Abstract
Recent work has shown that primary cilia are essential for Hh signaling during mammalian development1–9. It is also known that aberrant Hedgehog (Hh) signaling can lead to cancer10, but the role of primary cilia in oncogenesis is not known. Cerebellar granule neuron precursors (GNPs) can give rise to medulloblastomas, the most common malignant brain tumor in children11,12. The primary cilium and Hh signaling are required for GNPs proliferation8,13–16. We asked whether primary cilia in GNPs play a role in medulloblastoma growth in mice. Genetic ablation of primary cilia blocked medulloblastoma growth when this tumor was driven by a constitutively active Smoothened (Smo), an upstream activator of Hh signaling. In contrast, removal of cilia was required for medulloblastoma growth by a constitutively active Gli2, a downstream transcription factor. Thus, primary cilia are required for, or inhibit medulloblastoma formation, depending on the initiating oncogenic event. Remarkably, presence or absence of cilia were associated with specific variants of human medulloblastomas; primary cilia were found in medulloblastomas with activation in HH or WNT signaling, but not in most medulloblastomas in other distinct molecular subgroups. Primary cilia could serve as a diagnostic tool and provide new insights into the mechanism of tumorigenesis.
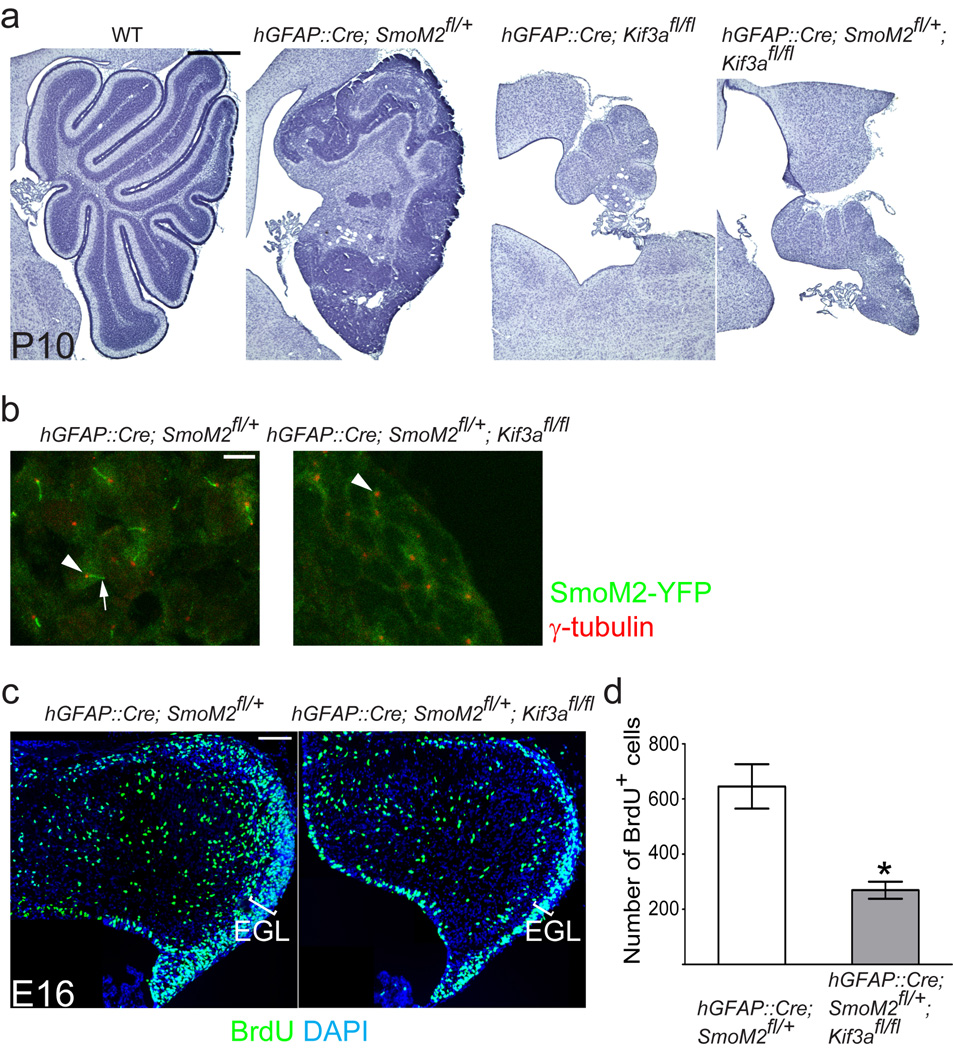
Abnormal activation of Hh signaling, through loss of Hh receptor, Patched1 (Ptch1), or activation of Smo induces medulloblastomas in mice17–22. To induce medulloblastoma we expressed constitutively active Smo (SmoM2) in GNPs using a human Glial Fibrillary Acidic Protein promoter-driven Cre (hGFAP::Cre)21. By postnatal day (P) 10, hGFAP::Cre; SmoM2fl/+ mice developed medulloblastoma (n = 7) (Fig. 1a). In these tumors, SmoM2 fused with yellow fluorescent protein localized to primary cilia (Fig. 1b). To investigate whether SmoM2-driven medulloblastoma formation requires primary cilia, we removed primary cilia from GNPs expressing SmoM2, using a conditional allele of Kif3a that encodes a subunit of the kinesin-II motor essential for ciliogenesis23–25. The removal of Kif3a and the consequent loss of cilia completely blocked tumorigenesis (n = 7). The cerebellum of the hGFAP::Cre; SmoM2fl/+; Kif3afl/fl mice resembled that of hGFAP::Cre; Kif3afl/fl mice (Fig. 1a), which fail to expand GNPs8,13. Loss of Ift88, another gene essential for ciliogenesis26,27 in hGFAP::Cre; SmoM2fl/+; Ift88fl/fl mice also blocked tumorigenesis driven by SmoM2 (Supplementary Fig. 1a).
Fig. 1. Kif3a is required for SmoM2-driven medulloblastoma formation.
(a) Hematoxylin stained sagittal sections of control and mutant cerebella. Expression of SmoM2 in GNPs using hGFAP::Cre induces medulloblastoma by P10. Removal of Kif3a, an essential gene for ciliogenesis, in SmoM2 expressing cells completely blocks tumorigenesis resulting in atrophic cerebella similar to those of hGFAP::Cre;Kif3afl/fl mice. (b) SmoM2-YFP is highly enriched in primary cilia (green, arrow) associated with the basal body (anti-γ-tubulin staining, shown in red, arrowhead). In hGFAP::Cre; SmoM2fl/+; Kif3afl/fl mice only the basal body is present (arrowhead). (c, d) BrdU incorporation (1h survival) at E16. GNPs proliferation in hGFAP::Cre; SmoM2fl/+; Kif3afl/fl is similar to that observed in hGFAP::Cre; Kif3afl/fl or wild type mice8. In contrast, already by E16, hGFAP::Cre; SmoM2fl/+ mice show an expanded EGL containing significantly more proliferating cells. *: P < 0.05. Scale bar = 0.5 mm (a), 5 µm (b) and 100 µm (c).
At E16, when the number of GNPs is not affected by removing cilia8, EGL was already expanded with many proliferating cells in hGFAP::Cre; SmoM2fl/+ mice but not in hGFAP::Cre; SmoM2fl/+; Kif3afl/fl mice (Fig. 1c,d), suggesting that SmoM2 requires Kif3a to initiate the aberrant GNP expansion and medulloblastoma. Similar requirement of cilia for SmoM2-driven expansion of GNPs was observed in the hippocampal dentate gyrus4.
Activated Smo converts Gli2 into a transcriptional activator and inhibits the formation of Gli3 repressors that form constitutively in the absence of Hh signaling28. We hypothesized that constitutively active Gli2 could induce medulloblastomas in the absence of primary cilia. To test this hypothesis we used CLEG2 mice that upon Cre-mediated recombination express a constitutively active Gli2 that lacks the N-terminal repressor domain (Gli2ΔN)29,30. Unexpectedly, none of hGFAP::Cre; CLEG2fl/+ mice (n = 14) developed medulloblastoma (Fig. 2) albeit two had a different type of tumors (see below). Surprisingly, unlike hGFAP::Cre; CLEG2fl/+ mice, removal of primary cilia in hGFAP::Cre; CLEG2fl/+; Kif3afl/fl mice induced medulloblastomas between P11 and P30 (n = 11) (Fig. 2). Tumors in hGFAP::Cre; CLEG2fl/+; Kif3afl/fl mice contained two types of cells frequently segregated into distinct zones: cells with darkly stained nuclei and lightly stained cytoplasm (type 1), and cells with large nuclei and highly eosinophilic cytoplasm (type 2) (Fig. 2b,c). Both cell types were actively proliferating (Fig. 2f) and expressed Gli1, suggesting active Hh signaling (Supplementary Fig. 2). The type 1 tumor cells were indistinguishable from those in SmoM2-driven medulloblastoma18,19,21 and reminiscent of classic medulloblastoma cells: small round cells with high nuclear-to-cytoplasmic ratio. Type 1 cells exhibited immunohistological characteristics of medulloblastoma similar to those in SmoM2 tumor: neuronal (Tuj1), glial (GFAP and Olig2), GNP (Pax6), and granule neuron (Zic1) markers (Supplementary Fig. 3). hGFAP::Cre; CLEG2fl/+; Kif3afl/fl and hGFAP::Cre; SmoM2fl/+ mice also had very similar gene expression profiles (Fig. 2h): up-regulation of Hh-responsive genes characteristic of medulloblastoma cells (Gli1, Gli2, Ptch1, Ptch2, Mycn, Ccnd2, Atoh1, and Bmi1). The expression of Gli3 and Wif1 were different between the two tumors likely due to the presence of type 2 tumor cells in hGFAP::Cre; CLEG2fl/+; Kif3afl/fl mice; type 2 tumor cells down-regulated Gli3 and up-regulated Wif1 (Fig. 2h and see below). Type 2 tumors lacked immunohistological characteristics of medulloblastoma markers (Supplementary Fig. 3), but expressed Sox2 (data not shown), suggesting that these are not medulloblastomas. Type 2 tumors were also found outside of cerebellum, suggesting that these cells have a different origin (Supplementary Fig. 4a). Both tumor types lacked primary cilia (Supplementary Fig. 5). Similar to loss of Kif3a, loss of Ift88 promoted tumor formation containing both type 1 and 2 tumor cells in hGFAP::Cre; CLEG2fl/+; Ift88fl/fl mice (Supplementary Fig. 1b).
Fig. 2. Kif3a suppresses Gli2ΔN-driven medulloblastoma formation.
(a–c) Hematoxylin and eosin (H&E) stained sagittal sections of control and mutant cerebella at P23. (a) hGFAP::Cre; CLEG2fl/+ mice have ectopic clusters of cells (arrows) without tumor formation. (b) All hGFAP::Cre; CLEG2fl/+; Kif3afl/fl mice develop medulloblastomas between P11 and P30. A green dotted line in b demarcates domains of the tumors containing type 1 and type 2 tumor cells (see text). (c) High magnification of red box in b, showing the two domains with different cell types. (d) Western blot analysis of Gli3 proteins isolated from P23 cerebella shows the decrease of Gli3 repressor (Gli3R) in hGFAP::Cre; CLEG2fl/+; Kif3afl/fl mice compared with wild-type and hGFAP::Cre; CLEG2fl/+ mice. Asterisk indicates a nonspecific band, which serves as a loading control. This banding pattern is similar to a previous report, which showed that this Gli3 antibody detects specific Gli3 bands that are absent in Gli3 mutants and non-specific bands38. (e,f) BrdU-labeling (1h survival) reveals that the ectopic clusters in hGFAP::Cre; CLEG2fl/+ mice are not proliferating (arrows), while tumors in hGFAP::Cre; CLEG2fl/+; Kif3afl/fl mice contained many proliferating cells. (g) hGFAP::Cre; CLEG2fl/+; Kif3afl/fl mice (P < 0.001) and hGFAP::Cre; CLEG2fl/+; Gli3Xt/+ mice (P < 0.02) die significantly earlier than hGFAP::Cre; CLEG2fl/+ mice. (h) Hh-responsive genes are up-regulated in both SmoM2- and Gli2ΔN-driven tumors (see text). *: P < 0.05. Scale bar = 0.5 mm (a,b,e,f), 10 µm (c).
hGFAP::Cre; CLEG2fl/+ mice developed only type 2 tumors later in life that were found throughout central nervous system (Supplementary Fig. 4b) and lived significantly longer than hGFAP::Cre; CLEG2fl/+; Kif3afl/fl mice (P < 0.0001, n = 19) (Fig. 2g). These tumors expressed high levels of Hh-responsive genes (Gli1, Gli2, Ptch1, Ptch2, Mycn, and Ccnd2) and Wnt-responsive genes (Axin2 and Wif1), but not genes associated with GNPs and medulloblastoma (Atoh120 and Bmi131) (Fig. 2h). Type 2 tumors in hGFAP::Cre; CLEG2fl/+ mice had primary cilia, excluding the possibility that they may have originated from cells that spontaneously lost primary cilia (Supplementary Fig. 5). Thus, loss of cilia is required for medulloblastoma development driven by Gli2ΔN, but not for type 2 tumors; yet, loss of cilia accelerated the development of the type 2 tumor (Fig. 2a–c,g).
The above results suggest that loss of primary cilia is required for Gli2ΔN-driven medulloblastoma development, which is in striking contrast to their requirement for SmoM2-driven tumorigenesis. One possible explanation for these opposite roles is that primary cilia are required not only for Smo function but also for Gli3 repressor formation2,6,7,9. Consistently, Gli3 repressor was dramatically reduced in hGFAP::Cre; CLEG2fl/+; Kif3afl/fl mice compared with wild-type or hGFAP::Cre; CLEG2fl/+ mice (Fig. 2d). Gli3 expression was also significantly decreased in type 2 tumors in hGFAP::Cre; CLEG2fl/+ mice (Fig. 2h), suggesting Gli3 could also have an inhibitory role in these tumors. Since Gli3 null mutants are embryonic lethal, we removed one copy of Gli3 in mice expressing Gli2ΔN (hGFAP::Cre; CLEG2fl/+; Gli3Xt/+) to test if Gli3 has an inhibitory effect on Gli2ΔN-driven tumorigenesis. hGFAP::Cre; CLEG2fl/+; Gli3Xt/+ mice developed type 2 tumors and died earlier in life than hGFAP::Cre; CLEG2fl/+ mice (P < 0.02, n = 18) (Fig. 2g and Supplementary Fig. 6). Thus, loss of one copy of Gli3 induces earlier development of the type 2 tumors in the presence of primary cilia, supporting our inference that Gli3 repressors may mediate the inhibitory function of primary cilia in tumorigenesis driven by Gli2ΔN. Interestingly, hGFAP::Cre; CLEG2fl/+; Gli3Xt/+ mice did not develop medulloblastoma, possibly due to the presence of one copy of Gli3. Additionally, loss of primary cilia could interfere with other signaling pathways that contribute to medulloblastoma development.
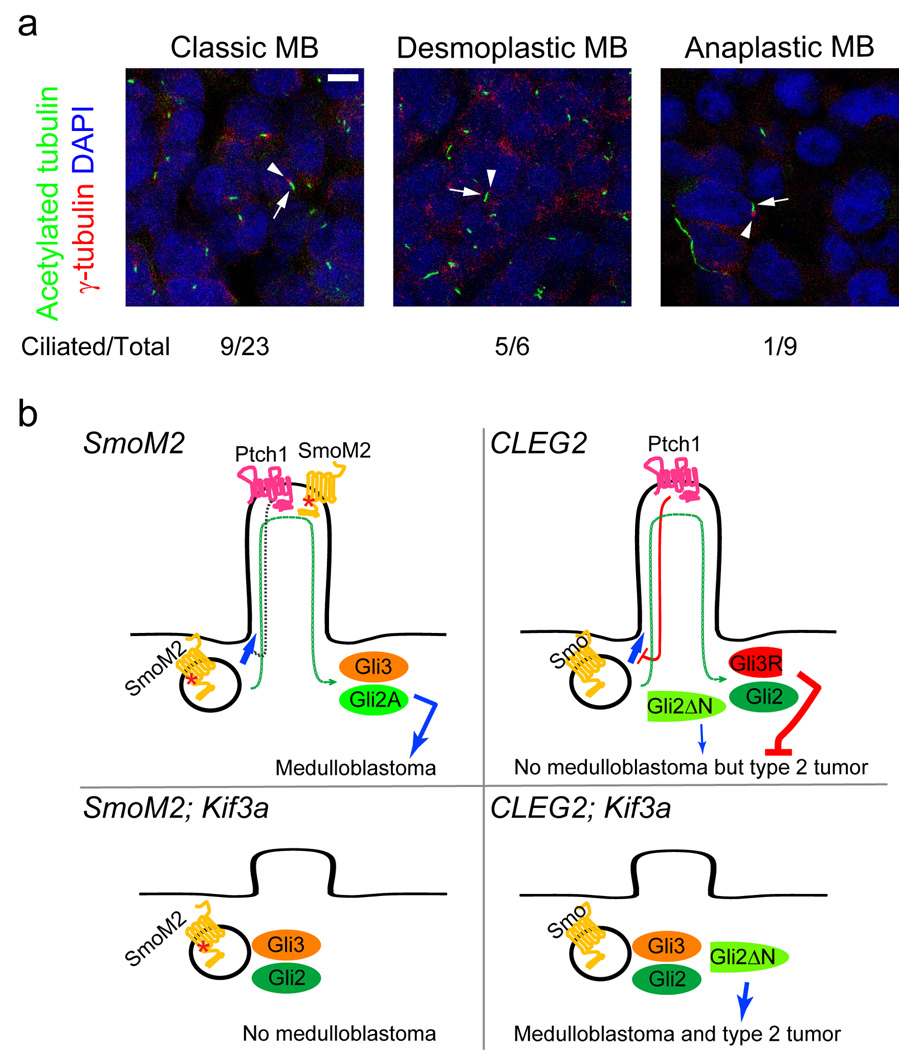
While SMOM2 mutations have been observed in one case of human medulloblastoma32, activating mutations in GLI transcription factors have not been described in medulloblastoma. Thus, our mouse models reveal important roles of primary cilia in tumorigenesis, but might not reflect on the behavior of this organelle in human cases. To determine if our observations in mice are relevant to human tumors, we analyzed 38 human medulloblastomas for the presence of cilia. Presence of primary cilia was significantly associated with desmoplastic medulloblastoma (P < 0.02); whereas the absence of cilia was significantly associated with anaplastic medulloblastoma (P < 0.05) (Fig. 3a and Table 1). Twenty-four of these samples were analyzed previously for gene expression profiling33. Remarkably, in these 24 samples, cilia were identified almost exclusively in tumors that have activation in either HH or WNT signaling (P < 0.0001, Table 1). Two of the four tumors with active HH signaling had inactivating mutations in PTCH133, which functions upstream of SMO and primary cilia5.
Fig. 3. Primary cilia are present in a subset of human medulloblastomas.
(a) Nine of twenty-three classic and five of six desmoplastic medulloblastomas have primary cilia in most cells. Eight of nine anaplastic medulloblastomas have few or no ciliated cells (shown is an example of non-ciliated anaplastic medulloblastoma). Cilia and basal bodies are stained by anti-acetylated tubulin (green, arrows) and anti-γ-tubulin (red, arrowheads), respectively. Scale bar = 5 µm. (b) Model summarizing results of dual roles of primary cilia in SmoM2- and Gli2ΔN-driven tumorigenesis. SmoM2 is insensitive to inhibition by Ptch1 and constitutively localizes to the cilia, where it activates Gli2 and inhibits Gli3 repressor (Gli3R) formation, leading to medulloblastoma (top left). Without the cilia, SmoM2 cannot activate downstream pathways; cerebellum remains small as in ciliary mutants8 and no tumors develop (bottom left). Gli2ΔN is constitutively active and independent of primary cilia, yet these mice do not develop medulloblastoma possibly due to thepresence of Gli3 repressor; however, these mice develop type 2 tumors later in life (top right). In the absence of cilia, Gli2ΔN induces medulloblastoma and type 2 tumors earlier in life. This may be due to the elimination of Gli3 repressor when cilia are removed (bottom right).
Table 1.
Primary cilia are present almost exclusively in medulloblastomas with either HH or WNT signaling activation.
Samples that were hard to be determined for the presence of cilia due to suboptimal staining are labeled as equiv.
Our results suggest that primary cilia can be permissive or inhibitory for tumor formation depending on the underlying oncogenic events (Fig. 3b). Interestingly, we found that subsets of human medulloblastomas have primary cilia, while others do not. Our mouse models predict both presence and absence of primary cilia in HH signaling-driven medulloblastomas, but all the observed medulloblastomas with high HH signaling activity had primary cilia. This suggests that medulloblastomas with high HH signaling in humans are largely due to mutations upstream of the primary cilia such as mutations in PTCH1. Oncogenic mutations in downstream molecules including GLI transcription factors, which would require loss of repressor activity mediated by the cilia, are rare11. A recent study identified rare amplifications of GLI1 and GLI2 genes in medulloblastomas34, but it is not known if these tumors are ciliated. Analyses of more medulloblastomas for both oncogenic mutations and presence/absence of primary cilia will be necessary to delineate the role of cilia in human medulloblastoma. Interestingly, all the observed human medulloblastomas with high WNT signaling had primary cilia. All these tumors had mutations in CTNNB1. It is possible that primary cilia are also involved in WNT signaling pathway35–37 or that oncogenic WNT activation requires concomitant signaling through primary cilia to induce medulloblastoma.
Our observations suggest that the presence of primary cilia be important for tumor diagnosis and treatment. HH signaling mutations found in medulloblastomas, loss of PTCH1 and SMO activation, act upstream of primary cilia2,4–9. Ciliated medulloblastomas showing HH signature would be susceptible to treatments that target primary cilia. The loss of cilia in subgroups of medulloblastomas suggests that tumor repressor functions mediated through primary cilia could serve as therapeutic targets to inhibit tumor growth. GLI3 is a strong candidate, but primary cilia could have multiple repressor activities. Understanding the role of primary cilia in normal and oncogenic signaling could lead to better understanding of tumor development and treatment not only in medulloblastomas, but also in other tumors (see Wong et al, this issue).
METHODS
Mice
All animal procedures were approved by the UCSF Institutional Animal Care and Use Committee. Genetically modified mice used in the study were described previously: hGFAP::Cre39, Kif3afl/fl 24, SmoM2-YFPfl/fl 19,Ift8826, CLEG229.
Tissue preparation and staining
Mice were injected with BrdU (50 mg kg−1) and perfused one hour later with 4% paraformaldehyde. The brains were postfixed overnight at 4°C. For frozen sections, brains were cryoprotected in 30% sucrose, embedded in OCT, and cut at 10 µm. For polyethylene glycol (PEG) sections, brains were dehydrated in an ethanol series, embedded in a 2:1 mixture of PEG 1000 and PEG 1500, and cut at 8 µm. For immunostaining, sections were incubated with primary antibodies overnight at 4°C followed by incubation with secondary antibodies at room temperature for two hours. Images were taken with an Olympus AX70 microscope, Leica SP2 or SP5 confocal microscopes, and processed using Adobe Photoshop.
Antibodies and reagents
The following primary antibodies were used: rat anti-BrdU (1:1000 Abcam), mouse anti-acetylated tubulin (1:1000 Sigma), anti-GFAP (1:1000 Chemicon), anti-Tuj1 (1:500 Covance), rabbit anti-γ-tubulin (1:1000 Sigma), anti-Zic (recognizes Zic1 and other Zic proteins, gift from Dr. R. Segal, Boston, 1:3000), anti-Olig2 (DF308, gift from Dr. C. Stiles, Boston, 1:20,000) anti-pericentrin (1:1000 Abcam), Goat anti-Pax6 (1:50 Santa Cruz) and chicken anti-GFP (1:500 Aves Labs). SmoM2-YFP was visualized by Anti-GFP antibody. Alexa Fluor-conjugated secondary antibodies (1:400 Molecular Probes) were used and DAPI (200 ng ml−1) was used for counter staining. Standard hematoxylin and eosin staining was used.
Gli1 in situ hybridization
Standard in situ protocol and antisense riboprobe from Gli1 cDNA (gift from A. Ruiz i Altaba) were used.
Western blot
Whole cerebella of P23 mice were lysed in RIPA buffer. Pellets were removed after centrifugation. Lysates were resolved in 7% SDS-PAGE gel. Rabbit anti-Gli3 antibody (1:500 Santa Cruz biotechnology, H280) was used for immunoblotting. This Gli3 antibody detects specific Gli3 bands that are absent in Gli3 mutants and non-specific bands38.
qRT-PCR
Total RNA was isolated from each tumor of an individual mouse (n = 3 per group) using RNeasy Mini Kit (Qiagen) and was reverse transcribed by Superscript III reverse transcriptase (Invitrogen). Quantitative PCR was performed on an Applied Biosystems 7900 quantitative PCR instrument using SYBR green. Transcript levels were normalized to the expression levels of Gapdh and measured relative to those in wild-type cerebellum. Primer sequences are listed in supplementary table 1.
Human medulloblastoma samples
Sections were prepared from formalin-fixed paraffin-embedded medulloblastomas archived in the Neurological Surgery Tissue Bank at UCSF or Neuropathology Laboratory at St. Jude Children’s Research Hospital. The St. Jude tumors were from a series reported previously33 and processed into tissue microarrays. Sections were cut at 10 µm (UCSF) or 6 µm (St. Jude) and were incubated in 0.01 M citrate buffer (pH 6.0) at 95°C for 30 minutes for antigen retrieval before immunostaining. The analysis to detect primary cilia was done blind as to the diagnosis or gene expression signature of the tumor. We used antibody to acetylated tubulin to detect primary cilia and antibody to γ-tubulin or pericentrin to detect the basal body.
Statistical analysis
We used Student’s t-test to compare BrdU+ cells in the embryonic EGL, Log-rank test to compare survival curves, Pair Wise Fixed Relocation Randomization test in Relative Expression Software Tool (REST©) to compare qRT-PCR results40, and Chi-square test to test the association of medulloblastoma subgroups and presence/absence of primary cilia.
Supplementary Material
ACKNOWLEDGEMENTS
We thank L.S. Goldstein at University of California San Diego for providing us with Kif3afl/fl mice; D. Rowitch at University of California San Francisco for SmoM2fl/+ mice; B. Yoder at University of Alabama Birmingham for Ift88fl/fl mice; C. Cowdrey and Neurological Surgery Tissue Bank at UCSF for human medulloblastoma samples; R. Segal and C. Stiles at Harvard University for anti-Zic and anti-Olig2 antibodies. We thank S. Wong, J. Reiter, D. Cano and S. Cervantes-Roldan for sharing unpublished data. We thank S. Vandenberg for helping with assessing the tumor types; J. Morris, K. Blaschke and M. Sachs for helping with qRT-PCR; R. Romero for technical assistance; D. Rowitch, J. Reiter, S Wong, R. Ihrie, S. Nader and T. Nguyen for comments on the manuscript. Y.-G. H. was, in part, supported by Mark Linder/American Brain Tumor Association Fellowship. The work was supported by grants from US National Institute of Health (NS28478 and HD32116) and a grant from the Goldhirsh foundation to A. A–B. Confocal microscopy at DERC Microscopy and Imaging Core was supported by an US National Institute of Health grant P30 DK063720.
REFERENCES
- 1.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 2.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science (New York, N.Y. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 4.Han YG, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 5.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 6.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development (Cambridge, England) 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 7.May SR, et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Developmental biology. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 8.Spassky N, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Developmental biology. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes & development. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 11.Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annual review of pathology. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- 12.Wechsler-Reya R, Scott MP. The developmental biology of brain tumors. Annu Rev Neurosci. 2001;24:385–428. doi: 10.1146/annurev.neuro.24.1.385. [DOI] [PubMed] [Google Scholar]
- 13.Chizhikov VV, et al. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. J Neurosci. 2007;27:9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development (Cambridge, England) 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 15.Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 16.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 17.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science (New York, N.Y. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 18.Hallahan AR, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer research. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 19.Mao J, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer research. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliver TG, et al. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development (Cambridge, England) 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- 21.Schuller U, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang ZJ, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 24.Marszalek JR, et al. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 25.Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haycraft CJ, et al. Intraflagellar transport is essential for endochondral bone formation. Development (Cambridge, England) 2007;134:307–316. doi: 10.1242/dev.02732. [DOI] [PubMed] [Google Scholar]
- 27.Murcia NS, et al. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development (Cambridge, England) 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- 28.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development (Cambridge, England) 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 29.Pasca di Magliano M, et al. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes & development. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roessler E, et al. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet. 2005;14:2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- 31.Leung C, et al. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- 32.Lam CW, et al. A frequent activated smoothened mutation in sporadic basal cell carcinomas. Oncogene. 1999;18:833–836. doi: 10.1038/sj.onc.1202360. [DOI] [PubMed] [Google Scholar]
- 33.Thompson MC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 34.Northcott PA, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nature genetics. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corbit KC, et al. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- 36.Gerdes JM, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nature genetics. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- 37.Simons M, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nature genetics. 2005;37:537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinelli DC, Fan CM. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes & development. 2007;21:1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhuo L, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 40.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic acids research. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.