Alignment is an essential component of contemporary protein NMR studies, especially for membrane proteins whose highly asymmetric structures are dominated by α-helices or β-sheets and are difficult to characterize based on short-range distance or intramolecular angle measurements. Solid-state NMR can take advantage of the immobilization and complete alignment of the protein in mechanically or magnetically aligned phospholipid bilayers.1 In contrast, weak alignment of detergent solubilized membrane proteins preserves the rapid molecular reorientation required for structure determination by solution NMR. Structure determination by solution NMR is complementary to that by X-ray diffraction of membrane proteins crystallized from the same types of detergent solutions, and it has the advantage that it can be performed on proteins that are resistant to crystallization.
Residual dipolar couplings (RDCs)2 are the principal source of structural constraints in solution NMR studies of membrane proteins because of the near-total absence of observable long-range NOEs, and the requirement of paramagnetic relaxation enhancement measurements for multiple mutations and chemical modifications.3 Unfortunately, the most widely used media for the alignment of globular proteins cannot be applied to membrane proteins, since they are immobilized by interactions with the long chain lipids in bicelles and the solubilizing detergents destroy the integrity of filamentous bacteriophages whose major coat protein is itself a membrane protein before particle assembly. These limitations led to the rapid adaptation of strained polyacrylamide gels4 as alignment media for membrane proteins. Presently, the limiting factor is obtaining a second alignment suitable for RDC measurements, since RDC data sets from two different protein alignments enable the direct analysis of the secondary structure with Dipolar Waves5 and the direct determination of the alignment tensors with λ-map plots6 without which the calculation of the three-dimensional structures can not proceed.
Although some success has been reported with the use of charged gels,7 additional alignment methods compatible with the detergents used to solubilize membrane proteins are needed. Binding of lanthanide ions to the membrane proteins, including those modified to interact specifically with metals, has been shown to induce alignment,8 however, there is always a concern about the effects of the paramagnetic metals or the requisite modifications. Recently, DNA-based liquid crystals have been shown to align detergent-solubilized membrane proteins.9
We describe here a general approach to obtaining a second alignment of membrane proteins that utilizes experimental conditions where fd bacteriophage remains intact in the presence of high concentrations (ca. 100 mM) of many of the detergents most commonly used to solubilize membrane protein, including short chain phospholipids, e.g., DHPC, isotropic bicelles (DMPC/DHPC mixtures), lyso lipids, e.g., LMPC, and dodecylphosphocholine (DPC) at pH ≥ 6.5. Measurements of RDCs for three different phage-aligned membrane proteins in DHPC micelles are illustrated in Figure 1. Notably, there is no detectable line broadening of the resonances under conditions that yield 1H-15N RDCs as large as 22 Hz.
Figure 1.
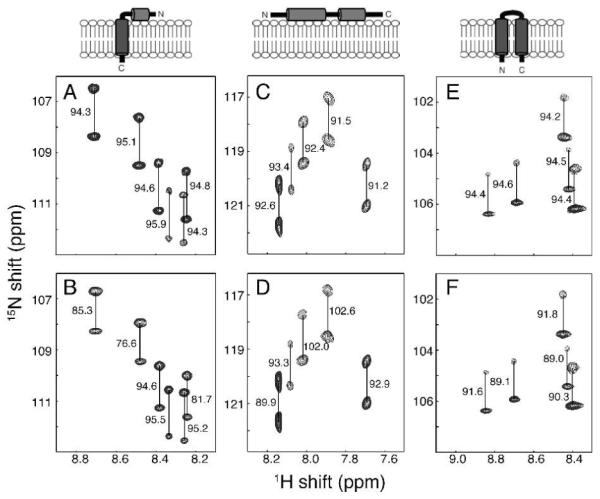
Representative regions of 1H-15N IPAP-HSQC spectra of isotropic (A, C and E) and aligned (B, D and F) samples. (A and B) Uniformly 15N labeled membrane-bound form of Pf1 coat protein10 in 100 mM DHPC, pH 6.7. (C and D) 15N-Ile labeled cytoplasmic domain of Vpu11 in 100 mM DHPC, 20 mM HEPES, pH 6.5. (E and F) Uniformly 15N-labeled MerFt12 in 100 mM DHPC, pH 6.5. The alignment resulted from the inclusion of 28 mg/ml, 13 mg/ml, and 30 mg/ml of fd bacteriophage in samples B, D, and F, respectively. The samples were maintained at 50°C for the measurements performed on a cryoprobe-equipped Bruker 600 MHz spectrometer. The measured values of the one-bond 1H-15N splittings are in Hz.
Figure 2 shows the Dipolar Waves fitted to the RDCs measured for the membrane-bound form of Pf1 coat protein weakly aligned in a compressed polyacrylamide gel (blue) and by added fd bacteriophage (red) plotted as a function of residue number. Dipolar Waves have exquisite structural resolution; they demonstrate the presence at residue 38 of a small kink in the hydrophobic transmembrane helix (residues 20-43), an interhelical region (residues 14-19), and an amphipathic in-plane helix (residues 5-13). A few residues at the N- and C- termini are mobile and unstructured. In order to calculate de novo the three-dimensional structure of a protein using only 1H-15N RDCs and no long-range NOEs, it is essential to have an accurate estimate of the order tensor, which is difficult to obtain for highly asymmetric proteins. As a result, this is an important application of the newly-introduced λ-maps, which analyze the distribution of RDCs from two alignments and extracts the order tensors without NMR resonance assignments or any structural information about the protein.6 The values of the alignment tensor’s magnitude and Rhombicity (R) are 11.44 Hz and 0.64 for the protein aligned by the presence of fd bacteriophage and 7.74 Hz and 0.21 for the protein aligned by a compressed gel, as shown in Figure 2.
Figure 2.
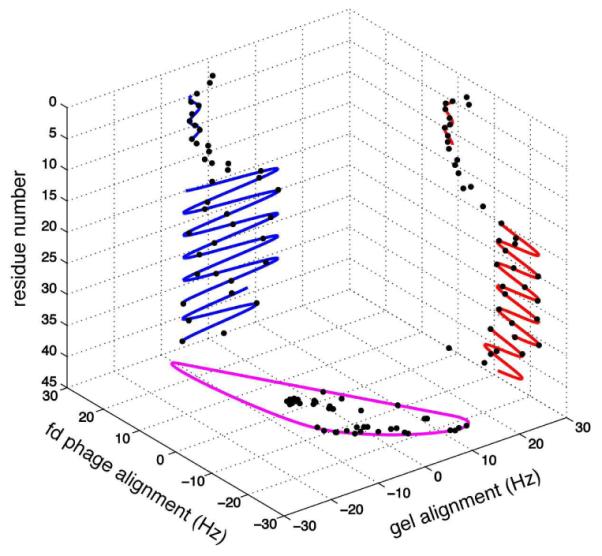
Cube correlation of Dipolar Waves and λ-map. Dipolar Waves fitted to the 1H-15N RDCs of membrane-bound form of Pf1 coat protein are shown for compressed polyacrylamide gel alignment (blue) and fd phage-induced alignment (red). λ-map estimate of the order tensors from both alignment media is shown in purple.
The order tensor information enabled structure calculations to be performed with the program XPLOR-NIH13 using the restraints from RDCs and dihedral angles as input.10 The helical regions of the protein are defined by Dipolar Waves and dihedral angles are derived from chemical shifts with the program TALOS.14 The simulated annealing step was performed at low temperature to impose RDC restraints with the XPLOR-NIH SANI potential.15 During the simulated annealing, the temperature was cooled from 300K to 25K in steps of 12.5K, with 10 ps of internal dynamics at each temperature, the RDC restraint force constant was ramped from 0.01 to 10.0 kcal Hz−2, while the dihedral angle restraint force constant was held at 300 kcal mol−1 rad−2 for helical regions. 20 out of 50 structures with RDC violations < 1.0 Hz and deviations of helical dihedral angles < 10° were accepted. These structures are shown in Figure 3; they form a unique fold with backbone rmsds of 1.950 Å (all residues), 0.361 Å (residues 5-13), 1.148 Å (residues 14-19) and 0.112 Å (residues 20-43), respectively. The interhelical residues contribute disproportionately to the large overall rmsd values because they have relatively few structural restraints.
Figure 3.
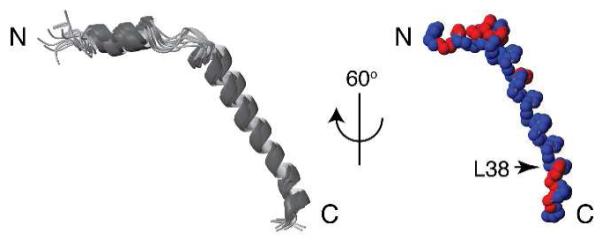
Superimposition of 15 calculated backbone structures of Pf1 coat protein (left) and 60° rotation of one structure to the vertical axis (right). A slight kink at residue 38 is indicated. Distribution of hydrophilic (red) and hydrophobic (blue) residues demonstrates the amphipathic character of the short N-terminal in-plane helix and the C-terminal region.
In combination, alignment of detergent solubilized membrane proteins with strained polyacrylamide gels and fd bacteriophage provides sufficient data for the use of Dipolar Waves and λ-maps, which is sufficient to characterize the secondary structure and to enable the calculation of their three-dimensional structures. The RDC data from multiple alignments are complementary to relaxation measurements for describing protein dynamics,16 such as that observed for the N-terminal amphipathic helix of the membrane-bound form of Pf1 coat protein.10
At pH > 6 added filamentous bacteriophage fd is compatible with many of the detergents used to solubilize membrane proteins for solution NMR studies of membrane proteins, and therefore serves as an alignment media. In combination with strained polyacrylamide gel alignment, Dipolar Waves can be used to directly assess the secondary structure and a λ-map extracts the order tensors for de novo structure calculation of membrane proteins without distance restraints.
Supplementary Material
Acknowledgement
We thank David Black for assistance. This research was supported by grants from the National Institutes of Health (S.J.O. and H.V.) and the National Science Foundation (H.V.) and utilized the Biomedical Technology Resource for NMR Molecular Imaging of Proteins at UCSD.
Footnotes
Supporting Information Available: Sample information, a table of RDC values and a scalar product between orientations of Pf1 coat protein. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Opella SJ, Marassi FM. Chem. Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2) (a).Bax A, Kontaxis G, Tjandra N. Methods Enzymol. 2001;339:127–74. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]; (b) Prestegard JH, Bougault CM, Kishore AI. Chem. Rev. 2004;104:3519–40. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- (3) (a).Iwahara J, Schwieters CD, Clore GM. J. Am. Chem. Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]; (b) Liang B, Bushweller JH, Tamm LK. J. Am. Chem. Soc. 2006;128:4389–97. doi: 10.1021/ja0574825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4) (a).Sass HJ, Musco G, Stahl SJ, Wingfield PT, Grzesiek S. J. Biomol. NMR. 2000;18:303–9. doi: 10.1023/a:1026703605147. [DOI] [PubMed] [Google Scholar]; (b) Tycko R, Blanco FJ, Ishii Y. J Am Chem Soc. 2000;122:9340–9341. [Google Scholar]; (c) Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. J. Biomol. NMR. 2001;21:377–82. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- (5).Mesleh MF, Lee S, Veglia G, Thiriot DS, Marassi FM, Opella SJ. J. Am. Chem. Soc. 2003;125:8928–35. doi: 10.1021/ja034211q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mukhopadhyay R, Miao X, Shealy P, Valafar H. J. Magn. Reson. 2009;198:236–47. doi: 10.1016/j.jmr.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7) (a).Jones DH, Opella SJ. J. Magn. Reson. 2004;171:258–69. doi: 10.1016/j.jmr.2004.08.022. [DOI] [PubMed] [Google Scholar]; (b) Cierpicki T, Bushweller JH. J. Am. Chem. Soc. 2004;126:16259–66. doi: 10.1021/ja046054g. [DOI] [PubMed] [Google Scholar]
- (8) (a).Ma C, Opella SJ. J. Magn. Reson. 2000;146:381–4. doi: 10.1006/jmre.2000.2172. [DOI] [PubMed] [Google Scholar]; (b) Feeny J, Birdsall B, Bradbury AF, Biekofsky RR, Bayley PM. J. Biomol. NMR. 2001;21:41–8. doi: 10.1023/a:1011924017938. [DOI] [PubMed] [Google Scholar]; (c) Kamen DE, Cahill SM, Girvin ME. J. Am. Chem. Soc. 2007;129:1846–7. doi: 10.1021/ja067089e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9) (a).Douglas SM, Chou JJ, Shih WM. Proc. Natl. Acad. Sci. U S A. 2007;104:6644–8. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lorieau J, Yao L, Bax A. J. Am. Chem. Soc. 2008;130:7536–7. doi: 10.1021/ja801729f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Lee S, Mesleh MF, Opella SJ. J. Biomol. NMR. 2003;26:327–34. doi: 10.1023/a:1024047805043. [DOI] [PubMed] [Google Scholar]
- (11).Marassi FM, Ma C, Gratkowski H, Straus SK, Strebel K, Oblatt-Montal M, Montal M, Opella S. J. Proc. Natl. Acad. Sci. U S A. 1999;96:14336–41. doi: 10.1073/pnas.96.25.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Howell SC, Mesleh MF, Opella S. J. Biochemistry. 2005;44:5196–206. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- (13).Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- (14).Cornilescu G, Delaglio F, Bax A. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- (15).Clore GM, Gronenborn AM, Tjandra N. J. Magn. Reson. 1998;131:159–62. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- (16).Yao L, Vogeli B, Torchia DA, Bax A. J. Phys. Chem. B. 2008;112:6045–56. doi: 10.1021/jp0772124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


