ± 0.004 | 9.4 ± 0.9 |
| MEF2 | 6 ± 1 | 0.089 ± 0.008 | 1.5 ± 0.3 | |
| Ubc9-K65A | MEF2P | 4.8 ± 0.3 | 0.161 ± 0.004 | 3.4 ± 0.2 |
| MEF2 | 9 ± 1 | 0.18 ± 0.01 | 2.0 ± 0.2 | |
| Ubc9 wild-type | Elk1 | 14 ± 2 | 0.066 ± 0.003 | 4.8 ± 0.7 |
| Ubc9-K59A | Elk1 | 21 ± 3 | 0.052 ± 0.004 | 2.4 ± 0.4 |
| Ubc9-K65A | Elk1 | 33 ± 7 | 0.068 ± 0.007 | 2.1 ± 0.5 |
We turned our attention next to the extended Elk1 SUMO consensus motif which includes a ψ-K-x-E SUMO conjugation site followed by three acidic residues (Fig. 1b). This motif was coined NDSM for negatively charged amino acid-dependent SUMO conjugation motif28. NDSM and PDSM sequences include negatively charged elements C-terminal to the ψ-K-x-E SUMO consensus site, thus acidic residues in NDSM substrates may substitute for the phosphorylated serine side chain in PDSMs to maintain a constitutively active motif for SUMO conjugation. Previous reports identified Ubc9 Lys59 and Arg61 as residues that contributed to recognition of NDSM within the Elk1 transcription factor28 insofar as substitution of Lys59 and Arg61 to glutamate diminished SUMO conjugation activity. These Ubc9 surfaces were also suggested to play a role in SUMO modification of MEF2A28.
We also found that Lys59 and Arg61 side chains did not contribute to PDSM discrimination insofar as Ubc9-K59A and Ubc9-R61A maintained higher levels of conjugation to the MEF2P substrate when compared to the MEF2 substrate (Fig. 3d, Table 1, Supplementary Figs. 1 and 6). These results are consistent with the fact that Lys59 and Arg61 are too far from the E2 substrate binding surface to facilitate simultaneous interaction with the ψ-K-x-E SUMO consensus site and phosphorylated serine residue even if the PDSM adopted a fully extended conformation (Fig. 2). Previous results utilized simultaneous glutamate substitution for Lys59 and Arg61 (ref. 28) and we predict that glutamate substitutions in these positions might substansially alter the properties of Ubc9 based on Arg61’s involvement in salt-bridging interactions with Glu41 and Glu78 and hydrogen bonding interactions with Asn39 (Fig. 2 and Supplementary Fig. 6 online). Together these results indicate that the basic surface on Ubc9 formed by lysines 65, 74 and 76 is important for PDSM discrimination of the MEF2A substrate in vitro (Fig. 3e).
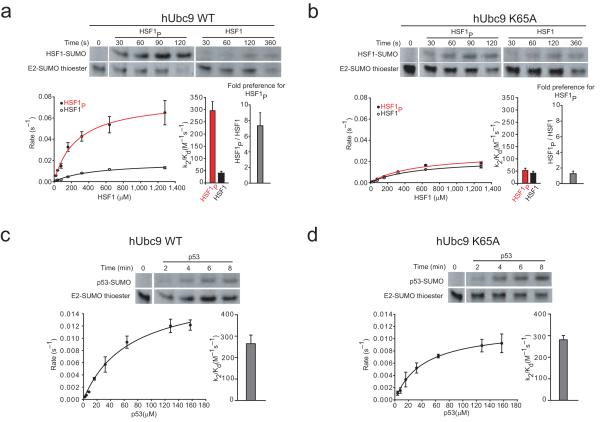
To test if this Ubc9 surface also plays a role in discriminating between phosphorylated and non-phosphorylated forms of another PDSM substrate we performed a similar kinetic analysis with model substrates for Heat Shock Factor 1 (HSF1; Fig. 1b), the primary transcription factor responsible for the transcriptional response to heat stress in mammalian cells. Similar to results observed for MEF2A, wild-type Ubc9 exhibited an 8-fold preference for phosphorylated HSF1 compared to the non-phosphorylated substrate (Fig. 4a and Table 1). Ubc9-K65A maintained nearly wild-type activity for non-phosphorylated HSF1, while losing its preference for phosphorylated HSF1. (Fig. 4b and Table 1). These results are similar to those obtained for MEF2 and MEF2P and suggest that Ubc9 Lys65 contributes to discrimination of PDSM phosphorylation status in vitro for both MEF2A and HSF1. It is important to note that MEF2A and HSF1 share no sequence conservation outside of their respective PDSM sequences (Fig. 1b), suggesting that the PDSM element is sufficient to mediate interactions with the E2.
Figure 4.
Ubc9 Lys65 is important for PDSM discrimination of HSF1 but not for the non-PDSM substrate p53. (a) Upper panel; insets showing examples of SDS-PAGE analysis and fluorescent detection for wild-type Ubc9 mediated SUMO conjugation to HSF1P and HSF1 at substrate concentrations of 80 μM. Lower panel; the plot on the left indicates initial rates versus substrate concentration and the bar chart shown on the right indicates the specificity constants (k2/Kd) for wild-type Ubc9 mediated SUMO conjugation to HSF1P (red) and HSF1 (black). (b) Similar data as in a, but for Ubc9-K65A. (c) Upper panel; insets showing examples of SDS-PAGE analysis and fluorescent detection for wild-type Ubc9 mediated SUMO conjugation to p53 at a substrate concentrations of 16 μM. Lower panel; the plot on the left indicates initial rates versus substrate concentration and the bar chart shown on the right indicates the specificity constant (k2/Kd) for wild-type Ubc9 mediated SUMO conjugation to p53. (d) Similar data as in c, but for Ubc9-K65A. Biochemical assays were conducted in triplicate. Error bars are ±1 standard deviation.
Our biochemical analysis revealed that wild-type Ubc9 and Ubc9-K65A have comparable rates of SUMO conjugation to MEF2A and HSF1 in their non-phosphorylated state (Figs. 1c, 3a, and 4b). Because the Lys65 side chain was not important for interactions with a non-phosphorylated PDSM substrate, we hypothesized that this side chain would not contribute to SUMO conjugation of substrates containing a simple SUMO consensus motif. To test this, we conducted single turnover SUMO conjugation assays with a non-PDSM substrate, in this case the C-terminal domain of p53. Comparison of the conjugation activities catalyzed by wild-type Ubc9 and Ubc9-K65A revealed similar rates of SUMO conjugation to p53 (refs. 11, 12) (Figs. 4c,d and Table 1). Thus, Ubc9-K65A had no discernable effect on SUMO conjugation to a non-PDSM containing substrate, or to MEF2A or HSF1 in their non-phosphorylated state.
Because PDSM and acidic NDSM SUMO conjugation motifs share similar negative electrostatic properties C-terminal to the SUMO ψ-K-x-E motif (Fig. 1b), we next compared Ubc9-K59A and Ubc9-K65A in conjugation reactions with a model Elk1 substrate to determine if these side chains contributed to interaction with an NDSM substrate (Methods and Supplementary Fig. 7 online). Our data revealed a mild 2-fold decrease in the specificity constant for Ubc9-K65A when compared to wild-type Ubc9 for SUMO conjugation to Elk1 (Table 2). When Ubc9-K59A was assessed, the specificity constant was similar to that obtained for Ubc9-K65A (Table 2). These data suggest that Lys65 contributes less to recognition of NDSM substrates such as Elk1 in comparison to its role in discriminating between phosphorylated states of either MEF2 or HSF1 PDSM substrates.
PDSM discrimination requires Ubc9 basic patch in vivo
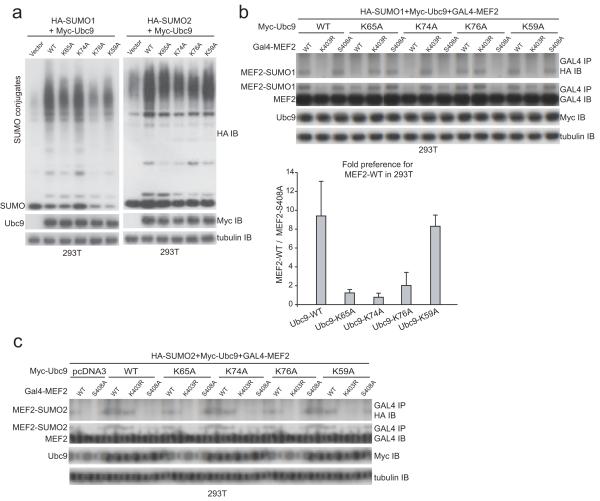
The effects of Ubc9 amino acid substitutions that abrogated PDSM discrimination in vitro were next explored in 293T cells through transfection assays through exogenous expression of HA-SUMO1 or HA-SUMO2, Myc-Ubc9 or respective mutant alleles, and GAL4-MEF2A (see Methods). In comparison to cells not expressing exogenous Ubc9, expression of the respective mutant Ubc9 proteins led to an increase in levels of SUMO conjugates independent of whether HA-SUMO1 or HA-SUMO2 was expressed (Fig. 5a). These data suggest that each Ubc9 isoform was capable of recognizing the bulk of SUMO substrates in the cell irrespective of the point mutation tested. Although some differences were observed in the levels of SUMO conjugates in cells expressing exogenous Myc-Ubc9 isoforms, expression of each Myc-Ubc9 mutant isoform led to higher levels of SUMO conjugates compared to cells expressing HA-SUMO1 or HA-SUMO2 in the presence of endogenous Ubc9 (vector control for expression of exogenous Myc-Ubc9; Fig. 5a).
Figure 5.
Amino acid side chains that constitute the basic surface on Ubc9 are important for PDSM discrimination of MEF2 in vivo. (a) SDS-PAGE analysis of whole-cell lysate obtained from 293T cells transfected with HA-SUMO1 (left panel) or HA-SUMO2 (right panel) and wild-type Myc-UBC9 or the indicated mutant isoforms immunoblotted using anti-HA, anti-Myc, or anti-tubulin antibodies (loading control). (b) Upper panel; SDS-PAGE analysis of GAL4 immunoprecipitates from 293T cells transfected with HA-SUMO1, indicated Myc-UBC9 alleles and GAL4-MEF2A immunoblotted with anti-HA and anti-GAL4 antibodies as well as anti-myc and anti-tubulin antibodies (loading controls). Lower panel; bar chart indicating ratios between the preference of SUMO modification for wild-type GAL4-MEF2A and GAL4-MEF2-S408A for wild-type and mutant Ubc9 isoforms obtained from three independent transfection (Methods) except for Myc-Ubc9-K59A which was calculated from two independent experiments due to technical difficulties. Ratios for SUMO conjugation (GAL4-MEF2A/GAL4-MEF2A-S408A) were 9.40 ± 3.67, 1.24 ± 0.35, 0.78 ± 0.43, 2.03 ± 1.39 and 8.30 ± 1.89 for Myc-Ubc9, Myc-Ubc9-K65A, Myc-Ubc9-K74A, Myc-Ubc9-K76A and Myc-Ubc9-K59A, respectively. Biochemical assays were conducted in triplicate. Error bars are ±1 standard deviation. (c) SUMO2 conjugation to MEF2A in 293T cells. GAL4 immunoprecipitates of 293T cells transfected with HA-SUMO2 and the indicated mutant isoforms of Myc-UBC9 and GAL4-MEF2A were immunoblotted with HA and GAL4 antibodies. Total lysates were immunoblotted with a myc antibody for Myc-Ubc9 and a tubulin antibody which served as a loading control for the SDS-PAGE analysis.
Previous studies demonstrated that approximately half of exogenously expressed GAL4-MEF2A is phosphorylated on Ser408 (ref. 23). In cells not expressing exogenous Myc-Ubc9, HA-SUMO1 conjugation to GAL4-MEF2A was enhanced by ~5-fold when compared to HA-SUMO1 conjugation to GAL4-MEF2A-S408A, a substrate incapable of phosphorylation at Ser408 (Fig. 1d). When exogenous Myc-Ubc9-WT was expressed, HA-SUMO1 conjugation was enhanced for both substrates. In this case, HA-SUMO1 conjugation was approximately 9-fold higher for GAL4-MEF2A in comparison to GAL4-MEF2A-S408A (Compare Fig. 1d with Fig. 5b).
We next assessed Myc-Ubc9-K65A, Myc-Ubc9-K74A, Myc-Ubc9-K76A and Myc-Ubc9-K59A for their ability to discriminate MEF2A PDSM phosphorylation status in vivo. Consistent with activities observed in vitro, expression of Myc-Ubc9-K65A and Myc-Ubc9-K74A led to selective loss of HA-SUMO1 conjugated GAL4-MEF2A when compared to GAL4-MEF2A-S408A, while expression of Myc-Ubc9-K76A maintained a ~2-fold preference for GAL4-MEF2A in comparison to GAL4-MEF2A-S408A (Fig. 5b). In contrast, Myc-Ubc9-K59A was comparable to Myc-Ubc9-WT with respect to its ability to discriminate between GAL4-MEF2A and GAL4-MEF2A-S408A.
To a first approximation, data obtained in vivo echo data obtained in vitro for Ubc9, Ubc9-K65A, Ubc9-K74A, Ubc9-K76A, and Ubc9-K59A, namely: 1) Ubc9-K65A lost the ability to discriminate between PDSM phosphorylation status but maintained the ability to conjugate SUMO at nearly wild-type levels to GAL4-MEF2A-S408A, 2) Ubc9-K74A lost the ability discriminate between substrates but had lower conjugation activity for both GAL4-MEF2A and GAL4-MEF2A-S408A, 3) Ubc9-K76A maintained a slight preference for GAL4-MEF2A in comparison to GAL4-MEF2A-S408A, and 4) the Ubc9-K59A side chain was not important for discriminating PDSM phosphorylation status. Because global defects were not observed for HA-SUMO1 conjugation in whole cell extracts (Fig. 5a), we suggest that defects observed for HA-SUMO1 conjugation to GAL4-MEF2A are due to the inability of the respective Ubc9 mutant isoform to discriminate MEF2 PDSM phosphorylation status.
We also examined whether Ubc9 mutant isoforms could discriminate MEF2A PDSM phosphorylation status in cells expressing exogenous HA-SUMO2. HA-SUMO2 conjugates of GAL4-MEF2A could not be detected in the presence of endogenous Ubc9 (Fig. 5c), similar to previous observations29. However, modification of GAL4-MEF2A by HA-SUMO2 was observed in the presence of exogenously expressed Myc-Ubc9-WT. Additionally, expression of the mutant Ubc9 isoforms Myc-Ubc9-K65A, Myc-Ubc9-K74A and Myc-Ubc9-K76A in the presence of HA-SUMO2 led to selective loss of HA-SUMO2 conjugated GAL4-MEF2A compared to GAL4-MEF2A-S408A. Myc-Ubc9-K59A maintained its ability to discriminate between GAL4-MEF2A and GAL4-MEF2A-S408A (Fig. 5c). Results obtained in transfection experiments with HA-SUMO2 and respective Myc-Ubc9 isoforms are consonant with results obtained with HA-SUMO1.
Because endogenous Ubc9 remains present in cells transfected with plasmids encoding respective Myc-UBC9 alleles, our data suggest that over-expression of Myc-Ubc9-K65A, Myc-Ubc9-K74A or Myc-Ubc9-K76A can suppress SUMO conjugation to GAL4-MEF2A by blocking the activities of endogenous Ubc9 by mass action (Myc-Ubc9 isoforms expressed at 5-10 fold higher levels in comparison to endogenous Ubc9; data not shown). As such, exogenously expressed Myc-Ubc9 may compete with endogenous Ubc9 for activation by the SUMO E1 or binding to its cognate E3, or by blocking endogenous Ubc9 from recognizing the phosphorylated MEF2 PDSM.
Ubc9 basic patch important for dendritic morphogenesis
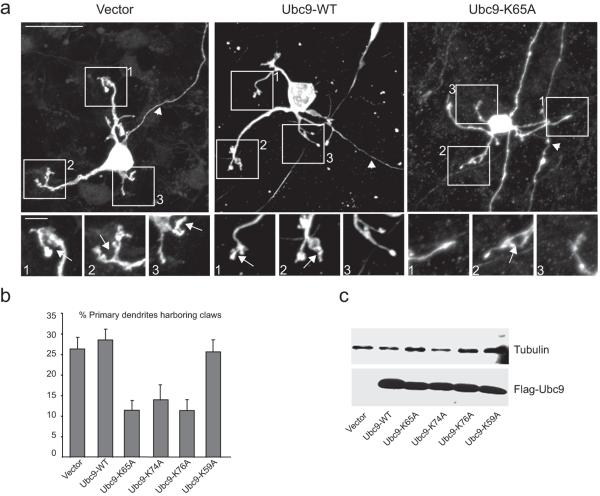
Phosphorylation-dependent SUMO conjugation to MEF2 is required for postsynaptic dendritic morphogenesis in the cerebellar cortex23,25. To determine whether Lys65, Lys74 or Lys76 side chains contribute to PDSM discrimination in vivo in morphological assays of postsynaptic dendritic claw development, we expressed exogenous Flag-Ubc9 and the respective mutant isoforms and assessed their effects on dendritic claw differentiation in the rat cerebellar cortex (Fig. 6). Cerebellar slices were prepared from postnatal day 9 (P9) or P10 rat pups and transfected four days later with the control pcDNA3 vector, Flag-UBC9-WT, Flag-UBC9-K65A, Flag-UBC9-K74A, Flag-UBC9-K76A or Flag-UBC9-K59A along with a GFP expression plasmid.
Figure 6.
Ubc9 mutants deficient in PDSM discrimination are also deficient for dendritic claw differentiation in the cerebellar cortex. (a) Representative confocal images of GFP-positive granule neurons expressing vector (left), Flag-Ubc9-WT (middle), or Flag-Ubc9-K65A (right). Arrows and arrowheads indicate dendritic claws and axons, respectively. Insets show higher-magnification views of ends of dendrites. Main scale bar is 20 microns; inset scale bar is 4 microns. Asterisk denotes an axon of a neuron that is not pictured. (b) Quantification of the effect of Ubc9 and Ubc9 mutants on dendritic claw differentiation in rat cerebellar slices. The number of dendritic claws was significantly reduced in granule neurons expressing Flag-Ubc9-K65A, Flag-Ubc9-K74A, and Flag-Ubc9-K76A as compared to those cells expressing Flag-Ubc9 WT-expressing neurons (p<0.001; ANOVA, followed by Bonferroni-Dunn post-hoc test). Vector, Flag-Ubc9-WT, and Flag-Ubc9-K59A expressing granule neurons were not markedly different from each other. (c) SDS-PAGE analysis and immunoblots using lysates obtained from 293T cells transfected with expression plasmids encoding Flag-UBC9 WT or indicated UBC9 mutant alleles. Error bars are ±1 standard deviation.
Cerebellar slices were fixed and subjected to immunohistochemistry using a GFP antibody to visualize granule neurons in the cerebellar cortex. Claws were frequently observed at the ends of granule neuron dendrites in cerebellar slices transfected with the control vector and Flag-UBC9-WT23,25 (Fig. 6a, left and middle panels). In contrast, cerebellar slices transfected with Flag-UBC9-K65A had markedly fewer granule neuron dendritic claws. Instead, many dendrites of granule neurons expressing FLAG-Ubc9-K65A frequently displayed bulbous or tapered ends (Fig. 6a, right panel). As with cerebellar slices transfected with Flag-UBC9-K65A, slices transfected with Flag-UBC9-K74A and Flag-UBC9-K76A also had significantly fewer granule neuron dendrites that harbored claws. Importantly, granule neurons in cerebellar slices transfected with Flag-UBC9-K59A had a similar number of dendritic claws as those in slices transfected with Flag-UBC9-WT or the control vector (Fig. 6b). As a control for effects of mutations on Flag-Ubc9 expression, 293T cells were also transfected with each expression plasmid, however no appreciable differences were observed (Fig. 6c).
As in our previous transfection experiments, endogenous Ubc9 remains present in neurons transfected with plasmids encoding respective UBC9 alleles. Thus, our data suggest that suppression of dendritic claw differentiation results from a dominant effect on the activities of endogenous Ubc9, either through blocking activation by the E1 or by blocking recognition of the phosphorylated MEF2 PDSM in cells over-expressing Flag-Ubc9-K65A, Flag-Ubc9-K74A and Flag-Ubc9-K76A. Interestingly, over-expression of Flag-Ubc9-WT or Flag-Ubc9-K59A had no detectable effect on dendritic claw differentiation suggesting that Ubc9 was not a rate-limiting factor during claw differentiation. Taken together, the high correlation (correlation coefficient >0.95) between results for Ubc9-catalyzed SUMO conjugation obtained in vitro and effects of Ubc9 mutants on dendritic claw differentiation in the cerebellar cortex suggest that Ubc9 Lys65, Lys74 and Lys76 play an important role in discriminating between phosphorylated and non-phosphorylated forms of MEF2A in vivo (Fig. 6b).
Discussion
Phosphorylation-dependent SUMO conjugation integrates two important signal transduction pathways. Using biochemical, structural, and biological analysis, we have demonstrated that a conserved basic batch on the Ubc9 surface is responsible for Ubc9’s ability to discriminate between phosphorylated and non-phosphorylated PDSM motifs within MEF2A and HSF1 in vitro and MEF2A in vivo. The present analysis offers a unique perspective on the evolution of SUMO conjugation. Instead of integrating substrate phosphorylation with E3-mediated interactions as is more commonly observed in the ubiquitin pathway, some SUMO substrates have developed an ability to engage the SUMO E2 at two sites by combining E2 surfaces responsible for recognition of the SUMO consensus motif with surfaces required for recognition of the phosphorylated serine in the PDSM consensus motif.
The phosphorylated serine residue is located just two residues downstream of the glutamate residue in the extended ψ-K-x-E-x-x-S-P SUMO motif and its position is highly conserved in other substrates containing PDSMs. Since the same basic surface on Ubc9 appears to be important for PDSM discrimination for both MEF2A (in vitro and in vivo) and HSF1 (in vitro), our data suggest that E2-mediated recognition of other PDSM substrates such as GATA-1 and PPARγ would occur a similar manner. The analysis presented herein for phosphorylation-dependent SUMO conjugation to MEF2A helps to establish a molecular mechanism for E2-dependent conjugation to PDSM-containing substrates that regulate diverse biological processes including synapse development and plasticity.
Supplementary Material
Acknowledgements
We thank the NMR staff, particularly Kaushik Dutta, at the New York Structural Biology Center (NYSBC, New York, NY) for assistance in NMR data collection and processing. We also thank Ary Shalizi and Ali Yunus for helpful discussions, and Nancy Arango for help with 293T transfection assays. NMR resources at NYSBC are supported by NIH P41 GM66354. The work was supported in part by NIH grants GM075695 (A.D.C.), NS041021 (A.B.), and GM065872 (C.D.L.).
Appendix
Methods
Cloning, expression and protein purification
We obtained human UBC9 as described31 and 15N-labeled hUbc9 was expressed in M9 minimal media with 5 g l-1 of 15N-NH4Cl (Cambridge Isotope Laboratories, Inc), purified, and concentrated to 10 mg ml-1 in 75 mM NaCl, 20 mM Hepes (pH 7.5) and 1 mM DTT, flash frozen in liquid nitrogen and stored at -80°C. Human E1, Nup358/RanBP2 IR1* (2631-2695), p53 (319-393) and SUMO1-Alexa Fluor 488 have been described11,32,33.
Mutations were introduced into human UBC9 using PCR (Quickchange, Stratagene). Ubc9 isoforms were expressed, purified and concentrated to ~5 mg ml-1 in 200 mM NaCl, 20 mM Tris-HCl (pH 8.0) and 1 mM DTT. MEF2, Elk1 (MicroChemistry Core of MSKCC) and HSF1 (NEP, Gardner, MA) were synthesized, dissolved in dH2O (5 mM). N- and C-terminal boundaries for the peptides are defined in Fig. 1b.
Plasmids for transfection were constructed using pEF-BOS or pCDNA3. N-terminal hemaglutinin (HA) SUMO1 was cloned into a modified pEF-BOS vector. SUMO1 was flanked 5’ by BamHI followed by the coding sequence (1-291bp), a stop codon and a 3’ Not1 site. All other plasmids were constructed using pCDNA3. N-terminal (HA) SUMO2 was flanked 5’ by BamHI followed by a consensus Kozak sequence, the coding sequence (1-288bp), the endogenous stop codon and a 3’ EcoRI site. Yeast galactose response gene 4-MEF2A (GAL4-MEF2A) fusions were described23. N-terminal Flag- and myc-UBC9 alleles were flanked 5’ by EcoRI, a consensus Kozak sequence, the coding regions, the endogenous stop codons and 3’ XhoI site.
NMR analysis
NMR spectra were collected at 30°C on a Bruker DRX600 equipped with triple resonance cryoprobes and z-axis pulsed field gradients (New York Structural Biology Center, New York, NY). 2D 1H-15N HSQC spectra were recorded using HSQC pulse programs implemented in TopSpin (Bruker). 1024 and 256 complex points were recorded for 1H and 15N dimensions and processed using NMRPipe34 and NMRView35. Ubc9 was diluted to 300 μM in 95% (v/v) H2O/5% (v/v) D2O buffer (75 mM NaCl, 20 mM Hepes, pH 7.5, and 1 mM βME). NMR samples with molar ratios of 15N-labeled Ubc9:peptide at 1:0, 1:1, 1:2, 1:4, and 1:8 were pre-mixed individually in 200-300 μL and adjusted to pH 7.5 if necessary. 1H-15N HSQC data were collected for Ubc9:MEF2P and Ubc9:MEF2 samples and data were collected overnight for 1:16 Ubc9:peptide molar ratios to confirm that titrations reached saturation. Backbone chemical shifts for hUbc9 (ref. 36) (BMRB 4132) guided assignment of peaks in our 1H-15N HSQC spectra facilitating assignment of 93% of non-proline residues of Ubc9 (Figure S4A). Chemical shift perturbations were measured for 300 μM Ubc9 with MEF2P or MEF2 at 0 μM, 300 μM, 600 μM, 1200 μM, 2400 μM, or 4800 μM. Differences between chemical shift perturbations were calculated from 1:8 Ubc9:MEF2 and 1:8 Ubc9:MEF2P. Combined normalized chemical shift changes (Δδ2) were calculated by: Δδ2 = (ΔδN)2 + (γH/γN)2(ΔδH)2, where ΔδX = ΔδX, complex - ΔδX, apo and ΔδX and γX are the chemical shift (in ppm) and gyromagnetic ratio for nucleus X37. Differences between normalized chemical shift changes between MEF2P and MEF2 titrations were calculated by Δδ2MEF2P - Δδ2MEF2.
Biochemical Assays
Multiple turnover assays were performed at 37°C with 200 nM E1, 2 μM E2 (Ubc9), 2 μM SUMO1, 5 mM MgCl2, 20 mM HEPES (pH 7.5), 1 mM ATP and 1 mM DTT, and 50 μM MEF2P, MEF2 or Elk1 substrate. Samples were removed at indicated time points (Supplementary Figs. 1 and 8), denatured, separated by SDS-PAGE (12% Bis-Tris gels, MES buffer (Invitrogen)), and detected with SYPRO Ruby (Bio-Rad). E2~SUMO thioester was prepared as described31,33. Single-turnover assays were performed at 23°C for MEF2, MEF2P, HSF1, HSF1P, and at 37°C for p53 by initiating reactions with E2~SUMO (0.7 μl; final concentration 15-30 nM) in a 50 μl reaction (50 mM NaCl, 20 mM HEPES (pH 7.5), 5 mM EDTA, 120 nM Alexa fluor 488 labeled SUMO1, 12 nM E1, 120 nM Ubc9 and MEF2, HSF1, p53 or Elk1 from 10 μM to 1.28 mM. Single-turnover reactions were performed at 4°C using IR1* at 60 nM with MEF2 from 0.05-25.6 μM and Elk1 from 0.4-51.2 μM. Samples were denatured in non-reducing SDS-PAGE buffer containing 4 M urea, separated by SDS-PAGE (4-12% Bis-Tris; MES buffer (Invitrogen)). Products were detected and images processed as described33. Rates were measured using 2-4 time points within the linear range of product formation. Error bars are ±1 standard deviation as calculated from three independent experiments. Apparent k2 and Kd were obtained by fitting data to a hyperbolic single rectangular two-parameter function (vi/[E]= k2[S]/(Kd+[S]) (SigmaPlot 9.0), where vi is the initial reaction rate, [E] is E2~SUMO concentration, k2 is the apparent rate constant, [S] is the substrate concentration and Kd is the apparent dissociation constant (Tables 1,2).
Immunoprecipitation, immunoblotting and cerebellar slice cultures
293T cells were transfected by DNA-calcium phosphate precipitation with plasmids encoding HA-SUMO, GAL4-MEF2A, Myc-UBC9 or respective mutant alleles. Cells were lysed in radioimmunoprecipitation (RIPA) buffer (150 mM NaCl; 10 mM Na2HPO4, pH 7.5; 50 mM NaF; 2 mM EDTA; 20 mM N-ethylmaleimide; 1 mM PMSF, 1% (v/v) Triton X-100; 1% (w/v) sodium deoxycholate; 0.1% (w/v) SDS). 5% of starting material was retained for detection of input proteins. The remainder was incubated with anti-GAL4 agarose (Santa Cruz) for 1h at 4°C, washed with RIPA (4X) and phosphate buffered saline (1X), suspended in LDS buffer (Invitrogen), analyzed by SDS-PAGE and western blotting using indicated antibodies. Fold preference for SUMO modification of GAL4-MEF2A/GAL4-MEF2A-S408A was calculated by imaging immunoblots with a Fujifilm LAS-3000 imager using GAL4-MEF2A-SUMO1 as a fraction of total GAL4-MEF2A and quantified using Multi-Guage v.2.02 (Fujifilm). Corresponding SUMO conjugated GAL4-MEF2-K403R lanes were used as background. Error bars are ±1 standard deviation as calculated from three independent experiments. Western analysis utilized hemaglutinin (HA) polyclonal and GAL4 monoclonal (Santa Cruz Biotechnology), anti-tubulin monoclonal (SIGMA, St. Louis, MO) and anti-Myc monoclonal antibodies (Monoclonal Antibody Core, MSKCC). Global SUMO conjugation was detected by transfecting 293T cells with UBC9 alleles and HA-SUMO1 or HA-SUMO2. Cells were lysed in LDS sample buffer (Invitrogen), sonicated, and analyzed as described above.
293T cells were transfected with Flag-UBC9-WT, Flag-UBC9-K65A, Flag-UBC9-K74A, Flag-UBC9-K76A or Flag-UBC9-K59A and immunoblotted with anti-Flag or anti-tubulin antibody (loading control). Rat cerebellar slices were prepared as described23 from postnatal day 9 (P9) or P10 rat pups, transfected four days later with control pcDNA3 vector, Flag-UBC9-WT, Flag-UBC9-K65A, Flag-UBC9-K74A, Flag-UBC9-K76A or Flag-UBC9-K59A along with a GFP expression plasmid, and subjected to immunohistochemistry with a GFP antibody after fixation four days after transfection. Microscopy was performed as described23,38. The percentage of primary dendrites per neuron that harbor dendritic claws was measured in transfected granule neurons in a blinded manner.
References
- 1.Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Geiss-Freidlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 3.Dasso M. Emerging roles of the SUMO pathway in mitosis. Cell Div. 2008;3:5. doi: 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Dye BT, Schulman BA. Structural mechanism underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 6.Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr. Opin. Struct. Biol. 2007;17:726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 8.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 9.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 10.Lin D, et al. Identification of a substrate recognition site on Ubc9. J. Biol. Chem. 2002;277:21740–21748. doi: 10.1074/jbc.M108418200. [DOI] [PubMed] [Google Scholar]
- 11.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 13.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 14.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA Is Mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Lin DY, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism & consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Mohideen F, Lima CD. SUMO Takes Control of a ubiquitin-specific protease. Mol. Cell. 2008;30:539–540. doi: 10.1016/j.molcel.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong X, et al. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron. 2003;38:33–46. doi: 10.1016/s0896-6273(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 19.Kang J, Gocke CB, Yu H. Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem. 2006;7:5. doi: 10.1186/1471-2091-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang XJ, Gregoire S. A recurrent phospho-sumoyl switch in transcriptional repression & beyond. Mol. Cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Hietakangas V, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohshima T, Koga H, Shimotohno K. Transcriptional activity of peroxisome proliferator-activated receptor gamma is modulated by SUMO-1 modification. J. Biol. Chem. 2004;279:29551–29557. doi: 10.1074/jbc.M403866200. [DOI] [PubMed] [Google Scholar]
- 23.Shalizi A, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 24.Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J. Neurosci. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalizi A, et al. PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J. Neurosci. 2007;27:10037–10046. doi: 10.1523/JNEUROSCI.0361-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregoire S, et al. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J. Biol. Chem. 2006;281:4423–4433. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita D, et al. The transactivating function of peroxisome proliferator-activated receptor gamma is negatively regulated by SUMO conjugation in the amino-terminal domain. Genes Cells. 2004;9:1017–1029. doi: 10.1111/j.1365-2443.2004.00786.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riquelme C, Barthel KKB, Liu X. SUMO-1 modification of MEF2A regulates its transcriptional activity. J. Cell. Mol. Med. 2006;10:132–144. doi: 10.1111/j.1582-4934.2006.tb00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delano WL. The PyMOL molecular graphics system. DeLano Scientific, San Carlos; California, USA: 2002. [Google Scholar]
- 31.Yunus AA, Lima CD. Purification & activity assays for Ubc9, the ubiquitin conjugating enzyme for the small ubiquitin-like modifier SUMO. Methods Enzymol. 2005;398:74–87. doi: 10.1016/S0076-6879(05)98008-7. [DOI] [PubMed] [Google Scholar]
- 32.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yunus AA, Lima CD. Purification of SUMO conjugating enzymes & kinetic analysis of substrate conjugation. Methods in Molecular Biology. SUMO Protocols. 2009;497:167–86. doi: 10.1007/978-1-59745-566-4_11. Ulrich, ed. (Humana Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–29. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–14. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 36.Liu Q, Shen B, Chen DJ, Chen Y. Backbone resonance assignments of human UBC9. J. Biomol. NMR. 1999;13:89–90. doi: 10.1023/a:1008364314328. [DOI] [PubMed] [Google Scholar]
- 37.Edwards TA, et al. Solution structure of the Vts1 SAM domain in the presence of RNA. J. Mol. Biol. 2006;356:1065–1072. doi: 10.1016/j.jmb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.