Abstract
Purpose: Accurate segmentation of lungs with severe interstitial lung disease (ILD) in thoracic computed tomography (CT) is an important and difficult task in the development of computer-aided diagnosis (CAD) systems. Therefore, we developed in this study a texture analysis-based method for accurate segmentation of lungs with severe ILD in multidetector CT scans.
Methods: Our database consisted of 76 CT scans, including 31 normal cases and 45 abnormal cases with moderate or severe ILD. The lungs in three selected slices for each CT scan were first manually delineated by a medical physicist, and then confirmed or revised by an expert chest radiologist, and they were used as the reference standard for lung segmentation. To segment the lungs, we first employed a CT value thresholding technique to obtain an initial lung estimate, including normal and mild ILD lung parenchyma. We then used texture-feature images derived from the co-occurrence matrix to further identify abnormal lung regions with severe ILD. Finally, we combined the identified abnormal lung regions with the initial lungs to generate the final lung segmentation result. The overlap rate, volume agreement, mean absolute distance (MAD), and maximum absolute distance (dmax) between the automatically segmented lungs and the reference lungs were employed to evaluate the performance of the segmentation method.
Results: Our segmentation method achieved a mean overlap rate of 96.7%, a mean volume agreement of 98.5%, a mean MAD of 0.84 mm, and a mean dmax of 10.84 mm for all the cases in our database; a mean overlap rate of 97.7%, a mean volume agreement of 99.0%, a mean MAD of 0.66 mm, and a mean dmax of 9.59 mm for the 31 normal cases; and a mean overlap rate of 96.1%, a mean volume agreement of 98.1%, a mean MAD of 0.96 mm, and a mean dmax of 11.71 mm for the 45 abnormal cases with ILD.
Conclusions: Our lung segmentation method provided accurate segmentation results for abnormal CT scans with severe ILD and would be useful for developing CAD systems for quantification, detection, and diagnosis of ILD.
Keywords: computer-aided diagnosis, lung segmentation, interstitial lung disease, texture analysis, co-occurrence matrix, computed tomography
INTRODUCTION
Computer-aided diagnosis (CAD) schemes for thoracic computed tomography (CT) are widely used to characterize, quantify, and detect numerous lung abnormalities, such as emphysema,1, 2 lung nodules,3, 4 and interstitial lung disease (ILD).5, 6, 7 An accurate lung segmentation method is always a critical first step in these CAD schemes and can significantly improve the performance level of these CAD schemes. Although manual or semiautomatic lung segmentation methods for CT images were used in some early CAD schemes,8, 9, 10 they are impractical for current CAD schemes because multidetector CT (MDCT) scanners can generate hundreds of CT slices for a patient. An automated lung segmentation method is needed for MDCT.
Various techniques based on CT values, such as thresholding,4, 11, 12 region growing,13, 14 and edge tracking,15, 16 were often employed for segmenting lungs. These methods become unreliable when the CT scans of lungs contain high attenuation patterns such as ILD. Haider et al.17 developed a region growing-based segmentation method followed by a manual revision for lungs with ILD. Uchiyama et al.6 segmented lungs with ILD by use of a combination of the gray-level thresholding technique and morphological operation; manual correction was further applied to the cases with consolidation patterns to obtain accurate lung segmentation results. Sluimer et al.18 employed a fully automated atlas-driven method for lung segmentation, in which a normal lung scan was elastically registered to, and utilized to guide the segmentation of a scan with severe ILD. Their approach was evaluated on ten CT scans with severe ILD and achieved a mean overlap rate of 82%. Prasad et al.19 segmented lungs by use of an adaptive thresholding technique guided by the curvature of ribs. Their segmentation method obtained a mean overlap rate of 87% for 44 CT scans with different disease patterns of scleroderma, asthma, and emphysema. Korfiatis et al.20 used statistical texture features and SVM classifier for distinction between abnormal lung regions and surrounding tissues. This method was evaluated on 22 HRCT scans with ILD patterns and achieved a high mean overlap rate of 95%.
The aim of our work is to develop a novel texture analysis-based method for the segmentation of lungs with severe ILD in MDCT scans. We first created texture-feature images, in which each pixel represents a specific texture-feature value. We then selected two effective texture features that well characterize abnormal lungs and provide very good distinction between abnormal lungs and surrounding tissues. From these texture-feature images, we can readily identify lung areas with severe ILD.
MATERIALS
IRB was obtained for this project. The database used in this study was collected from the University of Chicago Medical Center between September 2004 and August 2005.21 Two radiologists first identified 134 MDCT cases with ILD based on clinical reports. They also subjectively rated the severity of the disease as mild, moderate, and severe. They further selected 45 CT scans, each containing (1) at least one of the four kinds of abnormal patterns, i.e., ground glass opacity (GGO), reticular, nodular, and honeycombing, and (2) at least one abnormal pattern with a severity level of moderate or severe. In addition, the radiologists added to the database 31 normal cases, where each slice was considered to be normal.
The CT scans of all the subjects were acquired by use of 64-row (Brilliance 64; Philips Medical Systems, the Netherlands), 40-row (Brilliance 40), or 16-row (Brilliance 16P) CT scanners with an x-ray tube voltage of 120–140 kVp and a radiographic exposure of 200–400 mA s. The slice thickness ranged from 1 to 3 mm. Each slice had a matrix size of 512×512 pixels and the pixel size ranged from 0.50 to 0.89 mm with a 12-bit gray level in Hounsfield unit (HU).
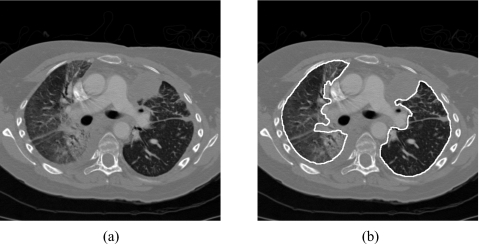
A reference standard for lungs was established to assess the accuracy of the lung segmentation method. To establish the reference standard in a manageable way, an expert chest radiologist (25 years of experience) first selected three slices for each CT scan based on the following criteria: an upper slice at the aortic arch level or above, a middle slice at the level of main bronchi or below, and a lower slice at the level of lower lobar bronchi or below. Radiologists at the University of Chicago Medical Center commonly use such three slices for diagnosis of ILD in clinical practice. A medical physicist used a LCD monitor to identify the lung regions in the three selected slices of each CT scan. The delineated lungs were confirmed or revised, if needed, by an expert chest radiologist (37 years of experience). Figure 1 shows a CT slice with severe ILD and the manually delineated lungs.
Figure 1.
(a) A CT slice with severe interstitial lung disease and (b) the manually delineated lungs.
METHODS
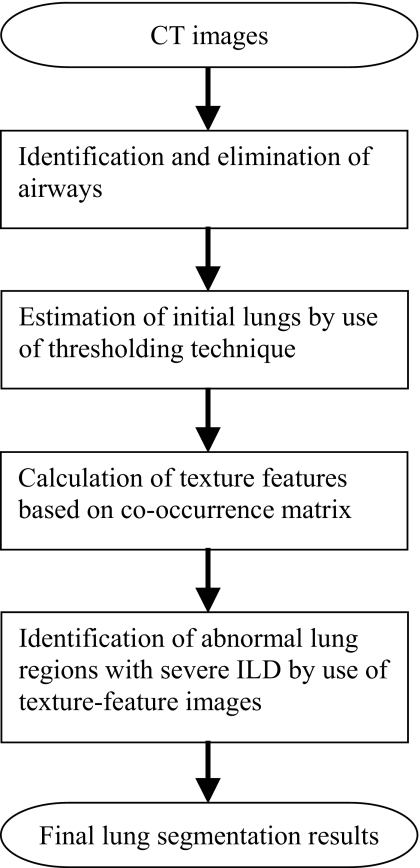
Figure 2 shows the overall scheme of the automated lung segmentation method. Our method first identifies and eliminates airways in each CT slice by use of pixel CT value and contiguity of the airways between adjacent slices. It then obtains an initial lung estimate that includes normal and mildly abnormal lung parenchyma by use of a CT value thresholding technique. It further calculates texture features for each pixel from the co-occurrence matrix and identifies the abnormal lung regions with severe ILD from the texture-feature images. Finally, it generates the final segmentation results by combing the initial lungs and the identified abnormal lung regions.
Figure 2.
The diagram of the automatic lung segmentation method.
Elimination of airways
Because the airways are filled with air only, they have very low CT values around −1000 HU in CT images. Therefore, we used a threshold of −900 HU to segment airways by indicating all the pixels with CT values lower than −900 HU as air-component pixels. With this technique, the trachea, bronchi, as well as some unwanted regions in lungs were identified as airways.
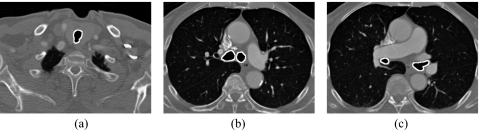
We then accurately identified airways from the air component pixels by use of the contiguity of airways. Generally, the trachea is the only significant air component in the beginning of a thoracic CT scan. Thus, we employed a 3D connected-component labeling algorithm in the first 30 slices to identify and retain the largest volume of air-component pixels that represents upper part of the trachea; other small connected components were eliminated as noise. Next, we used the identified trachea as a seed and traced the entire airways from an upper slice to a lower slice through the CT scan. In each slice, if an air-component pixel is adjacent to an airway pixel in the previous slice, the air-component pixel was added to the airway. Finally, we employed a morphological opening operation to separate the airways and unwanted regions and a 3D connected-component labeling algorithm to retain the largest volume as the airways. The segmented airways were then removed from the CT images to prevent interference in lung segmentation. The solid curves in Fig. 3 show the segmented airways in an upper slice, a middle slice, and a lower slice of a CT scan that were identified by our airway segmentation method.
Figure 3.
The segmentation of airways in (a) an upper, (b) a middle, and (c) a lower slices in a CT scan.
Estimation of initial lungs
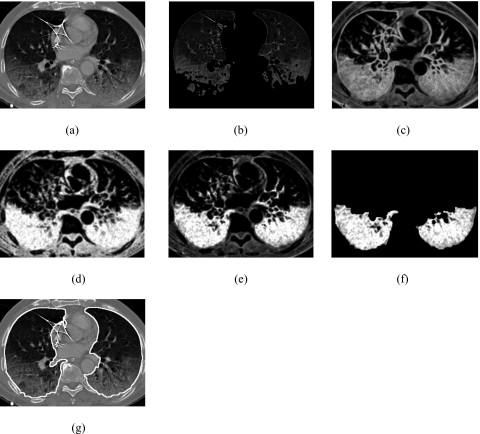
After removing airways from the CT images, we employed a CT value thresholding technique to estimate initial lungs. The initial lungs contain normal lung regions and mildly abnormal lung regions. In our previous study,4 we used a threshold of −400 HU to segment the lungs for nodule detection. Because many of the ILD patterns exhibited increased attenuation, in this study, we investigated the effect of different thresholds ranging from −400 HU to −200 HU with an increment of 50 HU and empirically selected a threshold of −300 HU to estimate initial lungs. The threshold was applied to the CT images to generate a 3D binary lung mask for each CT scan. The resulting binary lung masks included unwanted background air pixels from outside of the body and some isolated pixels with low CT values caused by noise or streak artifacts in surrounding tissue. To remove the unwanted pixels, we first applied a 2D connected-component labeling algorithm to each slice of the 3D binary lung masks to identify the connected components. If a connected component is connected to image edges, we considered it a background area and removed it. Next, we applied a 3D connected-component labeling algorithm to the 3D binary lung mask to identify the two largest components. If the largest component had a volume that was at least ten times larger than that of the second largest component, the two lungs were considered being connected and we retained the largest component as the initial lungs. Otherwise, the two lungs were not connected and we retained the two largest components as the initial lungs. Figures 4a, 4b show a CT image of a patient with severe ILD and the initial lungs, respectively. The lung regions with very severe ILD pattern were missed by the thresholding technique and resulted in poor segmentation of the initial lungs in Fig. 4b.
Figure 4.
(a) Original CT image of lungs with severe interstitial lung disease, (b) initial estimate of the lungs, (c) entropy image, (d) inverse difference moment image, (e) combined feature image, (f) the identified lung regions with severe interstitial lung disease, and (g) the final lung segmentation result obtained by combing the initial estimate and the lung regions with severe ILD, followed by a postprocessing to fill the holes in the segmented lung regions.
Identification of abnormal lung regions with severe interstitial lung disease
We found that the severe ILD patterns generally have rich texture (frequent spatial changes in CT values) compared to surrounding soft tissue. Such texture information can be used to identify the abnormal lung regions. We employed a 3D co-occurrence matrix (COM) to analyze the texture features of all the pixels in the CT images, except the background air pixels and airway pixels, to identify abnormal lung regions. From a volume of interest (VOI) of 15×15×15 pixels centered at each pixel, we determined a COM and calculated the following five COM texture features for the pixel: (1) energy, (2) entropy, (3) contrast, (4) maximum probability, and (5) inverse difference moment.22
An element of a COM was defined by the number of pairs of pixel values separated by a given distance along a specific direction in a VOI.22 We compared the performance levels for lung segmentation using COMs with four different distances of 1, 2, 4, and 8 pixels and found that the performance levels for these distances were comparable; thus, we used only a COM that was constructed with a distance of 1 pixel. Because the ILD patterns extended in all directions nearly homogenously, they can be considered nondirectional. Therefore, we first constructed three COMs for the x, y, and z directions, and then determined a combined COM by calculating the mean value of the corresponding elements in the three COMs.
In order to investigate the usefulness of COM features and select proper features for the segmentation of lung regions with severe ILD, we created five feature images, in which each pixel represents a feature value of energy, entropy, contrast, maximum probability, or inverse difference moment of COM. We found that the severe ILD patterns have very large entropy values compared to normal lung parenchyma, and thus can be easily identified in the entropy image, Fig. 4c. However, the lung borders also have large entropy values in Fig. 4c. Because the inverse difference moment emphasizes small changes in pixel values between pixel pairs, the lung borders, which have large changes, have a relatively low value in the inverse difference moment image, Fig. 4d. Therefore, we generated a new combined feature image, Fig. 4e, by multiplying the values of the corresponding pixels in the entropy image, Fig. 4c, and inverse difference moment image, Fig. 4d. This combined feature further enhanced the lung regions with severe ILD and suppressed most of the borders between lungs and surrounding tissue.
We empirically applied a fixed threshold of 600 to the combined feature images in Fig. 4e to identify the abnormal lung regions that were missed in the initial lungs in Fig. 4b and employed a 2D connected-component labeling technique to eliminate unwanted regions that were smaller than 10 mm in diameter. By doing this, we created the abnormal lung regions with severe ILD in Fig. 4f and added them to the initial lungs in Fig. 4b to generate the combined segmentation result in Fig. 4g.
Postprocessing for filling “holes” in the lungs
The segmented lungs may contain holes caused by blood vessels and other structures with high CT values. These holes were automatically identified and filled by use of a 2D connected-component labeling technique in each CT slice. The peak of the diaphragm, however, often resulted in a similar hole in the segmented lungs. In order to exclude the pixels that belonged to the diaphragm, we did not fill holes that exceeded 10% of the area of the segmented lungs in slices located in the lower half of the CT scan.
Evaluation of the segmentation method
We employed an overlap rate of lungs, a volume agreement of lungs, a mean absolute distance (MAD), and a maximum absolute distance (dmax) of lung borders as performance metrics to assess the accuracy of our lung segmentation method. The overlap rate is defined as the ratio of the intersection to the union between the automatically segmented lungs and the reference lungs in a CT image. The value of the overlap rate ranges from 0%, no overlap between the two lungs, to 100%, a perfect overlap. We employed the volume agreement to evaluate the accuracy in the size of segmented lungs. The volume agreement is defined as one minus the ratio of the absolute difference in the volume between the automatically segmented lungs and reference lungs to the volume of reference lungs. The typical value of the volume agreement ranges from 0% to 100%. The larger the agreement value, the closer the two volumes of the automatically segmented lung and the reference lung. To assess the shape accuracy of a segmented lung, the MAD is calculated by measuring the average distance from all points on the border of the automatically segmented lung to that of the reference lung. The smaller the MAD, the closer the points on the border of automatically segmented lung and that of the reference lung. To assess the local discrepancy between an automatically segmented lung and a reference lung, the maximum absolute distance dmax between the border of automatically segmented lung and that of reference lung was calculated. The mean value for each of the above four metrics for a case is defined as the average of the corresponding metric in the three selected slices.
RESULTS
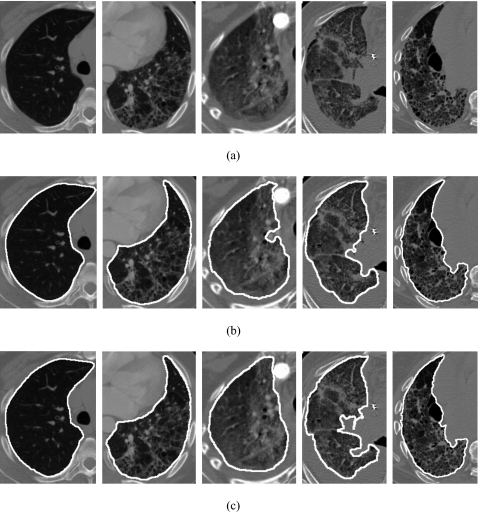
Figure 5 shows (a) original CT images of a normal lung, an abnormal lung with moderate ILD, and three abnormal lungs with severe ILD, (b) lung segmentation results, and (c) the corresponding reference standard. It is apparent that we obtained accurate lung segmentation results for the normal lungs as well as the abnormal lungs with severe ILD.
Figure 5.
(a) Original CT images of a normal lung, an abnormal lung with moderate interstitial lung disease, and three abnormal lungs with severe interstitial lung disease, (b) segmentation results, and (c) their corresponding reference standard.
Table 1 shows the mean, standard deviation, and range of the overlap rate, volume agreement, MAD, and dmax for our segmentation method. The (1) mean overlap rate, (2) mean volume agreement, (3) mean MAD, and (4) mean dmax for the segmentation of normal cases, abnormal cases, and all cases were (1) 97.7%, 96.1%, and 96.7%, (2) 99.0%, 98.1%, and 98.5%, (3) 0.66, 0.96, and 0.84 mm, and (4) 9.59, 11.71, and 10.84 mm, respectively. By use of two-tailed t-tests for unpaired data, we found that there were significant differences in the mean overlap rate (p<0.0001), mean volume agreement (p=0.003), mean MAD (p<0.0001), and mean dmax (p=0.03) between the normal lungs and abnormal lungs. The difference itself was quite small and, overall, the performance level for the abnormal lungs was quite high. However, for a small number of severely abnormal cases, the segmentation results may still be poor.
Table 1.
Accuracy of segmentation for normal lungs, abnormal lungs, and all the lungs in terms of overlap rates, volume agreement, mean absolute distance, and maximum absolute distance.
| Normal | Abnormal | All | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overlap (%) | Agreement (%) | MAD (mm) | dmax (mm) | Overlap (%) | Agreement (%) | MAD (mm) | dmax (mm) | Overlap (%) | Agreement (%) | MAD (mm) | dmax (mm) | |
| Mean | 97.7a | 99.0b | 0.66c | 9.59d | 96.1a | 98.1b | 0.96c | 11.71d | 96.7 | 98.5 | 0.84 | 10.84 |
| SD | 0.6 | 0.6 | 0.21 | 3.97 | 2.3 | 1.8 | 0.44 | 4.28 | 2.0 | 1.5 | 0.39 | 4.26 |
| Min | 96.3 | 97.4 | 0.36 | 4.14 | 89.7 | 90.0 | 0.35 | 5.39 | 89.7 | 90.0 | 0.35 | 4.14 |
| Max | 98.7 | 99.7 | 1.35 | 22.28 | 98.7 | 99.8 | 2.30 | 23.10 | 98.7 | 99.8 | 2.30 | 23.10 |
p<0.0001.
p=0.003.
p<0.0001.
p=0.03.
Table 2 shows the performance metrics for the abnormal lungs with moderate and severe ILD. The (1) mean overlap rate, (2) mean volume agreement, (3) mean MAD, and (4) mean dmax for the lungs with moderate and severe ILD were (1) 95.8% and 96.2%, (2) 97.4% and 98.4%, (3) 0.92 and 0.98 mm, and (4) 11.24 and 11.89 mm, respectively. There were no significant differences in the mean overlap rates (p=0.65), mean volume agreement (p=0.20), mean MAD (p=0.66), and mean dmax (p=0.66) between lungs with moderate and severe ILD.
Table 2.
Accuracy of segmentation for lungs with moderate and severe ILD in terms of overlap rates, volume agreement, mean absolute distance, and maximum absolute distance.
| Moderate | Severe | |||||||
|---|---|---|---|---|---|---|---|---|
| Overlap (%) | Agreement (%) | MAD (mm) | dmax (mm) | Overlap (%) | Agreement (%) | MAD (mm) | dmax (mm) | |
| Mean | 95.8a | 97.4b | 0.92c | 11.24d | 96.2a | 98.4b | 0.98c | 11.89d |
| SD | 2.5 | 2.7 | 0.44 | 4.64 | 2.2 | 1.1 | 0.44 | 4.18 |
| Min | 90.0 | 90.0 | 0.35 | 5.50 | 89.7 | 94.4 | 0.40 | 5.39 |
| Max | 98.7 | 99.8 | 1.85 | 20.10 | 98.3 | 99.7 | 2.30 | 23.10 |
p=0.65.
p=0.20.
p=0.66.
p=0.66.
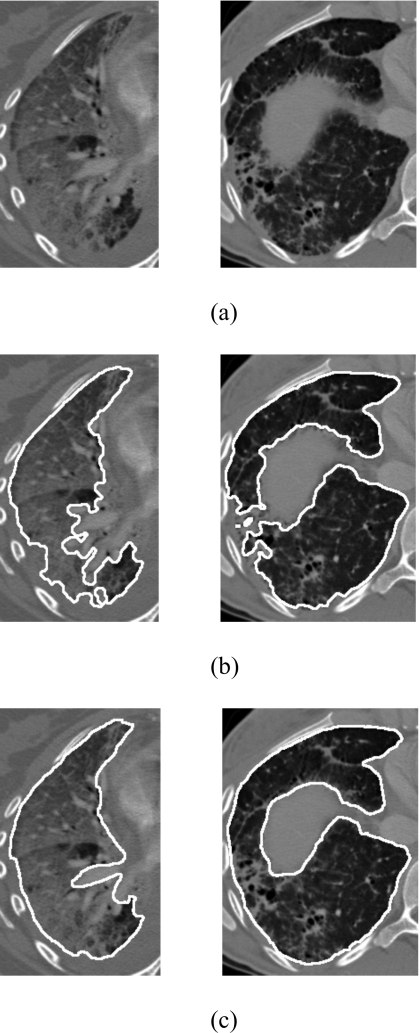
We found that the segmentation results were poor for some abnormal lungs. Figure 6 shows the lung segmentation results for the two poorest lungs in our database. The mean overlap rate, mean volume agreement, mean MAD, and mean dmax were 89.7%, 94.4%, 2.30 mm, and 23.10 mm for the first case, and 90.0%, 90.0%, 1.85 mm, and 14.01 mm for the second case.
Figure 6.
(a) CT images of two abnormal lungs with severe and moderate ILD, (b) poor segmentation results, and (c) their reference standards. The mean overlap rates, mean volume agreement, mean MAD, and dmax were 89.7%, 94.4% 2.30 mm, and 23.10 mm for the first case and 90.0%, 90.0%, 1.85 mm, and 14.01 mm for the second case.
DISCUSSION
Because the normal lungs generally have low density, we obtained good segmentation results for normal lungs by solely using the thresholding technique. In contrast, most of the abnormal lungs with severe ILD have increased density, thus, the thresholding technique lacks power for the identification of abnormal regions and the texture-analysis step is needed. In this study, we employed texture feature images to identify the abnormal lung parenchyma with severe ILD. In fact, if we excluded the texture-analysis step and segmented lungs by use of a CT value thresholding technique alone with a threshold of −300 HU, the mean overlap rate would degrade from 96.1% (range of 89.7%–98.7%) to 94.2% (range of 71.6%–98.7%), the mean volume agreement from 98.1% (range of 90.0%–99.8%) to 95.8% (range of 73.5%–99.8%), the mean MAD from 0.96 mm (range of 0.35–2.30 mm) to 1.51 mm (range of 0.35–5.78 mm), and mean dmax from 11.71 mm (range of 5.39–23.10 mm) to 15.48 mm (range of 5.39–29.85 mm) for the 45 abnormal CT scans.
We anticipated that the thresholding technique alone would work well for normal and mildly abnormal cases, and that the texture analysis would improve the performance of lung segmentation primarily for severe ILD cases. To verify our anticipation, we defined the “very severe” ILD cases as those with an overlap rate less than 93.0% for initial lungs (obtained by thresholding alone). We visually confirmed that these were indeed very severe ILD cases. The mean overlap rate, volume agreement, MAD, and dmax for the ten very severe ILD cases were 84.7%, 86.3%, 3.21 mm, and 22.89 mm for initial lungs and were improved considerably to 92.7%, 96.4%, 1.53 mm, and 15.45 mm by use of texture analysis step. The performance level for each of the ten cases was improved. On the other hand the mean overlap rate, volume agreement, MAD, and dmax for the 35 “not very severe” ILD cases were 96.9%, 98.4%, 1.03 mm, and 13.67 mm for initial lungs and were improved only slightly to 97.0%, 98.6%, 0.80 mm, and 10.64 mm by use of texture analysis step. Therefore, our texture analysis step can considerably improve the segmentation results of very severe abnormal cases and can only slightly improve those of not very severe ILD cases, because thresholding alone works well for them already.
It is intuitive to think that the segmentation results for the lungs with moderate ILD would be better than that for the lungs with severe ILD. However, we did not find significant difference in performance levels between cases with moderate ILD and severe ILD. As described in the Results section, the increased attenuation of abnormalities led to difficulty in segmentation of abnormal lungs. However, the attenuation is not the only factor that affects the lung segmentation results; the location and distribution of the disease patterns are also key factors. The image on the right side of Fig. 6a represents a lung with moderate ILD. Its segmentation result in Fig. 6b was poor because the lung contained a small portion of very high attenuation pattern in peripheral lung. If this small abnormal region was located completely inside the lung, then it would not constitute a problem.
Segmentation of lungs plays an import role in thoracic CAD schemes and has great impact on the overall performance level of a CAD scheme. Inaccurate lung segmentation may miss some lung parenchyma with severe abnormalities and may incorrectly include some nonlung structures as part of the lungs. A CAD scheme based on inaccurate lung segmentation thus will obtain a lower sensitivity and a lower specificity than that based on an accurate lung segmentation method. In this study, our lung segmentation method achieved good segmentation results for most of the cases. However, the segmentation results were poor for a small fraction of lungs with very severe ILD. In the future, others may develop a method to automatically identify and improve the poor segmentation results for lungs with very severe ILD. In some interactive CAD schemes, end users such as radiologists may also manually identify and refine the poor segmentation results for lungs based on their subjective judgment.
Our segmentation method achieved high performance level based on metrics of overlap rate, volume agreement, and MAD, but not as good based on the maximum absolute distance dmax. Our segmentation method employed a CT value thresholding technique to segment initial lungs for normal and mildly abnormal lung parenchyma. The thresholding technique may miss some “normal” structures inside the lungs, such as blood vessels near the mediastinum. Sometimes, these normal structures are treated as part of lungs by physicists and radiologists when they create reference lungs. Missing these normal structures by computer is not important in clinical practice, but creates a large dmax value (low performance), even though overall segmentation results are very good. Thus, we believe that dmax should be used only as a secondary metric because it measures the performance based only on a single point on the contour of a lung. The overlap rate, volume agreement, and MAD measure the overall accuracy of the segmentation results, and thus are more reliable performance metrics for assessing the lung segmentation methods.
When the gray-level information alone is not able to distinguish abnormal lungs from adjacent normal soft tissues, the use of shape constraints derived from normal lung atlas18 or surrounding structures19 could be beneficial. The mean overlap rates were 82% and 87%, respectively, for an atlas-driven lung segmentation method18 and a rib-curvature guided segmentation method.19 These overlap rates were relatively low probably because (1) the databases used in these two studies included many extremely difficult cases and (2) the usefulness of shape constraints were reduced for these extremely severe lungs. As with our method, Korfiatis et al. also employed a texture analysis-based lung segmentation method20 and obtained a mean overlap rate of 95%, which is comparable to ours.
The lung segmentation is an initial step in many CAD systems for detection of lung diseases; a fast and simple lung segmentation algorithm thus would be a considerable advantage for the development and application of the subsequent CAD systems. Our segmentation method was implemented in C (gcc 4.1.2) programming language. When we ran our segmentation method on a workstation server (8 GB RAM and 2.5 GHz Intel Xeon CPU), the processing time for each slice was approximately 5 s. Korfiatis et al. implemented their texture analysis-based segmentation method in MATLAB and ran it on a PC (2 GB RAM and Intel Core 2 Duo CPU); the processing time for each slice was approximately 20 s.20
CONCLUSIONS
We developed an automatic segmentation method for lungs with severe ILD in MDCT scans. The proposed method is composed of the elimination of airways, the estimation of initial lungs, and the identification of abnormal lung regions with severe ILD by use of texture-feature images. The experiments on a database of 45 abnormal cases with severe ILD and 31 normal cases indicated that our segmentation method achieved a high accuracy for the segmentation of lungs with ILD and would be very useful for developing computer-aided diagnosis schemes for ILD.
ACKNOWLEDGMENTS
This work was supported by USPHS Grant No. CA113870. The authors thank Drs. Yongkang Nie and Carl E. Ravin for their helpful discussion and technique assistance, Dr. W. Paul Segars for his software to delineate lung outlines, and Karen Wilson for improving the manuscript. CAD technologies developed by Qiang Li and his colleagues have been licensed to companies including R2 Technologies, Riverain Medical Group, Deus Technologies, Median Technology, Mitsubishi Space Software Co., General Electric Corporation, and Toshiba Corporation. It is the policy of Duke University and the University of Chicago that investigators disclose publicly actual or potential significant financial interests that may appear to be affected by research activities.
References
- Uppaluri R., Mitsa T., Sonka M., Hoffman E., and McLennan G., “Quantification of pulmonary emphysema from lung computed tomography images,” Am. J. Respir. Crit. Care Med. 156, 248–254 (1997). [DOI] [PubMed] [Google Scholar]
- Xu Y., Sonka M., McLennan G., Guo J., and Hoffman E., “MDCT-based 3-D texture classification of emphysema and early smoking related lung pathologies,” IEEE Trans. Med. Imaging 25, 464–475 (2006). 10.1109/TMI.2006.870889 [DOI] [PubMed] [Google Scholar]
- S.ArmatoIII, Giger M., and MacMahon H., “Automated detection of lung nodules in CT scans: Preliminary result,” Med. Phys. 28, 1552–1561 (2001). 10.1118/1.1387272 [DOI] [PubMed] [Google Scholar]
- Li Q., Li F., and Doi K., “Computerized detection of lung nodules in thin-section CT images by use of selective enhancement filters and an automated rule-based classifier,” Acad. Radiol. 15, 165–175 (2008). 10.1016/j.acra.2007.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppaluri R., Hoffman E., Sonka M., Hartley P., Hunninghake G., and Mclennan G., “Computer recognition of regional lung disease patterns,” Am. J. Respir. Crit. Care Med. 160, 648–654 (1999). [DOI] [PubMed] [Google Scholar]
- Uchiyama Y., Katsuragawa S., Abe H., Shiraishi J., Li F., Li Q., Zhang C., Suzuki K., and Doi K., “Quantitative computerized analysis of diffuse lung disease in high-resolution computed tomography,” Med. Phys. 30, 2440–2454 (2003). 10.1118/1.1597431 [DOI] [PubMed] [Google Scholar]
- Sluimer I., Waes P., Viergever M., and Ginneken B., “Computer-aided diagnosis in high resolution CT of the lungs,” Med. Phys. 30, 3081–3090 (2003). 10.1118/1.1624771 [DOI] [PubMed] [Google Scholar]
- Denison D., Morgan M., and Miller A., “Estimation of regional gas and tissue volumes of the lung in supine man using computed tomography,” Thorax 41, 620–628 (1986). 10.1136/thx.41.8.620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalender W., Fichte H., Bautz W., and Skalej M., “Semiautomatic evaluation procedures for quantitative CT of the lung,” J. Comput. Assist. Tomogr. 15, 248–255 (1991). [DOI] [PubMed] [Google Scholar]
- Zagers R., Vrooman H., Aarts N., Stolk J., Schultze Kool L., Voorthuisen E., and Reiber J., “Quantitative analysis of computed tomography scans of the lungs for the diagnosis of pulmonary emphysema: A validation study of a semiautomated contour detection technique,” Invest. Radiol. 30, 552–562 (1995). 10.1097/00004424-199509000-00008 [DOI] [PubMed] [Google Scholar]
- Kemerink G., Lamers R., Pellis B., Kruize H., and Engelshoven J., “On segmentation of lung parenchyma in quantitative computed tomography of the lung,” Med. Phys. 25, 2432–2439 (1998). 10.1118/1.598454 [DOI] [PubMed] [Google Scholar]
- S.ArmatoIII and Sensakovic W., “Automated lung segmentation for thoracic CT: impact on computer-aided diagnosis,” Acad. Radiol. 11, 1011–1021 (2004). 10.1016/j.acra.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Brown M., McNitt-Gray M., Mankovich N., Goldin J., Hiller J., Wilson L., and Aberle D., “Method for segmenting chest CT image data using an anatomical model: preliminary results,” IEEE Trans. Med. Imaging 16, 828–839 (1997). 10.1109/42.650879 [DOI] [PubMed] [Google Scholar]
- Hu S., Hoffman E., and Reinhardt J., “Automatic lung segmentation for accurate quantitation of volumetric x-ray CT images,” IEEE Trans. Med. Imaging 20, 490–498 (2001). 10.1109/42.929615 [DOI] [PubMed] [Google Scholar]
- Keller J., Edwards F., and Rundle R., “Automatic outlining of regions on CT scans,” J. Comput. Assist. Tomogr. 5, 240–245 (1981). 10.1097/00004728-198104000-00015 [DOI] [PubMed] [Google Scholar]
- Hedlund L., Anderson R., Goulding P., Beck J., Effmann E., and Putman C., “Two methods for isolating the lung area of a CT scan for density information,” Radiology 144, 353–357 (1982). [DOI] [PubMed] [Google Scholar]
- Haider C., Bartholmai B., Holmes D., Camp J., and Robb R., “Quantitative characterization of lung disease,” Comput. Med. Imaging Graph. 29, 555–563 (2005). 10.1016/j.compmedimag.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Sluimer I., Prokop M., and Ginneken B., “Toward automated segmentation of the pathological lung in CT,” IEEE Trans. Med. Imaging 24, 1025–1038 (2005). 10.1109/TMI.2005.851757 [DOI] [PubMed] [Google Scholar]
- Prasad M., Brown M., Ahmad S., Abtin F., Allen J., Costa I., Kim H., Mcnitt-Gray M., and Goldin J., “Automatic segmentation of lung parenchyma in the presence of disease based on curvature of ribs,” Acad. Radiol. 15, 1173–1180 (2008). 10.1016/j.acra.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Korfiatis P., Kalogeropoulou C., Karahaliou A., Kazantzi A., Skiadopoulos S., and Costaridou L., “Texture classification-based segmentation of lung affected by interstitial pneumonia in high-resolution CT,” Med. Phys. 35, 5290–5302 (2008). 10.1118/1.3003066 [DOI] [PubMed] [Google Scholar]
- Wang J., Li F., Doi K., and Li Q., “A novel scheme for detection of diffuse lung disease in MDCT by use of statistical texture features,” Proc. SPIE 7260, 726039-1–726039-8 (2009). 10.1117/12.811635 [DOI] [PubMed] [Google Scholar]
- Haralick R., “Statistical and structural approaches to texture,” Proc. IEEE 67, 786–804 (1979). 10.1109/PROC.1979.11328 [DOI] [Google Scholar]