Abstract
Purpose
Evidence has accumulated in recent years suggestive of a genetic basis for a susceptibility to the development of radiation injury following cancer radiotherapy. The purpose of this study was to assess whether patients with severe radiation-induced sequelae (RIS, i.e. NCI/CTCv3.0 grade ≥3) display both a low capacity of radiation-induced CD8 lymphocyte apoptosis (RILA) in vitro and possess certain single nucleotide polymorphisms (SNPs) located in candidate genes associated with the response of cells to radiation.
Experimental Design
DNA was isolated from blood samples obtained from patients (n = 399) included in the Swiss prospective study evaluating the predictive effect of in vitro RILA and RIS. SNPs in the ATM, SOD2, XRCC1, XRCC3, TGFB1, and RAD21 genes were screened in patients who experienced severe RIS (group A, n = 16 ) and control subjects who did not manifest any evidence of RIS (group B, n = 18).
Results
Overall, 13 and 21 patients were found to possess a total of <4 and ≥4 SNPs in the candidate genes. The median (range) RILA in group A was 9.4% (5.3–16.5) and 94% (95%CI: 70–100) of the patients (15/16) had ≥4 SNPs. In group B, median (range) RILA was 25.7% (20.2– 43.2) and 33% (95%CI: 13–59) of patients (6/18) had ≥4 SNPs (p < 0.001).
Conclusions
The results of this study suggest that patients with severe RIS possess 4 or more SNPs in candidate genes and low radiation-induced CD8 lymphocyte apoptosis in vitro.
Keywords: Apoptosis; CD8-Positive T-Lymphocytes; metabolism; Case-Control Studies; Fibrosis; Genetic Predisposition to Disease; Genotype; Humans; Neoplasms; genetics; pathology; radiotherapy; Polymorphism, Single Nucleotide; Radiation Injuries; diagnosis; Radiation Tolerance; adverse effects; Risk; Time Factors; Treatment Outcome
Radiation injury may develop months to years following radiotherapy (RT) with the manifestation of diverse pathological lesions such as fibrosis, necrosis, atrophy, and vascular damages (1). In recognition of the large number of people who survive for many years following a cancer diagnosis, the National Cancer Institute has identified cancer survivorship and the development of adverse late sequelae resulting from the use of therapeutic modalities, including RT, as an important area that has been under-researched (2).
A variety of patient, tumor, treatment, cellular, and molecular factors contribute to the variability in severity of normal tissue reactions exhibited after RT. In addition, it has been hypothesized that individual intrinsic radiosensitivity may influence the development of adverse radiation responses (3). Therefore, efforts have been made to develop assays capable of predicting susceptibility for the development of radiation injury that would allow customization of RT protocols on an individual basis (3, 4). By doing so, it has been estimated that a significant improvement in the therapeutic index could be achieved. In the clinic, two complementary approaches have been explored with encouraging results: the lymphocyte assay (4) and single nucleotide polymorphism (SNP) screening (3).
Lymphocyte apoptosis was developed as a rapid tool for characterization of normal tissue radiosensitivity (5–8), particularly due to the ease of blood collection in a standardized, patient-convenient manner. In a prospective study, blood samples were obtained from 399 patients treated with curative intent and tested using CD8 lymphocyte apoptosis following an X-ray dose of 8 Gy. Apoptosis was assessed by associated condensation of DNA in lymphocytes. The 2-year cumulative incidence for grade 2 or 3 late toxicity was 70, 32, and 12 percent for patients with absolute changes in CD8 T-lymphocyte apoptosis of ≤16, 16–24, and >24 percent, respectively (4).
Consistent with these results are those reported by Svensson et al. (9) who examined the expression of a series of genes associated with radiation response in radiotherapy patients with or without late radiation toxicity. It was reported that the majority of the discriminative genes and gene sets belonged to the ubiquitin, apoptosis, and stress signaling networks. In addition, it has been reported that possession of variants in genes, the products of which play a role in radiation response, may be associated with the development of adverse effects after RT (3). Candidate genes that possess SNPs associated with RIS include ATM, TGFB1, XRCC1, XRCC3, SOD2, and RAD21. The selection of these specific six genes for SNP screening was based upon previous results indicating that these genes possess SNPs associated with the development of adverse effects from RT (3, 10–14).
Here we present evidence that patients with severe RIS display both a low radiation-induced CD8 lymphocyte apoptosis (RILA) in vitro and possess SNPs located in candidate genes associated with the response of cells to radiation.
Materials and Methods
Blood samples
Between April 1998 and October 2001, a total of 399 patients with miscellaneous cancers were included in the KFS 00539-9-1997/SKL 00778-2-1999 prospective study evaluating the predictive value of CD4 and CD8 T-lymphocyte apoptosis on the development of radiation-induced late side effects (4). Among them, twenty-eight patients (7%) developed grade 3 sequelae. These latter patients experienced 32 grade 3 late side effects: 14 subcutaneous, 5 skin, 9 salivary gland, 1 brain, 1 pharynx, 1 ear and 1 mucous membrane. At the 2-year visit, 24 patients maintained the same grade 3 late toxicity. Grade 3 late toxicities were mainly observed for patients with breast (9%), and head and neck (16%) cancers. These patients were invited to participate in the Gene-Pare project(3) for genetic profile evaluations (see details below). Sixteen patients (group A) accepted this new study approved by the Lausanne University Ethical Committee and came for a medical visit where informed consent were signed before sending blood samples to the Mount Sinai hospital in New York (Pr Rosenstein) for genetic screening. Details of all patients were described (4) but briefly, 9/16 (56%) and 6/16 (38%) breast and head and neck-cancer patients, respectively, experienced grade 3 subcutaneous fibrosis. One patient (6%) treated for a localized meningioma presented a severe intracerebral edema with long term treatment of corticoids.
The second step was to include “control” patients (group B) from this trial treated for breast and head and neck cancers and meningioma without any long term clinical signs of radiation-induced toxicities. We stopped accrual when a “matched” sample size was obtained in comparison with group A, i.e. 10 breast, 7 head and neck and 1 meningioma. The selection of the “matched” patients was made from the entire population included initially in the prospective trial. Just before the genetic screening but after identifying patients who suffered from severe side-effects, we selected patients who did not present any radiation-induced sequelae. The selection was made according to the type and location of the tumor, initial tumor size, total radiation dose, irradiated volume, concurrent tamoxifen (breast) or cisplatin (head-and-neck) administration. No statistical difference was found between the two groups.
Blood samples were sent to the Mount Sinai hospital in New York for genetic screening. The human investigations were performed after approval by the local institutional review board.
Toxicity assessments
Toxicities were graded according to the RTOG/EORTC system in all patients by the same physician without knowledge of any translational studies so as not to bias the evaluation (15). The timing of late side effects from 6 weeks post RT up to 2 years corresponds to the time of observation of the worst late toxicity grade. The exposure to concomitant RT chemotherapy (head and neck cancer patients), concomitant hormonotherapy and post RT chemotherapy (mainly breast cancer patients) as well as all technical RT modalities (irradiated volume, total dose, dose at the surface of the skin, dose fractionation) were also taken into account to evaluate late side effects (4, 16).
Genetic screening
The lymphocyte isolation, DNA extraction and denaturing high performance liquid chromatography (DHPLC) procedures were performed as previously described (12). All PCR products were subjected to either DHPLC or the Surveyor nuclease assay using a Transgenomic WAVE High Sensitivity Nucleic Acid Fragment Analysis System. PCR primers for the DNA amplicons encompassing the SNPs of interest were designed using the genomic sequence obtained from NCBI (http://www.ncbi.nlm.nih.gov), and the online primer design program Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The SNPs in ATM were identified using DHPLC whereas the SNPs in all other candidate genes were detected using the Surveyor nuclease assay which was performed as described (17–20).
Briefly, a mixture was added to each 25 μl PCR reaction product consisting of 0.5 μl 10X buffer, 1 μl Surveyor nuclease W, 1 μl Enhancer W, and 2.5 μl ddH2O (Transgenomic Surveyor kit). The reaction was incubated at 42°C for 20 minutes. At this time, the reaction was terminated by adding 5 μl of a mixture of 3 μl Stop Buffer and 2 μl ddH2O. For each primer set, a gradient was chosen using Navigator software (Transgenomic) based on the size of the PCR fragment. The “DS multiple fragments” setting was used at 45°C with the addition of the dye WAVE-HS1 (Transgenomic). The fragment length minimum was set to 25 bp, and the fragment length maximum was set to roughly 50–75 bp larger than the size of the fragment. The resulting traces were examined for the presence of multiple peaks suggestive of heterozygosity for the SNP, comparing the sizes against a low molecular weight DNA ladder (New England Biolabs). To identify patients who may be homozygous for the minor allele for each SNP, an equal amount of DNA derived from a subject known to be homozygous for the major allele was mixed with each DNA sample prior to the PCR to create a potentially heterozygous SNP if that patient had been homozygous for the minor allele. Any samples for which the chromatogram differed from that obtained for DNA homozygous for the major allele were subjected to DNA sequencing using an ABI BigDye Terminator Version 3.1 Ready Reaction Cycle Sequencing kits and an ABI 3730xl DNA Analyzer. All analyses were blinded to treatment, side-effects, and RILA yield.
Radiation-induced CD8 lymphocyte apoptosis
As described (4, 5, 8), heparinized whole blood collected before starting RT was diluted 1:10 in RPMI 1640 medium containing 20 percent fetal bovine serum, irradiated with 0- and 8- Gy, and incubated for 48 h. The cells were then labeled with FITC-conjugated anti-CD8 monoclonal antibodies, red blood cells were lysed, and the DNA of the remaining cells stained with propidium iodide (PI). Samples were measured using a FACScan flow cytometer and data analysis performed using CellQuest software (Becton-Dickinson, San Jose, CA). Apoptotic CD8 T-lymphocytes were defined as those cells staining positively for their cell-type-specific antibodies, and displaying reduced PI fluorescence and cell size. These cells were previously demonstrated to be apoptotic using the TUNEL assay (8). Data from at least 10,000 cells/sample were acquired.
In our earlier study (4), a decreased percentage of grade 2 or more late toxicity was observed for increasing values of CD4 and CD8. No grade 3 side effects were observed for patients with CD4 apoptosis >15% and CD8 apoptosis >24%. A multivariate ROC analysis for CD4 and CD8 was performed among the patients who were evaluated at 2 years, or who experienced a grade ≥2 before 2 years. The area under the ROC curve was greater for CD8 (0.827 [95% CI: 0.78– 0.87]) than for CD4 (0.714 [95% CI: 0.66–0.77]). The addition of CD4 into the model did not contribute significantly in separating the two groups (AUC = 0.84). Therefore, we decided to take into account only CD8 results for the present study. In the prospective trial (4), all apoptosis yields were blindly collected by a technician before final analyses.
Statistical Methods
Fisher’s exact test was used for the comparison of 2x2 contingency tables. An odd’s ratio (OR) was estimated from the probabilities of developing grade ≥3 toxicity between groups A and B. 95% confidence intervals for probabilities were obtained from the binomial distribution. Youden’s index associated with ROC curves was obtained from sensitivity (Se) and specificity (Sp) as Se+Sp−1. This index was used to determine the best cut-off point for the number of SNPs. Median values for continuous variables were compared between groups with the Wilcoxon non parametric test.
Results
Genotype distributions and severe radiation-induced sequelae
The genotype distributions are depicted in Figures 1 and 2. A putative model for estimation of fibrosis risk based on multiple SNPs was used as has been described (13). Overall, 13 and 21 patients were found to possess a total of <4 and ≥4 SNPs in the candidate genes screened (ATM, TGFβ, SOD2, XRCC1, XRCC3, RAD21). Among the 16 patients who experienced grade ≥3 sequelae (group A), 15 (94%, 95%CI: 70–100%) carried ≥4 SNPs compared to 6/18 (33%, 95%CI: 13–59%) patients without any clinical signs of RT-induced late toxicity (p < 0.001).
Fig. 1.
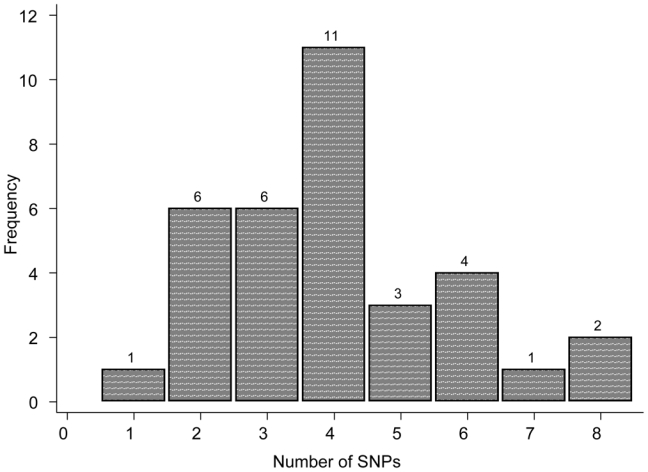
Distribution of the number of Single Nucleotide Polymorphisms (SNPs) in the study population. The candidate genes were selected from previous studies as ATM, SOD2, TGFb, XRCC3, XRCC1, RAD21. Thirteen and 21 patients presented less than three and four or more SNPs, respectively.
Fig. 2.
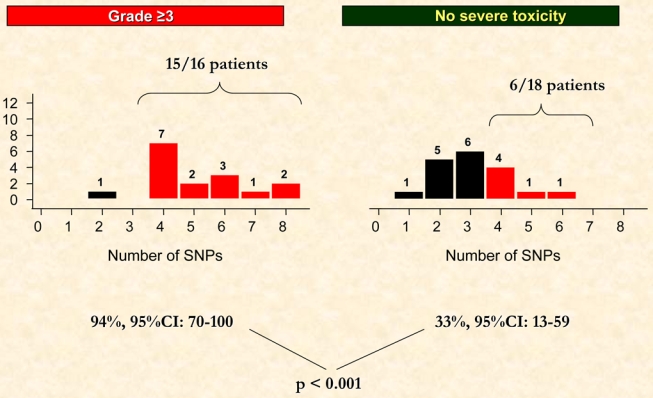
Distribution of the number of Single Nucleotide Polymorphisms (SNPs) according to radiation-induced late effects (grade ≥3 vs grade <3). Ninety-four percent (15/16) patients presented ≥4 SNPs in the grade ≥3 toxicity group compared to 33% (6/18) in the “control” group without any severe clinical radiation-induced toxicity (p <0.001).
The probability for the development of grade ≥3 toxicity (Figure 3) was significantly higher (OR 9.3, 95% CI: 1.4–62, p = 0.003) for patients with ≥4 SNPs (0.71, 95%CI: 0.48–0.89) compared to those with <4 SNPs (0.08, 95%CI 0.01–0.36). In terms of sensitivity and specificity, as can be seen from Figure 2, the cut-off at 4 or more SNPs gave the largest values for Youden’s index with sensitivity (true positives) = 94% (15/16) and specificity (true negatives) = 67% (12/18) corresponding to an index of 0.61. At the 4 or more cut-off value, the positive predictive value (PPV) was 71% and the negative predictive value (NPV) was 92%. Few patients (10/34) had 5 or more SNPs in our study. Youden’s index was lower and equal to 0.39 (sensitivity = 50% and specificity = 89%) and PPV = 80% and NPV = 67%.
Fig. 3.
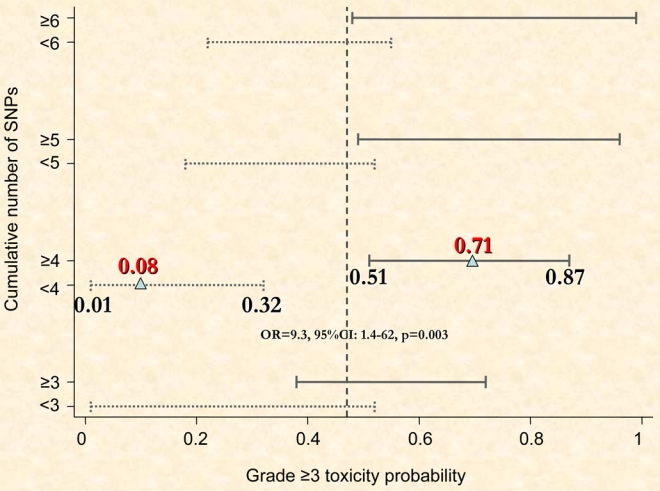
The probability of grade 3 or more toxicity as a function of the cumulative number of Single Nucleotide Polymorphisms (SNPs). Four and more SNPs are strongly correlated with the risk of grade ≥3 toxicity; OR, odds ratio = 9.3, 95%CI: 1.4–62, p = 0.003).
Taken individually, the total numbers of SNPs detected in ATM 5557 G>A (number of patients, n=15), SOD2 47 T>C (n=22), TGFB1 -509 C>T (n=18) and 870 T>C (n=24), XRCC1 1321 G>A (n=15), XRCC3 1075 C>T (n=18), and RAD21 1625 T>C (n=10) were greater in group A compared with group B (Table 1).
Table 1.
Number of patients presenting Specific Single Nucleotide Polymorphisms (SNPs) according to both groups (A, grade ≥ 3 late effects and B, no severe late effects)
| Number of patients (%) |
||
|---|---|---|
| Type of SNPs | Grade ≥ 3 late effects group | No severe late effects group |
| n = 16 | n = 18 | |
| ATM 5557 G>A | 9 (56%) | 6 (33%) |
| SOD2 47 T>C | 13 (81%) | 9 (50%) |
| TGFB1 -509 C>T | 11 (69%) | 7 (39%) |
| TGFB1 870 T>C | 15 (94%) | 9 (50%) |
| XRCC1 1321 G>A | 8 (50%) | 7 (39%) |
| XRCC3 1075 C>T | 9 (56%) | 9 (50%) |
| RAD21 1625 T>C | 7 (44%) | 3 (17%) |
Low radiation-induced CD8 lymphocyte apoptosis (RILA), number of SNPs, and radiation-induced sequelae
Grade 3 late toxicities were mainly observed for patients with breast (9%), and head and neck (16%) cancers with an inverse relationship between the incidence of late side effects and percent CD8 apoptosis. The median (range) RILA in group A was 9.4% (5.3–16.5) and 94% (95%CI: 70–100) of patients (15/16) had ≥4 SNPs.
In our control population (group B), median (range) RILA was 25.7% (20.2–43.2) and 33% (95%CI: 13–59) of patients (6/18) had ≥4 SNPs (p < 0.001, Figure 2).
Discussion
Normal-tissue radiobiology has achieved notable advances mainly through an improved mechanistic understanding of molecular pathogenesis (21). Radiation-induced tissue remodeling is a complex process driven by intercellular communication via cytokines and growth factors, which are induced during the radiation response of participating cells (22).
Mechanistically, the early steps of radiation-induced fibrosis are similar to those of a wound-healing response characterized by an upregulation of cytokines with proinflammatory recruitment of cells within the surrounding irradiated tissue (21). Transforming growth factor-β (TGFβ) is secreted in a latent form and may be activated directly in the extracellular space by a triggering event such as ionizing radiation (23). The second key process characteristic of radiation fibrogenesis is damage of endothelial cells (24), which activates the apoptosis death program and also leads to release of pro-fibrotic cytokines, mainly TGFβ (21).
We reported that radiation-induced T-lymphocyte apoptosis can be predictive for grade 2 and 3 late effects (P <0.0001). Patients with grade 3 late effects showed CD8 radiation-induced apoptosis significantly (P <0.0001) below the median (4).
Regarding the relationship between genetic variation and clinical radiosensitivity (3, 13), we screened for SNPs in candidate genes that are associated with the response of cells to radiation; ATM, TGFβ1, SOD2, XRCC3, XRCC1, and RAD21. It has been reported (8) that AT patients, who are homozygous for mutations in ATM, display a low apoptotic response of CD4 and CD8 lymphocytes. Therefore, it is plausible that SNPs in genes which encode proteins involved with apoptosis may affect the apoptotic response in those cells.
Recent clinical investigations have suggested that fibrosis risk is not only dependent on the type of the SNPs but also on the number of ‘risk alleles’ (14). The results reported in the present article reinforce this hypothesis and indicate that possession of multiple SNPs associated with radiosensitivity correlates with an increased probability for developing severe RIS.
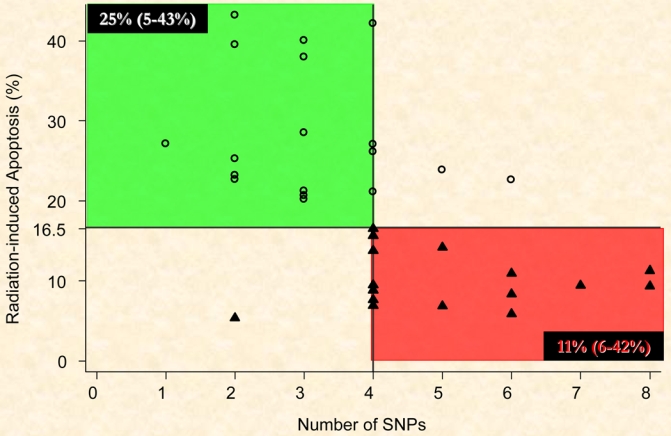
Of particular note may be the inverse correlation between RILA and the number of SNPs in the candidate genes screened, whose products are involved in both fibrosis and apoptosis following irradiation (3, 9, 11, 25). It is our hypothesis that radiation-induced late effects are exacerbated in tissues with reduced apoptosis since a diminished apoptotic cell death response may enhance the probability for induction the cytokine cascade induced by radiation that result in a pro-inflammatory response resulting in long-term adverse responses. We should be able to verify this hypothesis in our on-going COHORT trial (NCI PDQ: NCT00208273) with 150 patients specifically designed to evaluate this association.
Further investigations are strongly warranted to identify more comprehensively the SNPs that are associated with RIS and may provide the basis for an assay to predict which patients are at greatest risk for the development of radiation injury, particularly in the current era of combined modality treatments (16, 26, 27).
Acknowledgments
Grateful appreciation is extended to Ms. Michèle Ben Sta and Eliane Cottin for their technical assistance.
Grant Support: This study was supported by US Department of the Army (grant DAMD 17-02-1-0503), the Swiss Cancer League (KFS 00539-9-1997 and SKL 00778-2-1999), the Programme Hospitalier de Recherche Clinique (PHRC) 2005 from the French “Institut National du Cancer” (INCa) and was included in the Gene-PARE project (Genetic Predictors of Adverse Radiotherapy Effects).
References
- 1.Stone HB, Coleman CN, Anscher MS, McBride WH. Effects of radiation on normal tissue: consequences and mechanisms. Lancet Oncol. 2003;4:529–36. doi: 10.1016/s1470-2045(03)01191-4. [DOI] [PubMed] [Google Scholar]
- 2.Aziz NM. Cancer survivorship research: state of knowledge, challenges and opportunities. Acta Oncol. 2007;46:417–32. doi: 10.1080/02841860701367878. [DOI] [PubMed] [Google Scholar]
- 3.Ho AY, Atencio DP, Peters S, et al. Genetic predictors of adverse radiotherapy effects: the Gene-PARE project. Int J Radiat Oncol Biol Phys. 2006;65:646–55. doi: 10.1016/j.ijrobp.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Ozsahin M, Crompton NE, Gourgou S, et al. CD4 and CD8 T-Lymphocyte Apoptosis Can Predict Radiation-Induced Late Toxicity: A Prospective Study in 399 Patients. Clin Cancer Res. 2005;11:7426–33. doi: 10.1158/1078-0432.CCR-04-2634. [DOI] [PubMed] [Google Scholar]
- 5.Crompton NE, Miralbell R, Rutz HP, et al. Altered apoptotic profiles in irradiated patients with increased toxicity. Int J Radiat Oncol Biol Phys. 1999;45:707–14. doi: 10.1016/s0360-3016(99)00256-4. [DOI] [PubMed] [Google Scholar]
- 6.Crompton NE, Ozsahin M. A versatile and rapid assay of radiosensitivity of peripheral blood leukocytes based on DNA and surface-marker assessment of cytotoxicity. Radiat Res. 1997;147:55–60. [PubMed] [Google Scholar]
- 7.Crompton NE, Shi YQ, Emery GC, et al. Sources of variation in patient response to radiation treatment. Int J Radiat Oncol Biol Phys. 2001;49:547–54. doi: 10.1016/s0360-3016(00)01477-2. [DOI] [PubMed] [Google Scholar]
- 8.Ozsahin M, Ozsahin H, Shi Y, Larsson B, Wurgler FE, Crompton NE. Rapid assay of intrinsic radiosensitivity based on apoptosis in human CD4 and CD8 T-lymphocytes. Int J Radiat Oncol Biol Phys. 1997;38:429–40. doi: 10.1016/s0360-3016(97)00038-2. [DOI] [PubMed] [Google Scholar]
- 9.Svensson JP, Stalpers LJ, Esveldt-van Lange RE, et al. Analysis of gene expression using gene sets discriminates cancer patients with and without late radiation toxicity. PLoS medicine. 2006;3:e422. doi: 10.1371/journal.pmed.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall EJ, Schiff PB, Hanks GE, et al. A preliminary report: frequency of A-T heterozygotes among prostate cancer patients with severe late responses to radiation therapy. The cancer journal from Scientific American. 1998;4:385–9. [PubMed] [Google Scholar]
- 11.Severin DM, Leong T, Cassidy B, et al. Novel DNA sequence variants in the hHR21 DNA repair gene in radiosensitive cancer patients. Int J Radiat Oncol Biol Phys. 2001;50:1323–31. doi: 10.1016/s0360-3016(01)01608-x. [DOI] [PubMed] [Google Scholar]
- 12.Iannuzzi CM, Atencio DP, Green S, Stock RG, Rosenstein BS. ATM mutations in female breast cancer patients predict for an increase in radiation-induced late effects. Int J Radiat Oncol Biol Phys. 2002;52:606–13. doi: 10.1016/s0360-3016(01)02684-0. [DOI] [PubMed] [Google Scholar]
- 13.Andreassen CN, Alsner J, Overgaard M, Overgaard J. Prediction of normal tissue radiosensitivity from polymorphisms in candidate genes. Radiother Oncol. 2003;69:127–35. doi: 10.1016/j.radonc.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Andreassen CN, Alsner J, Overgaard J, et al. TGFB1 polymorphisms are associated with risk of late normal tissue complications in the breast after radiotherapy for early breast cancer. Radiother Oncol. 2005;75:18–21. doi: 10.1016/j.radonc.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Cox JD, Stetz J, Pajak TF. Toxicity criteria of the Radiation Therapy Oncology Group (RTOG) and the European Organization for Research and Treatment of Cancer (EORTC) Int J Radiat Oncol Biol Phys. 1995;31:1341–6. doi: 10.1016/0360-3016(95)00060-C. [DOI] [PubMed] [Google Scholar]
- 16.Azria D, Gourgou S, Sozzi WJ, et al. Concomitant use of tamoxifen with radiotherapy enhances subcutaneous breast fibrosis in hypersensitive patients. Br J Cancer. 2004;91:1251–60. doi: 10.1038/sj.bjc.6602146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu P, Shandilya H, D’Alessio JM, O’Connor K, Durocher J, Gerard GF. Mutation detection using Surveyor nuclease. BioTechniques. 2004;36:702–7. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- 18.Bannwarth S, Procaccio V, Paquis-Flucklinger V. Surveyor Nuclease: a new strategy for a rapid identification of heteroplasmic mitochondrial DNA mutations in patients with respiratory chain defects. Hum Mutat. 2005;25:575–82. doi: 10.1002/humu.20177. [DOI] [PubMed] [Google Scholar]
- 19.Shi R, Otomo K, Yamada H, Tatsumi T, Sugawara I. Temperature-mediated heteroduplex analysis for the detection of drug-resistant gene mutations in clinical isolates of Mycobacterium tuberculosis by denaturing HPLC, SURVEYOR nuclease. Microbes and infection/Institut Pasteur. 2006;8:128–35. doi: 10.1016/j.micinf.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Janne PA, Borras AM, Kuang Y, et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006;12:751–8. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- 21.Bentzen SM. Preventing or reducing late side effects of radiation therapy: radiobiology meets molecular pathology. Nat Rev Cancer. 2006;6:702–13. doi: 10.1038/nrc1950. [DOI] [PubMed] [Google Scholar]
- 22.Rodemann HP, Blaese MA. Responses of normal cells to ionizing radiation. Semin Radiat Oncol. 2007;17:81–8. doi: 10.1016/j.semradonc.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence DA. Latent-TGF-beta: an overview. Molecular and cellular biochemistry. 2001;219:163–70. doi: 10.1023/a:1010819716023. [DOI] [PubMed] [Google Scholar]
- 24.Milliat F, Francois A, Isoir M, et al. Influence of endothelial cells on vascular smooth muscle cells phenotype after irradiation: implication in radiation-induced vascular damages. Am J Pathol. 2006;169:1484–95. doi: 10.2353/ajpath.2006.060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moullan N, Cox DG, Angele S, Romestaing P, Gerard JP, Hall J. Polymorphisms in the DNA repair gene XRCC1, breast cancer risk, and response to radiotherapy. Cancer Epidemiol Biomarkers Prev. 2003;12:1168–74. [PubMed] [Google Scholar]
- 26.Azria D, Pelegrin A, Dubois JB, Mirimanoff RO, Ozsahin M. Radiation Therapy and Tamoxifen: Concurrent or Sequential? It’s No Longer the Question! J Clin Oncol. 2005;23:4239–41. doi: 10.1200/JCO.2004.00.8623. [DOI] [PubMed] [Google Scholar]
- 27.Azria D, Rosenstein BS, Ozsahin M. Radiation-induced side-effects with or without systemic therapies: Primetime for prediction of individual radiosensitivity! Int J Radiat Oncol Biol Phys. 2008 doi: 10.1016/j.ijrobp.2008.03.057. in press. [DOI] [PubMed] [Google Scholar]


