Abstract
Defensins are highly basic cationic peptides that are important components of the innate and adaptive immune response pathways. In addition, these peptides are involved in CD8+ T cell response to HIV-1, increased pulmonary infection risk among cystic fibrosis patients, upregulated levels of HNP-5 for patients with ulcerative colitis and Crohn’s disease, monitoring HNP-3 levels as a tumor classification scheme for cutaneous T cell lymphomas, and have promise in the pharmaceutical field as a new class of antibiotics. Here we present a parallel assay for the α (HNP1-3) and β (HBD1-2) classes of defensins in saliva that are naturally observed in the concentration range of 1 ng/mL to 10 μg/mL. The method utilizes solid phase extraction of saliva samples combined with liquid chromatography-tandem mass spectrometry to identify and quantitate defensin targets. The approach involves limited sample manipulation and is easily amenable to automation. The saliva samples analyzed are derived from a large cohort study focused on examining the role of polymorphisms in genes of innate and adaptive immunity in modulating the response to vaccination for two gastrointestinal tract infections: typhoid and cholera. The α-defensin levels observed range from 1 to 10 μg/mL and correlate well with known active concentrations against a wide variety of pathogens. The observed concentration range for β-defensins was between the detection limit and 33 ng/mL and had a sensitivity level that was comparable to immunoassay-based detection. This method is easily adapted for use in a clinical immunology setting and can be modified for other biological matrices. This assay will facilitate examination of the production, secretion, and regulation of defensin peptides in a direct fashion in order to coordinate levels of these compounds with gender, age, response to vaccination, gene copy number, and oral health.
Keywords: peptides, defensins, tandem mass spectrometry, innate immunity, adaptive immunity
Introduction
One of the major routes of pathogen exposure in humans occurs through the oral cavity1, 2. In response to potential pathogen infection, there exists a variety of different mediators present in human saliva that are involved in host defense3. Antimicrobial peptides present in saliva are key components of the innate immune system that function as a primary defense against infection4. The majority of these peptides that have activity against bacteria and fungi are small (3–5 kD), cationic, and amphiphilic. One group of these peptides is the defensins5–7, which can be classified into two different forms: α- and β-defensins8. The active form of α-defensins found in saliva are a series of highly conserved peptides coded for by genes located on chromosome eight that differ by a single amino acid residue present on the N-terminus as shown in table 1. Originally isolated from myeloid neutrophil cells in the 1980’s, these peptides are commonly referred to as human neutrophil peptides9, 10. Expression of the 30 amino acid residue HNP-1 and HNP-3 peptides is derived from two separate genes on chromosome eight, while HNP-2 is a posttranslational modification of HNP-1 and/or HNP-3 involving cleavage of the N-terminal alanine (the specific protease or prtoeases involved in this process is unknown). All three of the forms found in human saliva contain six cysteine residues that are linked through disulfide bridges at cysteine residue positions 1–6, 2–4, and 3–5.
Table 1.
Sequences and masses of defensin peptides.
| Peptide | Sequence | Monoisotopic Mass |
|---|---|---|
| Human Neutrophil α–Defensin-1 (HNP-1) | ACYCRIPACIAGERRYGTCIYQGRLWAFCC | 3445.557 |
| Human Neutrophil α–Defensin-2 (HNP-2) | CYCRIPACIAGERRYGTCIYQGRLWAFCC | 3374.520 |
| Human Neutrophil α–Defensin-3 (HNP-3) | DCYCRIPACIAGERRYGTCIYQGRLWAFCC | 3489.547 |
| Human β–defensin 1 (HBD-1) | DHYNCVSSGGQCLYSACPIFTKIQGTCYRGKAKCCK | 3931.774 |
| Human β–defensin-2 (HBD-2) | GIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKP | 4331.179 |
Although the β-defensins (originally isolated from human neutrophils in 1993)11 found in saliva (HBD-1 and HBD-2) have a similar β-sheet confirmation, disulfide linkage scheme (1–5, 2–4, 3–6), and close chromosome location as α-defensins, the amino acid sequences are markedly different12 (see table 1). Interestingly of the approximately 50 known forms of β-defensins, these peptides of 35–50 residues in length contain only eight conserved amino acids six of which are the cysteine residues involved in disulfide bridge formation. It was soon discovered the β-defensins were not only involved in innate immunity but also played a role in immune response as well. For example, expression of human β-defensin-2 (HBD-2) is upregulated in response to inflammatory mediators (TNF-α, IL-1β, various Gram-negative and positive bacteria, and lipopolysaccharide). β-Defensins have also exhibited chemotactic activity by binding to the CCR6 receptor on dendritic and T cells, as well as mast cells, and macrophages12.
Defensins have activity against a wide variety of microbes (Gram-positive and negative bacteria), fungal, and even some viral targets13. The major mechanism of antimicrobial activity of all defensin peptides (which adopt a β-sheet confirmation) is thought to occur through interaction with the membrane of the invading microbe resulting in a release of the cell contents. In addition to their traditional role in innate immunity, defensins may play a role in CD8+ T cell response to HIV-1 infection14–17 and indicate increased pulmonary infection risk among cystic fibrosis patients18. Upregulated levels of HNP-5 have been observed in patients with ulcerative colitis and Crohn’s disease19, and the assay of HNP-3 levels has also been employed as a tumor classification scheme for cutaneous T cell lymphomas20. There has also been interest in defensins by the pharmaceutical field as potential novel antibiotics21, 22.
Therefore, a reliable analytical method that covers all classes of defensins in a single assay would be a valuable tool. Previous methods for assay of defensins have included semiquantiative Western blot analysis23, 24, relative expression of mRNA levels25, ELISA-based approaches26, 27, RIA28, 29, immunohistochemistry30, flow cytometric analysis31, liquid chromatography (LC) and LC-MS (liquid chromatography mass spectrometry)32, 33. The approaches with the highest sensitivity include ELISA- and RIA-based approaches34, 35; however, these techniques have limited dynamic range and can be subject to interferents that can affect binding specificity. The Western blot approach is also subject to limited dynamic range and is only semiquantitative at best. Microarray techniques are excellent for measuring relative expression levels but cannot differentiate the α-defensin human neutrophil protein-2 (HNP-2) from human neutrophil protein-3 (HNP-3) because HNP-2 is a posttranslationally modified form of HNP-1 and/or HNP-3. Currently published LC and LC-MS methods33, 36 are subject to limited specificity due to interferents that co-elute with the peptide of interest using photometric detectors or other compounds present that have the same mass as the desired peptide targets. In addition if sample complexity has not been reduced significantly, then low signal-to-noise ratios may result that will limit the overall sensitivity of the method especially when monitoring multiply charged precursor ions between m/z 400–700. Matrix assisted laser desorption ionization time-of-flight techniques (MALDI-TOF) have also been employed successfully for the analysis of defensins 27, 32, 33. Jørgensen et al. have reported the addition of n-octylglucoside for the absolute quantification (0.4 to 40 ng) of the β-defensins from human eye biopsy samples37. Also, Thompson et al. have used a combination of the MALDI-TOF approach with tandem mass spectrometry for relative quantitation of HNP1-3 using tracheal aspirates labeled with d0 and d3 acrylamides38. Although effective for specialized applications, the drawbacks of these MS based approaches are the limited dynamic range, the need for independent confirmation of defensin identity, and the complexity of the sample preparation and analysis schemes.
Tandem mass spectrometry (MS/MS), on the other hand, could provide a highly specific and sensitive approach for direct quantitation of a wide variety of defensin peptides found in saliva. By coupling solid phase extraction and reversed phase LC, we can effectively determine the levels of all the known defensin peptides found in saliva including the α-defensins (HNP-1, HNP-2, and HNP-3) in the physiologically relevant μg/mL range, and the β-defensins (HBD-1 and HBD-2) in the low ng/mL range (see sequences and molecular weights in table 1) in a single LC-MS/MS based assay. The defensin concentration values obtained from this assay are being applied to a larger study whose objective is to understand the role of polymorphisms in genes of innate and adaptive immunity in modulating the response to vaccines for two gastrointestinal tract infections: typhoid and cholera. The mechanisms of action of these two vaccines are expected to be different; hence, this study has the potential of providing a broader understanding of immune response and its relation to genetic polymorphisms. This research is an opportunity to study, in detail, the responses longitudinally to vaccination (using laboratory assays on longitudinally-collected biospecimens) in a large ethnic population in Kolkata, India. The focus of this publication is on the development of a comprehensive assay of defensin peptides that will be used to determine the biological impact of these molecules as outlined above.
Materials and Methods
Chemicals and Reagents
The synthetic standards Human Neutrophil Defensin-1 (HNP-1), Human Neutrophil Defensin-2 (HNP-2), Human Neutrophil Defensin-3 (HNP-3), Human β-Defensin-1 (HBD-1), and Human β-Defensin-2 (HBD-2) were obtained from New England Peptide, Inc., Gardner, MA, USA. Acetonitrile, isopropanol, and water (HPLC grade) were obtained from Burdick and Jackson, Muskegon, MI, USA. Formic acid was obtained from Pierce, Rockford, IL, USA. All other reagents were obtained from Sigma Aldrich, St. Louis, MO, USA.
Instrumentation and Equipment
Solid-phase extraction was carried out using a Supelco Visiprep 24 DL manifold (Sigma Aldrich, St. Louis, MO, USA), with an Edwards model E2M8 vacuum pump (BOC Edwards, West Sussex, UK) using Strata-X 60mg/3mL cartridges (Phenomenex, Torrance, CA, USA). Sample evaporation was carried out using a model EV2012S evaporator (Glas-Col, Terra Haute, IN, USA). Corrosion from the metal guides located on the underside of the manifold lid caused problems with analyte recovery and sample contamination. Therefore, the metal guides were removed from the manifold lid. Primary standards were weighed using a Mettler-Toledo, Inc. (Columbus, OH, USA) model XS 105 balance. HPLC separation was performed using an Agilent Technologies (Santa Clara, CA, USA) 1200 series HPLC system, with a G1312A binary pump, G1379B inline degasser, G1367B temperature controlled wellplate autosampler, and G1316A temperature controlled column compartment. The HPLC column was a Zorbax SB-C3, 2.1×150mm, 5μm particle (Agilent Technologies, Santa Clara, CA, USA). Tandem MS analysis was performed using a linear ion trap LTQ mass spectrometer (Thermo Fisher Scientific Corp., San Jose, CA, USA). Exploratory tandem MS experiments were also carried out on all defensin standards using an Applied Biosystems (Foster City, CA, USA) API 4000 triple quadrupole mass spectrometer.
Sample Preparation
Stock solutions of standards were prepared by weighing the neat peptide, at a nominal concentration of 1 mg/mL in 1:1 acetonitrile : water. All containers used for samples and standards, whether before or after extraction, were polypropylene. All such containers were treated to prevent adsorptive loss of analytes, by incubating them with a 1 mg/mL aqueous lysozyme solution for two hours. The solution was then removed, and the containers rinsed with an equal volume of water and allowed to air dry. All standards, QCs, and blanks were prepared from the stock solutions in a surrogate saliva matrix containing 0.84 mg/mL bovine serum albumin and 4.9 mg/mL (84 mM) of sodium chloride. Standards, QCs, blanks, and unknown samples were mixed 1:1 with 6M guanidine HCl, buffered to pH 8 with 50mM tris buffer, and then treated with 10mM dithiothreitol (DTT) for 1 hour at 37°C to reduce any disulfide bonds. Treated samples were then extracted by SPE.
For SPE, cartridges were conditioned with 2 mL acetonitrile followed by 2 mL of 1% (v/v) aqueous acetic acid. Following sample loading, the cartridges were washed with 2mL of 1% acetic acid. The analyte was eluted with 0.5mL of 35:65 acetonitrile : 1% aqueous acetic acid, followed by 0.5mL of 70:30 acetonitrile : 1% acetic acid, followed by 0.5mL of 70:25:5 isopropanol : acetic acid : water. For a given sample, all eluent was collected in a single tube, held at 40°C and evaporated under a stream of dry nitrogen. Dry samples were then reconstituted in 20:80 acetic acid : water containing bradykinin as an internal standard at 100 ng/mL.
An eight-point calibration curve was prepared, with concentrations ranging from 0.034 to 8.6 μg/mL for the α-defensins (0.034, 0.086, 0.17, 0.34, 0.69, 1.7, 3.4, and 8.6 μg/mL), and from 1.2 to 310 ng/mL for the β-defensins (1.2, 3.1, 6.2, 12, 25, 62, 120, and 310 ng/mL). The QC standards were prepared and analyzed at two concentration levels. The low QC level contained 0.34 μg/mL of the α-defensins and 12 ng/mL of the β-defensins. The high QC level contained 3.4 μg/mL of the α-defensins and 120 ng/mL of the beta defensins. Each QC sample was analyzed after every ten (or fewer) samples.
HPLC-MS/MS Analysis
LC-MS/MS experiments were performed using a Thermo-Electron (San Jose, CA) LTQ mass spectrometer with an Agilent 1200 HPLC system. Chromatographic separation was conducted using a Zorbax SB-C3, 2.1×150mm, 5 mm particle column (Agilent Technologies, Santa Clara, CA, USA). Mobile phase “A” was 99:1 water : isopropanol with 0.1% formic acid. Mobile phase “B” was 85:5:10 acetonitrile : isopropanol : water with 0.1% formic acid. The flow rate was 300 μL/min. The gradient was begun at 5% “B”, and was increased linearly to 70% “B” over 13 minutes, followed by a return to 5% “B” over 0.5 minutes. The injection volume was 80μL. A system wash, consisting of a 1-minute step to 100% B accompanied by two injection valve cycles, was used after the initial gradient cycle to minimize carryover. MS analysis was by positive ion ESI MS/MS. MS/MS conditions for analytes, given in table 2, were optimized by infusing standard solutions of the peptides in linear form.
Table 2.
Tandem mass spectrometry parameters (LTQ Classic) for the defensin peptides.
| Peptide | Charge State | Precursor Ion (m/z) | Product Ion (m/z) | Collision Energy (%) | Activation Time (ms) | Product Ion Identification |
|---|---|---|---|---|---|---|
| HNP-1 | +5 | 690.9 | 806.9 | 24 | 10 | b28 (+4) |
| HNP-2 | +5 | 676.7 | 789.0 | 32 | 10 | b27 (+4) |
| HNP-3 | +5 | 699.7 | 817.9 | 26 | 10 | b28 (+4) |
| HBD-1 | +6 | 657.0 | 716.3 | 35 | 10 | y19 (+3) &/or b20 (+3) |
| HBD-2 | +7 | 620.6 | 694.9 | 25 | 10 | y39 (+6) |
Results and Discussion
Tandem Mass Spectrometry
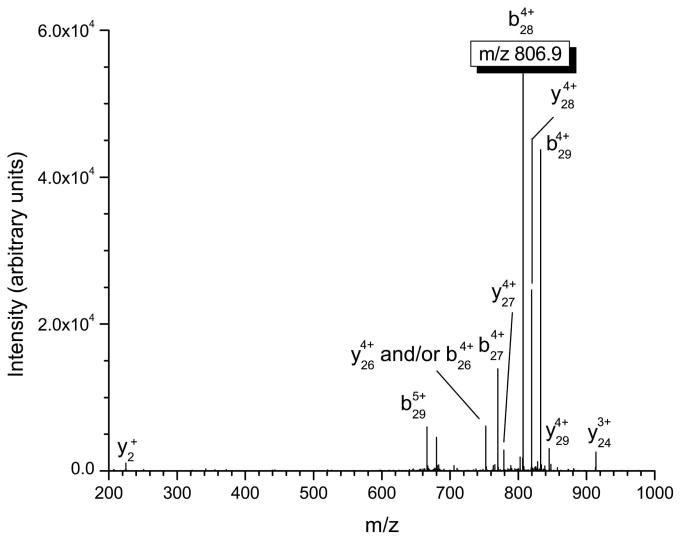
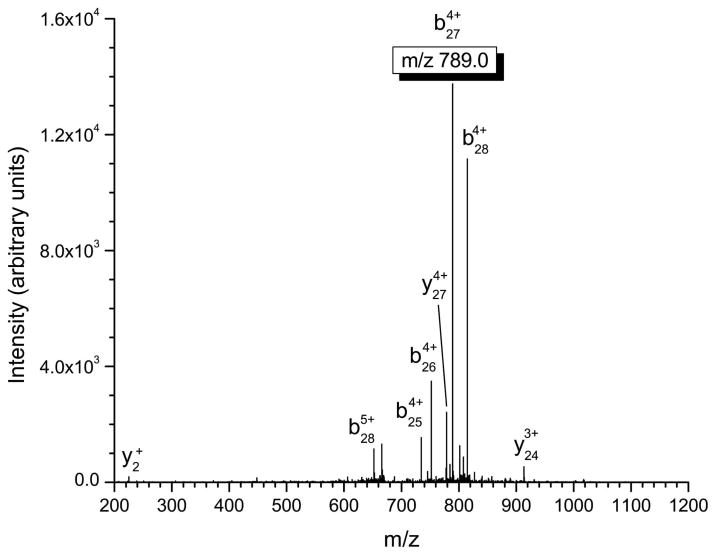
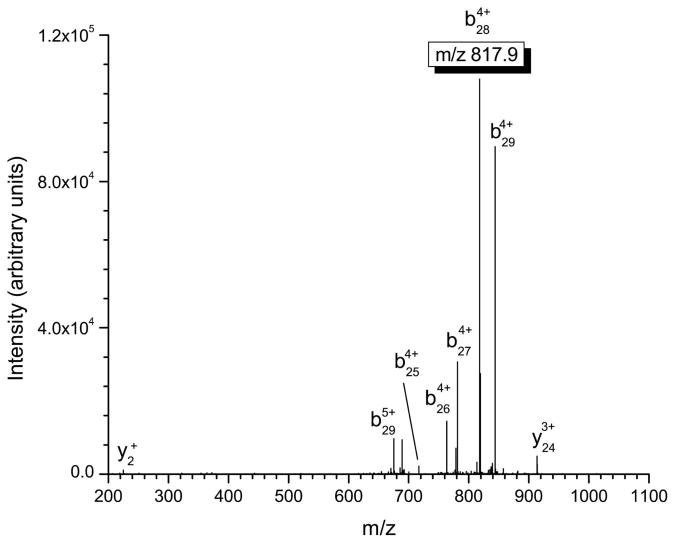
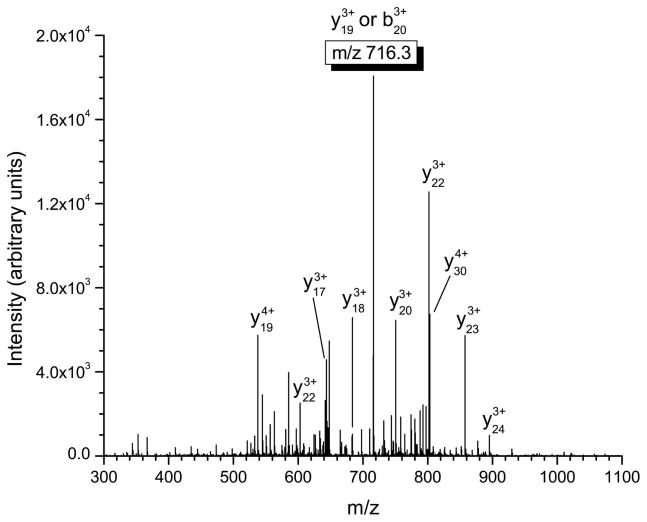
Several important structural characteristics of defensin peptides need to be considered when performing tandem mass spectrometry experiments. First, the disulfide linkages present in these peptides are not amenable to fragmentation via standard low-energy collisional activation and must be reduced in order to produce appropriate product ions. Second, from a sensitivity and signal-to-noise ratio standpoint, the most intense product ion should be used for quantitation in tandem mass spectrometry. Finally, the product ions chosen for quantitation should incorporate the differences in the α-defensins at the N-terminus in order to maximize the specificity of the approach. This consideration is not important for β-defensins where sequence conservation is limited to only eight residues. In our experiments, b-type ions that retain charge on the N-terminus of the α-defensin peptide were chosen for quantitation (see table 2) due to the fact they were the most intense product ions observed and provide a high degree of specificity for the assay. The MS/MS spectra of HNP-1-3 are shown in figures 1a–1c, with the corresponding multiply-charged b-type product ions highlighted. Since the linear ion trap used in these studies operates at unit mass resolution, assignment of the product ions was calculated by determining all the m/z values of b- and y-type ions with charge states up to the charge state of the precursor ion. The most intense product ion observed is a cleavage on the C-terminal side of alanine in position 28 for HNP-1 and HNP-3, and at position 27 for HNP-2 (counting from the free N-terminus). This corresponds to the 4+ charge state b-type product ions listed in table 2.
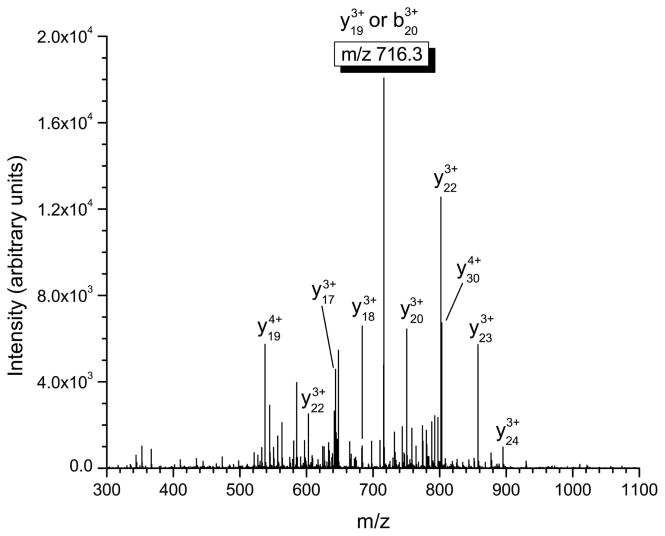
Figure 1.
Mass spectrometric fragmentation patterns of the five defensin targets for the saliva assay. Due to the highly basic nature of these peptides, fragmentation is limited to only a few dissociation routes which results in improved signal-to-noise for absolute quantitation. The product ions used for quantitative purposes are highlighted in the box. a) HNP-1 b) HNP-2 c) HNP-3 d) HBD-1 e) HBD-2.
Figures 1d and 1e show the tandem mass spectra of HBD-1 and HBD-2. For all defensins the spectra shown were obtained from a 0.34 μg/mL standard directly infused into the linear ion trap. For HBD-1, the observed product ion is derived from either the C-terminal cleavage of the phenylalanine residue at position 20 or the N-terminal cleavage of the proline residue at position 18. These product ions have the same nominal mass and cannot be distinguished using the linear ion trap. Therefore the product ion for quantitation is either a b20 or y19 ion with a 3+ charge state. The primary cleavage site for HBD-2 is observed on the N-terminal side of the glycine located at residue 3, resulting in the formation of the y39 product ion that is used for quantitation.
The fragmentation of the precursor peptide ions to diagnostically relevant quantitation ions is critically important in this assay. The limited fragmentation observed for the α-defensins is consistent with the highly basic nature of these large peptides. In instances where the number of charges (5 in this example) just exceeds (or is equal to) the total number of arginine residues (4 per molecule) for a collisional-activation experiment39, 40, limited fragmentation is observed due to the high gas-phase basicity of the guanidinium group of arginine that restricts the mobility of protons during the dissociation step. This has a distinct advantage in quantitation where the ion signal can be concentrated primarily into a single diagnostically relevant fragment (see figure 1 and table 2). Quadrupole ion trapping devices are ideal for this type of experiment in that the ion-activation step is performed on the millisecond timescale via multiple collisions with the helium bath gas41, 42. These multiple collisions slowly and efficiently push the ion past its critical energy of dissociation, where the ion is thought to dissociate at a rate much greater than the ion activation event43. Similar tandem MS experiments performed on a triple quadrupole instrument (quadrupole collision cell), showed a much higher degree of scattering (reduction in product to precursor ion signal) and significantly reduced fragmentation efficiency. This is due primarily to the fact that the residence time of any given ion in the collision cell is on the microsecond timescale with fewer collisions with the background gas. Therefore, all the factors effecting energy transfer necessary to produce fragmentation (e.g. impact parameter, velocity of the ion, and size of the neutral atom) must be met in fewer higher energy collisions in the triple quadrupole in order to produce and transmit fragments to Q3. Thus the difference in the dissociation behavior of highly stable precursor species (with localized areas of high gas-phase basicity) is highly dependent on the ion activation method.
Similar behavior is observed for highly basic peptides that contain a series of lysine residues. This is the case for HBD-1 where an abundant series of y-type ions ranging from y19 (ion used for quantitation) to y24 is the result of collisional activation of the +6 charge state precursor ion. Since there are seven basic residues on HBD-1 (including the N-terminus) and the C-terminus of the peptide is populated with three lysine residues, the observed y-ion series is expected (similar to what is seen for incomplete tryptic digest products). For the case of HBD-2 there are nine basic residues (including the N-terminus) that include a total of two arginine amino acids. The almost exclusive formation of the y39 6+ charge state ion from the 7+ precursor ion at m/z 620.6 is interesting in that very little other fragmentation is observed for this charge state of HBD-2. The larger size, two arginine residues compared to one for HBD-1 and a higher number of proline residues could all contribute to the limited fragmentation observed compared to HNP-1. It should also be noted that for both α- and β-defensins, the use of product ions with a higher m/z value than the precursor greatly reduces the chance of chromatographic interference, by eliminating all singly charged ions as potential interferences.
The internal standard chosen for these studies was the neuropeptide bradykinin. Bradykinin is a nine-residue, basic peptide (pI = 12.0) that has been commonly used in mass spectrometry studies and has similar physiochemical properties as the defensin targets. The peptide is not present in human saliva and can be obtained inexpensively in sufficient quantities for large scale sample studies. In addition, this peptide is well separated from the defensin targets where it can be monitored in a separate mass window and does not limit duty cycle time for quantitation.
Sample Preparation Factors
Due to the highly basic nature (and thus the mechanism of action) of the defensin peptides, it was observed that these molecules had limited recovery and linearity issues over their normal physiological concentration ranges. This is primarily due to the fact that defensins can have strong surface interactions with polymers and can be lost easily in the sample preparation process. Carryover and response problems were initially believed to be a result of adsorption onto silanols in the LC column; however, the problems were also observed with a polymeric column (data not shown). Several derivitization schemes were tested in an attempt to mitigate adsorption onto surfaces in the flow path because of polar or ion-exchange interactions. Alkylation of cysteines with iodoacetamide did not improve response or carryover behaviors, and additionally resulted in a loss of signal because of the inefficiencies of the reaction. Digestion of the analytes with the enzymes trypsin, chymotrypsin, and Glu-C were also separately tested, with the goal of employing a digestion product (smaller peptide) specific to each analyte as a quantitation surrogate32. Here, we did not consistently observe specific digestion products that would differentiate HNP-1, HNP-2, and HNP-3 from one another (data not shown). Additionally, we wanted to minimize the amount of time spent on sample preparation and therefore did not pursue proteolytic digestion to the full extent.
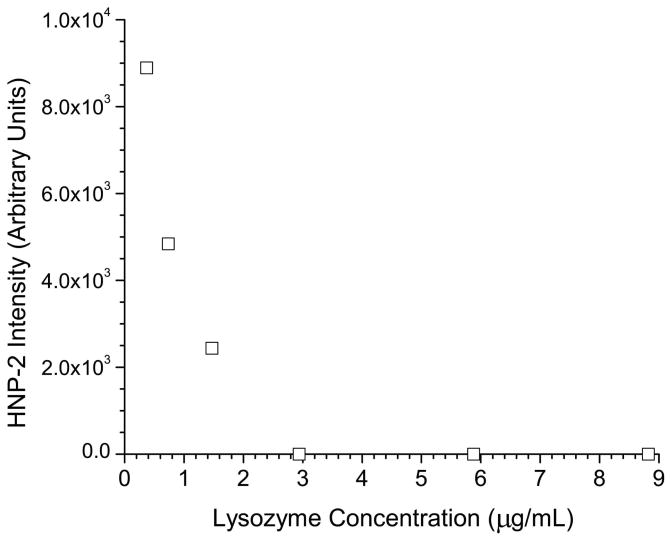
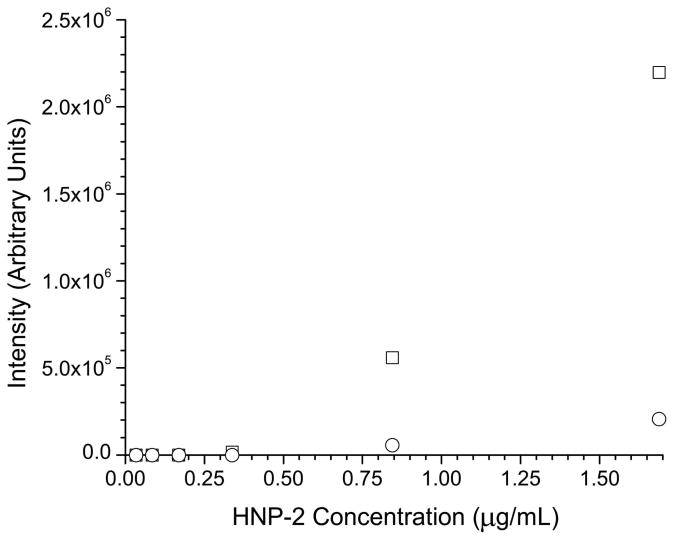
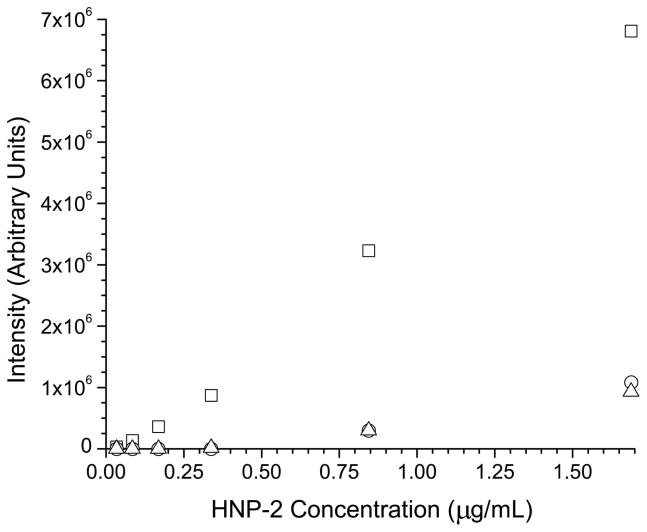
It was then observed that injection of larger volumes of low concentration standards produced a significantly lower response than injecting, theoretically, the same mass injected on column from a higher concentration standard (data not shown). Thus, the cause of the problem was determined to be adsorption of the peptides onto the polypropylene vial and tube surfaces encountered during sample storage. In order to correct for the adsorption process, several strategies were evaluated to minimize sample losses. One approach involving the use of carrier proteins has proven to be an effective way to minimize sample losses on container surfaces. Lysozyme was first tried as a carrier protein due to its similar pI (9.3 compared to 10.2 for the average defensin) to the defensin peptides, availability, and low cost. Some defensin peptides did show an improvement in signal intensity by adding lysozyme in concentrations ranging from 0.1 to 1 mg/mL. However, some defensins such as HNP-2, demonstrated a marked decrease in signal as a function of the addition of a carrier protein as shown in figure 2a. When comparing two calibration curves for HNP-2 between standards (one with and without 1 mg/mL of lysozyme) shown in figure 2b, signal suppression was clearly observed for the HNP-2 containing the lysozyme carrier protein. At the high-standard level (1.7 μg/mL), the signal suppression observed for the carrier protein standard was approximately ten fold. For both the calibration curves, little or no signal was observed on the low end of the curve (levels 0.034, 0.086, 0.17, 0.34 μg/mL), indicating that adsorption of the peptide to surfaces or some form of matrix ionization effect was the major cause of signal loss. To validate this hypothesis, a second set of experiments used centrifuge tubes that were washed with a 1 mg/mL solution of lysozyme and were allowed to dry at room temperature. Standards were then prepared as described and a calibration curve was run over the 0.034–1.7 μg/mL range. The calibration curve obtained was compared directly to curves run with untreated vials and lysozyme treated vials that were filled with calibration standards and then stored at (4° C) for 48 hrs. The results, shown in figure 2c, show a linear calibration of HNP-2 for the treated vials, indicating the use of a lysozyme carrier solution directly affects the instrument response. The curves obtained for the untreated and 48 hr treated vials are similar, demonstrating that treating the vials is only effective in improving response for a limited time frame.
Figure 2.
a) Response of HNP-2 as a function of the concentration of the carrier protein lysozyme. b) Individual calibration curves for HNP-2 with (○) and without (□) a 1 mg/mL of the carrier protein lysozyme present c) Calibration curves for HNP-2 using lysozyme treated vials (□), treated vials after 48 hours (△), and untreated vials (○).
Calibration and Chromatography
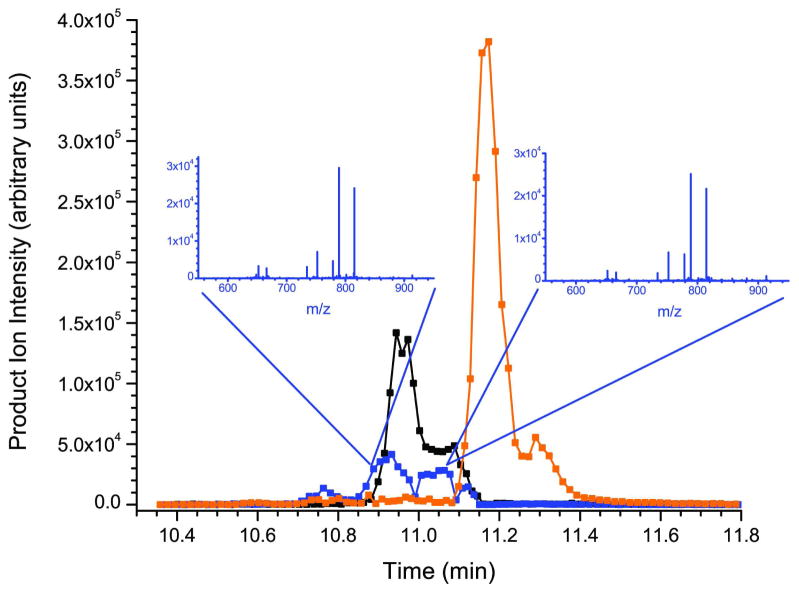
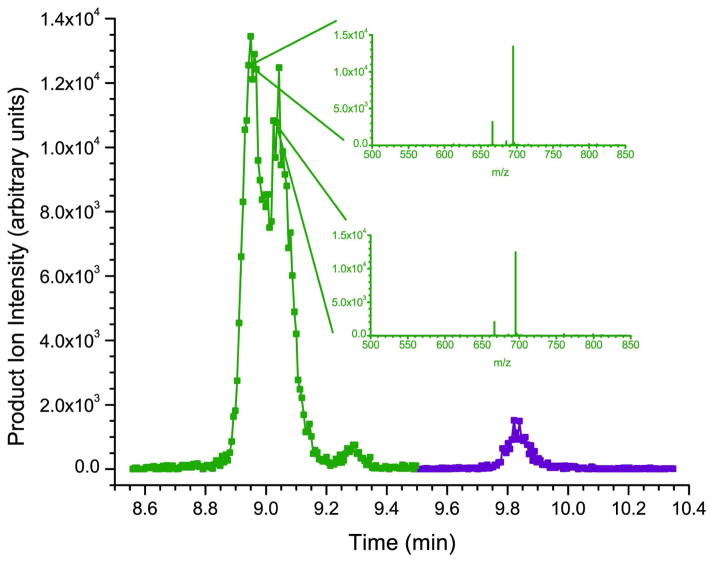
The chromatographic profiles of the three forms of the synthesized α- and two forms of β-defensins are shown in figures 3a and 3c. Initially it was thought that the multiple peaks for each species represented different synthesized peptides. However, under carefully controlled chromatographic conditions this translates to the separation of the various forms of the peptides (diastereoisomers) that are side products of solid-phase peptide synthesis. Solid-phase methods of peptide synthesis (Fmoc/tBu) were employed by the manufacturer due to their efficiency. Although significant progress has been made in the field, there are drawbacks to the approach that must be accounted for when considering strategies for elucidating the structure of the synthesized peptide and its impurities. Aggregation, enantiomerization, and various side reactions can confound the synthesis process and contribute substantial impurities to any peptide preparation. For example, aggregation is typically the most common failure process in solid-phase synthesis. This effect can occur with as little as five amino acid residues, and can lead to lower yields through failure of the deprotection or acylation reactions. Enantiomerization can occur via several mechanisms, including ring opening of the oxazolone intermediate via deprotonation, through coupling where chiral changes can occur due to removal and reattachment of the acidic proton on the α-carbon (particularly during coupling), and on cysteine residues upon removal of the Nα-Fmoc and during the coupling reaction and is hypothesized to be the major issue here44.
Figure 3.
a) Chromatographic profile of the product ion intensity for synthetic HNP-1, HNP-2, and HNP-3. The inset shows the tandem mass spectrum of the peptide at two different points across the peak for HNP-2. b) Chromatographic profile of the product ion intensity for naturally occurring (saliva sample) HNP-1, HNP-2, and HNP-3. c) Chromatographic profile of the product ion intensity for synthetic HBD-1, and HBD-2. The inset shows the tandem mass spectrum of the peptide at two different points across the peak for HBD-2. d) Chromatographic profile of the product ion intensity for naturally occurring (saliva sample) HBD-1, and HBD-2.
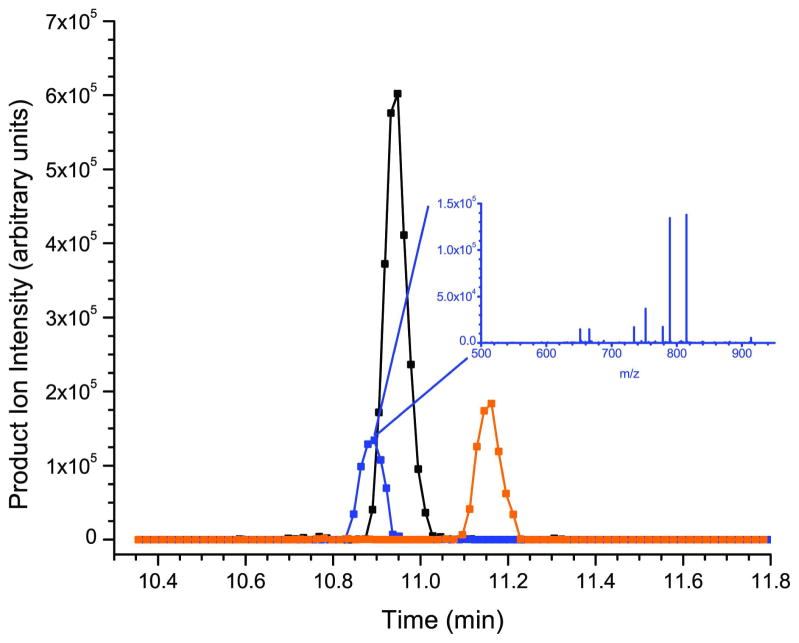
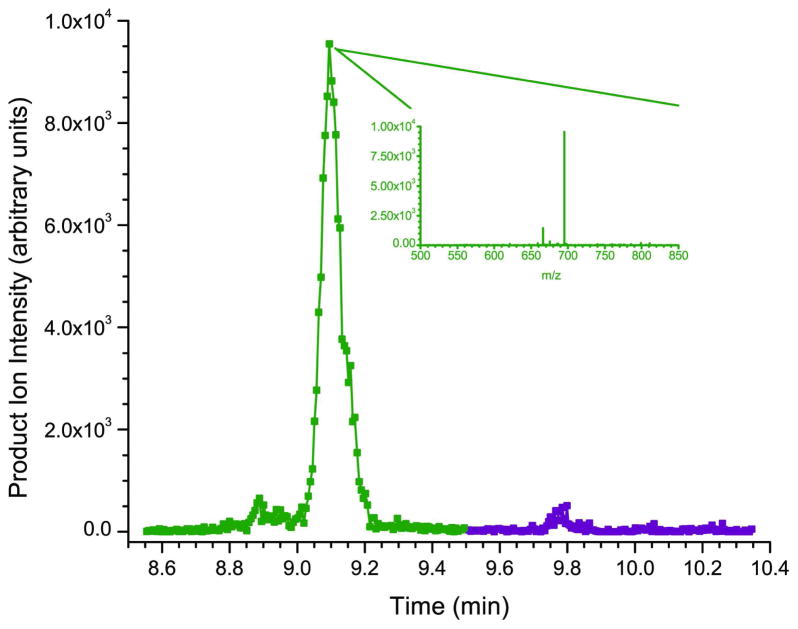
To partially test this hypothesis, the tandem mass spectrum for each of the three standards was examined on all points across the asymmetric peaks. In each case, the tandem mass spectra were identical, suggesting that the multiple peaks in question have the exact same sequence (see the HNP-2 and HBD-1 example in the insets of figure 3a and 3c respectively) but could have a different conformation. For comparison, figures 3b and 3d show the chromatographic profiles of the α- and β-defensins extracted from human saliva. Here the defensin peptides are present as single peaks as would be expected for naturally occurring peptides. One way to address the peak shape issue concerning the synthesized peptides in the future is to express the recombinant forms of the α- and β-defensins in the appropriate genetic systems. This has recently been accomplished for the α-defensins and HBD-2 in an E. coli expression system45, 46.
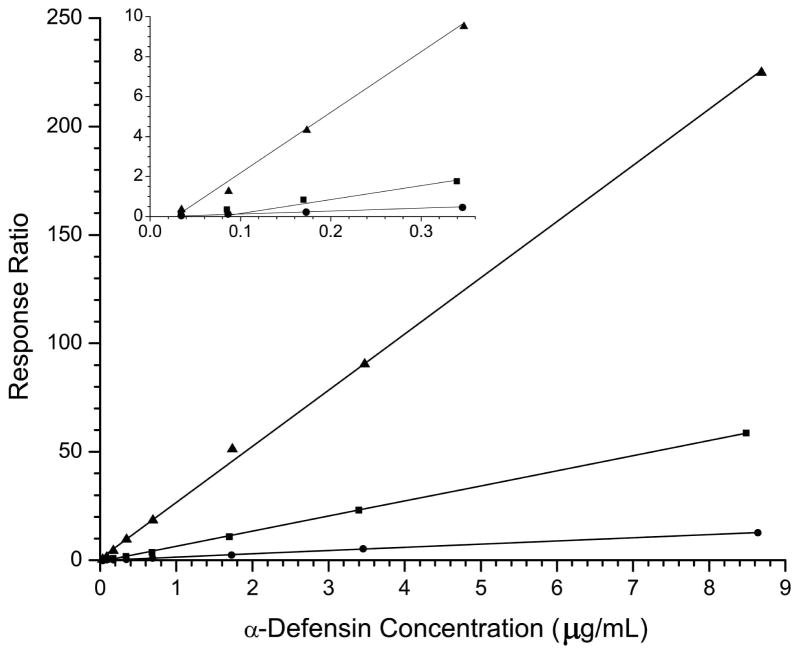
Calibration curves for the α-defensins are shown in figure 4a. The observed differences in the slopes of the calibration curves increase from HNP-2, to HNP-1, to HNP-3 with a corresponding increase in response from 4.8 going from HNP-2 to HNP-1 and an 18-fold increase going from HNP-2 to HNP-3. Although the response varies widely for the three targets, appropriate adjustments of the automatic gain control software allow for reproducible responses across the physiological concentration range of interest where 9–11 data points are obtained across the peaks of interest (see figure 3b) from biologically relevant samples. The three α-defensins elute in an approximate 0.5 minute time window between 10.8 and 11.3 minutes. During this elution period only the putatitive precursor ions specific to the three α-defensins are subject to fragmentation in order to maximize the sensitivity and minimize the duty cycle of the experiment.
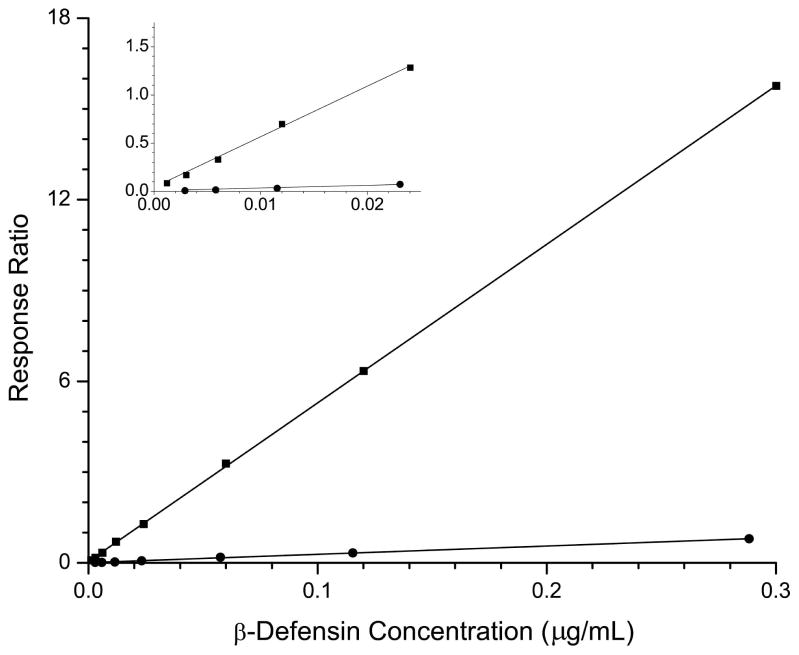
Figure 4.
a) Calibration curves for HNP-1 (■), HNP-2 (●), and HNP-3 (▲). In the inset is shown an expanded view of the low concentration region. b) Calibration curves for HBD-1 (●) and HBD-2 (■). The inset shows an expanded view of the low concentration region.
Figure 4b shows the calibration curves for β-defensins HBD-1 and -2. HBD-2 has a 21-fold increase in response (as measured by the slope of the calibration curve) over that of HBD-1. Approximately 25 data points across the peak are obtained for the β-defensins, which elute in their own mass window. The increased sampling allows for much better line fitting and calibration for the β-defensins at the low end of the curve compared to the corresponding α-defensins (see the insets of figures 4a and 4b).
Analysis of Saliva Samples
The assay was applied to an original study design with 1000 participants derived from the Muslim and Hindu populations in Kolkata, India. This prospective study is part of the Population Genetics Program sponsored by the National Institute of Allergy and Infectious Disease (NIAID) Program. The goal is to evaluate the defensin levels in participants on day 0 and 3 post vaccination and correlate these with gender, age, cytokine/chemokine levels, response to vaccination with typhoid, and gene copy polymorphisms. In this initial study, we analyzed 612 saliva samples from the larger population both at days 0 and 3 and put in place a variety of quality control checks to ensure reproducibility. For analysis of the typhoid saliva samples (0.1–10 μg/mL for the α-defensins and 1–100 ng/mL for the β-defensins), known calibration values could not drift more than 20% from their true concentration. In addition, QA/QC samples were run at both low and high levels for all analytes and were analyzed at three separate times during the analysis of any sample set. Any QA/QC level that was outside 20% of its true value made the analysis of a given sample set invalid (e.g., analysis had to be repeated). Random replicate injections were performed for every sample set with an average cv value of less than 3% for all analytes. Repeatability of the extraction process was also checked for each of the five defensin analytes. Of the 612 samples analyzed from the typhoid study (responders and nonresponders to the vaccine), 10% were randomly chosen for re-extraction and analysis. The extraction variability experiments were carried out on a large sample aliquot removed from the main sample. Here multiple extractions were performed on different days to test the reproducibility. This was done to separate the variability associated with extraction efficiency and homogenization of the original sample. Using this approach to the study the observed extraction variability was as follows: HNP-1 3.6%, HNP-2 8.3%, HNP-3 5.3%, HDB-1 3.0%, and HBD-2 8.0% for the coefficient of variation (cv). If the repeat extractions were performed on the original random samples without removing a specified aliquot, then the range of the cv values increased in some cases to ~60%. It should be noted that many subjects in the study population have a compromised oral health status which can add to the complexity of the sample. Homogenization of some of these samples was particularly challenging due to the presence of a wide variety of particulates, emulsions, mucous, and limited amounts of blood contamination.
In order to determine the extraction efficiency the method of standard addition (e.g. due to the presence of endogenous defensins in the sample) was employed on saliva samples from the study population. The samples chosen for this process represented moderate complexity considering the presence of particulates, blood contamination, mucous and other interferents, and contained the majority of all the defensin targets in the sample. The calibration curves were analyzed over five different concentrations defined by: 1.00c, 1.25c, 1.50c, 1.75c, and 2.00c, where c represents the original concentration as determined by the method described herein. The percent recoveries, as determined by this approach, ranged between 50 and 135%. The large range associated with these results were attributed to the homogenization of the original samples as described above.
In table 3, representative data (randomly selected) is shown from the field study. The values are reported in μg/mL, which is the accepted format in the current literature. The majority of the values obtained for HNP-1 ranged between 0.1 to 10 μg/mL, and correlated well with previous values reported in the literature36. HNP-2 levels were found to be less than HNP-1 levels, ranging from approximately 0.02 to 6.0 μg/mL. For HNP-3, observed concentrations ranged from a high of several μg/mL down to “not detected” (ND). This is consistent with the observation that some individuals lack the gene for the production of HNP-3. Overall, the percentage of samples quantitated in the 0.1–10 μg/mL range for HNP1-3 were 87%, 82%, and 13% respectively. Samples found to be above the upper limit of quantitation were diluted and reanalyzed. The observed levels of β-defensins were from ND to ~39 ng/mL and ND to ~33 ng/mL for HBD-1 and -2, respectively. For HBD-1 and -2, the observed concentrations correlate well with a recent report using a new immunoassay that reported levels in saliva in the low ng/mL range47. HBD-2 levels have been reported in the literature to be up-regulated as result of increased TNF-α and ILB-1 levels, and this too is consistent with our observations. It is not the intention of this manuscript to discuss the biological aspects of defensins and their role in innate immunity. However, it is important to point out the utility and reproducibility of this approach as a clinical tool for potentially monitoring biomarkers of susceptibility, evaluating oral and dental health, and studying basic immunological processes.
Table 3.
Representative results from the typhoid field study.
| Sample ID | HNP-1 (μg/mL) | HNP-2 (μg/mL) | HNP-3 (μg/mL) | HBD-1 (μg/mL) | HBD-2 (μg/mL) |
|---|---|---|---|---|---|
| 977-3 | 2.13 | 1.64 | ND | ND | 0.00820 |
| 296-0 | 20.1 | 13.7 | 1.75 | ND | 0.00206 |
| 35-3 | 0.980 | 0.0622 | 0.0601 | ND | ND |
| 148-3 | > ULOQ | 16.2 | 1.33 | ND | ND |
| 46-0 | 2.63 | 1.72 | 0.189 | ND | ND |
| 1010-3 | 2.43 | 1.15 | ND | ND | < LLOQ |
| 806-0 | 2.04 | 1.62 | 0.366 | ND | ND |
| 35-3* | 0.910 | 0.127 | < LLOQ | ND | ND |
| 966-3 | 2.77 | 1.88 | 0.189 | ND | < LLOQ |
| 1068-3 | 7.06 | 8.49 | ND | ND | 0.00141 |
| 144-3 | 1.56 | 1.51 | 0.593 | ND | 0.00379 |
| 690-3 | 3.43 | 1.95 | 0.219 | ND | < LLOQ |
| 909-0 | 5.25 | 6.07 | 0.805 | ND | < LLOQ |
| 966-3* | 2.89 | 2.09 | 0.191 | ND | 0.00135 |
| 50-0 | 1.43 | 1.22 | 0.283 | ND | 0.00171 |
| 70-0 | 4.68 | 5.15 | 0.747 | 0.00871 | < LLOQ |
| 1018-3 | 4.93 | 2.70 | ND | ND | 0.00175 |
| 246-0 | 5.05 | 1.60 | 0.292 | 0.0117 | 0.0329 |
| 918-3 | 3.90 | 2.72 | 0.268 | ND | ND |
| 406-3 | 2.93 | 1.92 | 0.232 | ND | < LLOQ |
| 261-3 | 2.34 | 2.16 | 0.198 | ND | < LLOQ |
| 1018-3* | 4.85 | 2.93 | ND | ND | 0.00175 |
| 291-0 | 5.84 | 6.19 | 0.414 | ND | ND |
| 128-0 | 5.25 | 5.41 | 0.415 | 0.00863 | 0.00196 |
| 320-3 | 4.46 | 2.80 | ND | ND | < LLOQ |
| 896-3 | 7.22 | 7.74 | 1.13 | 0.00825 | 0.00174 |
| 920-3 | 3.88 | 2.67 | 0.226 | ND | ND |
| 22-3 | 2.70 | 2.61 | 0.204 | ND | < LLOQ |
| 192-0 | 5.69 | 3.28 | 0.399 | ND | 0.00283 |
| 320-3* | 4.37 | 2.14 | < LLOQ | ND | < LLOQ |
| 766-0 | 3.39 | 2.51 | ND | ND | 0.00290 |
| 662-0 | 4.71 | 3.29 | 0.372 | ND | ND |
| 1035-0 | 5.51 | 3.79 | ND | 0.00565 | < LLOQ |
| 1072-0 | 8.03 | 9.81 | 0.968 | 0.0226 | 0.00315 |
| 197-0 | 39.2 | 18.2 | 1.80 | ND | < LLOQ |
| 729-3 | 5.14 | 3.91 | 0.514 | ND | 0.0110 |
| 17-3 | 2.53 | 1.81 | 0.398 | ND | 0.00146 |
| 1008-3 | 2.14 | 1.91 | 0.537 | ND | < LLOQ |
| 171-3 | 8.16 | 11.7 | 2.72 | 0.00889 | 0.00237 |
| 232-3 | 5.12 | 5.79 | 0.680 | ND | 0.00151 |
Indicates a duplicate sample that was extracted and run two times.
>ULOQ value above the upper limit of quantitation
<LLOQ value below the lower limit of quantitation.
Conclusions
Here we present a robust, mass spectrometric assay for the analysis of defensin peptides in saliva. This assay can detect HNP-1, -2, and -3, as well as HBD-1 and -2, at known physiological levels in a single assay (0.1–10 μg/mL and 1–100 ng/mL respectively). The assay employs solid phase extraction and can be used on any saliva sample, independent of the amount of particulate, ionic strength, or presence of any other potential interferents. In addition, this assay could be easily adapted to detect α- or β-defensins in a variety of other matrices including blood, urine, sputum, or tissue samples (e.g intestinal lining).
Compared to previous mass spectrometry based approaches that involve multiple instruments (for quantitation and validation of identity) and comprehensive sample preparation, the approach presented here is highly specific, more sensitive, and easier to execute than previously published methods. The sensitivity obtained rivals that of standard immunoassay based formats (low ng/mL range), and is far less susceptible to interferents. An area of focus for future studies will involve improvements in the homogenization process of saliva samples with compromised oral health status. In addition the use of recombinantly expressed defensin standards45, 46 will be explored to reduce the analysis cost, limit the need for manual inspection of the calibration curves, and improve reproducibility. We are currently expanding this approach to evaluate its utility for the detection of other basic cationic peptides involved in host defense including histatins, cathelicidin (LL-37), and small proline rich proteins.
Acknowledgments
The authors wish to acknowledge NIH-NIAID for funding of this research under contract number HHSN266200400067C. In addition, we wish to recognize Partha P. Majumder and colleagues at the Indian Statistical Institute for performing the saliva sample collection.
References
- 1.Abiko Y, Saitoh M. Current Pharmaceutical Design. 2007;13:3065–3072. doi: 10.2174/138161207782110417. [DOI] [PubMed] [Google Scholar]
- 2.Abiko Y, Saitoh M, Nishimura M, Yamazaki M, Sawamura D, Kaku T. Medical Molecular Morphology. 2007;40:179–184. doi: 10.1007/s00795-007-0381-8. [DOI] [PubMed] [Google Scholar]
- 3.Dale BA, Fredericks LP. Current Issues in Molecular Biology. 2005;7:119–133. doi: 10.1093/jac/dki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowdish DME, Davidson DJ, Scott MG, Hancock REW. Antimicrobial Agents And Chemotherapy. 2005;49:1727. doi: 10.1128/AAC.49.5.1727-1732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganz T. Nature Reviews Immunology. 2003;3:710. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 6.Ganz T. Combinatorial Chemistry & High Throughput Screening. 2005;8:209. doi: 10.2174/1386207053764594. [DOI] [PubMed] [Google Scholar]
- 7.Ganz T, Lehrer RI. Pharmacology & Therapeutics. 1995;66:191. doi: 10.1016/0163-7258(94)00076-f. [DOI] [PubMed] [Google Scholar]
- 8.Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. Cellular and Molecular Life Sciences. 2006;63:1294–1313. doi: 10.1007/s00018-005-5540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganz T, Selsted ME, Szklarek D, Harwig SSL, Daher K, Bainton DF, Lehrer RI. Journal of Clinical Investigation. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selsted ME, Harwig SSL, Ganz T, Schilling JW, Lehrer RI. Journal of Clinical Investigation. 1985;76:1436–1439. doi: 10.1172/JCI112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selsted ME, Tang YQ, Morris WL, McGuire PA, Novotny MJ, Smith W, Henschen AH, Cullor JS. Journal Of Biological Chemistry. 1993;268:6641–6648. [PubMed] [Google Scholar]
- 12.Kluver E, Adermann K, Schulz A. Journal of Peptide Science. 2006;12:243–257. doi: 10.1002/psc.749. [DOI] [PubMed] [Google Scholar]
- 13.Klotman ME, Chang TL. Nature Reviews Immunology. 2006;6:447–456. doi: 10.1038/nri1860. [DOI] [PubMed] [Google Scholar]
- 14.Chang TL, Klotman ME. Aids Reviews. 2004;6:161–168. [PubMed] [Google Scholar]
- 15.Cole AM, Cole AL. American Journal of Reproductive Immunology. 2008;59:27–34. doi: 10.1111/j.1600-0897.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- 16.Garzino-Demo A. Current Pharmaceutical Design. 2007;13:163–172. doi: 10.2174/138161207779313696. [DOI] [PubMed] [Google Scholar]
- 17.Lama J, Planelles V. Retrovirology. 2007:4. doi: 10.1186/1742-4690-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soong LB, Ganz T, Ellison A, Caughey GH. Inflammation Research. 1997;46:98. doi: 10.1007/s000110050114. [DOI] [PubMed] [Google Scholar]
- 19.Fahlgren A, Hammarstrom S, Danielsson A, Hammarstrom ML. Clinical And Experimental Immunology. 2003;131:90. doi: 10.1046/j.1365-2249.2003.02035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escher N, Spies-Weisshart B, Kaatz M, Melle C, Bleul A, Driesch D, Wollina U, Von Eggeling F. European Journal of Cancer. 2006;42:249–255. doi: 10.1016/j.ejca.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 21.Gordon YJ, Romanowski EG, McDermott AM. Current Eye Research. 2005;30:505–515. doi: 10.1080/02713680590968637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke DJ, Campopiano DJ. Biochemical Society Transactions. 2006;34:251. doi: 10.1042/BST20060251. [DOI] [PubMed] [Google Scholar]
- 23.Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, Tack BF, McCray PB. Infection And Immunity. 1999;67:2740. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, Gallo RL, Leung DYM. New England Journal Of Medicine. 2002;347:1151. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 25.Song JJ, Chae SW, Woo JS, Lee HM, Jung HH, Hwang SJ. Annals Of Otology Rhinology And Laryngology. 2007;116:235. doi: 10.1177/000348940711600312. [DOI] [PubMed] [Google Scholar]
- 26.Melle C, Ernst G, Schimmel B, Bleul A, Thieme H, Kaufmann R, Mothes H, Settmacher U, Claussen U, Halbhuber KJ, von Eggeling F. Gastroenterology. 2005;129:66–73. doi: 10.1053/j.gastro.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Panyutich AV, Panyutich EA, Krapivin VA, Baturevich EA, Ganz T. Journal of Laboratory and Clinical Medicine. 1993;122:202–207. [PubMed] [Google Scholar]
- 28.Hiratsuka T, Nakazato M, Date Y, Ashitani J, Minematsu T, Chino N, Matsukura S. Biochemical and Biophysical Research Communications. 1998;249:943–947. doi: 10.1006/bbrc.1998.9239. [DOI] [PubMed] [Google Scholar]
- 29.Ihi T, Nakazato M, Mukae H, Matsukura S. Clinical Infectious Diseases. 1997;25:1134–1140. doi: 10.1086/516075. [DOI] [PubMed] [Google Scholar]
- 30.Lu Q, Jin LJ, Darveau RP, Samaranayake LP. Journal Of Periodontal Research. 2004;39:221. doi: 10.1111/j.1600-0765.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- 31.Klut ME, Whalen BA, Hogg JC. European Journal Of Haematology. 2000;64:114. doi: 10.1034/j.1600-0609.2000.90069.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhou L, Huang LQ, Beuerman RW, Grigg ME, Li SFY, Chew FT, Ang L, Stern ME, Tan D. Journal Of Proteome Research. 2004;3:410. doi: 10.1021/pr034065n. [DOI] [PubMed] [Google Scholar]
- 33.Pisano E, Cabras T, Montaldo C, Piras V, Inzitari R, Olmi C, Castagnola M, Messana I. European Journal Of Oral Sciences. 2005;113:462. doi: 10.1111/j.1600-0722.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 34.Panyutich AV, Voitenok NN, Lehrer RI, Ganz T. Journal Of Immunological Methods. 1991;141:149. doi: 10.1016/0022-1759(91)90141-2. [DOI] [PubMed] [Google Scholar]
- 35.Ihi T, Nakazato M, Mukae H, Matsukura S. Clinical Infectious Diseases. 1997;25:1134. doi: 10.1086/516075. [DOI] [PubMed] [Google Scholar]
- 36.Mizukawa N, Sugiyama K, Ueno T, Mishima K, Takagi S, Sugahara T. Oral Surgery Oral Medicine Oral Pathology Oral Radiology And Endodontics. 1999;87:539. doi: 10.1016/s1079-2104(99)70130-7. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen M, Bergkvist KSG, Welinder KG. Analytical Biochemistry. 2006;358:295–297. doi: 10.1016/j.ab.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 38.Thompson L, Turko I, Murad F. Molecular Immunology. 2006;43:1485. doi: 10.1016/j.molimm.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Stephenson JL, McLuckey SA. Analytical Chemistry. 1998;70:3533–3544. doi: 10.1021/ac9802832. [DOI] [PubMed] [Google Scholar]
- 40.Stephenson JL, McLuckey SA, Reid GE, Wells JM, Bundy JL. Current Opinion in Biotechnology. 2002;13:57–64. doi: 10.1016/s0958-1669(02)00285-9. [DOI] [PubMed] [Google Scholar]
- 41.Louris JN, Cooks RG, Syka JEP, Kelley PE, Stafford GC, Todd JFJ. Analytical Chemistry. 1987;59:1677–1685. [Google Scholar]
- 42.McLuckey SA. Journal of the American Society for Mass Spectrometry. 1992;3:599–614. doi: 10.1016/1044-0305(92)85001-Z. [DOI] [PubMed] [Google Scholar]
- 43.McLuckey SA, Goeringer DE. Journal of Mass Spectrometry. 1997;32:461–474. [Google Scholar]
- 44.Chan WCWPD, editor. Fmoc Solid Phase Peptide synthesis. Oxford University Press; New York: 2000. [Google Scholar]
- 45.Pazgier M, Lubkowski J. Protein Expression And Purification. 2006;49:1. doi: 10.1016/j.pep.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Xu ZN, Peng L, Zhong ZX, Fang XM, Cen PL. Biotechnology Progress. 2006;22:382–386. doi: 10.1021/bp0502680. [DOI] [PubMed] [Google Scholar]
- 47.Ghosh SK, Gerken TA, Schneider KM, Feng ZM, McCormick TS, Weinberg A. Clinical Chemistry. 2007;53:757–765. doi: 10.1373/clinchem.2006.081430. [DOI] [PubMed] [Google Scholar]