Abstract
SUMO proteases catalyze two reactions, deconjugation of SUMO from substrates and processing of precursor SUMO isoforms to prepare SUMO for conjugation. The SUMO protease family includes two members in yeast (Ulp1 and Ulp2) and as many as six members in human (SENP1–3, SENP5–7). SENP/Ulp proteases each contain conserved C-terminal domains that catalyze protease activity. The C-terminal protease domains exhibit unique specificities during SUMO processing and deconjugation in vitro. While there are many available reagents to assess these activities, including fusion proteins and chemically modified SUMO isoforms, our studies have indicated that the composition of substrates C-terminal to the scissile bond can substantively influence the activity of the protease. As such, we have relied extensively on assays that utilize endogenous substrates, namely wild-type SUMO precursors and SUMO conjugated substrates. In this chapter, we will describe methodological details for purification and characterization of SUMO precursors, SUMO conjugated substrates, and SUMO proteases. We will also describe methods for kinetic analysis of SUMO deconjugation and maturation using endogenous substrates.
1. Introduction
SUMO is a member of the ubiquitin (Ub) and ubiquitin-like (Ubl) family. Post-translational attachment of SUMO to target proteins occurs through an enzymatic cascade analogous to the ubiquitin conjugation cascade (E1-E2-E3 enzymes), ultimately resulting in formation of an isopeptide bond between the Ub/Ubl C-terminal residue and substrate lysine residue (1,2). While yeast includes one SUMO ortholog named Smt3, mammals contain at least four SUMO family members. SUMO-2 and SUMO-3 share greater than 96% sequence identity to each other in their processed forms, although each share only 43% and 42% identity to SUMO-1, respectively. SUMO-4 is more similar to SUMO-2/3, but it remains unclear whether SUMO-4 forms SUMO conjugates (3). We utilize UniProtKB/Swiss-Prot nomenclature for human SUMO isoforms 1–4.
The steady state level of a particular SUMO conjugated substrate is regulated by maintaining balance between SUMO conjugation and SUMO deconjugation. SUMO deconjugation occurs through the action of SUMO (SENP/Ulp) proteases. These enzymes are composed of at least two domains, an N-terminal domain which directs subcellular localization and a conserved C-terminal catalytic domain which shares similarity to other papain-like cysteine proteases (4). SENP/Ulp proteases catalyze two essential activities. The first involves SUMO precursor maturation in a reaction that entails proteolysis and removal of amino acids C-terminal to the conserved SUMO diglycine motif. The second proteolytic activity entails SUMO deconjugation from proteins, releasing both the target lysine and SUMO for subsequent rounds of conjugation. These two activities share a common catalytic mechanism, although the substrates differ insomuch as processing involves hydrolysis of an α-linked peptide bond and deconjugation catalyzes hydrolysis of the ε-linked lysine isopeptide bond (Figure 1).
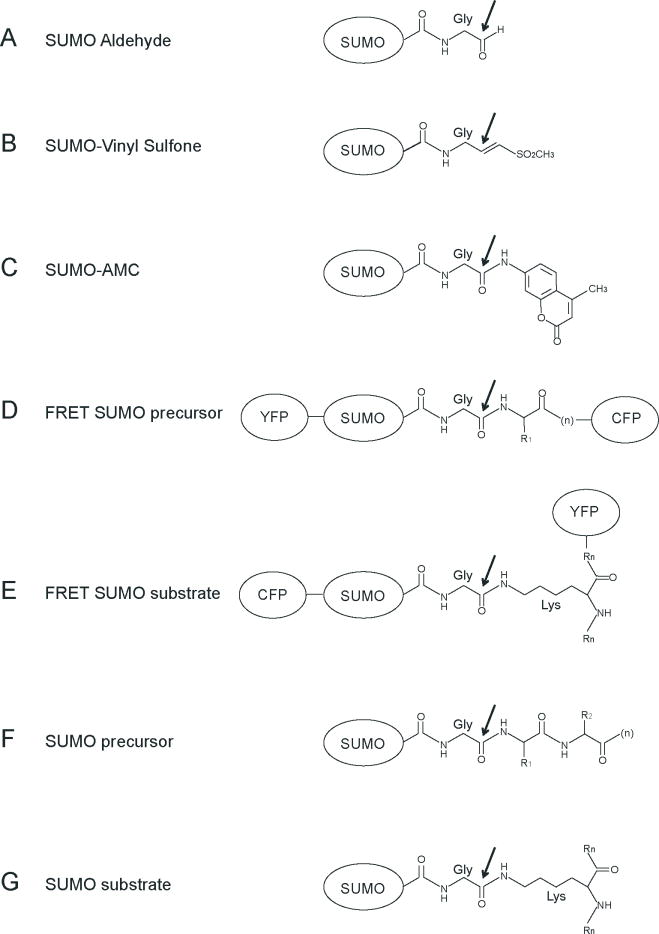
Figure 1. SUMO variants and substrates used in proteolytic assays.
A) SUMO with its C-terminal glycine reduced to aldehyde. B) SUMO with its C-terminal glycine modified with vinyl sulfone. C) SUMO with its C-terminal glycine modified with 7-Amido-4-methylcoumarin. D) Fusions in a single polypeptide of the SUMO precursor with the yellow fluorescent protein (YFP) at the N-terminus and the cyan fluorescent protein (CFP) at the C-terminus. E) Fusion of SUMO with the cyan fluorescent protein (CFP) at the N-terminus forming an isopeptide bond with a lysine of a SUMO substrate fused to yellow fluorescent protein (YFP). F) Endogenous SUMO precursor substrate. G) Endogenous SUMO substrate, with its C-terminal glycine forming an isopeptide bond with a lysine of a substrate. Black arrow indicates the site of nucleophilic attack and cleavage by the catalytic cysteine of the protease.
The six human Ulp/SENP protease family members identified thus far are termed SENP1–3 and SENP5–7 (4,5). While their individual physiological roles remain somewhat obscure, recent reports support the hypothesis that SENP family members participate in non-redundant cellular functions. For example, mouse SENP1 is required during embryonic development (6), while knockdown of SENP5 inhibited cell proliferation and exhibited defects in nuclear morphology, revealing essential roles during mitosis and/or cytokinesis (7). The diversity of cellular functions for SENP proteases is supported by the observation that SENP proteases exhibit distinct subcellular localization patterns (8). It is believed in most instances that non-conserved N-terminal domains direct subcellular localization, and that subcellular localization contributes in part to SENP function by restricting protease activity to distinct areas within the cell.
Recent studies have revealed differences in the ability of some SENP catalytic domains to catalyze maturation of SUMO precursors. While SENP1 and SENP2 can hydrolyze all three SUMO precursors, SENP2 exhibits a preference for SUMO-2 > SUMO-1 > SUMO-3 (9,10) while SENP1 exhibits different specificities during processing (SUMO-1 > SUMO-2 > SUMO-3) (11,12). For SENP2, substrate preferences were correlated to differences in polypeptide composition C-terminal to the scissile peptide bond in respective SUMO isoforms while for SENP1 and SENP2, substrate preferences could be explained by differences in affinity between the protease domain and principle interaction surfaces for respective SUMO isoforms (9–12). Recent analysis of SENP6 and SENP5 revealed that each preferred substrates containing isopeptide bonds, and both exhibited a preference for SUMO-2/3, either as a conjugated substrate or as chains (7, 8).
Kinetic analysis has provided unique insights into substrate specificity for SENP1 and SENP2, albeit with substantive differences in observed Kcat and Km values (10, 12). While these differences may be attributable to indirect methodologies used to extract kinetic parameters from the experimental data, it is also possible that utilization of chemical or genetic modifications C-terminal to the SUMO scissile peptide bond might interfere with accurate assessment of protease activity. For instance, ubiquitin or SUMO-AMC (7-amido-4-methylcoumarin) modifications are commonly used substrates in proteolytic assays because they are easily followed by spectrofluorometric analysis (13) (Figure 1). Ubiquitin aldehyde or vinyl sulfone derivatives are also commonly used as these C-terminal modifications trap intermediates of the proteolytic reaction (14). Another useful methodology employs FRET (fluorescence resonance energy transfer), in this case GFP and YFP are linked to both ends of the substrate and changes in fluorescence are monitored during proteolytic cleavage. This method is amenable to applications involving peptide and isopeptide cleavage assays, but is dependent on suitable FRET signal between donor and acceptor positions within the substrate (12). While these reagents have been used to determine Km or Ki values during proteolysis, they do not represent physiological substrates (13, 14).
In this chapter we describe procedures to express and purify SENP family members and methods to extract kinetic parameters for SENP protease activities under steady-state conditions using full-length SUMO isoforms and substrates that do not contain chemical modifications or fusion to foreign proteins.
2. Materials
2.1. Protein expression and purification
Luria-Bertani (LB) media: 10 g bacto-tryptone, 10 g sodium chloride, 5 g bacto-yeast extract in 1 L water.
Super Broth (SB) media: 32 g bacto-tryptone, 20 g bacto-yeast extract, and 5 g sodium chloride in 1 L water.
Antibiotics: Ampicilin: 50 mg/mL in water, filter sterilized. Kanamycin: 50 mg/mL in water, filter sterilized. Chloramphenicol: 34 mg/ml in ethanol.
Plasmids: pET-28b (Novagen).
PCR: Primers (Invitrogen); Pfu turbo polymerase (Strategene) or Deep Vent polymerase (New England Biolabs); human cDNA (Clontech).
Bacterial strains: Escherichia coli BL21(DE3) RIL Codon Plus (Stratagene).
Isopropyl-beta-D-thiogalactopyranoside (IPTG): 1M in water, filter sterilized.
Fermentation: BioFlo-3000 fermentor equipped with a 14 L vessel (New Brunswick).
Thrombin (SIGMA): 1 U/μl (0.33 μg/μl) in 20 mM Tris-HCl pH 8.0, 350 mM NaCl, 1 mM β-mercaptoethanol (BME).
Ulp1 protease catalytic domain (amino acids 403–621): 3 mg/ml in 20 mM Tris-HCl pH 8.0, 350 mM NaCl, 1 mM BME, 10% glycerol at −80°C.
Suspension Buffer: 20% sucrose, 20 mM Tris-HCl pH 8.0, and 1 mM BME.
Lysis Buffer: 20% sucrose, 20 mM Tris-HCl pH 8.0, 1 mM BME, 350 mM NaCl, 20 mM imidazole, 10 μg/ml DNAse, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1% IGEPAL CA-630, and 20 μg/ml lysozyme.
NTA-Ni Superflow resin (Qiagen).
Buffer A: 20 mM Tris-HCl pH 8.0, 350 mM NaCl, 1 mM BME, and 20 mM imidazole.
Buffer B: 20 mM Tris-HCl pH 8.0, 350 mM NaCl, 1 mM BME, and 400 mM imidazole.
Buffer C: 20 mM Tris-HCl pH 8.0, 200 mM NaCl, and 1 mM BME.
Buffer D: 20 mM Tris-HCl pH 8.0, 50 mM NaCl, and 1 mM BME.
Buffer E: 20 mM Tris-HCl pH 8.0, 1 M NaCl, and 1 mM BME.
Buffer F: 20 mM Tris-HCl pH 8.0, 350 mM NaCl, and 1 mM BME.
AKTA-FPLC (GE Healthcare), equipped with gel filtration (Superdex-75 26/60 and Superdex-200 26/60) and ion exchange columns (Mono-Q 10/10 and Mono-S 10/10) (See Note 1).
Centricon or Centriprep micro-filtration devices (Amicon) with appropriate molecular weight cut-offs (10 kDa or 30 kDa).
2.2 SUMO-deconjugation assays and SDS-polyacrylamide gel electrophoresis (SDS-PAGE)
Buffer I: 20 mM Hepes pH 7.5, 5 mM MgCl2, 0.1 % Tween-20, 50 mM NaCl, 1 mM DTT and 2 mM ATP.
Buffer II: 25 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% Tween-20 and 2 mM DTT.
NuPAGE system for SDS-PAGE analysis with MES or MOPS running buffer (Invitrogen).
4–12% polyacrylamide gradient gels.
12% polyacrylamide gels.
SYPRO Ruby (Bio-Rad).
SYPRO fixing and destaining solution: 7% acetic acid, 10% methanol.
2.3. Integration and data analysis
Gel-Doc apparatus (Bio-Rad) for gel visualization under UV irradiation.
Image processing and data integration: Quantity-One software (Bio-Rad).
Raw data processing: EXCEL (Microsoft).
Data and regression analysis: SigmaPlot 9.0 software (Systat Software, Inc).
3. Methods
The C-terminal SENP/Ulp catalytic protease domains consist of approximately 220 residues and exhibit homology throughout the SENP/Ulp family. The first identified SENP/Ulp family member was Ulp1 (15), and the first structure of a SENP/Ulp catalytic domain was elucidated through x-ray structure determination of the yeast Ulp1 catalytic domain in complex with Smt3-aldehyde (16). We have utilized the structure of the Ulp1 catalytic domain in conjunction with sequence alignments to design PCR primers for respective catalytic domains for each member of the human SENP/Ulp protease family (for alignments, see Reverter and Lima, 2004). Details of this process and expression of a representative SENP family member will be addressed below.
To analyze processing reactions for human SUMO family members, we have utilized native proteins and SUMO isoforms that exclude the native C-terminal stop codon and include a C-terminal hexahistidine affinity tag (Figure 1). Based on our previous data with SENP2, protease specificity was substantially influenced by only 2–3 amino acids immediately C-terminal to the scissile peptide bond. In this instance, use of SUMO precursors with C-terminal hexahistidine sequences located after the endogenous stop codon appeared equivalent in processing reactions using SENP2 when compared to endogenous SUMO isoforms. Use of a C-terminal tag is especially useful during analysis of the SUMO-2 precursor as it includes a very short C-terminal native sequence (GG-VY) that complicates analysis because of the difficulty associated with resolving SUMO-2 substrates from SUMO-2 products by conventional SDS-PAGE electrophoresis. With that said, it remains a distinct possibility that other SUMO proteases may be inhibited or affected by inclusion of C-terminal affinity tags, so it is best to utilize comparative assays with both native and tagged SUMO isoforms during initial characterization (10).
Deconjugation reactions utilize SUMO isoforms which are conjugated to a substrate via a covalent isopeptide bond between the SUMO C-terminal glycine and substrate lysine (Figure 1). For kinetic analysis, we have utilized the C-terminal domain of human RanGAP1 since it can be readily conjugated to any SUMO isoform in quantities sufficient for further analysis. We also know this substrate to be a monomer in solution, simplifying kinetic analysis. While current structural and functional data suggest that the RanGAP1 substrate does not interact specifically with the protease catalytic domain, it does engage in contacts to the protease surface (10, 12). As such, it is important to consider that SUMO substrates can interact with the protease domain and may thus affect kinetic analysis.
3.1. Preparation of SENP catalytic domains
Expression constructs containing catalytic domains for SENP/Ulp family members were defined by the structures of the catalytic domains from Ulp1, SENP1 and SENP2 (9, 10, 12, 15). Primers were designed to amplify respective coding regions for human SENP family members using PCR and human cDNA. Use of a low error PCR polymerase is recommended.
Using SENP2 as an example, primers were designed to include NheI and HindIII restriction sites 5′ and 3′ to the coding regions to facilitate ligation into pET-28b. Use of the NheI site encodes a N-terminal thrombin-cleavable hexahistidine tag fused to the SENP2 catalytic domain (364–489). Stable clones were obtained by transforming into E. coli DH5α with ligation products. Clones containing suitable inserts were verified by DNA sequencing. This procedure has been repeated for other SENP/Ulp family members and has resulted in successful isolation of Ulp1, Ulp2 from yeast genomic DNA and SENP1, SENP2, SENP3, SENP5, SENP6 and SENP7 from human cDNA.
Bacterial strains suitable for protein expression were obtained by transforming respective plasmids into E. coli BL21 DE3 codon plus strains that contained the T7 polymerase and a ColE1-compatible plasmid that encodes additional copies of rare tRNA genes to enhance expression of recombinant polypeptides.
To induce protein expression, bacterial cultures were grown by fermentation at 37°C to OD600=0.8 and induced by addition of IPTG to a final concentration of 0.5–1.0 mM. Cultures were then incubated for 3 to 4 hours at 30°C. Cultures were harvested by centrifugation (7000 × g) and the supernatant discarded.
Cell pellets were suspended in Suspension Buffer. Cell pellets can be stored at this stage for later use by snap freezing the suspended pellets in liquid nitrogen. Cell suspensions were equilibrated in Lysis Buffer, and cells were disrupted by sonication. Cell debris was removed by centrifugation (40000 × g).
The supernatant was applied to Ni-NTA resin equilibrated with Buffer A. To elute His6-SENP2 from the Ni-NTA resin, a step gradient with Buffer B was applied to the chromatography column. Peak fractions containing His6-SENP2 were dialyzed against Buffer C overnight at 4°C in the presence of thrombin at a 1:1000 (w:w) thrombin to protein ratio. To ensure that thrombin cleavage was complete, the reaction was analyzed by SDS-PAGE before proceeding to the next step.
To purify SENP2, the dialyzed mixture was passed through a 0.2 μm filter and applied to a gel filtration column (Superdex-200) equilibrated with Buffer F (see Note 1 & 2). Fractions were analyzed by SDS-PAGE and those containing SENP2 were pooled, equilibrated to Buffer D, and applied to a cation exchange matrix (Mono-S). SENP2 was eluted from the cation exchange resin by application of a salt gradient from Buffer D to 50% Buffer E. Fractions were again analyzed by SDS-PAGE and those containing SENP2 were pooled, concentrated to 10 mg/ml, snap frozen in liquid nitrogen, and stored at −80 for future use. Purification protocols for other SENP/Ulp family members employ similar chromatography methods. Expression levels for recombinant Ulp1, SENP1, and SENP2 are high and approach 30 mg of purified material per liter of starting culture.
3.2. Cloning and preparation of SUMO precursors fused to C-terminal hexahistidine tags
DNA encoding full-length SUMO-1, SUMO-2, and SUMO-3 precursors was isolated by PCR from human cDNA as described above.
Plasmids containing suitable SUMO DNA fragments for recombinant protein expression were obtained by engineering NcoI and XhoI restriction sites into the upstream and downstream primers, respectively, but excluding the native stop codon to encode a polypeptide fused to a C-terminal hexahistidine sequence to facilitate purification by metal-affinity chromatography. These DNA fragments were prepared and ligated into pET-28b. Gene sequences were confirmed by DNA sequencing.
DNA plasmids were transformed into appropriate E. coli expression strains and protein expression was induced as described above.
Cell pellets were suspended, lysed, and prepared as described in Step 3.1.5 and proteins were purified via Ni-NTA agarose chromatography (see above).
Fractions were analyzed by SDS-PAGE and those fractions containing SUMO were buffer exchanged by overnight dialysis into Buffer D, and applied to an anion exchange matrix (Mono-Q). SUMO isoforms were eluted by application of a salt gradient from Buffer D to 50% Buffer E. Fractions were analyzed by SDS-PAGE and those that contained SUMO were pooled and applied to a gel filtration column (Superdex-75) equilibrated in Buffer F.
To prepare SUMO isoforms for large scale conjugation reactions, preSUMO was subjected to proteolysis using Ulp1 (16) at a protease to substrate ratio of 1:1000 (w:w). Trace quantities of Ulp1 were removed by applying the reaction to an anion exchange resin (Mono-Q) followed by elution using a salt gradient from Buffer D to 50% Buffer E over 12 column volumes. Fractions were analyzed by SDS-PAGE and those containing SUMO were pooled and applied to a gel filtration column equilibrated in Buffer F (Superdex-75). Fractions were again analyzed by SDS-PAGE and those that contained SUMO were pooled, concentrated to 10 mg/ml, snap frozen in liquid nitrogen, and stored at −80°C.
3.3. Cloning and preparation of native SUMO precursors fused to N-terminal hexahistidine tags
This protocol is similar to that described for purification of C-terminal hexahistidine tagged SUMO isoforms (see above) but the cloning strategy differs. In this case, full-length SUMO precursors containing native C-terminal tails and respective stop codons were produced by introducing an N-terminal hexahistidine tag which can be removed by proteolysis using thrombin, leaving behind three non-native N-terminal amino acid residues (Gly-Ser-His-).
To produce the desired native SUMO isoform precursors, coding DNA for respective SUMO isoforms was ligated into pET-28b using NheI and XhoI restriction sites which were engineered in upstream and downstream primers, respectively.
Plasmids were transformed into expression strains as described above.
Cell pellets were suspended, lysed, and prepared as described in Step 3.1.5 and proteins were purified via Ni-NTA agarose chromatography (see above).
The N-terminal His-tag was removed by overnight treatment of the His6-SUMO fusion with thrombin at a final thrombin to protein ratio of 1:1000 (w:w). The native SUMO isoform was purified by applying this mixture to anion exchange resin (Mono-Q) and eluting with a salt gradient from Buffer D to 50% Buffer E. Fractions containing SUMO were pooled and further purified by gel filtration chromatography (Superdex-75) as described above.
3.4. Preparation of SUMO conjugated substrates
SUMO conjugated substrates are prepared using SUMO conjugation enzymes in conjunction with mature SUMO and respective substrates (see Chapter on SUMO conjugation in this volume (Yunus and Lima) and 18). It is advantageous to purify and isolate SUMO conjugated substrates by conventional chromatography methods to avoid potential artifacts generated by remnants of the affinity tags used for initial purification. We have developed preparative conjugation reactions for human SUMO substrates that include IkBα, P53, 10-mer P53 peptide, and RanGAP1. For our studies on deconjugation, we have principally utilized SUMO isoforms conjugated to the C-terminal RanGAP1 domain (9,10).
Preparative quantities of the RanGAP1 C-terminal domain can be produced in E. coli by conventional protein expression methods (17). RanGAP1 conjugation reactions were performed at 37°C in Buffer I containing 150 nM SUMO E1, 100 nM Ubc9 (E2), 16 μM of the desired SUMO isoform and 8 μM RanGAP1. The reaction typically requires 2–3 hours to complete. The extent of conjugation was evaluated by SDS-PAGE before proceeding to the next step.
The mixture is filtered, concentrated (if necessary), and purified by gel filtration chromatography (Superdex-75) as described above. In this instance, RanGAP1-SUMO can be readily separated from free SUMO and RanGAP1 since it migrates with an apparent molecular weight approximately twice that of its constituents.
3.5. Analysis of processing and deconjugation reactions
Processing reactions (carboxyl-terminal hydrolase activity) included 10 μM native SUMO-1, -2 and -3 precursors in Buffer II.
SUMO precursors are incubated with 10 nM SENP proteases (for SENP2) which corresponds to a protease:substrate ratio of 1:1000 (w:w). The reaction is incubated at 23°C, 30°C or 37°C in Buffer II.
Samples are removed at time points ranging from seconds to hours, stopped by addition of SDS-PAGE loading buffer, and analyzed by SDS-PAGE.
Gels are fixed in SYPRO solution and stained using SYPRO-Ruby for 3 hours. Gels are de-stained in SYPRO solution for 30 minutes prior to visualization (see Note 3).
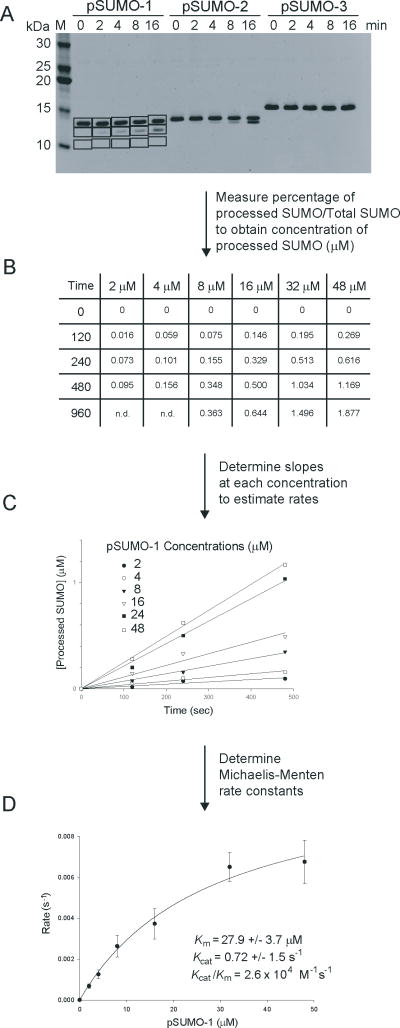
Protein bands were visualized in the gel by UV irradiation. Protein quantities were determined by 2-dimensional integration using a Gel-Doc apparatus and associated software. Figure 2 depicts an example for quantifying SUMO processing reactions.
Deconjugation reactions utilized 3 μM SUMO-modified RanGAP1 and 5 nM SENP protease (SENP2) at 23°C, 30°C or 37°C in Buffer II.
Samples are removed at time points ranging from a few seconds to several minutes, stopped by addition of SDS-PAGE loading buffer, and analyzed by SDS-PAGE.
Gels are fixed in SYPRO solution and stained using SYPRO-Ruby for 3 hours. Gels are de-stained in SYPRO solution for 30 minutes prior to visualization.
Gels were visualized and analyzed as described in Step 5 above. Figure 3 depicts an example utilized for quantifying SUMO deconjugation reactions.
Figure 2. Detection and kinetic analysis for SUMO processing reactions.
A) SDS-PAGE for a processing reaction with preSUMO-1, preSUMO-2 and preSUMO-3 stained with SYPRO. Bands were quantified with Bio-RAD Quantity One software. Boxes indicate integrated areas for estimation of protein levels and representative background levels. B) Table indicating the concentration of processed SUMO at different times for a range of different SUMO concentrations. C) Linear representation of the SUMO processing reaction from table in B. Slopes represent SUMO processing rates at different substrate concentrations. D) Michaelis-Menten kinetics was applied to SUMO processing rates in C to extract kinetic parameters. Data was obtained in triplicate to determine standard deviations and error bars.
Figure 3. Detection and kinetic analysis for SUMO deconjugating reactions.
A) SDS-PAGE for SUMO deconjugation reaction at 2, 4, 8 and 16 μM RanGAP1-SUMO-1 concentrations stained with SYPRO. Bands were quantified with Bio-RAD Quantity One software. Boxes indicate integrated areas for estimation of protein levels and representative background levels. B) Table listing concentrations of released SUMO at different times for a range of different RanGAP1-SUMO concentrations. C) Linear representation of the SUMO deconjugation reaction from table in B. Slopes represent SUMO deconjugation rates at different substrate concentrations. D) Michaelis-Menten kinetics was applied to SUMO deconjugation rates in C to extract kinetic parameters. Data was obtained in triplicate to determine standard deviations and error bars.
3.6. Kinetic analysis under multiple turnover conditions
Catalytic parameters for processing and deconjugation were determined under steady state multiple turnover conditions using Michaelis-Menten kinetics by establishing conditions that enable rate measurements in the linear range during product formation. For reactions containing the SENP2 catalytic domain, preliminary titration analysis indicated that the respective binding constants were in the low μM range, thus enabling analysis by conventional SDS-PAGE electrophoresis and SYPRO-Ruby staining which is capable of detecting a broad range of protein concentrations (from 1–1000 ng/band) with sensitivity comparable to that obtained using silver stain. Using this method, we have extracted rate constants for deconjugation of RanGAP1-SUMO-1 and RanGAP1-SUMO-2/3 and for processing of preSUMO-1, −2, and −3 (10) (Figure 2 and 3).
During processing and deconjugation, initial reaction velocities were determined by titrating the substrate (from 40 nM to 96 μM) using SENP2 concentrations of 5 nM or 0.5 nM for processing or deconjugation reactions, respectively. The length for each respective time course was empirically determined to obtain initial rate velocities in the linear range.
Three to four time points were measured for each substrate concentration to obtain estimates of reaction velocity in a linear range (Figures 2 & 3). The respective bands were quantified by integrating the intensities contained within respective boxed areas (see Note 4). Numerical values were extracted for each band and analyzed to ensure that increasing substrate concentrations resulted in a commensurate increase in initial rate velocity at each substrate concentration (Figures 2B, 2C, 3B, 3C).
The slope of each line represents an initial reaction velocity (see Note 5). These results are plotted against respective substrate concentrations (Figures 2D & 3D). To obtain rate parameters, these data are fitted to a hyperbolic 2-parameter, single rectangular Michaelis-Menten function (υ=Vmax [S]/Km+[S]) using SigmaPlot to derive rate constants (kcat=Vmax/[E], and Km=Michaelis-Menten constant). Data was measured in triplicate to obtain standard deviations and respective error bars (indicated at ±1 standard deviation).
4. Notes
To ensure reproducibility and to protect chromatography columns from undo wear and tear, all chromatographic steps were performed using filtered buffer solutions prepared from MilliQ (Millipore) water or the equivalent. All buffers should were degassed under vacuum for at least 1 hour prior to use.
All protein purification was conducted at 4°C to avoid degradation and/or aggregation. All protein preparations were passed through a 0.2 μm filter prior to application to chromatography media. All proteins were flash-frozen in liquid nitrogen prior to storage at −80°C.
SYPRO-Ruby staining is a reasonable method to quantify protein bands, but its sensitive detection limits can give rise to slow migrating bands in the gel. Duplicate gels are recommended for each experiment to ensure reproducibility.
Integration of signal requires proper background selection. We select individual background boxes adjacent to each band of interest.
To obtain kinetic parameters Km and Kcat, it is important to determine velocities at substrate concentrations at least ten-fold higher than Km to ensure that the reaction is nearing saturation. For Km values in the low nM range, SYPRO staining is not recommended due to its sensitivity limits. In this instance, one may utilize immunoblotting and detection with appropriate antibodies raised against SUMO or the desired substrate (18).
References
- 1.Hershko A, Ciechanover A. The Ubiquitin System. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Saitoh H, Pu RT, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. TIBS. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 3.Towerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KMT. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 4.Melchior F, Schergaut M, Pichler A. SUMO: ligases, isopeptidases and nuclear pores. TIBS. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi T, et al. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol Cell Biol. 2005;25:5171–82. doi: 10.1128/MCB.25.12.5171-5182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Bacco A, et al. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007 May 11; doi: 10.1016/j.tibs.2007.05.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 9.Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human SENP2 and a SENP2-SUMO complex. Structure. 2004;12:1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Reverter D, Lima CD, editors. Nat Struct Mol Biol. Vol. 13. 2006. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates; pp. 1060–1068. [DOI] [PubMed] [Google Scholar]
- 11.Shen LN, Dong C, Liu H, Naismith JH, Hay RTT. The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem J. 2006;397:279–288. doi: 10.1042/BJ20052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen L, Tatham MH, Dong C, Zagorska A, Naismith JH, Hay RT. SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat Struct Mol Biol. 2006;13:1069–1077. doi: 10.1038/nsmb1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang LC, Melandri FD, Stein RL. Kinetic and Mechanistic Studies on the Hydrolysis of Ubiquitin C-Terminal 7-Amido-4-Methylcoumarin by Deubiquitinating Enzymes. Biochemistry. 1998;37:1868 –1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 14.Hemelaar J, Borodovsky A, Kessler BM, Reverter D, Cook J, et al. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol Cell Biol. 2004;24:84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 16.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 17.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 18.Yunus AA, Lima CD. Purification and activity assays for Ubc9, the ubiquitin conjugating enzyme for the small ubiquitin-like modifier SUMO. Methods in Enzymology. 2005;398:74–87. doi: 10.1016/S0076-6879(05)98008-7. [DOI] [PubMed] [Google Scholar]