Abstract
The purpose of this study was to investigate the effect of a noise injection method on the “overfitting” problem of artificial neural networks (ANNs) in two-class classification tasks. The authors compared ANNs trained with noise injection to ANNs trained with two other methods for avoiding overfitting: weight decay and early stopping. They also evaluated an automatic algorithm for selecting the magnitude of the noise injection. They performed simulation studies of an exclusive-or classification task with training datasets of 50, 100, and 200 cases (half normal and half abnormal) and an independent testing dataset of 2000 cases. They also compared the methods using a breast ultrasound dataset of 1126 cases. For simulated training datasets of 50 cases, the area under the receiver operating characteristic curve (AUC) was greater (by 0.03) when training with noise injection than when training without any regularization, and the improvement was greater than those from weight decay and early stopping (both of 0.02). For training datasets of 100 cases, noise injection and weight decay yielded similar increases in the AUC (0.02), whereas early stopping produced a smaller increase (0.01). For training datasets of 200 cases, the increases in the AUC were negligibly small for all methods (0.005). For the ultrasound dataset, noise injection had a greater average AUC than ANNs trained without regularization and a slightly greater average AUC than ANNs trained with weight decay. These results indicate that training ANNs with noise injection can reduce overfitting to a greater degree than early stopping and to a similar degree as weight decay.
Keywords: artificial neural networks, overfitting, regularization, early stopping, weight decay, BANN, noise injection, jitter
INTRODUCTION
Artificial neural networks (ANNs) are frequently used in computer-aided detection and diagnosis (CAD) applications.1, 2 ANNs are popular because they are capable of modeling complicated classification decision boundaries from training data (of which the diagnostic truth status is known in every case) with minimal supervision or explicit modeling.3, 4
To model complex decision boundaries, ANNs must be flexible, but this flexibility can also result in “overfitting.”5 Overfitting occurs when the classification algorithm learns to classify the training data better than the population of cases at large (i.e., the algorithm does not generalize well to the population of cases from which the training dataset was sampled).
Regularization attempts to avoid overfitting by using a flexible model with constraints on the values that model parameters can take, usually through the addition of a penalty term.5, 6 Bayesian ANNs (BANNs) (which are closely related to weight decay) and early stopping are two widely used regularization methods that favor models with smooth decision boundaries.7, 8, 9
A third method of regularization, called noise injection, penalizes complex models indirectly by adding noise to the training dataset.5, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 However, to our knowledge, noise injection has not been compared to the more common methods of BANN and early stopping. The purpose of this work was to compare the effect of noise injection to BANNs and early stopping.
METHODS
We conducted several simulation studies and a study of a breast ultrasound (US) dataset in a CAD application. The idealized simulation studies allowed us to study the effect of regularization in greater depth and the US study provided an example of real-world application. In the first simulation study, we studied ANNs of common complexity in terms of the number of hidden nodes and training iterations; in the second simulation study, we studied a highly complex ANN; and in the third simulation study, we evaluated a method for automatically selecting a critical noise parameter: the standard deviation of the noise kernel. ANN performance was evaluated with receiver operating characteristic (ROC) analysis,20, 21, 22 using the nonparametric area under the ROC curve (AUC) as a summary index.
Simulation study datasets
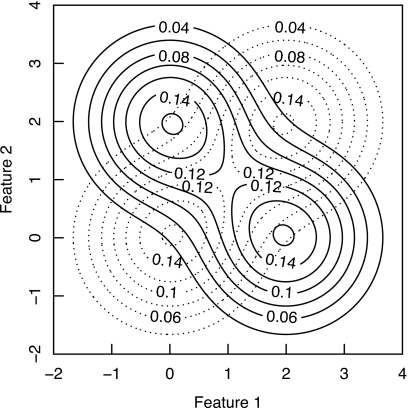
We simulated datasets from a two-dimensional exclusive-or (XOR) population,3 which requires a nonlinear decision boundary to achieve an AUC value greater than 0.5. We chose this problem because it has already been shown theoretically that weight decay and noise injection perform identically in classification problems that require linear decision boundaries.10 The XOR population was constructed with four two-dimensional Gaussian distributions with equal covariance matrices in a two-dimensional feature space. Two Gaussian distributions were centered at (0, 0) and (x, x), respectively, and they represented the “normal” class, whereas another pair of Gaussian distributions was centered at (x, 0) and (0, x), respectively, and they represented the “abnormal” class. The ideal observer’s AUC value23, 24 depends on the separation x and the covariance matrices of the four Gaussian distributions. We set the covariance matrix of each of the four Gaussian distributions to be a 2-by-2 identity matrix and set x to 2.0, thus giving rise to a classification problem with an ideal-observer AUC value of 0.83. A contour plot of the XOR population is shown in Fig. 1.
Figure 1.
The XOR population. Normal cases were drawn from the dotted-line probability density and abnormal cases were drawn from the solid-line probability density. The lines depict isopleths of the probability densities.
We created training datasets of various sizes: 50, 100, and 200 total cases with half of each dataset being normal and the other half being abnormal cases. We chose small training datasets because ANNs exhibit overfitting more often with small training datasets. Overfitting can also occur with large training datasets when the feature space is large, or the underlying distribution is complex, which is probably the case in many real-world applications. We repeated each experiment 500 times with independently drawn training datasets for training datasets of 50 and 100 total cases, 100 times for training datasets of 200 total cases (because the results were less variable), and report here the summary results. An independently drawn validation dataset of 2000 cases (1000 normal and 1000 abnormal cases) was used to evaluate all ANNs.
Breast ultrasound dataset
The breast ultrasound dataset has been described elsewhere.25 The goal was to differentiate malignant from benign breast lesions. The dataset contained 157 malignant lesions and 969 benign lesions (1126 total cases). In the previous work,25 a BANN with four input nodes and five hidden nodes was used and the BANN was trained and tested using leave-one-out cross validation to obtain an AUC value of 0.90. We trained ANNs on this dataset and found no evidence of overfitting. To simulate a situation in which the ANNs do overfit, we randomly and independently sampled 500 sets of 50 cases and 500 sets of 100 cases from this dataset (each subset of cases consisted of half cancer and half benign cases) and tested the ANNs on the subset of training cases with the .632+ bootstrap AUC estimate. We will describe the .632+ bootstrapping method later.
We did not evaluate the method of early stopping with this breast US dataset. Given the single dataset that was available in this experiment rather than the independently drawn training and test datasets that were available in the simulation studies, we would have to measure the ANN performance with the .632+ bootstrap AUC estimate and use it to decide when to stop ANN training. Doing so would bias the results. In the simulation studies, we decided when to stop ANN training based on the .632+ bootstrapping results and evaluated the ANN performance on the independent validation dataset.
ANN implementation
We trained all ANNs by minimizing the cross-entropy error function26 with a conjugate gradient algorithm. The ANNs in the first and third simulation studies had a single hidden layer with six hidden nodes and were trained to 500 iterations. With very large training datasets (500, 1000, and 5000 cases in 100 repeated experiments), this ANN architecture was able to achieve nearly ideal-observer AUC performance on the validation dataset (0.81, 0.81, and 0.82, respectively). Therefore, the small size of the training datasets was the main reason why the ANNs did not perform as well as the ideal observer in simulation results that we report below. The ANN in the second simulation study also had a single hidden layer but with 20 hidden nodes and was trained to 1500 iterations. This was a more flexible ANN configuration, which allowed us to observe overfitting to a greater degree. The ANN in the US study had a single hidden layer with five hidden nodes, the same as that of the previous study.25 The ANNs in the simulation studies had two input nodes and the ANN in the US study had four input nodes. All ANNs had a single output node. We did not vary the random initialization of the ANN weights because we found that the variability in our results from different training datasets was much greater than that from different ANN initial weights, which is known to cause variability in ANN output values.27 All ANNs were implemented using the NETLAB toolkit for MATLAB.26
Noise injection
The method of noise injection refers to adding “noise” artificially to the ANN input data during the training process. Jitter is one particular method of implementing noise injection. With this method, a noise vector is added to each training case in between training iterations. This causes the training data to “jitter” in the feature space during training, making it difficult for the ANN to find a solution that fits precisely to the original training dataset, and thereby reduces overfitting of the ANN. This effect has been shown both in theory13, 14 and in practice.5, 19 The noise vector is typically drawn from some probability density function, known as a “kernel.”14 We used a zero-mean Gaussian kernel and updated the noise vector independently and nonincrementally before every training iteration.
Because at every training iteration the ANN “sees” a slightly different training dataset caused by the added noise, the noisy trajectory of the AUC value of the ANN at various training iterations reflects both the incremental convergence of the ANN toward its final training performance and an effect of the added noise. Because our primary interest is in the training progression of the ANN, we applied a running average of 30 iterations (selected empirically) on the ANN weights when analyzing the AUC values of the ANN as a function of training iterations.
The standard deviation of the noise kernel affects the results of noise injection and ANN performance. Holmström and Koistinen14 described a method for automatically determining an appropriate value of this standard deviation based on cross validation and a broad assumption that the training dataset was drawn from a continuous underlying population. Let us use f1(xi) to denote the probability density in the feature space of the underlying population associated with training case xi of class 1. Let us further use f1,σ(xi) to denote an estimate of this probability density obtained by summing over the noise kernels (denoted by Kσ) of standard deviation σ and centered on every training case of class 1. If we now remove case xi from the calculation of f1,σ(xi), the result is an estimate of f1(xi) from all training cases of class 1 except for case i,
| (1) |
where n is the total number of training cases of class 1. A similar expression can be written for class 2: f2,σ(x−i). Let us create a likelihood expression to represent a cross-validation probability estimate based on the entire training dataset (assuming that all training cases are independent),
| (2) |
where m is the total number of training cases of class 2. It was shown14 that by maximizing Eq. 2, one can select an appropriate σ value to reduce overfitting.
To evaluate this method of automatically selecting an appropriate σ value for the noise kernel, we compared the results obtained with this method and those with incrementing σ in a stepwise fashion. To implement the automatic method, we determined the σ value that maximized L(σ) by calculating L(σ) for values of σ ranging from 0.01 to 1.50 in intervals of 0.01. This calculation took a few seconds for each training dataset.
BANNs and weight decay
A modified error function, , where E(w) is the cross-entropy error function and α is a parameter that controls the weight of the penalty against large values of ANN weights, is used to train both ANNs with weight decay and BANNs.4, 8, 9, 28 BANNs use the Bayes’ rule to estimate an appropriate α value automatically (some of the assumptions involved may not be valid for small training datasets),8, 9, 26 whereas weight decay requires the user to select an α value.
We used both BANN and weight decay in the simulation studies with training datasets of 200 total cases. However, we used only weight decay with training datasets of 50 and 100 total cases because with these datasets the BANNs failed to converge in their estimate of α. To manually select an α value, we trained ANNs with α values ranging between 0.01 and 4.0, repeated the experiment independently 50 times, and chose the α value that maximized the .632+ bootstrapping AUC value (calculated on the training dataset only) averaged across the replicated experiments.29, 30 This α value was then used as a fixed value in all ANN training experiments. We used the method of .632+ bootstrapping because we did not want to bias the results by involving the validation dataset during training. However, there was still potential bias in our method because, in practice, independent replication of the experiment would not be possible in estimating the α value. By replicating the experiment, we reduced the inherent variability in the estimate of the α value, which could have improved the ANN performance and caused a bias in favor of weight decay.
Early stopping
With the method of early stopping, ANN training is stopped before the training error is minimized.31 Typically, an independent test dataset is used to monitor the ANN performance during training, based on which an appropriate point is selected to stop the ANN training. However, withholding training cases for testing is not an efficient use of the data for small training datasets. We used the method of .632+ bootstrapping,7, 29, 30 which allows all cases be used for training.
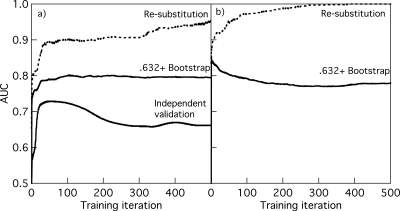
Figure 2a shows how the AUC values of ANNs varied with training iterations for three testing methods that could be used to determine when to stop training for the XOR experiment. The testing method of resubstitution (test the ANN on the training dataset) produced the greatest AUC values. Obviously, this is a biased validation method.32 The method of independent validation (test the ANN on our independent validation dataset) produced the smallest AUC values. This method is also biased for training purposes because the validation dataset should not be used to decide when to stop training. However, the independent validation results are a good surrogate of the ANN performance on the underlying XOR population because the validation dataset of 2000 total cases is large. The method of .632+ bootstrapping produced intermediate AUC values. We used this method for ANN training and stopped the training at the iteration that maximized the .632+ bootstrapping AUC values. For example, in Fig. 2a, we stopped ANN training after 105 training iterations based on the .632+ bootstrapping results. The AUC value of the ANN on the validation dataset (as surrogate of ANN performance on the underlying population) at 105 training iterations is close to the highest AUC value at 60 training iterations (which is not necessarily attainable from the training dataset alone) and clearly greater than that at 500 training iterations with overtraining. Figure 2b shows the AUC values for the two testing methods available for the breast ultrasound experiment. We did not have an independent validation dataset in the breast ultrasound experiment.
Figure 2.
Two ANNs’ AUC values obtained with three training and evaluation methods as a function of the training iterations for (a) simulated XOR dataset of 50 training cases, and (b) breast ultrasound dataset of 50 training cases. The results of .632+ bootstrapping were obtained from 50 bootstrapping samples. The results of independent validation for the XOR data were obtained from an independent validation dataset of 2000 cases not used in any way in training. The breast ultrasound data did not have an independent validation dataset. The standard deviations of estimates of AUC, calculated over different training datasets, were approximately 0.04 for the resubstitution and independent validation estimates, and 0.06 for the .632+ bootstrapping estimates.
Data analysis
We calculated the AUC values of ANNs on the validation dataset as a surrogate measure of the ANN performance on the underlying population. Except for ANNs trained with early stopping, which were not stopped at a fixed training iteration, all ANNs were trained to a large number of training iterations (e.g., 500). A running average of 30 training iterations was used in all ANNs to reduce the noisy trajectory of their performance as a function of training iterations. The average and standard deviation of the AUC values were calculated from 500 or 100 repetitions of each ANN training experiment. Therefore, the average AUC values and standard deviations describe the distribution of observed AUC values over different training datasets. Uncertainties in the average AUC value [95% confidence intervals (CIs)] were estimated with a normal approximation and uncertainties in the standard deviation in the AUC value were estimated from 10 000 bootstrapping samples. Statistical hypothesis testing (i.e., p values) was not used in analyzing our results because in our simulation studies we could have simulated a sufficiently large number of trials to obtain statistical significance for arbitrarily small differences.
For clarity of presentation, we define two measures of relative change in the AUC value: The loss and gain in the AUC value. Both of these measures are defined in terms of ANN performance on the validation dataset. We define the loss in AUC due to overtraining as the difference between the highest AUC value on the validation dataset and the AUC value on the validation dataset without regularization after a large number (e.g., 500) of training iterations. For example, in Fig. 2a, the loss in AUC due to overtraining is the difference between the highest AUC value at 60 iterations and the AUC value at 500 iterations: 0.729−0.661=0.068. We define the gain in AUC as the difference between the AUC value on the validation dataset obtained with regularization and the AUC value on the validation dataset without regularization after a large number (e.g., 500) of training iterations. For example, in Fig. 2a, the gain in AUC due to the method of early stopping is 0.722−0.661=0.061 at 105 iterations. Therefore, the loss represents the magnitude of overfitting, and the gain represents the recovery of the loss with regularization. We further define the ratio of the gain to the loss as the percent recovery. In Fig. 2a, this ratio is 0.061∕0.068=89.7%. Note that this percent recovery is relative to what early stopping could have achieved and, therefore, the percent recovery from early stopping cannot be greater than 100%, whereas the percent recovery from noise injection and weight decay is not limited by 100%—a regularization method could improve ANN training beyond the recovery of loss in performance from overtraining. Uncertainty in the percent recovery was estimated from 10 000 bootstrapping samples and presented as 95% CIs.
RESULTS
Simulation study results of absolute differences
The simulation results in terms of absolute differences are summarized in Table 1. In the first simulation study, ANNs trained on datasets with 50 total cases and without regularization had an average AUC value of 0.723 and a standard deviation of 0.043. Training ANNs with noise injection increased the average AUC value to 0.756 and reduced the standard deviation to 0.037. Training ANNs with weight decay and early stopping also improved the average AUC values to 0.742 and 0.740, respectively, but weight decay increased the standard deviation to 0.050, whereas early stopping did not change the standard deviation. Therefore, training ANNs with noise injection resulted in a greater average and smaller standard deviation in the AUC values than those of the alternative methods.
Table 1.
Comparison of the absolute performance of the ANN training methods in the simulation studies.
| No regularization | Noise injection | Weight decay | BANNa | Early stopping | ||
|---|---|---|---|---|---|---|
| 50 training cases | Average AUC [95% CI] | 0.723 [0.719, 0.727] | 0.756 [0.751, 0.758] | 0.742 [0.738, 0.746] | 0.740 [0.737, 0.744] | |
| AUC standard deviation [95% CI] | 0.043 [0.038, 0.048] | 0.037 [0.033, 0.041] | 0.050 [0.045, 0.055] | 0.041 [0.035, 0.046] | ||
| 50 training cases, complex ANNsb | Average AUC [95% CI] | 0.694 [0.685, 0.703] | 0.758 [0.755, 0.761] | 0.745 [0.735, 0.754] | 0.748 [0.740, 0.757] | |
| AUC standard deviation [95% CI] | 0.044 [0.036, 0.052] | 0.034 [0.031, 0.038] | 0.048 [0.037, 0.060] | 0.043 [0.032, 0.053] | ||
| 100 training cases | Average AUC [95% CI] | 0.762 [0.760, 0.765] | 0.785 [0.784, 0.787] | 0.784 [0.782, 0.786] | 0.770 [0.768, 0.772] | |
| AUC standard deviation [95% CI] | 0.028 [0.026, 0.030] | 0.017 [0.016, 0.019] | 0.020 [0.019, 0.022] | 0.023 [0.021, 0.025] | ||
| 200 training cases | Average AUC [95% CI] | 0.788 [0.785, 0.790] | 0.799 [0.797, 0.801] | 0.797 [0.795, 0.799] | 0.800 [0.798, 0.802] | 0.790 [0.788, 0.792] |
| AUC standard deviation [95% CI] | 0.012 [0.011, 0.014] | 0.009 [0.008, 0.010] | 0.009 [0.007, 0.011] | 0.009 [0.007, 0.010] | 0.010 [0.008, 0.011] |
The BANNs failed to converge for training datasets with 50 and 100 cases.
These ANNs had 20 hidden nodes and were trained to 1500 training iterations, whereas all other ANNs had 6 hidden nodes and were trained to 500 training iterations. The results were calculated at the 1485th and 485th training iteration, respectively.
In the second simulation study, the highly complex ANNs trained without regularization (1485 training iterations) had an average AUC value of 0.694 and standard deviation of 0.044. Therefore, these more complex ANNs exhibited greater overfitting. Training the ANNs with noise injection, weight decay, and early stopping increased the average AUC values to 0.758, 0.745, and 0.748, respectively. Training the ANNs with noise injection reduced the standard deviation only slightly, to 0.034, and training the ANNs with weight decay and early stopping did not change the standard deviation (0.048 and 0.043, respectively). Therefore, for the more complex ANNs weight decay and early stopping performed similarly, and noise injection performed best.
All ANNs trained on the larger datasets of 100 total cases improved in performance and reduced in the differences between the various training methods (Table 1). The differences in the average AUC values between all training methods were of the order of 0.02, which is small in terms of practical importance. However, all regularization methods increased the average and decreased the standard deviation in AUC values with nonoverlapping 95% CIs. Noise injection and weight decay increased the average AUC value to 0.785 and 0.784, respectively, whereas early stopping increased the average AUC value to 0.770, and they reduced the standard deviation to 0.017, 0.020, and 0.023, respectively. Therefore, noise injection and weight decay performed similarly and both outperformed early stopping.
Further increase in the number of training cases to 200 total cases reduced the differences in the average AUC values of ANNs trained with the various methods to the order of 0.005, which are of little practical value (Table 1). However, performance was still improved with noise injection, weight decay, and BANNs. The improvement in the average AUC value from noise injection was similar to that from weight decay and BANNs.
Percent recovery results
Table 2 summarizes the gain in AUC from regularization, the loss of AUC from overfitting, and the percent recovery (i.e., the ratio of gain to loss). For training datasets of 50 cases, the average loss in AUC due to overfitting in the first simulation study was 0.036 (Table 2), or approximately 80% of the standard deviation in AUC of 0.043 (Table 1). Noise injection provided an average gain of 0.032 and a percent recovery of 87%. Weight decay and early stopping provided smaller average gains of 0.019 and 0.017, respectively, and percent recoveries of 53% and 48%, respectively. Training the more complex ANNs on 50 total cases in the second simulation study, noise injection, weight decay, and early stopping produced percent recoveries of 90%, 71%, and 76%, respectively. Therefore, noise injection performed better than weight decay and early stopping.
Table 2.
Comparison of the relative gain in performance from regularization and loss due to overfitting of the ANN training methods in the simulation studies.
| No regularization | Noise injection | Weight decay | BANN | Early stopping | ||
|---|---|---|---|---|---|---|
| 50 training cases | Average gaina or lossb [95% CI] | −0.036 [−0.034, −0.039] | 0.032 [0.028, 0.035] | 0.019 [0.015, 0.023] | 0.017 [0.015, 0.020] | |
| Percent recovery [95% CI] | 87% [80%, 94%] | 53% [44%, 61%] | 48% [43%, 52%] | |||
| 50 training cases, complex ANNsc | Average gaina or lossb [95% CI] | −0.071 [−0.065, −0.077] | 0.064 [0.058, 0.070] | 0.051 [0.043, 0.058] | 0.054 [0.048, 0.061] | |
| Percent recovery [95% CI] | 90% [84%, 96%] | 71% [63%, 79%] | 76% [69%, 83%] | |||
| 100 training cases | Average gaina or lossb [95% CI] | −0.019 [−0.017, −0.021] | 0.023 [0.021, 0.025] | 0.021 [0.020, 0.023] | 0.008 [0.006, 0.009] | |
| Percent recovery [95% CI] | 121% [114%, 129%] | 114% [107%, 120%] | 40% [35%, 45%] | |||
| 200 training cases | Average gaina or lossb [95% CI] | −0.006 [−0.005, −0.008] | 0.011 [0.009, 0.013] | 0.009 [0.008, 0.011] | 0.012 [0.010, 0.014] | 0.002 [0.001, 0.003] |
| Percent recovery [95% CI] | 176% [148%, 204%] | 146% [126%, 166%] | 194% [165%, 223%] | 31% [15%, 47%] |
Gain=the difference in the AUC value between ANNs trained with regularization and the ANNs trained without regularization at the 485th training iteration (1485th training iteration for the more complex ANNs).
Loss=the difference between the maximum AUC value and the AUC value at the 485th training iteration (1485th training iteration for the more complex ANNs) for ANNs trained without regularization.
These ANNs had 20 hidden nodes and were trained to 1500 training iterations, whereas all other ANNs had 6 hidden nodes and were trained to 500 training iterations. The results were calculated at the 1485th and 485th training iteration, respectively.
As the number of training cases increased from 50 to 200 total cases, noise injection produced increasing percent recoveries of 87% (90% for the more complex ANNs), 121%, and 176%. Therefore, noise injection recovered essentially all of the loss in AUC from overfitting, and sometimes achieved better performance than possible without regularization. The percent recoveries from early stopping were essentially constant but smaller than those from noise injection: 48% (76% for the more complex ANNs), 40%, and 31%, respectively. The percent recoveries from weight decay increased with larger training datasets, from 53% (71% for the more complex ANNs) to 114% and to 146%, respectively. BANNs achieved a percent recovery of 194% on the training dataset with 200 cases.
Breast ultrasound results
Table 3 summarizes the breast ultrasound results. On datasets of 50 total cases, ANNs trained without regularization had an average AUC value of 0.801. Noise injection and weight decay increased the average AUC value to 0.849 and 0.838, respectively. The standard deviation of training the ANNs without regularization was 0.065, and it was reduced to 0.056 with noise injection and 0.058 with weight decay. On datasets of 100 total cases, the AUC values improved with all methods. ANNs trained without regularization attained an average AUC value of 0.807. Noise injection and weight decay increased the average AUC values to 0.856 and 0.851, respectively. The standard deviation of training ANNs without regularization was 0.047, and it was reduced to 0.039 and 0.040, respectively, with noise injection and weight decay. Therefore, both regularization methods increased the average AUC values and reduced standard deviations.
Table 3.
Comparison of the absolute performance of the ANN training methods in the breast ultrasound study.
| No regularizationa | Noise injection | Weight decay | ||
|---|---|---|---|---|
| 50 training cases | Average AUCb [95% CI] | 0.801 [0.795. 0.807] | 0.849 [0.844, 0.853] | 0.838 [0.833, 0.843] |
| AUC standard deviation [95% CI] | 0.065 [0.061, 0.069] | 0.056 [0.052, 0.060] | 0.058 [0.054, 0.062] | |
| 100 training cases | Average AUCb [95% CI] | 0.807 [0.803, 0.811] | 0.856 [0.853, 0.860] | 0.851 [0.847, 0.854] |
| AUC standard deviation [95% CI] | 0.047 [0.044, 0.050] | 0.039 [0.037, 0.042] | 0.040 [0.038, 0.043] |
These ANNs had five hidden nodes and were trained to 500 training iterations. The results were calculated at the 485th training iteration.
Performance measured by the .632+ bootstrap AUC values.
Automatic selection of the noise kernel standard deviation
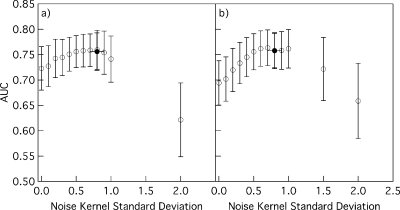
From 100 independent replications of the experiment, the average value of the noise kernel standard deviation σ selected by maximizing Eq. 2 for datasets of 50 training cases was 0.80 with a standard deviation of 0.10 (less than the standard deviation of each feature in our XOR population, which was 1.4). The ANNs trained with noise injection and the automatically selected σ values achieved an average AUC value of 0.756 with a standard deviation of 0.037. In comparison, Fig. 3 shows the results of manually incrementing σ from 0 to 2.0. The average AUC value without regularization (i.e., a σ value of 0) was 0.723 and 0.694, respectively, for the typical and more complex ANNs, both with a standard deviation of approximately 0.043. As the σ value increased, the AUC values of the ANNs of typical complexity increased, reaching a plateau in the range of σ=0.5–0.8 with an average AUC value of approximately 0.76 and standard deviation of 0.03, and then decreased. The more complex ANNs showed a similar trend and attained similar AUC values in the range of σ=0.6–1.0. Therefore, the σ values selected by the automatic selection method were within a wide range of near optimal values.
Figure 3.
Average ANN performance measured on the independent validation dataset when the ANNs were trained with noise injection of various noise kernel standard deviation values. Empty circles represent the average AUC values and error bars represent one standard deviation. Filled circles represent the average AUC values of ANNs trained with noise standard deviation values estimated by maximizing Eq. 2, the vertical error bars represent one standard deviation in the AUC values, and the horizontal error bars represent one standard deviation in the selected noise kernel standard deviation values. The ANNs in (a) had 6 hidden nodes and their performance was measured at the 485th training iteration; the ANNs in (b) had 20 hidden nodes and their performance was measured at the 1485th training iteration.
DISCUSSION
In this study we compared the effect of noise injection, weight decay, and early stopping on the overfitting problem of ANNs. We showed that training ANNs with noise injection reduces overfitting and produces greater AUC values with smaller standard deviations compared with training ANNs without regularization. Weight decay performed similarly: ANNs trained with weight decay attained greater AUC values with smaller standard deviations. Early stopping also increased the AUC values and reduced the standard deviation, but to a lesser extent than the other two methods. The complexity of the ANN model did not appear to affect the absolute performance from the three methods. We also found that the algorithm14 for selecting the noise kernel standard deviation value σ was effective in reducing overfitting. The automatically selected σ values, with an average of 0.80, was within the optimal range of 0.5–0.8 for the ANNs of typical complexity and 0.6–1.0 for the more complex ANNs and produced near optimal AUC values. Our results agree qualitatively with other studies.5, 11, 14 However, direct comparison is difficult because other studies used the classification error rate as the measure of classification performance, whereas we used the more general and more meaningful AUC values as the measure of classification performance.
Our results show both statistical significance and potentially practical significance. The practical significance of an increase in the AUC value of 0.02 or 0.04 depends on the context of the classification problem. We have found, from a survey of the medical literature, that reported improvements in the AUC value from new diagnostic technologies are rarely greater than 0.1, of which 0.04 is 40%.33 In this context, the improvement is not negligible. Further, our study shows that the improvement in the AUC value from noise injection is comparable to, or better than, that from the methods of weight decay (or Bayesian ANN) and early stopping. Early stopping and Bayesian ANN are methods known to be effective for reducing overtraining. Therefore, our results support the conclusion that noise injection is at least as effective as these known methods for reducing overtraining. Finally, our results are statistically significant because the standard deviations that we report (Tables 1, 3) include variation in ANN performance due to different training datasets, whereas the improvement in AUC represents an average improvement due to the different training methods. In other words, the standard deviation values in the AUC are not a measure of variations due to only each training method, and we should not judge the magnitude of the average improvement in terms of that variability. Rather, we conducted our statistical analysis based on the 95% CIs, which are calculated from the standard errors, not the standard deviations.
A potential advantage of noise injection that we did not study is the possibility of incorporating prior information of the training data into the ANN training process. For example, if the values of a particular feature were smaller than the values of other features, then one could assign a smaller noise kernel standard deviation value for this particular feature than those of other features. From a Bayesian viewpoint, the noise kernels can be interpreted as “prior information” of the data. BANNs apply a specific constraint on ANN weights via prior distributions but do not have a mechanism to do so on the training data. Wright et al.17, 18 interpreted noise injection from a Bayesian viewpoint, but more research is needed. In this study we assumed no specific prior knowledge of the training data.
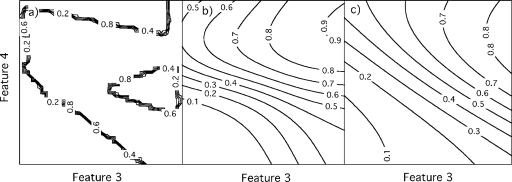
Figure 4 illustrates differences in the classification decision boundaries between ANNs trained with no regularization, with noise injection, and with weight decay in terms of two features from the US study. ANNs trained without regularization had decision boundaries with sharp corners, local extrema, and strong gradients (i.e., decision boundaries that are close to each other). Training ANNs with noise injection removed local extrema, reduced the number and curvature of corners, and reduced the gradients of the decision boundaries. Training ANNs with weight decay produced decision boundaries that were similar to those attained with noise injection, but generally with smaller gradients. These differences partially explain the small performance differences between ANNs trained with noise injection and those trained with weight decay, and the larger performance differences between ANNs trained with noise injection and those trained with no regularization.
Figure 4.
Contour plots showing classification decision boundaries for (a) ANNs trained without regularization (i.e., overfitting), (b) ANNs trained with noise injection, and (c) ANNs trained with weight decay. The ANNs were trained on a dataset of 50 cases drawn from the breast US dataset, and are shown for fixed values of features 1 and 2.
In this study we were unable to use BANNs on small datasets because the BANNs failed to estimate the α value automatically. While this problem has been addressed in the literature,9 the proposed BANN model is more complicated, more computationally costly, and less frequently used than the BANN model that we used in this study.8
In conclusion, we have shown that training ANNs with noise injection using zero-mean Gaussian noise kernels and automatically selected standard deviation values can reduce overfitting. Our simulation studies and the breast US study showed that training ANNs with noise injection outperformed training ANNs with early stopping and produced results comparable to or sometimes better than weight decay and BANNs. Furthermore, noise injection reduces overfitting with a different mechanism than weight decay and BANNs and noise injection can be used as an alternative to BANNs for training ANNs. The possibility of using noise injection in situations where specific prior information about the training data can be incorporated into the injected noise is appealing and warrants further study.
ACKNOWLEDGMENTS
This work was supported in part by the National Cancer Institute of the National Institutes of Health through Grant Nos. R01 CA92361, R21 CA93989, S10 RR021039, and P30 CA14599 and by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health through Grant No. R01 EB000863 (Kevin S. Berbaum, PI). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of any of the supporting organizations. The authors thank Charles Metz, Ph.D., and Patrick LaRiviere, Ph.D., for their help and suggestions.
References
- Wu Y., Doi K., Metz C. E., Asada N., and Giger M. L., “Simulation studies of data classification by artificial neural networks: Potential applications in medical imaging and decision making,” J. Digit Imaging 6, 117–125 (1993). [DOI] [PubMed] [Google Scholar]
- Jiang Y. et al. , “Malignant and benign clustered microcalcifications: Automated feature analysis and classification,” Radiology 198, 671–678 (1996). [DOI] [PubMed] [Google Scholar]
- Bishop C. M., Neural Networks for Pattern Recognition (Oxford University Press, New York, 1995). [Google Scholar]
- Kupinski M. A., Edwards D. C., Giger M. L., and Metz C. E., “Ideal observer approximation using Bayesian classification neural networks,” IEEE Trans. Med. Imaging 20, 886–899 (2001). 10.1109/42.952727 [DOI] [PubMed] [Google Scholar]
- Sietsma J. and Dow R. J. F., “Creating artificial neural networks that generalize,” Neural Networks 4, 67–79 (1991). 10.1016/0893-6080(91)90033-2 [DOI] [Google Scholar]
- Geman S., Bienenstock E., and Doursat R., “Neural networks and the bias∕variance dilemma,” Neural Comput. 4, 1–58 (1992). 10.1162/neco.1992.4.1.1 [DOI] [Google Scholar]
- Hastie T., Tibshirani R., and Friedman J. H., The Elements of Statistical Learning: Data Mining, Inference, and Prediction (Springer, New York, 2001). [Google Scholar]
- MacKay D. J. C., Bayesian Methods for Adaptive Models (California Institute of Technology, Pasadena, 1992). [Google Scholar]
- Neal R. M., Bayesian Learning for Neural Networks (Springer, New York, 1996). [Google Scholar]
- Sarle W. S., Neural Network FAQ. Retrieved September 9, 2008. (website: ftp://ftp.sas.com/pub/neural/FAQ.html).
- An G. Z., “The effects of adding noise during backpropagation training on a generalization performance,” Neural Comput. 8, 643–674 (1996). 10.1162/neco.1996.8.3.643 [DOI] [Google Scholar]
- Bishop C. M., “Training with noise is equivalent to Tikhonov regularization,” Neural Comput. 7, 108–116 (1995). 10.1162/neco.1995.7.1.108 [DOI] [Google Scholar]
- Grandvalet Y., Canu S., and Boucheron S., “Noise injection: theoretical prospects,” Neural Comput. 9, 1093–1108 (1997). 10.1162/neco.1997.9.5.1093 [DOI] [Google Scholar]
- Holmström L. and Koistinen P., “Using additive noise in back-propagation training,” IEEE Trans. Neural Netw. 3, 24–38 (1992). 10.1109/72.105415 [DOI] [PubMed] [Google Scholar]
- Matsuoka K., “Noise injection into inputs in backpropagation learning,” IEEE Trans. Syst. Man Cybern. 22, 436–440 (1992). 10.1109/21.155944 [DOI] [Google Scholar]
- Raviv Y. and Intrator N., in Combining Artificial Neural Nets: Ensemble and Modular Multi-net Systems, edited by Sharkey A. J. C. (Springer, New York, 1999), p. 298. [Google Scholar]
- Wright W. A., “Bayesian approach to neural-network modeling with input uncertainty,” IEEE Trans. Neural Netw. 10, 1261–1270 (1999). 10.1109/72.809073 [DOI] [PubMed] [Google Scholar]
- Wright W. A., Ramage G., Cornford D., and Nabney I., “Neural network modelling with input uncertainty: Theory and application,” J. VLSI Sig. Proc. Syst. 26, 169–188 (2000). 10.1023/A:1008111920791 [DOI] [Google Scholar]
- Zur R. M., Jiang Y., and Metz C. E., “Comparison of two methods of adding jitter to artificial neural network training,” Proceedings of CARS (Elsevier, Chicago, 2004), pp. 886–889.
- Pepe M. S., The Statistical Evaluation of Medical Tests for Classification and Prediction (Oxford University Press, New York, 2003). [Google Scholar]
- Metz C. E., “ROC methodology in radiologic imaging,” Invest. Radiol. 21, 720–733 (1986). 10.1097/00004424-198609000-00009 [DOI] [PubMed] [Google Scholar]
- Wagner R. F., Metz C. E., and Campbell G., “Assessment of medical imaging systems and computer aids: A tutorial review,” Acad. Radiol. 14, 723–748 (2007). 10.1016/j.acra.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Egan J. P., Signal Detection Theory and ROC-Analysis (Academic, New York, 1975). [Google Scholar]
- Metz C. E. and Pan X., “’Proper binormal ROC curves: Theory and maximum-likelihood estimation,” J. Math. Psychol. 43, 1–33 (1999). 10.1006/jmps.1998.1218 [DOI] [PubMed] [Google Scholar]
- Drukker K., Gruszauskas N. P., Sennett C. A., and Giger M. L., “Breast US computer-aided diagnosis workstation: Performance with a large clinical diagnostic population,” Radiology 248, 392–397 (2008). 10.1148/radiol.2482071778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabney I., NETLAB: Algorithms for Pattern Recognitions (Springer, New York, 2002). [Google Scholar]
- Jiang Y., “Uncertainty in the output of artificial neural networks,” IEEE Trans. Med. Imaging 22, 913–921 (2003). 10.1109/TMI.2003.815061 [DOI] [PubMed] [Google Scholar]
- Lampinen J. and Vehtari A., “Bayesian approach for neural networks-review and case studies,” Neural Networks 14, 257–274 (2001). 10.1016/S0893-6080(00)00098-8 [DOI] [PubMed] [Google Scholar]
- Sahiner B., Chan H. P., and Hadjiiski L., “Classifier performance estimation under the constraint of a finite sample size: resampling schemes applied to neural network classifiers,” Proceedings of IJCNN (IEEE, Orlando, 2007), pp. 1762–1766. [DOI] [PMC free article] [PubMed]
- Yousef W. A., Wagner R. F., and Loew M. H., “Comparison of non-parametric methods for assessing classifier performance in terms of ROC parameters,” Proceedings of the 33rd AIPR Workshop (IEEE, Washington, 2004), pp. 190–195.
- Sarle W. S., “Stopped training and other remedies for overfitting,” Proceedings of the 27th Symposium on the Interface: Computer Science and Statistics (Interface Foundation of North America, Pittsburgh, 1995), pp. 352–360.
- Chan H. P., Sahiner B., Wagner R. F., and Petrick N., “Classifier design for computer-aided diagnosis: Effects of finite sample size on the mean performance of classical and neural network classifiers,” Med. Phys. 26, 2654–2668 (1999). 10.1118/1.598805 [DOI] [PubMed] [Google Scholar]
- Shiraishi J., Pesce L., Metz C. E., and Doi K., “On experimental design and data analysis in receiver operating characteristic (ROC) studies: Lessons learned from papers published in Radiology from 1997 to 2006,” Radiology (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]