Abstract
Background
Different iron transport systems evolved in Gram-negative bacteria during evolution. Most of the transport systems depend on outer membrane localized TonB-dependent transporters (TBDTs), a periplasma-facing TonB protein and a plasma membrane localized machinery (ExbBD). So far, iron chelators (siderophores), oligosaccharides and polypeptides have been identified as substrates of TBDTs. For iron transport, three uptake systems are defined: the lactoferrin/transferrin binding proteins, the porphyrin-dependent transporters and the siderophore-dependent transporters. However, for cyanobacteria almost nothing is known about possible TonB-dependent uptake systems for iron or other substrates.
Results
We have screened all publicly available eubacterial genomes for sequences representing (putative) TBDTs. Based on sequence similarity, we identified 195 clusters, where elements of one cluster may possibly recognize similar substrates. For Anabaena sp. PCC 7120 we identified 22 genes as putative TBDTs covering almost all known TBDT subclasses. This is a high number of TBDTs compared to other cyanobacteria. The expression of the 22 putative TBDTs individually depends on the presence of iron, copper or nitrogen.
Conclusion
We exemplified on TBDTs the power of CLANS-based classification, which demonstrates its importance for future application in systems biology. In addition, the tentative substrate assignment based on characterized proteins will stimulate the research of TBDTs in different species. For cyanobacteria, the atypical dependence of TBDT gene expression on different nutrition points to a yet unknown regulatory mechanism. In addition, we were able to clarify a hypothesis of the absence of TonB in cyanobacteria by the identification of according sequences.
Background
Filamentous cyanobacteria contain molecular machines for oxygenic photosynthesis under all growth conditions [1]. These machines, as well as those involved in respiration and nitrogen metabolism, depend on non-proteinaceous cofactors such as iron [2,3]. The level of iron found in cyanobacteria is generally one order of magnitude higher than in non-photosynthetic bacteria [4] and represents about 0.1% of their biomass [5]. Even though iron and copper are required for the function of respiratory and photosynthetic complexes, their intracellular level has to be tightly controlled as these ions pose a risk of oxidation [3]. Therefore, the uptake of iron is highly regulated in order to avoid intoxication. On the other hand, it is hypothesized that iron limitation might have been one of the selective forces in the evolution of cyanobacteria [6], and one might speculate that those cyanobacteria with the most efficient iron uptake systems might have had an evolutionary advantage. To enhance iron uptake, eubacteria secrete low-molecular-weight iron chelators (siderophores) under iron-limiting conditions to complex environmental iron [7]. The siderophore-iron complexes are bound by receptor proteins (TonB-dependent transporters, TBDTs) in the outer membrane which are composed of a transmembrane β-barrel domain, a so-called plug domain and a periplasmic exposed TonB box. The siderophore-iron is subsequently transferred to the cytoplasm by transport proteins in the cytoplasmic membrane [8,9]. This process is dependent on TonB which provides the energy required for the translocation of siderophore-iron complexes across the outer membrane [10]. In order to facilitate this translocation, the periplasmic domain of TonB interacts with the TonB box of the loaded TBDT. It is proposed that TonB exerts a pulling force on the TonB box and, thereby, partially unfolds the plug domain enabling the translocation of the siderophore into the periplasmic space [11]. Several TBDTs have been identified. Beside the ones for iron transport [12,13], TBDTs for nickel [14], disaccharides (for sucrose SuxA; [15], for maltose MalA; [16]), oligo- (CsuF; [17]), polysaccharides (SusC; [18]) or large degradation products of proteins (RagA; [19]) are described. The most intensively studied function of TBDTs is the iron uptake in Gram-negative bacteria. Three large classes are defined, namely transferrin-/lactoferrin-binding proteins, porphyrin and siderophore transporters [20]. In addition to the transport of iron across the outer membrane by TBDTs, an additional ferric iron uptake system is postulated, but the corresponding outer membrane receptor has not yet been identified [21]. The TBDTs TbpA (transferring-binding protein A) and LbpA (lactoferrin-binding protein A) facilitate the uptake of iron from transferrin/lactoferrin, respectively; the uptake is also assisted by the lipoproteins TbpB and LbpB which face the extracellular side [22]. The porphyrin-transporting TBDTs include HasR, HgbA, HmbR (heme; [12,22]) and BtuB which transports the cobalt-complexing vitamin B12 (cobalamin [23]). Heme uptake is especially important in bacterial pathogens, where various heme-containing compounds are utilized [13]. The siderophore TBDTs are further sub-classified according to their substrate - that is the chemical nature of the siderophore they bind. Siderophores belong inter alia to hydroxamates, catecholates, phenolates, citrates or combinations thereof [9]. For example, the siderophore transporters FepA, ViuA and IroN recognize catecholates, FhuA, FoxA and FhuE hydroxamate and FecA citrate.
The iron uptake system in cyanobacteria is not well understood. For the non-filamentous cyanobacterium Synechocystis sp. PCC 6803 the TBDTs encoded by sll1206, sll1406, sll1409 and slr1490 were partially characterized [24,25]. For filamentous cyanobacteria such as Anabaena sp. PCC 7120 (also termed Nostoc sp. PCC 7120) only siderophore secretion [26-28], and the influence of enhanced or reduced iron levels on the growth [29-32], were investigated. Anabaena sp. PCC 7120 secretes the hydroxamate-type siderophore schizokinen, allegedly the only siderophore secreted [26,27]. Only recently, a TBDT encoded by schT (alr0397) involved in the uptake of schizokinen was identified. The expression of the gene schT (alr0397) was mildly increased under a shortage of Fe3+. A schT knock-out mutant showed a moderate phenotype of iron starvation, and the characterization of its siderophore-dependent iron uptake demonstrated the function of schT as a TonB-dependent schizokinen transporter [33].
To learn more about iron transport systems in general and in cyanobacteria particularly we searched for genes coding for TBDTs based on previously experimentally characterized TBDTs. Subsequently, we assigned putative substrates for so far uncharacterized TBDTs, according to their sequence similarity to already known TBDTs. We observed a substantial difference in the number of TBDT genes in the analysed cyanobacteria. The expression pattern of the TBDT genes in Anabaena sp. PCC 7120 is analysed with respect to iron, copper and nitrogen availability.
Results and discussion
Classification of TonB-dependent transporters
Ninety-eight TBDTs and the (putative) substrates (for example, metallophores or sugars) were extracted from the published literature (see Additional file 1) [14-19,22,34-124]. In order to classify the TBDTs with unknown substrates, we first searched for putative TBDTs in 686 sequenced genomes. We identified 4600 putative TBDTs in 347 species (see Additional file 2). Compared to previously published bioinformatic analyses [15,125], we identified fewer sequences in the species which had been analysed in the past due to a more stringent cutoff (not shown). More specifically, within the species analysed by Koebnik, we selected seven sequences not previously identified, but did not consider 103 sequences [125]. A similar ratio was found when analysing the number of sequences selected by us from the species analysed by Blanvillain et al. (+22/-142; [15]), who selected 3020 sequences which resulted in a discrepancy of about 5%.
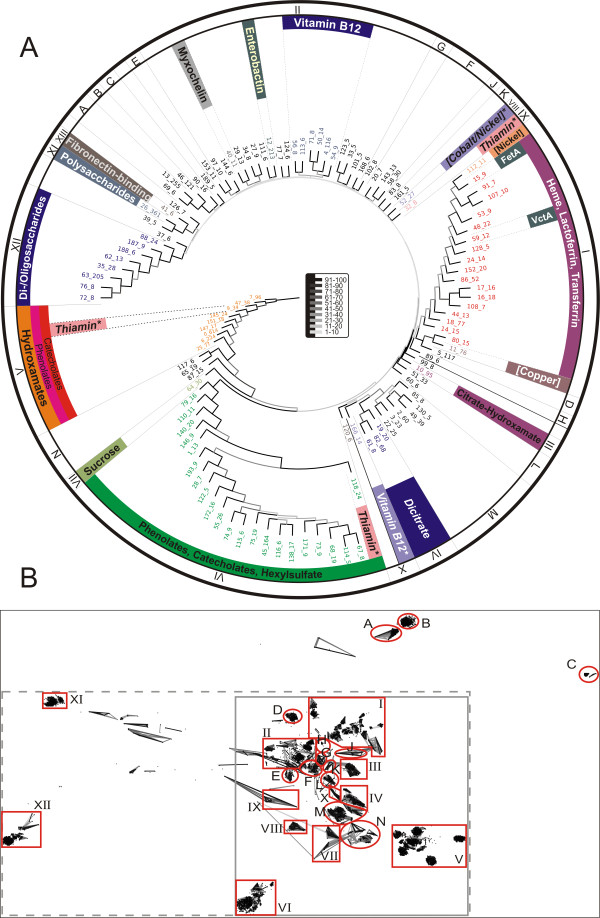
We subsequently performed a cluster analysis of the identified sequences of putative TBDTs (see Methods) leading to 195 clusters with at least two sequences. Figure 1A shows the consensus tree used to highlight 'regions' on the two-dimensional sequence landscape. A region is marked by roman numerals if the substrate for at least one TBDT in this region is experimentally verified (expTBDTs) or predicted (pTBDTs), and marked by upper case letters if no substrate TBDT in the region is known (Figure 1). Figure 1B shows the expTBDT regions I-VII, XI, XII and XIII and the pTBDTs regions VIII, IX and X together with the uncharacterized regions A-N. Figure 2 shows an enlarged version of the dashed rectangle in Figure 1B. The colours describe the substrate that binds to the corresponding TBDTs. Figure 2 (bottom) shows a magnification of the expTBDTs regions, where the numbers refer to sequences with a known substrate (Additional file 1, [14-19,22,34-124]). In the following, we characterize the regions according to the 98 TBDTs that have been experimentally verified or predicted (Additional file 1, [14-19,22,34-124]).
Figure 1.
Clustering of the sequences of putative TonB-dependent transporters (TBDTs). The sequences found by the described genome-wide searches were analysed by CLANs as depicted. (A) shows the consensus tree of the pair-wise mean cluster distances. The branches are coloured according to their respective bootstrap value in shades of grey as indicated by the legend in the middle of the tree. The numbers at each leaf are of the format 'x_y', where 'x' is the cluster number and 'y' the number of sequences belonging to this cluster. We have further indicated the transported substrates and the regions as shown in Figure 1B are marked by I to XII and A to N. Brackets indicate that the metal ion is known, but the metallophore has not yet been identified. An asterisk marks predicted substrates. (B) shows the result of two-dimensional clustering in CLANS. The regions from Figure 1A are marked by red polygons (containing at least a single exp/pTBDT) and red circles (no functionally characterized TBDT). Sequences with a high similarity (P-value < 10-90) are connected by lines coloured in shades of grey (the darker the smaller the P-value). The regions shown in Figure 2 (grey dashed line) and Figure 3 (grey dashed-dotted line) are highlighted.
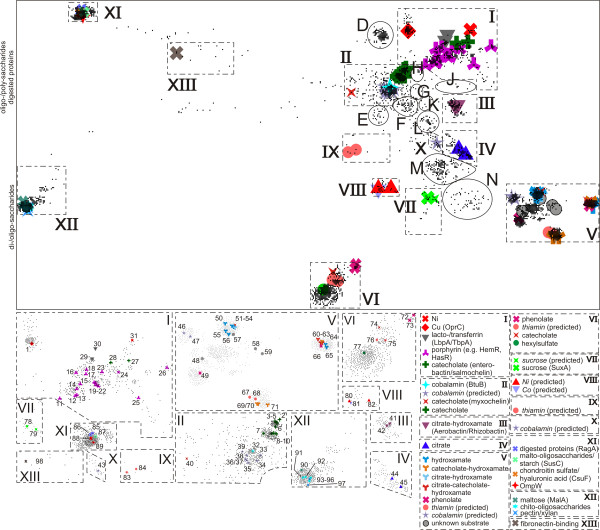
Figure 2.
Distribution of the sequences of characterized (experimental/predicted) TonB-dependent transporters (TBDTs). The grey dashed frame from Figure 1B is shown here containing expTBDTs and pTBDTs, which are marked by colored symbols and a number, which corresponds to the numbering in column 1 (see Additional file 1, [14-19,22,34-124]). Symbols are used to enhance the readability of the figures and are explained on the bottom right. The dashed frames are shown in a magnified view on the bottom. Circles define regions without functionally characterized TBDTs. For regions XI and XII the substrates are indicated on the left. The region numbering is explained in the text.
Region I
This consists of 19 clusters with at least five sequences and 10 of them have assigned functions which include porphyrin, lacto-/transferrin and nickel transporters (Figure 2). Cluster 11 contains the sequence of the copper chelate binding protein OprC (sequence 1, for references see Additional file 1, [14-19,22,34-124]). A group of clusters (15, 17, 18, 59, 86 and 107; sequences 11-28) are comprised of heme-transporting (HmbR) proteins. Remarkably, the two enterobactin (catecholate; see Additional file 1, [14-19,22,34-124]) transporters VctA (cluster 59; sequence 28) and FetA (cluster 15; sequence 27) are located within the porphyrin group. This finding corroborates the observation that VctA and FetA are, supposedly, involved in transporting porphyrin [126,127]. In cluster 16, the LbpA or TbpA proteins are found (sequences 29, 30). There is also a small cluster (112) which contains nickel-transporting TBDTs with a single expTBDT (sequence 31, FrpB4).
Region II
This contains 20 clusters, three of them represented by expTBDTs (cluster 4, 12, 40). In cluster 4 the experimentally confirmed cobalt-complexing vitamin B12 transporter BtuB (sequence 32) is present. However, in the same cluster, and in clusters 160 and 165, predicted BtuBs were identified (sequences 33-39). Cluster 12 contains IrgA, BfrA or IroN sequences (No. 2-10) transporting enterobactin, DHBS (catecholate) or salmochelin (glycosylated catecholate). In addition, a myxochelin (catecholate) transporter (sequence 40, cluster 40) occurs in region II.
Region III
Cluster 10 is defined by sequences of the aerobactin/rhizobactin (Citrate-hydroxamate; see Additional file 1, [14-19,22,34-124]) transporters IutA and RhtA (sequence 41 and 42).
Region IV
The largest cluster in region IV (No. 82) contains the sequences of the ferric rhizoferrin (carboxylate) transporter RumA and the diferric dicitrate transporter FecA (sequences 44 and 45).
Region V (sequences 46-71)
This consists of nine clusters with three (clusters 0, 6, 7) of them containing expTBDTs and two pTBDTs (clusters 9, 25). This region mainly contains transporters for hydroxamate-type siderophores, such as desferrioxamine (hydroxamate; cluster 0, sequences 50, 51, 56), ferrichrome (hydroxamate; cluster 0, sequence 55), pseudobactin A (citrate-catecholate-hydroxamate; cluster 6, sequences 62, 63), pyochelin (phenolate; cluster 6, sequence 66), or anguibactin (catecholate-hydroxamate; cluster 7, sequences 69-71). Interestingly, the proteins for which sequences are found in cluster 9 (sequence 67, 68) are predicted to transport thiamin, whereas proteins 46 and 47 (cluster 25) are predicted to transport vitamin B12. The latter appears to be a false prediction as judged from the large distance to cluster 4 containing the experimentally confirmed BtuB. Setting an even lower P-value (1E-100 instead of 1E-90) as the threshold for defining the clusters in CLANS leads to cluster 0 splitting up in the upper part with all hydroxamate-type TBDTs (including all cyanobacterial TBDTs of cluster 0) and the lower part containing phenolate-transporting TBDTs and VciA, which has been shown to transport neither heme, vibriobactin, enterobactin, ferrioxamine B, aerobactin nor shizokinen [77].
Region VI
This region represents transporters for phenolates, catecholates or hexylsulfate and contains several clusters. A hexylsulfate transporting TBDT (sequence 77) can be found in cluster 45, a vibriobactin (catecholate) transporter (sequence 74) in cluster 140 and proteins transporting yersiniabactin (phenolate; sequences 72, 73) in cluster 79. As already observed in region V, we also detected two sequences (75, 76) in cluster 118 that are putative thiamin transporters.
Region VII (cluster 67)
Cluster 67 contains SuxA (sequence 78), an experimentally verified sucrose transporter. Please note, that sequence 79 has been predicted to transport sucrose [15]. The prediction was based on the co-localization of the corresponding gene with the transcriptional regulator ScrR. Thus, our bioinformatic analysis provides additional evidence for the functional characterization.
Region VIII (cluster 52)
This region contains predicted nickel and cobalt TBDTs with unknown metallophore specificity and no representative of the expTBDTs.
Region IX
This consists of eight sequences in one cluster (No. 32), where two of the eight are putative thiamin transporters. However, proteins assigned as thiamin transporters were also found in regions V (sequences 67, 68, cluster 7) and VI (sequences 75 and 76, cluster 118). Their genes are co-localized on the genome with a cytoplasmic membrane transporter for thiamin (PnuT, [128]), however, the functional assignment remains to be proven.
Region X
This contains a TBDT predicted to transport cobalt-complexing vitamin B12 (sequence 43, cluster 166). However, it is far away from the BtuB cluster in region II (Figure 2). Hence, the assigned function should be experimentally confirmed.
Region XI
The region is clearly separated from the rest and contains cluster 26. The experimentally characterized TBDTs include oligosaccharide (CsuF, sequence 88), polysaccharide transporters (SusC, sequence 87) and transporters for degradation products of proteins (RagA, sequences 85-86). While many taxa are represented by sequences in the region I-X, region XI consists almost exclusively of bacteroidetes with the exception of one δ-proteobacterial sequence (gi|108757959, Myxococcus xanthus). Thus, sequences in this region may indicate a special adaptation of these organisms, which may be due to their lifestyle. Bacteroidetes are involved in food digestion in the intestinal tract of mammals. Hence a specific TBDT class for the uptake of substrates provided by the host seems plausible.
Region XII
This also appears as an outlier (Figure 2). It contains eight clusters (35, 62, 63, 72, 76, 88, 187 and 188) and only one expTBDT MalA (sequence 90, cluster 63) that transports maltodextrin. Seven pTBDTs are said to transport xylan, pectin or chito-oligosaccharides (No. 91-97; for references see Additional file 1, [14-19,22,34-124]), where sequences 91-96 belong to cluster 63 and sequence 97 to cluster 72. It appears that this region is composed of di- and oligosaccharide transporters. In line with this notion, the δ-proteobacterial TBDTs are from Myxobacteria (Myxococcus xanthus, Sorangium cellulosum), which are found on decaying plant material consuming their saccharides. Most of the sequences in this region stem from α- and γ-proteobacteria (18.4%, 76%) and a few bacteroidetes, δ- and β-proteobacteria taxa.
Region XIII
Positioned between region XI and the crowded area on the right side, this region is defined by a fibronectin-binding TBDT (sequence 98, cluster 41). As in most of the sequences in region XI, the sequences of this region consist almost exclusively of bacteroidetes. The close proximity of regions XI and XIII is consistent with the observed interaction of the TBDT with a glycoprotein.
Other regions
For regions I to XIII we were in the lucky position of being able to infer at least putative functions to ~3700 sequences. (The putative annotation can be viewed at http://www.cibiv.at/TBDT.) However, from the 4600 sequences from GenBank ~900 sequences remain in regions A-N, where we were unable to assign any function (Figure 1). While we cannot discuss potential substrates for clusters in regions A-N, we can at least point to some regions that show a peculiar taxonomic composition. In regions A and B sequences from mostly γ- (74%) and α-proteobacteria (19%), but also a few β-proteobacterial (5%) and bacteroidetes (1.5%), are present. Region C contains exclusively γ-proteobacterial sequences.
Classification of TonB-dependent transporters in cyanobacteria
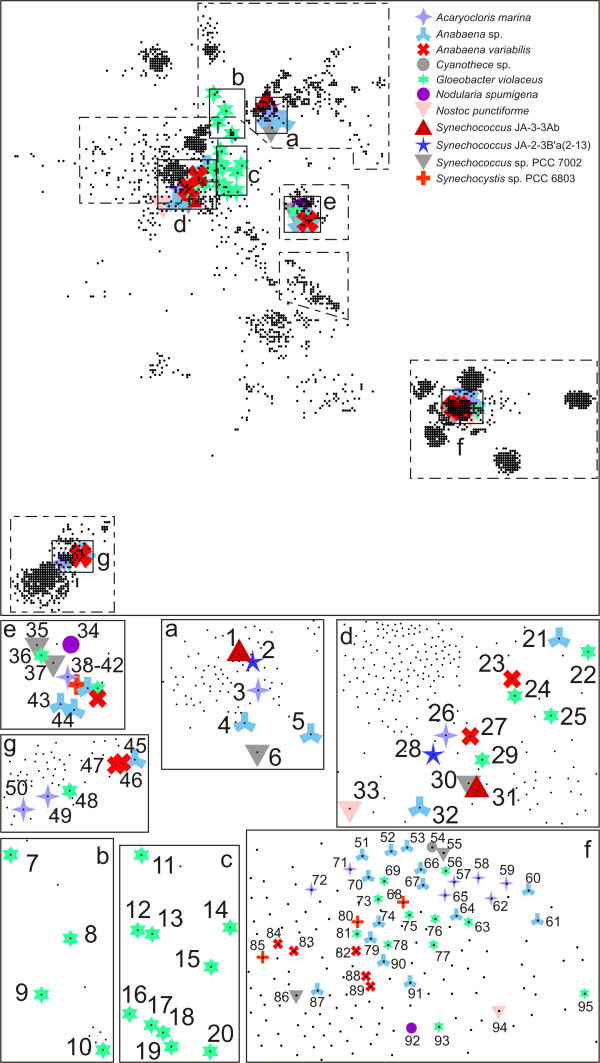
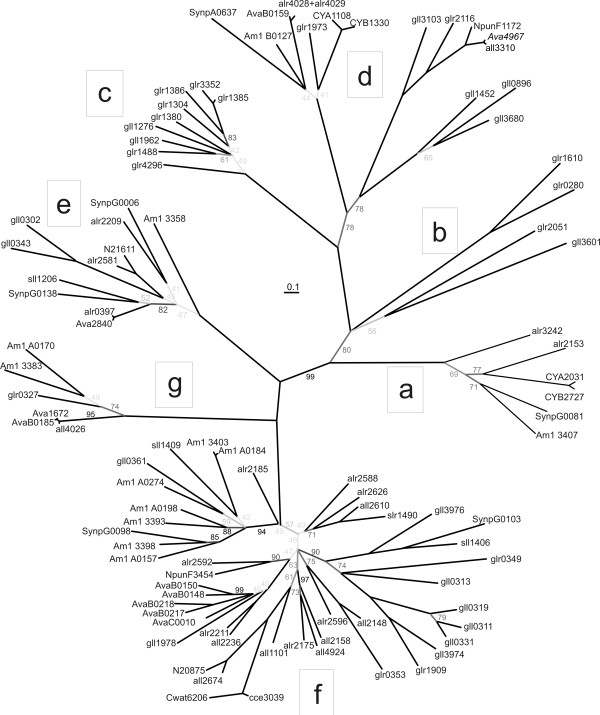
One of our aims was the identification and classification of cyanobacterial TBDTs. Hence we searched for sequences of putative TBDTs in 32 cyanobacterial genomes (proteins listed according to their accession code (Table 1, column 1). We additionally extracted the automated annotation from GenBank (Table 1, column 2). At present, this annotation is mostly limited to CirA, FhuE or BtuB. Hence, we analysed the location of cyanobacterial sequences on the CLANS plot (Figure 3 shows the section of Figure 1B indicated by a grey box). All cyanobacterial TBDTs belong to regions with experimentally characterized TBDTs (see Figure 2 and dashed frames in Figure 3). To further confirm the classification determined with CLANS we also constructed a phylogenetic tree for the cyanobacterial sequences (Figure 4). Seven 'subtrees' (a-f) were identified and mapped to regions I-X.
Table 1.
Sequences used for the phylogenetic analysis of cyanobacteria
| Code | Old | New | Spot | Database code | Species |
| Am1 B0127 | BtuB | BtuB | 26 | gi|158339996 | Acaryochloris marina MBIC11017 |
| Am1 3407 | CirA | HutA | 3 | gi|158336543 | |
| Am1 3358 | CirA | IutA | 38 | gi|158336494 | |
| Am1 A0170 | CirA | ViuA | 50 | gi|158339820 | |
| Am1 3383 | CirA | ViuA | 49 | gi|158336519 | |
| Am1 A0184 | Fiu | FhuA | 58 | gi|158339834 | |
| Am1 A0274 | FhuE | FhuA | 57 | gi|158339535 | |
| Am1 A0198 | - | FhuA | 71 | gi|158339845 | |
| Am1 A0157 | FhuE | FhuA | 59 | gi|158339807 | |
| Am1 3393 | FhuE | FhuA | 72 | gi|158336529 | |
| Am1 3398 | - | FhuA | 65 | gi|158336534 | |
| Am1 3403 | Fiu | FhuA | 62 | gi|158336539 | |
| Ava_B0148 | FhuE | FhuA | 89 | gi|75812430 | Anabaena variabilis ATCC 29413 |
| Ava_B0150 | CirA | FhuA | 88 | gi|75812432 | |
| Ava_B0159 | BtuB | FhuA | 82 | gi|75812441 | |
| Ava_B0185 | CirA | ViuA | 46 | gi|75812467 | |
| Ava_B0217 | Fiu | BtuB | 23 | gi|75812499 | |
| Ava_B0218 | CirA | IutA | 42 | gi|75812500 | |
| Ava_C0010 | FhuE | BtuB | 27 | gi|75812671 | |
| Ava_1672 | CirA | ViuA | 47 | gi|75907894 | |
| Ava_2840 | CirA | FhuA | 83 | gi|75909052 | |
| Ava_4967 | BtuB | FhuA | 84 | gi|75911163 | |
| cce3039 | CirA | FhuA | 54 | gi|172037952 | Cyanothece sp. CCY0110 |
| glr0280 | CirA | ? | 7 | gi|37519849 | Gloeobacter violaceus PCC7421 |
| gll0302 | CirA | IutA | 41 | gi|37519871 | |
| gll0311 | - | FhuA | 81 | gi|37519880 | |
| gll0313 | - | FhuA | 56 | gi|37519882 | |
| gll0319 | - | FhuA | 63 | gi|37519888 | |
| glr0327 | CirA | ViuA | 48 | gi|37519896 | |
| gll0331 | FecA | FhuA | 69 | gi|37519900 | |
| gll0343 | CirA | IutA | 36 | gi|37519912 | |
| glr0349 | FhuE | FhuA | 95 | gi|37519918 | |
| glr0353 | - | FhuA | 76 | gi|37519922 | |
| gll0361 | FhuE | FhuA | 78 | gi|37519930 | |
| gll0896 | FepA | BtuB | 22 | gi|37520465 | |
| gll1276 | OMC | BtuB | 20 | gi|37520845 | |
| glr1304 | OMC | BtuB | 17 | gi|37520873 | |
| glr1380 | - | - | - | gi|37520949 | |
| glr1385 | - | BtuB | 18 | gi|37520954 | |
| glr1386 | CirA | BtuB | 15 | gi|37520955 | |
| gll1452 | BtuB | BtuB | 25 | gi|37521021 | |
| glr1488 | - | BtuB | 12 | gi|37521057 | |
| glr1610 | - | ? | 8 | gi|37521179 | |
| glr1909 | FhuE | FhuA | 73 | gi|37521478 | |
| gll1962 | - | BtuB | 16 | gi|37521531 | |
| glr1973 | BtuB | BtuB | 29 | gi|37521542 | |
| gll1978 | Fiu | FhuA | 93 | gi|37521547 | |
| glr2051 | FepA | ? | 10 | gi|37521620 | |
| glr2116 | BtuB | BtuB | 24 | gi|37521685 | |
| gll3103 | BtuB | BtuB | 11 | gi|37522672 | |
| glr3352 | OMCa | BtuB | 19 | gi|37522921 | |
| gll3601 | BtuB | BtuB | 14 | gi|37523170 | |
| gll3680 | BtuB | ? | 9 | gi|37523249 | |
| gll3974 | FhuE | FhuA | 77 | gi|37523543 | |
| gll3976 | CirA | FhuA | 75 | gi|37523545 | |
| glr4296 | CirA | BtuB | 13 | gi|37523865 | |
| NpunF1172 | - | BtuB | 33 | gi|186681644 | Nostoc punctiforme PCC73102 |
| NpunF3454 | - | FhuA | 94 | gi|186683610 | |
| all1101 | - | FhuA | 52 | gi|17228596 | Anabaena sp. PCC 7120 |
| all2148 | FhuE | FhuA | 66 | gi|17229640 | |
| all2158 | FhuE | FhuA | 53 | gi|17229650 | |
| all2236 | Fiu | FhuA | 87 | gi|17229728 | |
| all2610 | CirA | FhuA | 67 | gi|17230102 | |
| all2674 | Fiu | FhuA | 70 | gi|17230166 | |
| all3310 | BtuB | BtuB | 21 | gi|17230802 | |
| all4026 | CirA | ViuA | 45 | gi|17231518 | |
| all4924 | FhuE | FhuA | 61 | gi|17232416 | |
| alr0397 | CirA | IutA | 40 | gi|17227893 | |
| alr2153 | CirA | HutA | 4 | gi|17229645 | |
| alr2175 | - | FhuA | 69 | gi|17229667 | |
| alr2185 | Fiu | FhuA | 51 | gi|17229677 | |
| alr2209 | CirA | IutA | 44 | gi|17229701 | |
| alr2211 | CirA | FhuA | 90 | gi|17229703 | |
| alr2581 | CirA | IutA | 43 | gi|17230073 | |
| alr2588 | CirA | FhuA | 74 | gi|17230080 | |
| alr2592 | FhuE | FhuA | 91 | gi|17230084 | |
| alr2596 | FhuE | FhuA | 64 | gi|17230088 | |
| alr2626 | - | FhuA | 79 | gi|17230118 | |
| alr3242 | CirA | HutA | 5 | gi|17230734 | |
| alr4028 +alr4029 |
- | BtuB | 32 | gi|17231520 gi|17231521 |
|
| sll1206 | IutA | IutA | 39 | gi|16329186 | Synechocystis sp. PCC6803 |
| sll1409 | FhuA | FhuA | 80 | gi|16329191 | |
| sll1406 | FhuA | FhuA | 85 | gi|16329194 | |
| slr1490 | FhuA | FhuA | 68 | gi|16329201 | |
| CYA_1108 | BtuB | BtuB | 31 | gi|86605797 |
Synechococcus sp. JA-3-3Ab |
| CYA_2031 | CirA | HutA | 1 | gi|86606671 | |
| CYB_1330 | BtuB | BtuB | 28 | gi|86608804 |
Synechococcus sp. JA-2-3B'a(2-13) |
| CYB_2727 | CirA | HutA | 2 | gi|86610153 | |
| SynpA0637 | BtuB | BtuB | 30 | gi|170077260 | Synechococcus sp. PCC 7002 |
| SynpG0081 | CirA | HutA | 6 | gi|170076551 | |
| SynpG0006 | CirA | IutA | 35 | gi|170076476 | |
| SynpG0138 | CirA | IutA | 37 | gi|170076608 | |
| SynPG0089 | FhuE | FhuA | 55 | gi|170076568 | |
| SynpG0103 | FhuA | FhuA | 86 | gi|170076573 | |
| Nspu20875 | CirA | FhuA | 92 | gi|119508873 |
Nodularia spumigena CCY9414 |
| Nspu21611 | CirA | IutA | 43 | gi|119509643 | |
| Cwat6206* | - | FhuA | 1 | gi|67920343 | Crocosphaera watsonii WH8501 |
The annotation of the sequences is indicated in column 1, the spot number according to Figure 3 is indicated in column 4; the initial annotation in the database is given in column 2; the classification according to Figure 3 using the name of a representative transporter of the related category is given in column 3; the accession code in column 5; and the source organism column 6.
a(OMC, outer membrane channel; '*', incomplete sequence; '?', no clear assignment possible).
Figure 3.
Distribution of TonB-dependent transporters (TBDTs) found in genomes of cyanobacteria. Cyanobacterial sequences of TBDTs are highlighted and the containing frames are enlarged at the bottom. Dashed boxes indicate the dimensions shown in Figure 1. The colour code shows the different species as indicated in the right corner. The numbers are according to Table 1.
Figure 4.
Distribution of cyanobacterial sequences. An alignment of sequences of TonB-dependent transporters listed in Table 1 was used to reconstruct a maximum likelihood phylogeny. Bootstrap values were calculated from 1000 phylogenetic trees. To indicate the probability of occurrence of an edge in these trees the edges are shown in shades of grey.
The six sequences in subtree 'a' belong to region I (Figure 3, 4) and show a relation to heme transporters such as HutA (Figures 1, 2, sequence 13). The sequences are found in Synechococcus sp., Acaryochloris marina and Anabaena sp. PCC 7120 (see new assignment in Table 1, column 3). Subtrees 'b' and 'c' contain only sequences from Gloeobacter violaceus. Subtree 'b' is within region I and is equidistant to enterobactin and heme transporters. Thereby, a clear assignment to a characterized TBDT family appears currently impossible. Subtree 'd' is close to the BtuB transporter cluster (region II) (Figure 1). In this region we find sequences from most of the analysed cyanobacteria (8 of 12), suggesting that transporters with similarity to BtuB are common. Subtree 'e' (Figure 3, 4) represents transporters, which can clearly be assigned as specific for aerobactin/rhizobactin (IutA-/RhtA-type). Subtree 'f' represents sequences of transporters with the closest relation to FhuA-type transporters of cluster 0. The sequences of subtree 'g' (cluster 1), closely related to ViuA, are probably transporters for catecholates. The sequences of subtree 'g' are also close to cluster 118, which contains putative thiamin transporters. Nevertheless, since the two putative thiamin transporters have not yet been experimentally confirmed, we consider these cyanobacterial TBDTs to be iron transporters of the ViuA-type.
Summarizing, the assignment of the cyanobacterial TBDTs to regions with functional characterization was successful with the exception of some TBDTs from Gloeobacter violaceus (subtrees 'b' and 'c'). Although BtuB-like transporters and hydroxamate-type metallophore transporters were found in cyanobacteria, we did not find FecA-type (diferric dicitrate) TBDTs, even though they occur in α-, β-, γ-, δ- and ε-proteobacteria, bacteroidetes and spirochaetes.
Identification of TBDTs in Anabaena sp. PCC 7120
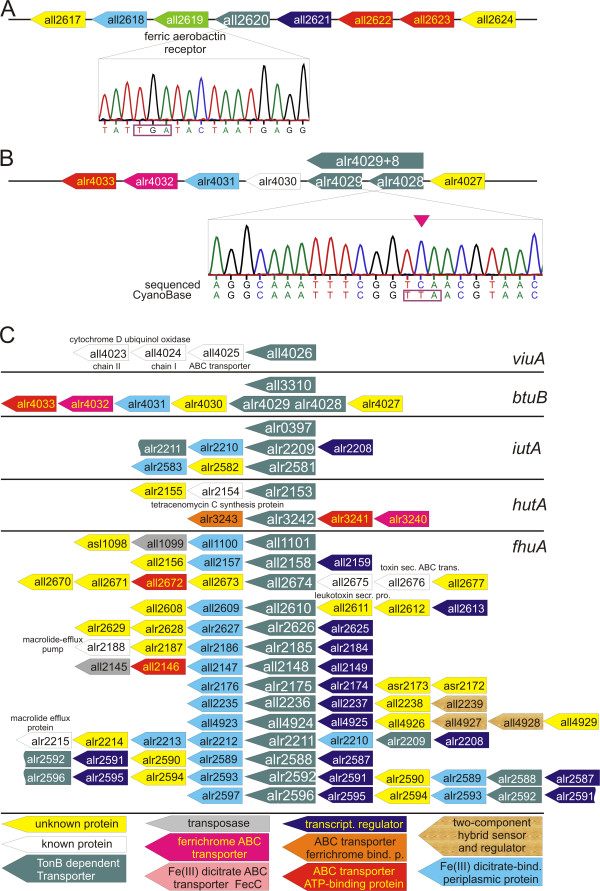
In order to explore the cyanobacterial TBDTs in more detail we analysed the full genome of Anabaena sp. PCC 7120. We identified 21 TBDT genes carrying the plug domain and β-barrel domain characteristic for TBDTs. In addition, we identified four genes (all2620, alr2179, all2578, alr4028) containing the plug domain of the TBDT, but an incomplete β-barrel domain. Downstream of all2620 (Figure 5A) and alr4028 (Figure 5B) a gene coding for the 'missing part' of the β-barrel domain is present (all2619 and alr4029, respectively). Consequently, we checked the stop codon separating the two gene pairs. We confirmed the stop codon between all2620 and all2619 (Figure 5A) and could not identify a frame shift in the sequence of the region 500 bp upstream or downstream of the stop codon. If All2620 is, indeed, part of a TBDT it has to form a heterodimer. A putative interaction partner would be All2619. It would, therefore, be interesting to investigate the existence of such complex and to understand whether it is just a remnant of a genetic accident which led to a split of the TBDT gene in all2620 and all2619. In contrast to all2620 and all2619, for alr4028 and alr4029 we found a T to C exchange in the sequence when comparing our results with that of the deposited sequence. Hence, we conclude that the stop codon does not exist and that the two genes alr4028 and alr4029 encode one protein. Therefore, 22 TBDTs exist in Anabaena sp. PCC 7120.
Figure 5.
The genomic structure of the loci coding for TonB-dependent transporters (TBDTs) in Anabaena sp. PCC 7120. For A-C, the genomic structure was excised from Cyanobase (http://bacteria.kazusa.or.jp/cyano/[148]) and the nomenclature of the colour code is given in C. White boxes indicate genes for which the name is given in the figure. (A) Shows the genomic orientation surrounding all2620 (top) and the sequencing profile of the region coding for the stop codon (in reverse orientation, bottom). (B) Shows the genomic orientation surrounding alr4028 (top) and the sequencing profile of the region coding for the stop codon (in reverse orientation, bottom). (C) The genomic organization surrounding the 22 other genes coding for TBDTs.
For 19 TBDTs the genomic organization suggests the integration of the gene in an operon (Figure 5C). Twelve TBDTs are directly positioned behind a gene coding for a (putative) transcriptional regulator (Figure 5C, violet), and most of the (putative) operon structures contain genes coding for proteins involved in iron transport. The gene coding for a ViuA-type transporter is in a putative operon with subunits of a cytochrome D ubiquinol oxidase, which is rather unexpected, because, to date, a relation between this oxidase and iron transport has not been reported (Figure 5C). Of the BtuB transporters one is a single gene (all3310), whereas the other (the gene which we confirmed and which is still annotated as alr4028/alr4029) is in a rather typical genomic environment, namely in front of three genes encoding the periplasmic and the plasma membrane localized iron transport machinery. The same holds true for the hutA-like gene alr3242. The other hutA-like gene (alr2153) is in a putative operon with a gene encoding a tetracenomycin C synthesis protein and a gene of unknown function. Again, the relation between the TBDT and the downstream genes are rather questionable.
Three genes are classified as iutA-like. alr0397 (schT) is single standing in the genome. Downstream of alr2581 we found two genes coding for an unknown protein and a dicitrate binding protein, respectively. Alr2209 is a component of a large genomic region (~14 kbp, alr2208-alr2215) containing upstream a transcription regulator and downstream a cluster with three genes coding for periplasmic dicitrate binding proteins and one fhuA-like gene (alr2211). Thirteen of the 14 fhuA-like genes are upstream of a gene coding for a protein annotated as dicitrate-binding. However, most of the genes found in the putative operons defined by the 14 fhuA-like genes encode for proteins of unknown function. Three of the fhuA-like genes (alr2588, alr2592, alr2596) are in the same chromosomal region. Upstream of these, a gene coding a transcription regulator and downstream a gene encoding a dicitrate binding protein are found. However, the phylogenetic analysis (Figure 4) argues against recent gene duplication.
Variations of the number of genes encoding TBDTs in cyanobacteria
The results presented in Figures 3, 4, 5 and Table 1 show that the number of TBDTs varies among cyanobacteria. We found 22 TBDTs in Anabaena sp. PCC 7120, 10 in Anabaena variabilis, six in Synechococcus sp. PCC 7002, four in Synechocystis sp. PCC 6803, 33 in Gloeobacter violaceus, but no TBDTs in the genomes of, for example, Prochlorococcus). This variation of the number of genes, however, does not reflect an elevated amount of outer membrane protein coding genes in Anabaena sp. PCC 7120, because such a variation is not found for other outer membrane proteins (Omp85, TolC, OstA and others; not shown). Furthermore, in a previous report, a correlation of the number of TBDTs to the number of open reading frames as, for example, for transporters in the cytoplasmic membrane [129] was not observed [128], which is supported by our analysis (not shown).
TBDTs are regulated by TonB proteins. Hence, the large number of TBDTs leads to the question of whether each TBDT is regulated individually or (at least a sub-population of the TBDTs) in concert by one TonB protein. We, therefore, screened the genomes for the presence of tonB (Table 2). One to three tonB genes were detected. Hence, the number of TBDTs largely exceeds the number of TonB proteins. Please note that we identified a TonB-like protein (Slr1484) in Synechocystis sp. PCC 6803, which corrects a previous statement excluding the presence of a TonB-like protein in this species [130].
Table 2.
TonB-like genes in cyanobacteria
| species | No. TonBs | Locus tag |
| Acaryochloris marina MBIC11017 | 2 | AM1_A0167, AM1_3413 |
| Anabaena variabilis ATCC 29413 | 1 | Ava_2295 |
| Crocosphaera watsonii WH 8501 | 1 | CwatDRAFT_6356 |
| Cyanothece sp. CCY0110 | 2 | CY0110_08196, CY0110_24616 |
| Gloeobacter violaceus PCC 7421 | 3 | glr1389, glr1815, glr2404 |
| Nodularia spumigena CCY9414 | 1 | N9414_10453 |
| Nostoc punctiforme PCC 73102 | 1 | Npun_F0783 |
| Anabaena sp. PCC 7120 | 1 | all5036 |
| Synechococcus sp. PCC 7002 | 2 | SYNPCC7002_G0090, SYNPCC7002_A2465 |
| Synechococcus sp. JA-3-3Ab | 1 | CYA_2030 |
| Synechococcus sp. JA-2-3B'a(2-13) | 1 | CYB_2726 |
| Synechocystis sp. PCC 6803 | 1 | slr1484 |
Expression of genes in Anabaena sp. coding for TBDTs
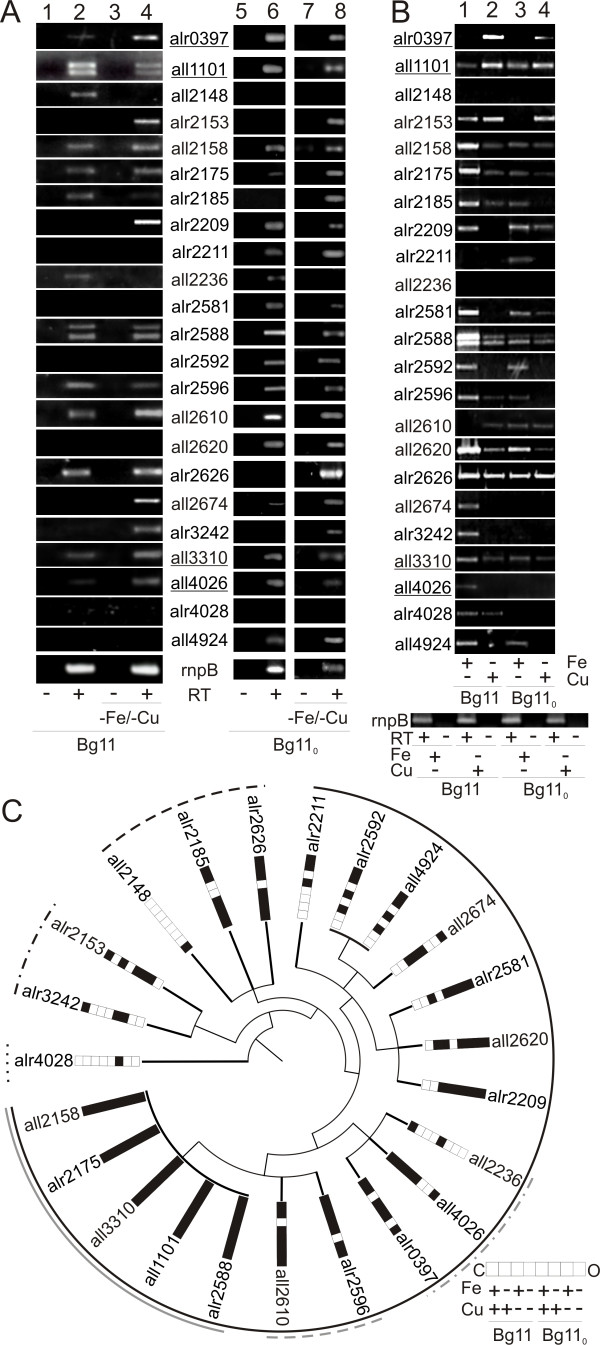
We analysed the gene expression of the 22 TDBT genes and of all2620, which only codes for the N-terminal portion of a TBDT (Figure 5A) in Anabaena sp. PCC 7120 (Figure 6). To this end, Anabaena sp. PCC 7120 was grown in normal medium (BG11), medium without iron (BG11-Fe), medium without copper (BG11-Cu) or medium lacking both (BG11-Fe-Cu). The presence of transcript was then determined by non-quantitative reverse transcription polymerase chain reaction (RT-PCR; primers are listed in Table 3). Iron and copper were chosen, because iron is known to be involved in the regulation of the gene expression of TBDTs and copper was recently found to induce an expression of a gene cluster involved in siderophore synthesis [131]. Remarkably, 13 TBDT-gene transcripts were present under normal growth conditions in such amounts that they could be amplified and visualized by RT-PCR (Figure 6A, lane 2; Figure 6C, grey lines and black dashed line). It should be noted that the absence of a transcript for the other genes might only reflect low transcript abundance. For 19 genes, we detected transcripts under Fe minus or/Cu minus conditions (Figure 6A lane 4, 6B, lane 1, 2). The analysis of the detection pattern revealed the following: (1) the genes all2148 and all2236, both hydroxamate-type TBDTs, were down-regulated upon iron and/or copper starvation compared to transcript levels under normal conditions; (2) the expression of seven genes (iutA-like genes alr2209 and alr2581, the btuB-like gene alr4028, the hutA-like gene alr3242 and the fhuA-like genes all2674, all4924 and alr2592) not detected under normal growth conditions is increased in response to copper, but not iron, limitation in the BG11 medium (Figure 6B, lane 1). This is notable, because, for four of these seven genes, the expression in the absence of one metal-ion (either Cu or Fe) is higher than in the absence of both iron and copper. One viuA-like gene (all4026) is expressed at a low level in BG11, but not in BG11 deficient of iron. An exclusive dependence of (upregulation of) expression in BG11 medium on iron limitation was only observed for alr0397 (iutA-like) and all2610 (fhuA-like).
Figure 6.
Iron-dependent expression of TonB-dependent transporters (TBDTs) of Anabaena sp. PCC 7120. (A) Anabaena sp. PCC 7120 were grown in media with (lane 1, 2, 5 and 6) and without a source of ferric ammonium citrate and copper (-Fe/-Cu, lane 3, 4, 7 and 8) as well as with (BG11, lane 1-4) and without a source of nitrogen (BG110, lane 5-8). RNA was isolated and RT-PCR was performed to visualize expression with (+) and without (-) addition of reverse transcriptase using primers listed in Table 3. The transcript of rnpB was amplified as control. (B)Anabaena sp. PCC 7120 were grown in media with (BG11, lane 1, 2) or without nitrogen source (BG110, lane 3, 4) in the absence of either copper sulfate (lane 1, 3) or ferric ammonium citrate (lane 2, 4) and RNA isolated from 50 ml cells of log phase cultures as described. RT-PCR was performed to visualize expression with and without addition of reverse transcriptase using primers listed in Table 3. (C) The results from A and B are visualized as bar code where black stands for detected expression, and white for no expression detected under conditions used. The order is given on the right (C stands for: pointing to the centre). The black outer circle marks all genes expressed in BG110, the black dashed line all genes expressed in BG11, the black dashed dotted line all genes not expressed without starvation and the dotted line the gene only weakly expressed upon copper limitation in BG11. The grey line on the outer rim indicates all genes always expressed, the grey dashed line on the outer rim all genes where expression was not detected under one condition and the grey dashed dotted line on the outer rim all genes which are expressed in BG11 and BG110, but not in all other media.
Table 3.
Primers used for reverse transcription polymerase chain reaction analysis and sequence confirmation.
| Use | Primer | Oligonucleotide sequence |
| RT-PCR | all1101-F | CGCTGCTCTATCGCCTGACTG |
| all1101-R | GCGTACTTTCCAGCGATTGTGC | |
| all2148-F | ACGGTGACGGGGGAAACAGGA | |
| all2148-R | ACTTCCACTCGCTCAATCGTCC | |
| all2158-F | CACCACCAGCAGAACCAACAGC | |
| all2158-R | CGATAAATCCACCTCACCACGG | |
| all2236-F | AATCGCGCGGCACTCTACCGT | |
| all2236-R | GGATAACTCCAATCCCACGAGC | |
| all2610-F | CAGTCATAGGAGAGGCGGGATT | |
| all2610-R | CATAGAGTACAGAAGCCGGTCC | |
| all2620-F | AAACTCCCTCAACCGCGCTGG | |
| all2620-R | GGAATCCGCGAACCATCGGC | |
| all2674-F | CCCAGAAACACCTACCGCAGAA | |
| all2674-R | AACTAGGTTGACGACCCCACCT | |
| all3310-F | TTTAGGCAACCCAGGCGGCAC | |
| all3310-R | GCATTATGCTCAAAGGTGACGCG | |
| all4026-F | GTGGTTTTTGTGGAGTGTGGGG | |
| all4026-R | CCATCAACTGGTGTGTCTTCCC | |
| all4924-F | CCCTACCAGAGGATCTGGGGA | |
| all4924-R | CTGTACCAACGGCTGGTAAGAC | |
| alr0397-F | TGCGTCGCGGGATTTGCGAAC | |
| alr0397-R | GGATAGTATTGACCCTGGGGTC | |
| alr2153-F | CCTCCCGTGGGATTAACTTTGG | |
| alr2153-R | CGCATCAGGGCCCACTCGA | |
| alr2175-F | GGTGTCCCGGCTGTTGGTACT | |
| alr2175-R | AGTACCTCAAACCTCTCTGGGC | |
| alr2185-F | CCCCGTCAGGTACTCGAAGAC | |
| alr2185-R | TCTTCCATGTCAAGCTAGGGGC | |
| alr2209-F | CCTACTCCCACACCCCCAAC | |
| alr2209-R | CTGTCTTGGTCAGGTCTTCGGG | |
| alr2211-F | CAAGATCGCCAAGTGGTGAGGC | |
| alr2211-R | GTGTTTCCAGGTAACAAGCCCC | |
| alr2581-F | GCGGGGACAGAAGGCAAATTTG | |
| alr2581-R | CGCCTAATTGTTCTGTACCCCC | |
| alr2588-F | CAGGTGAGGCGGGATTACCTG | |
| alr2588-R | ACTCCCCCTGGTTCTAGTTGTC | |
| alr2592-F | CCCCAAAGCCAGTGGAGGGA | |
| alr2592-R | CCCCTAGCGGCTCGAACATTG | |
| alr2596-F | CGCCTGTGCGCGATATTCCTG | |
| alr2596-R | CAGGCGCAACAAATACCCGTTC | |
| alr2626-F | GGCGTTCAACCAGGGGGAGT | |
| alr2626-R | CTAAACCAGGTCTGGGTATGGC | |
| alr3242-F | CCCGACGTGATAGTGGGTCAC | |
| alr3242-R | CTGGGGGAATCCGGCTGCAT | |
| rnpB-F | AGGGAGAGAGTAGG GTTGG | |
| rnpB-R | GGTTTACCGAGCCAGTACCTCT | |
| Sequence confirmation | all2619-20 F | CTCCCATTTCTCCGAAGCTG |
| all2619-20 R | CAACGCTGGGGCCAACATAG | |
| alr4028-29 F | CTATGGACTTAACCAACAAAGCATTC | |
| alr4028-29 R | CTTCTCTGGTTTAAGGTCAGGATTAC | |
Finally, we investigated the expression pattern of TDBTs under conditions enforcing heterocyst formation by growth in medium without a nitrogen source (BG110). We again analysed the amount of transcript in the four different media. Strikingly, in BG110 medium 17 genes are expressed (Figure 6A, lane 6, Figure 6C, black hemicircle) but seven of them are not expressed in BG11. Moreover, we found four genes - alr2153 and alr3242 (hutA-like),alr2626 and alr2185 (fhuA-like) - for which a transcript was detected only under additional metal starvation (BG110 -Fe, -Cu or -Fe/-Cu). Remarkably, all2620 is expressed under all conditions without a nitrogen source, which suggests that all2620 is not a pseudogene. In general, one can conclude that not only metal starvation but also nitrogen starvation induces transcription of TBDT-encoding genes in Anabaena sp. PCC 7120.
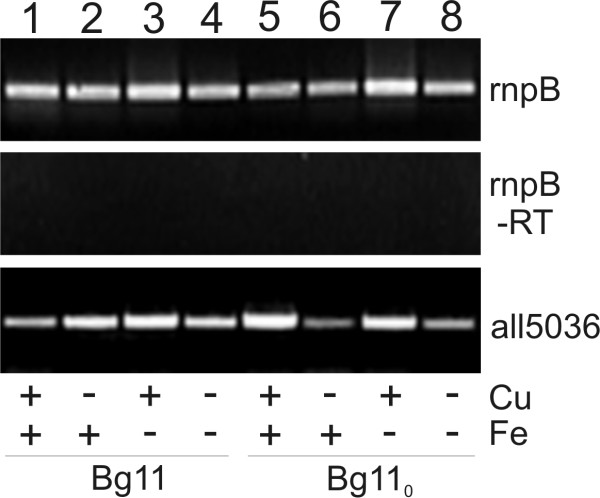
As only all5036 encodes a TonB-like protein in Anabaena sp. PCC 7120 we analysed its expression under the conditions outlined (Figure 7). As expected, all5036 transcript can be detected under all conditions tested. Assuming that the function of all identified TBDTs in Anabaena sp. depends on TonB, All5036 is required for iron homeostasis in general.
Figure 7.
Expression of all5036 encoding the only TonB in Anabaena sp. PCC 7120. Anabaena sp. PCC 7120 were grown as for experiments shown in Figure 6 in BG11 (lane 1-4) or BG110 (lane 5-8) with and without a source of ferric ammonium citrate and/or copper and reverse transcription (RT) polymerase chain reaction was performed in the absence (-RT) or presence of reverse transcriptase on isolated RNA using primers for rnpB and all5036 (listed in Table 3) to visualize expression.
Conclusion
By clustering ~4,600 TBDTs we found that they group by their substrate and not according to their taxonomy with the exception of regions IX, XI, XIII and C. The latter are specific for sequences from bacteroidetes and γ-proteobacteria, respectively. Hence, the transported molecule dominates the sequence variation among TBDTs. According to the occurrence of expTBDTs within clusters, we were able to assign a tentative substrate for almost two-thirds of the analysed sequences. We have developed a website for a further detailed inspection of the clustering of individual sequences http://www.cibiv.at/TBDT. Here, the individual clusters or sequences can be highlighted based on the presentation in Figure 2. However, the current assignment has to be viewed with care as Schauer and colleagues pointed out that further substrates might be discovered in future [128], which will then be introduced into the web interface. We identified several clusters of TBDTs with putatively so far unknown substrates. Further research on a few candidate proteins of each of these clusters would be of great interest, as it would significantly advance the knowledge on substrate uptake by bacteria on the protein level and it might also reveal new potential drug targets.
Large differences to previously suggested classifications were not observed for iron-transporting TBDTs. Generally, our approach resembles previous classifications of TBDTs according to their substrates based on a smaller number of sequences and a phylogenetic tree reconstruction [53,80,82,111,112], but the positioning of the IutA and of the ViuA sequences differs with respect to distances previously proposed [82,112]. In contrast to the report by LeVier and Guerinot who placed ViuA between the lactoferrin and transferrin recognizing transporters [82], we found that ViuA (sequence 74, Region VI) clearly clusters with FyuA (sequence 72) sequences. This discrepancy might reflect the fact that: (i) more sequences of TBDTs are available nowadays; and (ii) the methodology to analyse sequence relationships has improved.
A deviation from this general picture was found for the predicted BtuBs, which are spread over a long stripe from regions II to V. Hence, BtuBs might show a similar diffuse distribution pattern like the heme and hydroxamate transporters (regions I and V, respectively). The predicted BtuBs might, therefore, transport substrates only structurally related to cobalt-complexing vitamin B12.
TBDTs in Anabaena sp
Based on database searches, we have identified 25 sequences with TonB-box signature [39] leading to 22 sequences coding for putative TBDTs in Anabaena sp. PCC 7120 (Figures 3, 4, 5). Strikingly, at least five different types of transporters are identified (FhuA, ViuA, IutA, BtuB and HutA type) and this number greatly exceeds the number of genes coding for TBDTs of almost all other (sequenced) cyanobacteria with the exception of Gloeobacter violaceus. In turn, this is the only species for which some of the TBDTs could not be functionally assigned. As already discussed, it has been hypothesized that iron limitation might have been one of the selective forces in evolution of cyanobacteria. Gloeobacter violaceus (33 TBDTs) was isolated in 1972 from a calcareous rock near the Vierwaldstättersee in Switzerland, whereas Anabaena variablis (previously Anabaena flos-aquae strain A-37; 10 TBDTs) was isolated in 1964 from fresh water of the Mississippi, USA. Both strains are considered non-symbiotic. G. violaceus is rather unique with respect to the absence of thylakoid structure and does not form filaments [132] like Synechocystis sp. PCC 6803 (4 TBDTs), which was isolated from fresh water in California and deposited in the Pasteur collection in 1968 http://www.crbip.pasteur.fr. Hence, the number of TBDTs does not correlate with filament or heterocyst formation. It might, however, correlate with the habitat from which the species were isolated, with respect to either species competition for iron or iron limitations in the environment per se. Therefore, symbiotic cyanobacteria such as Nostoc punctiforme may possibly contain a rather low number of TBDTs because iron is provided by the host. Unfortunately, to the best of our knowledge, the source of Anabaena sp. strain PCC 7120 - formerly named Nostoc muscorum ISU ([133]; further synonyms are Anabaena sp. ATCC 27893, Nostoc sp. strain PCC 7120) - is unknown and it is considered to be a 'free living cyanobacterium'. The observation that this cyanobacterium is susceptible to viruses isolated from the Lake Mendota, Dane County, Wisconsin, USA, [133] might suggest that a similar environment was its place of isolation. This would be in line with an original natural habitat of Anabaena sp. PCC 7120 that contained rather limited iron sources, because it has been reported that the iron concentration in rivers is higher than in lakes ([134]). The variety of TBDT classes found in Anabaena sp. rather agrees with iron limited environmental conditions. The only TBDT type which could not be identified in the analysed cyanobacterial species, in general, and, thereby, also in Anabaena sp. PCC 7120, is the FecA-type (diferric dicitrate) which can be found in many other bacteria. To date, schizokinen is the only confirmed siderophore which is secreted by Anabaena sp. PCC 7120 [27] and, recently, its transporter was identified [33]. However, additional siderophores are secreted by Anabaena sp. [33,131], but they have not yet been characterized. Nevertheless, other interpretations for the variable number of TBDTs might still be possible.
The environment influences the expression of TBDT genes in Anabaena sp
In line with iron limitation in the native environment, several differential expression regulation regimes have been observed. For instance, six out of 14 genes encoding hydroxamate recognizing FhuA-like transporters are expressed under (almost) all tested conditions (Figure 6C, grey and grey dashed line, Table 1). The same holds true for one BtuB-like transporter encoded by all3310, which is in accordance with its identification in a proteome analysis of cells grown under standard conditions [135,136]. Interestingly, the other BtuB-like transporter encoded by the joint gene all4028/all4029 is only expressed under iron-limiting conditions (Figure 6C, black dotted line). Furthermore, the iutA-like genes are always expressed under nitrogen-limiting conditions, whereas hutA-like genes are only expressed upon metal starvation (Figure 6C, black dashed dotted line). Also, for the gene encoding the schizokinen transporter SchT (Alr0397) only a moderate and intermediate influence of iron starvation on expression was observed [33]. The gene encoding the only putative catecholate transporter (All4026) appears to be expressed under non-limiting conditions as well as after nitrogen starvation. To our surprise, we did not observe a transcript under iron limitation but under copper limitation in BG11 or in the absence of both metals in BG11 and BG110. Such a clear relation to copper starvation was detected for four FhuA-type transporters as well (Figure 6C). The relation between the expression of genes encoding for TBDTs in Anabaena sp. and copper agrees with the recent observation that genes involved in siderophore production are also induced by copper starvation [131]. Nevertheless, the components of the network regulating the expression of TBDT encoding genes still need to be identified. Even though a complex network of TBDTs was discovered, only a single TonB protein was found in 58% of Gram-negative bacteria [137]. The gene is expressed under all tested conditions and, hence, it has to be considered as a master 'regulator' of the large group of TBDTs.
Methods
Identification of TonB-dependent transporters
Ninety-eight TBDT sequences were extracted from the NCBI database after extensive literature search. For 67 of them, experimental data is available, but for four of them the substrate is still unknown. Information on predicted substrates for the remaining 27 is available (see Additional file 1, [14-19,22,34-124]). These predictions are based on co-localization with genes of a specific metabolic pathway or on co-regulation by either transcription factors or a riboswitch [128]. Moreover, we downloaded 686 completely sequenced eubacterial genomes from the NCBI ftp server ftp://ftp.ncbi.nih.gov/genomes/Bacteria/ that were available in June 2008. In order to locate putative TBDTs in the genomes, we searched for open reading frames containing the TBDT β-barrel domain and the plug domain. To this end, we used hmmsearch (hidden Markov model search) from the hmmer package http://hmmer.janelia.org/ and the profile hidden Markov models PF00593 and PF07715 provided by the PFAM database [138,139]. The hmmsearch output-files were parsed considering only hits with an E-value < 10-10. We used only sequences for further analysis that resulted in a significant hit for both domains.
Phylogenetic analysis and clustering
The 97 cyanobacterial TBDT sequences were aligned with MAFFT [140] and a maximum likelihood tree was constructed with IQPNNI v3.3.b4 [141]. As a substitution model we selected VT [142] with gamma-distributed rate heterogeneity. Support values were calculated from 1000 bootstrap replicates. The consensus tree was reconstructed with Tree-Puzzle v5.2 [143] applying the majority consensus rule. The program CLANS [144] was used to cluster the 4648 putative TBDTs detected in the complete genomes, and to visualize their degree of similarity. In CLANS we set the cut off such that only P-values < 10-10 obtained by pairwise BLASTs were used for the CLANS-clustering. In the context of this manuscript, we use the term 'cluster' to refer to an aggregation of sequences. Each sequence in a cluster has at least one correspondent within the cluster with a BLAST p-value < 10-90 leading to 195 clusters with at least two elements.
To further elucidate the relationship of the 195 clusters, we ran CLANS 100 times with a random initial configuration of the sequences in 3d space. In each run we determined the cluster centres and computed pair-wise distances between the centres. With the PHYLIP package v3.68 [145] we constructed a neighbour-joining tree for the resulting 100 distance matrices and we inferred the majority rule consensus tree with support values for the splits in the consensus tree.
Genome loci of TonB-dependent transporters in Anabaena sp. PCC 7120
The annotations of genes upstream and downstream of the TBDT loci, shown in Figure 5, were done manually.
Analysis of the operon structure
Genomic DNA of Anabaena sp. was isolated as described [146]. The intergenic sequences between all2619 and all2620 and between alr4028 and alr4029, respectively, and additional ~250 bp inside each flanking gene were amplified with 5' Prime PCR Extender Polymerase (5' Prime, Hamburg, Germany) according to the manufacturer's protocol. The PCR product was cloned into pCR2.1 (Invitrogen, Karlsruhe, Germany), transformed into DH5α (GibcoBRL, Eggenstein, Deutschland) and the resulting plasmids purified for sequencing.
RNA isolation and analysis
Total RNA was isolated from 50 ml cells of log phase cultures (OD750 ≈ 2) as described [147]. RT-PCRs were performed according to the protocol of the Invitrogen SuperScript® III First-Strand Synthesis System for Random Hexamer Primers (Invitrogen, Carlsbad, USA). The used oligonucleotides are listed in Table 3.
Abbreviations
Hmmsearch: hidden Markov model search; LbpA: lactoferrin-binding protein A; RT-PCR: reverse transcription polymerase chain reaction; TbpA: transferrin-binding protein A; TBDT: TonB-dependent transporter; expTBDT: experimentally characterized TBDT; pTBDT: TBDT with predicted substrate.
Authors' contributions
The idea and strategy were developed by ES and AvH. OM and SS performed database searches and bioinformatic analyses. Laboratory experiments were performed by KN. ES drafted the manuscript, which was finalized by AvH and ES. All authors read and approved the final manuscript.
Supplementary Material
List of experimentally characterized TBDTs and of TBDTs with predicted substrate used for classification.
Number of TBDTs detected in analyzed genomes.
Acknowledgments
Acknowledgements
Financial support from the Deutsche Forschungsgemeinschaft (DFG, SFBTR1-C7, SFBTR1-A10) and the Volkswagenstiftung to ES, from Wiener Wissenschafts-, Forschung- und Technologiefonds (WWTF) and Deutsche Forschungsgemeinschaft (DFG, SPP 1174) to AvH is acknowledged.
Contributor Information
Oliver Mirus, Email: o.mirus@bio.uni-frankfurt.de.
Sascha Strauss, Email: sascha.strauss@univie.ac.at.
Kerstin Nicolaisen, Email: kerstin.nicolaisen@gmx.de.
Arndt von Haeseler, Email: arndt.von.haeseler@univie.ac.at.
Enrico Schleiff, Email: schleiff@bio.uni-frankfurt.de.
References
- Adams DG, Duggan PS. Heterocyst and akinete differentiation in cyanobacteria. New Phytol. 1999;144:3–33. [Google Scholar]
- Kustka A, Carpenter EJ, Sanudo-Wilhelmy SA. Iron and marine nitrogen fixation: progress and future directions. Res Microbiol. 2002;153:255–262. doi: 10.1016/s0923-2508(02)01325-6. [DOI] [PubMed] [Google Scholar]
- Shcolnick S, Keren N. Metal homeostasis in cyanobacteria and chloroplasts. Balancing benefits and risks to the photosynthetic apparatus. Plant Physiol. 2006;141:805–810. doi: 10.1104/pp.106.079251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Aurora R, Pakrasi HB. Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol. 2004;135:1666–1673. doi: 10.1104/pp.104.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger PA, Tirol A, Ardales S, Watanabe I. Chemical composition of cultures and natural samples of N2-fixing blue-green algae from rice fields. Biol Fertil Solis. 1986;2:131–146. [Google Scholar]
- Ting CS, Rocap G, King J, Chisholm SW. Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 2002;10:134–142. doi: 10.1016/s0966-842x(02)02319-3. [DOI] [PubMed] [Google Scholar]
- Ferreira F, Straus NA. Iron deprivation in cyanobacteria. J Appl Phyc. 1994;6:199–210. [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Gumbart J, Wiener MC, Tajkhorshid E. Mechanics of FORCE PROPAGATION in TonB-dependent outer membrane transport. Biophys J. 2007;93:496–504. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TE, Tari LW, Vogel HJ. Structural biology of bacterial iron uptake systems. Curr Top Med Chem. 2001;1:7–30. doi: 10.2174/1568026013395623. [DOI] [PubMed] [Google Scholar]
- Lee BC. Quelling the red menace: haem capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- Schauer K, Gouget B, Carrière M, Labigne A, de Reuse H. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol Microbiol. 2007;63:1054–1068. doi: 10.1111/j.1365-2958.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denancé N, Vasse J, Lauber E, Arlat M. Plant carbohydrate scavenging through tonb-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS ONE. 2007;2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer H, Herrmann C, Kammer W, Schwarz G, Nordheim A, Braun V. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J Bacteriol. 2005;187:8300–8311. doi: 10.1128/JB.187.24.8300-8311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Yu MC, Reeves AR, Salyers AA. Identification and characterization of a Bacteroides gene, csuF, which encodes an outer membrane protein that is essential for growth on chondroitin sulfate. J Bacteriol. 1995;177:3721–3727. doi: 10.1128/jb.177.13.3721-3727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, D'Elia JN, Frias J, Salyers AA. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol. 1996;178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano K, Murakami Y, Nishikawa K, Sakakibara J, Shimozato K, Yoshimura F. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J Med Microbiol. 2007;56:1536–1548. doi: 10.1099/jmm.0.47289-0. [DOI] [PubMed] [Google Scholar]
- Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. Feo-transport of ferrous iron into bacteria. Biometals. 2006;19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- Perkins-Balding D, Ratliff-Griffin M, Stojiljkovic I. Iron transport systems in Neisseria meningitidis. Microbiol Mol Biol Rev. 2004;68:154–171. doi: 10.1128/MMBR.68.1.154-171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AD, Deisenhofer J. TonB-dependent receptors-structural perspectives. Biochim Biophys Acta. 2002;1565:318–332. doi: 10.1016/s0005-2736(02)00578-3. [DOI] [PubMed] [Google Scholar]
- Katoh H, Hagino N, Grossman AR, Ogawa T. Genes essential to iron transport in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2001;183:2779–2784. doi: 10.1128/JB.183.9.2779-2784.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, McIntyre LM, Sherman LA. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2003;132:1825–1839. doi: 10.1104/pp.103.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson FB, Neilands JB. Siderochromes in cyanophyceae: isolation and characterization of schizokinen from Anabaena sp. J Phycol. 1976;12:44–48. [Google Scholar]
- Goldman SJ, Lammers PJ, Berman MS, Sanders-Loehr J. Siderophore-mediated iron uptake in different strains of Anabaena sp. J Bacteriol. 1983;156:1144–1150. doi: 10.1128/jb.156.3.1144-1150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SE, Stuart J, Sanders-Loehr J. Induction of siderophore activity in Anabaena spp. and its moderation of copper toxicity. Appl Environ Microbiol. 1987;53:917–922. doi: 10.1128/aem.53.5.917-922.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massalski A, Laube VM, Kushner DJ. Effects of cadmium and copper on the ultrastructure of Ankistrodesmus braunii and Anabaena 7120. Microbial Ecol. 1981;7:183–189. doi: 10.1007/BF02032499. [DOI] [PubMed] [Google Scholar]
- Guikema JA, Sherman LA. Organization and function of chlorophyll in membranes of cyanobacteria during iron starvation. Plant Physiol. 1983;73:250–256. doi: 10.1104/pp.73.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutber GN, Hutson KG, Rogers LJ. Effect of iron deficiency on levels of two ferredoxins and flavodoxin in a cyanobacterium. FEMS Microbiol Lett. 1977;1:193–196. [Google Scholar]
- Latifi A, Jeanjean R, Lemeille S, Havaux M, Zhang CC. Iron starvation leads to oxidative stress in Anabaena sp. strain PCC 7120. J Bacteriol. 2005;187:6596–6598. doi: 10.1128/JB.187.18.6596-6598.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaisen K, Moslavac S, Samborski A, Valdebenito M, Hantke K, Maldener I, Muro-Pastor AM, Flores E, Schleiff E. Alr0397 is an outer membrane transporter for the siderophore schizokinen in Anabaena sp. strain PCC 7120. J Bacteriol. 2008;190:7500–7507. doi: 10.1128/JB.01062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama H, Nakae T. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiol. 1996;142:2137–2144. doi: 10.1099/13500872-142-8-2137. [DOI] [PubMed] [Google Scholar]
- Beall B, Sanden GN. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiol. 1995;141:3193–3205. doi: 10.1099/13500872-141-12-3193. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Neilands JB. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem Biophys Res Commun. 1970;38:989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Johnson Z, Vasil ML. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiol. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- Dean CR, Poole K. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J Bacteriol. 1993;175:317–324. doi: 10.1128/jb.175.2.317-324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundrigan MD, Kadner RJ. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB. J Biol Chem. 1986;261:10797–10801. [PubMed] [Google Scholar]
- Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci USA. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bister B, Bischoff D, Nicholson GJ, Valdebenito M, Schneider K, Winkelmann G, Hantke K, Süssmuth RD. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals. 2004;17:471–481. doi: 10.1023/b:biom.0000029432.69418.6a. [DOI] [PubMed] [Google Scholar]
- Carswell CL, Rigden MD, Baenziger JE. Expression, purification, and structural characterization of CfrA, a putative iron transporter from Campylobacter jejuni. J Bacteriol. 2008;190:5650–5662. doi: 10.1128/JB.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau CD, Konisky J. Evolutionary relationship between the TonB-dependent outer membrane transport proteins: nucleotide and amino acid sequences of the Escherichia coli colicin I receptor gene. J Bacteriol. 1989;171:1041–1047. doi: 10.1128/jb.171.2.1041-1047.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Dihydroxybenzoylserine - a siderophore for E. coli. FEMS Microbiol Lett. 1990;67:5–8. doi: 10.1016/0378-1097(90)90158-m. [DOI] [PubMed] [Google Scholar]
- Goldberg MB, Boyko SA, Butterton JR, Stoebner JA, Payne SM, Calderwood SB. Characterization of a Vibrio cholerae virulence factor homologous to the family of TonB-dependent proteins. Mol Microbiol. 1992;6:2407–2418. doi: 10.1111/j.1365-2958.1992.tb01415.x. [DOI] [PubMed] [Google Scholar]
- Beall B, Hoenes T. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiol. 1997;143:135–145. doi: 10.1099/00221287-143-1-135. [DOI] [PubMed] [Google Scholar]
- Mey AR, Payne SM. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol Microbiol. 2001;42:835–849. doi: 10.1046/j.1365-2958.2001.02683.x. [DOI] [PubMed] [Google Scholar]
- Henderson DP, Payne SM. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazoy R, Osorio CR, Toranzo AE, Lemos ML. Isolation of mutants of Vibrio anguillarum defective in haeme utilisation and cloning of huvA, a gene coding for an outer membrane protein involved in the use of haeme as iron source. Arch Microbiol. 2003;179:329–338. doi: 10.1007/s00203-003-0529-4. [DOI] [PubMed] [Google Scholar]
- Lewis LA, Gray E, Wang YP, Roe BA, Dyer DW. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I, Hwa V, de Saint Martin L, O'Gaora P, Nassif X, Heffron F, So M. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- Srikumar R, Mikael LG, Pawelek PD, Khamessan A, Gibbs BF, Jacques M, Coulton JW. Molecular cloning of haemoglobin-binding protein HgbA in the outer membrane of Actinobacillus pleuropneumoniae. Microbiol. 2004;150:1723–1734. doi: 10.1099/mic.0.27046-0. [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica : similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung JM, Jones HA, Perry RD. The hmu locus of Yersinia pestis is essential for utilization of free haemin and haem-protein complexes as iron sources. Mol Microbiol. 1996;20:725–739. doi: 10.1111/j.1365-2958.1996.tb02512.x. [DOI] [PubMed] [Google Scholar]
- Torres AG, Payne SM. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1997;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- Mills M, Payne SM. Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect Immun. 1997;65:5358–5363. doi: 10.1128/iai.65.12.5358-5363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope LD, Yogev R, Müller-Eberhard U, Hansen EJ. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CE, Olsen B, Elkins C. Cloning and characterization of tdhA, a locus encoding a TonB-dependent heme receptor from Haemophilus ducreyi. Infect Immun. 1998;66:4254–4262. doi: 10.1128/iai.66.9.4254-4262.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoffe S, Ghigo JM, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano K, Luke NR, Howlett AJ, Campagnari AA. Identification of a conserved Moraxella catarrhalis haemoglobin-utilization protein, MhuA. Microbiol. 2005;151:1151–1158. doi: 10.1099/mic.0.27820-0. [DOI] [PubMed] [Google Scholar]
- Carson SD, Klebba PE, Newton SM, Sparling PF. Ferric enterobactin binding and utilization by Neisseria gonorrhoeae. J Bacteriol. 1999;181:2895–2901. doi: 10.1128/jb.181.9.2895-2901.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Wyckoff EE, Oglesby AG, Rab E, Taylor RK, Payne SM. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect Immun. 2002;70:3419–3426. doi: 10.1128/IAI.70.7.3419-3426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas GD, Sparling PF. Characterization of lbpA, the structural gene for a lactoferrin receptor in Neisseria gonorrhoeae. Infect Immun. 1995;63:2958–2967. doi: 10.1128/iai.63.8.2958-2967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen CN, Biswas GD, Tsai J, Paruchuri DK, Thompson SA, Sparling PF. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to TonB-dependent outer membrane receptors. J Bacteriol. 1992;174:5788–5797. doi: 10.1128/jb.174.18.5788-5797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller K, Kadner RJ. Nucleotide sequence of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J Bacteriol. 1985;161:904–908. doi: 10.1128/jb.161.3.904-908.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Silakowski B, Kunze B, Nordsiek G, Blöcker H, Höfle G, Müller R. The myxochelin iron transport regulon of the myxobacterium Stigmatella aurantiaca Sg a15. Eur J Biochem. 2000;267:6476–6485. doi: 10.1046/j.1432-1327.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- Kunze B, Bedorf N, Kohl W, Höfle G, Reichenbach H. Myxochelin A, a new iron-chelating compound from Angiococcus disciformis (Myxobacterales). Production, isolation, physico-chemical and biological properties. J Antibiot (Tokyo) 1989;42:14–17. doi: 10.7164/antibiotics.42.14. [DOI] [PubMed] [Google Scholar]
- Krone WJA, Stegehuis F, Koningstein G, van Doorn C, Roosendaal B, de Graaf FK, Oudega B. Characterization of the pColV-K30 encoded cloacin DF13/aerobactin outer membrane receptor protein of Escherichia coli ; isolation and purification of the protein and analysis of its nucleotide sequence and primary structure. FEMS Microbiol Lett. 1985;26:153–161. [Google Scholar]
- Gibson F, Magrath DI. The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim Biophys Acta. 1969;192:175–184. doi: 10.1016/0304-4165(69)90353-5. [DOI] [PubMed] [Google Scholar]
- Lynch D, O'Brien J, Welch T, Clarke P, Cuiv PO, Crosa JH, O'Connell M. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J Bacteriol. 2001;183:2576–2585. doi: 10.1128/JB.183.8.2576-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persmark M, Pittman P, Buyer JS, Schwyn B, Gill PR, Neilands JB. Isolation and structure of rhizobactin 1021, a siderophore from the alfalfa symbiont Rhizobium meliloti 1021. J Am Chem Soc. 1993;115:3950–3956. [Google Scholar]
- Kühn S, Braun V, Köster W. Ferric rhizoferrin uptake into Morganella morganii : characterization of genes involved in the uptake of a polyhydroxycarboxylate siderophore. J Bacteriol. 1996;178:496–504. doi: 10.1128/jb.178.2.496-504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel H, Metzger J, Freund S, Jung G, Boelaert JR, Winkelmann G. Rhizoferrin -- a novel siderophore from the fungus Rhizopus microsporus var. rhizopodiformis. BioMetals. 1991;4:238–243. [Google Scholar]
- Pressler U, Staudenmaier H, Zimmermann L, Braun V. Genetics of the iron dicitrate transport system of Escherichia coli. J Bacteriol. 1988;170:2716–2724. doi: 10.1128/jb.170.6.2716-2724.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AD, Chakraborty R, Smith BS, Esser L, Helm D van der, Deisenhofer J. Structural basis of gating by the outer membrane transporter FecA. Science. 2002;295:1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]
- Mey AR, Wyckoff EE, Hoover LA, Fisher CR, Payne SM. Vibrio cholerae VciB promotes iron uptake via ferrous iron transporters. J Bacteriol. 2008;190:5953–5962. doi: 10.1128/JB.00569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Vasil ML. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa : cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CD, Rinehart KL, Jr, Moore ML, Cook JC., Jr Pyochelin: novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1981;78:4256–4260. doi: 10.1073/pnas.78.7.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Hantke K. Ferrioxamine uptake in Yersinia enterocolitica : characterization of the receptor protein FoxA. Mol Microbiol. 1992;6:1309–1321. doi: 10.1111/j.1365-2958.1992.tb00852.x. [DOI] [PubMed] [Google Scholar]
- Keller-Schierlein W, Prelog V. Stoffwechselprodukte von Actinomyceten. 29. Mitteilung. Die Konstitution des Ferrioxamins D1. Helv Chim Acta. 1961;44:709–713. [Google Scholar]
- LeVier K, Guerinot ML. The Bradyrhizobium japonicum fegA gene encodes an iron-regulated outer membrane protein with similarity to hydroxamate-type siderophore receptors. J Bacteriol. 1996;178:7265–7275. doi: 10.1128/jb.178.24.7265-7275.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage C, Franza T, Expert D. Analysis of the Erwinia chrysanthemi ferrichrysobactin receptor gene: resemblance to the Escherichia coli fepA-fes bidirectional promoter region and homology with hydroxamate receptors. J Bacteriol. 1996;178:1227–1231. doi: 10.1128/jb.178.4.1227-1231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persmark M, Expert D, Neilands JB. Isolation, characterization, and synthesis of chrysobactin, a compound with siderophore activity from Erwinia chrysanthemi. J Biol Chem. 1989;264:3187–3193. [PubMed] [Google Scholar]
- Alice AF, López CS, Lowe CA, Ledesma MA, Crosa JH. Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243. J Bacteriol. 2006;188:1551–1566. doi: 10.1128/JB.188.4.1551-1566.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HM, Chaowagul W, Sokol PA. Siderophore production by Pseudomonas pseudomallei. Infect Immun. 1991;59:776–780. doi: 10.1128/iai.59.3.776-780.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol PA, Darling P, Lewenza S, Corbett CR, Kooi CD. Identification of a siderophore receptor required for ferric ornibactin uptake in Burkholderia cepacia. Infect Immun. 2000;68:6554–6560. doi: 10.1128/iai.68.12.6554-6560.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan H, Freund S, Beck W, Jung G, Meyer JM, Winkelmann G. Ornibactins--a new family of siderophores from pseudomonas. Biometals. 1993;6:93–100. doi: 10.1007/BF00140109. [DOI] [PubMed] [Google Scholar]
- Coulton JW, Mason P, Cameron DR, Carmel G, Jean R, Rode HN. Protein fusions of beta-galactosidase to the ferrichrome-iron receptor of Escherichia coli K-12. J Bacteriol. 1986;165:181–192. doi: 10.1128/jb.165.1.181-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalkin A, Forrester JD, Templeton DH. Crystal and molecular structure of ferrichrome A. Science. 1964;146:261–263. doi: 10.1126/science.146.3641.261. [DOI] [PubMed] [Google Scholar]
- Llamas MA, Sparrius M, Kloet R, Jiménez CR, Vandenbroucke-Grauls C, Bitter W. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J Bacteriol. 2006;188:1882–1891. doi: 10.1128/JB.188.5.1882-1891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Locht C. Expression of the putative siderophore receptor gene bfrZ is controlled by the extracytoplasmic-function sigma factor BupI in Bordetella bronchiseptica. J Bacteriol. 2001;183:2910–2917. doi: 10.1128/JB.183.9.2910-2917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito B, Aldon D, Barberis P, Boucher C, Genin S. A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol Plant Microbe Interact. 2002;15:109–119. doi: 10.1094/MPMI.2002.15.2.109. [DOI] [PubMed] [Google Scholar]
- Morris J, Donnelly DF, O'Neill E, McConnell F, O'Gara F. Nucleotide sequence analysis and potential environmental distribution of a ferric pseudobactin receptor gene of Pseudomonas sp. strain M114. Mol Gen Genet. 1994;242:9–16. doi: 10.1007/BF00277342. [DOI] [PubMed] [Google Scholar]
- Teintze M, Leong J. Structure of pseudobactin A, a second siderophore from plant growth promoting Pseudomonas B10. Biochemistry. 1981;20:6457–6462. doi: 10.1021/bi00525a026. [DOI] [PubMed] [Google Scholar]
- Poole K, Neshat S, Krebes K, Heinrichs DE. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J Bacteriol. 1993;175:4597–4604. doi: 10.1128/jb.175.15.4597-4604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philson SB, Llinás M. Siderochromes from Pseudomonas fluorescens. II. Structural homology as revealed by NMR spectroscopy. J Biol Chem. 1982;257:8086–8090. [PubMed] [Google Scholar]
- Bitter W, Marugg JD, de Weger LA, Tommassen J, Weisbeek PJ. The ferric-pseudobactin receptor PupA of Pseudomonas putida WCS358: homology to TonB-dependent Escherichia coli receptors and specificity of the protein. Mol Microbiol. 1991;5:647–655. doi: 10.1111/j.1365-2958.1991.tb00736.x. [DOI] [PubMed] [Google Scholar]
- Koster M, Vossenberg J van de, Leong J, Weisbeek PJ. Identification and characterization of the pupB gene encoding an inducible ferric-pseudobactin receptor of Pseudomonas putida WCS358. Mol Microbiol. 1993;8:591–601. doi: 10.1111/j.1365-2958.1993.tb01603.x. [DOI] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J Bacteriol. 1999;181:5958–5966. doi: 10.1128/jb.181.19.5958-5966.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio T, Tanaka N, Hiratake J, Katsube Y, Ishida Y, Oda J. Isolation and structure of the novel dihydroxamate siderophore alcaligin. J Am Chem Soc. 1988;110:8733–8734. [Google Scholar]
- Sauer M, Hantke K, Braun V. Sequence of the fhuE outer-membrane receptor gene of Escherichia coli K12 and properties of mutants. Mol Microbiol. 1990;4:427–437. doi: 10.1111/j.1365-2958.1990.tb00609.x. [DOI] [PubMed] [Google Scholar]
- Atkin CL, Neilands JB. Rhodotorulic acid, a diketopiperazine dihydroxamic acid with growth-factor activity. I. Isolation and characterization. Biochemistry. 1968;7:3734–3739. doi: 10.1021/bi00850a054. [DOI] [PubMed] [Google Scholar]
- Charlang G, Ng B, Horowitz NH, Horowitz RM. Cellular and extracellular siderophores of Aspergillus nidulans and Penicillium chrysogenum. Mol Cell Biol. 1981;1:94–100. doi: 10.1128/mcb.1.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhungana S, White PS, Crumbliss AL. Crystal structure of ferrioxamine B: a comparative analysis and implications for molecular recognition. J Biol Inorg Chem. 2001;6:810–818. doi: 10.1007/s007750100259. [DOI] [PubMed] [Google Scholar]
- Ankenbauer RG, Quan HN. FptA, the Fe(III)-pyochelin receptor of Pseudomonas aeruginosa : a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. J Bacteriol. 1994;176:307–319. doi: 10.1128/jb.176.2.307-319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J Biol Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- Actis LA, Tolmasky ME, Farrell DH, Crosa JH. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J Biol Chem. 1988;263:2853–2860. [PubMed] [Google Scholar]
- Wuest WM, Sattely ES, Walsh CT. Three siderophores from one bacterial enzymatic assembly line. J Am Chem Soc. 2009;131:5056–5057. doi: 10.1021/ja900815w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey CW, Tomaras AP, Connerly PL, Tolmasky ME, Crosa JH, Actis LA. The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiol. 2004;150:3657–3667. doi: 10.1099/mic.0.27371-0. [DOI] [PubMed] [Google Scholar]
- Koebnik R, Hantke K, Braun V. The TonB-dependent ferrichrome receptor FcuA of Yersinia enterocolitica : evidence against a strict co-evolution of receptor structure and substrate specificity. Mol Microbiol. 1993;7:383–393. doi: 10.1111/j.1365-2958.1993.tb01130.x. [DOI] [PubMed] [Google Scholar]
- Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica : a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Drechsel H, Stephan H, Lotz R, Haag H, Zähner H, Hantke K, Jung G. Structure elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs Annalen. 1995;10:1727–1733. [Google Scholar]
- Fetherston JD, Lillard JW, Jr, Perry RD. Analysis of the pesticin receptor from Yersinia pestis : role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterton JR, Stoebner JA, Payne SM, Calderwood SB. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J Bacteriol. 1992;174:3729–3738. doi: 10.1128/jb.174.11.3729-3738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths GL, Sigel SP, Payne SM, Neilands JB. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem. 1984;259:383–385. [PubMed] [Google Scholar]
- Kahnert A, Kertesz MA. Characterization of a sulfur-regulated oxygenative alkylsulfatase from Pseudomonas putida S-313. J Biol Chem. 2000;275:31661–31667. doi: 10.1074/jbc.M005820200. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J Bacteriol. 2006;188:317–327. doi: 10.1128/JB.188.1.317-327.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P, Linker A, Lippman V, Meyer K. The structure of chondroitin sulfate B from studies with Flavobacterium enzymes. J Biol Chem. 1960;235:3066–3069. [PubMed] [Google Scholar]
- Atkins ED, Sheehan JK. The molecular structure of hyaluronic acid. Biochem J. 1971;125:92. doi: 10.1042/bj1250092pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B, Dalwadi H, Gordon LK, Landers C, Bruckner D, Targan SR, Braun J. Molecular cloning of a Bacteroides caccae TonB-linked outer membrane protein identified by an inflammatory bowel disease marker antibody. Infect Immun. 2001;69:6044–6054. doi: 10.1128/IAI.69.10.6044-6054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Rodionov DA, Li X, Laikova ON, Gelfand MS, Zagnitko OP, Romine MF, Obraztsova AY, Nealson KH, Osterman AL. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J Biol Chem. 2006;281:29872–29885. doi: 10.1074/jbc.M605052200. [DOI] [PubMed] [Google Scholar]
- Karlsen S, Hough E. Crystal structures of three complexes between chito-oligosaccharides and lysozyme from the rainbow trout. How distorted is the NAG sugar in site D? Acta Crystallogr D Biol Crystallogr. 1995;51:962–978. doi: 10.1107/S0907444995005105. [DOI] [PubMed] [Google Scholar]
- Pauer H, Ferreira Ede O, dos Santos-Filho J, Portela MB, Zingali RB, Soares RM, Domingues RM. A TonB-dependent outer membrane protein as a Bacteroides fragilis fibronectin-binding molecule. FEMS Immunol Med Microbiol. 2009;55:388–395. doi: 10.1111/j.1574-695X.2009.00532.x. [DOI] [PubMed] [Google Scholar]
- Koebnik R. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 2005;13:343–347. doi: 10.1016/j.tim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Beucher M, Sparling PF. Cloning, sequencing, and characterization of the gene encoding FrpB, a major iron-regulated, outer membrane protein of Neisseria gonorrhoeae. J Bacteriol. 1995;177:2041–2049. doi: 10.1128/jb.177.8.2041-2049.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Payne SM. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol Microbiol. 2001;42:835–849. doi: 10.1046/j.1365-2958.2001.02683.x. [DOI] [PubMed] [Google Scholar]
- Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: do we only see the 'tip of the iceberg'? Trends Biochem Sci. 2008;33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Ren Q, Paulsen IT. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput Biol. 2005;1:e27. doi: 10.1371/journal.pcbi.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Parmryd I, Nilsson F, Persson AL, Pakrasi HB, Andersson B, Norling B. Proteomics of Synechocystis sp. strain PCC 6803: identification of plasma membrane proteins. Mol Cell Proteomics. 2002;1:956–966. doi: 10.1074/mcp.m200043-mcp200. [DOI] [PubMed] [Google Scholar]
- Jeanjean R, Talla E, Latifi A, Havaux M, Janicki A, Zhang CC. A large gene cluster encoding peptide synthetases and polyketide synthases is involved in production of siderophores and oxidative stress response in the cyanobacterium Anabaena sp. strain PCC 7120. Environ Microbiol. 2008;10:2574–2585. doi: 10.1111/j.1462-2920.2008.01680.x. [DOI] [PubMed] [Google Scholar]
- Guglielmi G, Cohen-Bazire G, Bryant DA. The structure of Gloeobacter violaceus and its phycobilisomes. Arch Microbiol. 1981;129:181–189. [Google Scholar]
- Adolph KW, Haselkorn R. Isolation and characterization of a virus infecting the blue-green alga Nostoc muscorum. Virology. 1971;46:200–208. doi: 10.1016/0042-6822(71)90023-7. [DOI] [PubMed] [Google Scholar]
- Dean WE, Neff BP, Rosenberry DO, Winter TC, Parkhurst R. The significance of ground water to the accumulation of iron and manganese in the sediments of two hydrologically distinct lakes in North-Central Minnesota: A geological perspective. Ground Water. 2003;41:951–963. [Google Scholar]
- Moslavac S, Bredemeier R, Mirus O, Granvogl B, Eichacker LA, Schleiff E. Proteomic analysis of the outer membrane of Anabaena sp. strain PCC 7120. J Proteome Res. 2005;4:1330–1338. doi: 10.1021/pr050044c. [DOI] [PubMed] [Google Scholar]
- Moslavac S, Reisinger V, Berg M, Mirus O, Vosyka O, Plöscher M, Flores E, Eichacker LA, Schleiff E. The proteome of the heterocyst cell wall in Anabaena sp. PCC 7120. Biol Chem. 2007;388:823–829. doi: 10.1515/BC.2007.079. [DOI] [PubMed] [Google Scholar]
- Chu BC, Peacock RS, Vogel HJ. Bioinformatic analysis of the TonB protein family. Biometals. 2007;20:467–483. doi: 10.1007/s10534-006-9049-4. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Finn R. Pfam: a domain-centric method for analyzing proteins and proteomes. Methods Mol Biol. 2007;396:43–58. doi: 10.1007/978-1-59745-515-2_4. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement and accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Vinh LS, von Haeseler A, Schmidt HA. pIQPNNI -- Parallel reconstruction of large maximum likelihood phylogenies. Bioinformatics. 2005;21:3794–3796. doi: 10.1093/bioinformatics/bti594. [DOI] [PubMed] [Google Scholar]
- Müller T, Vingron M. Modeling amino acid replacement. J Comput Biol. 2000;7:761–776. doi: 10.1089/10665270050514918. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. Tree-Puzzle: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Cai YP, Wolk CP. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares A, Muro-Pastor AM, Fillat MF, Herrero A, Flores E. Constitutive and nitrogen-regulated promoters of the petH gene encoding ferredoxin:NADP+ reductase in the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 1999;449:159–164. doi: 10.1016/s0014-5793(99)00404-4. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kohara M, Matsumoto M, Matsuno A, Muraki A, Nakazaki N, Shimpo S, Sugimoto M, Takazawa M, Yamada M, Yasuda M, Tabata S. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2001;8:205–213. doi: 10.1093/dnares/8.5.205. 227-253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of experimentally characterized TBDTs and of TBDTs with predicted substrate used for classification.
Number of TBDTs detected in analyzed genomes.