Abstract
Genetic diversity within a population of the southern plains woodrat was examined using DNA sequences (967 base pairs [bp]) obtained from the control or d-loop region of the mitochondrial genome. One hundred fourteen individuals from 10 collection sites were assigned to 42 haplotypes. Haplotype diversity values were moderate to high (0.974 overall and ranged from 0.524 to 0.964 across collecting sites), whereas nucleotide diversity values were low (0.008 overall and ranged from 0.001 to 0.010 across sites), indicating that this population possesses a high number of closely related haplotypes. Seventy-nine percent of the genetic variability was partitioned within groups that corresponded to the collecting sites. In addition, 13 samples from Texas, New Mexico, and Mexico were included as references for evaluating the evolutionary history of haplotypes. Nested clade analysis revealed that restricted gene flow with isolation by distance in conjunction with contiguous range expansion was responsible for the observed pattern of genetic diversity. A test of neutrality supported the diagnosis of restricted gene flow, but failed to support contiguous range expansion due solely to population growth. Examination of the spatial distribution of the haplotypes indicated that most haplotypes were restricted to a single collecting site; however, a small number of haplotypes were found at 2 or more sites. A phylogenetic analysis indicated that some haplotypes (28.6%) were restricted to the study area whereas the remaining haplotypes occupied a broader geographic region.
Keywords: control region, d-loop, genetic diversity, Neotoma micropus, population genetics, southern plains woodrat
Woodrats (genus Neotoma) are a principal host of Whitewater Arroyo arenavirus (WWAV—Fulhorst et al. 1996, 2001, 2002; Kosoy et al. 1996), with isolates reported from 4 species: Neotoma albigula (changed to N. leucodon by Edwards et al. 2001), N. cinerea, N. mexicana, and N. micropus. Specific rodents (usually 1 or 2 closely related species) are the principal hosts of WWAV and other arenaviruses for which natural host relationships have been characterized. The dominant feature of arenaviruses is their ability to establish chronic infections in their respective principal rodent hosts. A recent study (Fulhorst et al. 2002) revealed extensive genetic diversity among 5 strains (isolates) of WWAV isolated from contemporary N. micropus (southern plains woodrat) captured on the Chaparral Wildlife Management Area in southern Texas. In that study, levels of genetic divergence among WWAV isolates ranged from 0.2% to 12.9%. Perhaps the most intriguing finding was that individuals collected less than 1 km from each other possessed a genetic divergence of 12.7%. These findings indicated that substantial genetic diversity could exist within a single host species over a relatively small geographic area.
Consequently, 2 hypotheses were proposed by Fulhorst et al. (2002) to explain the observed pattern of viral genetic diversity in the host population. The coexistence of these multiple genetic variants of WWAV on the Chaparral Wildlife Management Area might have been a consequence of commingling of allopatric populations of N. micropus, each associated with a distinct WWAV genotype. Alternatively, the different genetic forms of the virus might have descended from a common ancestor and diverged in situ (in this population) over an extended period of time.
Knowledge of genetic diversity within this population of N. micropus is required to elucidate the source(s) of genetic diversity within WWAV on the Chaparral Wildlife Management Area. Two approaches can be used to examine genetic diversity in this context. Diversity can be determined from several loci (such as microsatellites) for each individual in a population. This approach results in a complex and perhaps unique genotype being estimated for each individual in the population. Alternatively, genetic diversity can be examined from a broader subdivision or cross-section of the population. For example, such a subdivision could correspond to maternal or paternal lineages. In order to examine genetic diversity in the context of the hypotheses outlined above, it seems prudent to 1st examine patterns of genetic diversity along maternal lineages. Although a complex and unique genotype can provide greater resolution in most cases, more general patterns of association might be overlooked. Examination of maternal lineages especially would be valuable if arenaviruses are transmitted vertically (mother to offspring).
The present study quantified and characterized the levels of genetic diversity in a population of N. micropus in south Texas, and attempted to elucidate the biological processes that might be the source of the genetic diversity in this population. To accomplish this, nucleotide sequences from the d-loop region (control region) of the mitochondrial genome were examined. The d-loop region was selected based on its rapid rate of sequence evolution (on average 4 times faster than mitochondrial protein coding regions—Pesole et al. 1999) and its utility in constructing maternal lineages (Rooney et al. 2001).
Materials and Methods
The research area (Chaparral Wildlife Management Area) primarily is a mixture of mesquite and acacia grasslands, described in detail by Fulhorst et al. (2002) and Suchecki et al. (2004), and currently is a wildlife management area for the state of Texas. Sampling of woodrats was conducted once per season (January, March, June, and October) during 2001 and 2002 and included 10 sampling sites. The sampling design included 2 phases, mark-recapture (3 sites), and excavation of middens (7 sites) with subsequent collection of all inhabitants. Woodrats included in this study were part of a prospective study on the ecology of WWAV and the natural history of woodrats on the Chaparral Wildlife Management Area.
Collection and processing of woodrats
The mark-recapture phase used a web-based design (Anderson et al. 1983) and encompassed 3 webs established approximately 3–4 km apart (Fig. 1). Webs were constructed at locations containing arenavirus positive animals reported by Fulhorst et al. (2002). Each web contained 16 equidistant spokes, with 20 Sherman traps (H. B. Sherman Trap Inc., Tallahassee, Florida) placed 5 m apart on each spoke (320 traps per web). Traps were baited with a mixture of birdseed and rolled oats, set at dusk, and checked at dawn. Webs were sampled for 3 consecutive nights each season. Captured individuals were measured, weighed, ear-punched, and given a unique number (TK number) cross-referenced to an individual-specific toe-clip pattern for identification purposes, and released at the site of initial capture.
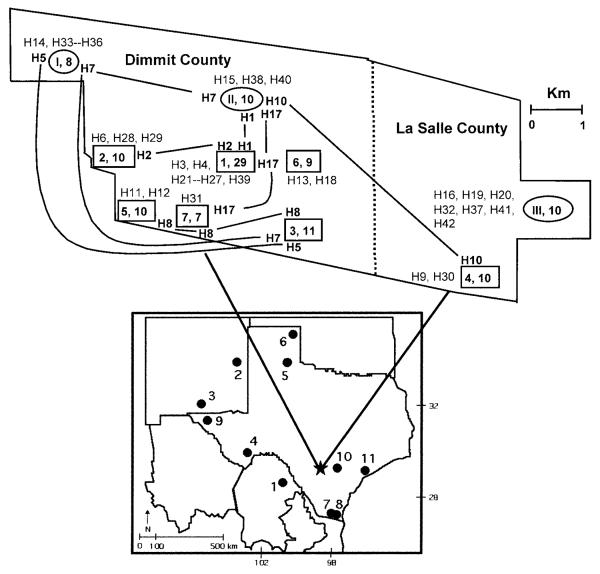
Fig. 1.
Map depicting collection sites at the Chaparral Wildlife Management Area. Web sites are denoted by roman numerals in ovals and midden sites are denoted by numbers in rectangles. In both instances, numerals and numbers are followed by sample size. Star represents the study site, and closed circles and numbers (see Appendix I) depict the reference samples. See text for description of web and midden sites. Haplotypes are represented by the letter “H” followed by the haplotype number. Boldface numbers indicate haplotypes that were shared between sites and plain numbers indicate haplotypes that were site specific. Lines depict sites sharing a particular haplotype.
The midden design involved sampling 7 sites, each represented by a 25 m radius circle as described by Suchecki et al. (2004). These sites were selected for their high density of middens (confirmed by visual inspection), but were not located closer than 500 m from other midden sites or webs in order to preserve sampling independence. The objective was to excavate middens in order to capture family units (females and their presumed offspring). Similar data were recorded as above; however, as the midden study was part of another ongoing research project, tissues (heart, kidney, muscle, lung, liver, and spleen) were obtained instead of toe and ear clips, and voucher specimens were prepared following animal care and use procedures approved by the American Society of Mammalogists (Animal Care and Use Committee 1998).
In addition, 13 individuals of N. micropus collected from Texas, New Mexico, and Mexico, and 1 sample of N. leucodon were included as reference and outgroup taxa for data analysis. Collection localities are provided in the Specimens Examined (Appendix I).
DNA sequencing
Two hundred eighty one woodrats were captured from the 3 webs and 177 from the 7 midden sites. Of these, a sub-sample containing 114 individuals (28 individuals from webs and 86 from midden sites) was selected for genotyping (Appendix I). Individuals comprising this subsample were chosen to represent age and sex structure of the captured woodrats, as well as to include individuals from all collection sites. The midden sample included 12 family units (12 females and 22 presumed offspring).
Toe and ear clips, tissues, and voucher specimens were deposited in the Vital Tissue Collection and the Recent Mammal Collection in the Natural Science Research Laboratory of the Museum of Texas Tech University and served as the DNA source in this study. Whole genomic DNA was isolated from the toe-clip, ear-clip, or liver samples using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minnesota). Polymerase chain reaction (PCR) primers specific to the d-loop region (reverse primer 2340-5, Castro-Campillo et al. 1999 and forward primer Nmic5′, TCCTCAAGGCATCAAGAAG) were used to amplify the entire d-loop region (967 bp) of the mitochondrial genome. The following thermal profile was used for amplification: an initial cycle of 93.5° C for 1 min, followed by 33 cycles of 93.5° C for 40 s, 49° C for 40 s, and 72° C for 2 min 40 s, and 72° C for 2 min. Purification of PCR products was done with a QIAquick kit (Quiagen, Inc., Chatsworth, California). Sequencing reactions used the 2 PCR primers described above and 2 internal cycle sequencing primers (1115 Reverse: ATGACCCTGAAGAARGAACCAG and 500 Forward: TCTCTTAATCTACCATCCTCCGTG). Both forward and reverse sequences were obtained using an automated ABI 310 or 3100 Avant automated sequencers (PE Applied Biosystems, Foster City, California). Sequences initially were aligned using Sequencher 3.1 software (Gene Codes, Ann Arbor, Michigan) and then adjusted manually. All sequences were deposited into GenBank (accession numbers AY338500–AY338613; AY496214–AY496225). GenBank and voucher specimen reference numbers are provided in Specimens Examined (Appendix I).
Data analysis
For this study, a mitochondrial haplotype was defined as a unique DNA sequence. Therefore, multiple individuals with identical d-loop sequences were considered to represent the same haplotype. In cases where haplotypes differed by a single nucleotide, chromatographs and resultant sequences were reexamined to verify the single nucleotide change.
Haplotype distribution and frequencies, and 2 standard genetic diversity indices, haplotype diversity (h—Nei 1987), and nucleotide diversity (π—Tajima 1983) were estimated using Arlequin 2.0 software (Schneider et al. 2000). An analysis of molecular variance (AMOVA—Excoffier et al. 1992; Weir 1996; Weir and Cockerham 1984) was performed to estimate the partitioning of genetic variation. Genetic differentiation among sites was measured using Wright's (1951, 1965) FST statistic. Individuals representing the reference samples and those comprising the family units were not included in these analyses.
The program TCS (version 1.12—Clement et al. 2000) was used to verify haplotype frequencies of haplotypes obtained from the study site and generate a network depicting the haplotype genealogy under the method of statistical parsimony (Templeton et al. 1992). The resulting network was converted into a series of nested clades (1-step to 5-step) following the procedure described by Templeton et al. (1987). A network was used to depict differences among haplotypes because of its appropriateness at intraspecific and population levels (Posada and Crandall 2001).
The geographic locations of collection sites were combined with nested clades and analyzed using the program GeoDis (Posada et al. 2000) to test for geographic association and possible processes generating the observed pattern of haplotype distribution. Clade distance (Dc: estimation of geographic variability represented in each clade), nested clade distance (Dn: description of geographic distribution of a clade in relation to other clades in the same nesting category), and interior-tip statistics (I–T: average interior distance minus average tip distance for each nesting level) were estimated. These parameters were used in conjunction with an inference key (Templeton et al. 1995) associated with GeoDis (Posada et al. 2000) to interpret results of the geographic distance analysis and test of interior versus tip clades. The null hypothesis for this test was that no association existed between haplotype genealogy and geographic distribution of clades.
Explanations for the patterns of genetic diversity inferred from the test of Templeton et al. (1995) were further evaluated using Fu's test of neutrality (FS—Fu 1996). Under this test, excessively negative values are indicative of an excess of recent mutations (Fu 1997). This test was performed using the software program Arlequin 2.0 (Schneider et al. 2000) and 2 methods of analysis. In the 1st analysis, individuals were grouped according to collection sites, whereas in the 2nd analysis individuals were grouped based on the hierarchy of nested clade (highest in this case).
A 2nd test, spatial autocorrelation, was used to evaluate the relationship between haplotypes (genetic distances) and geographic distribution. This test was performed using the software package GenA1Ex v.5 (Peakall and Smouse 2001) with 1,000 bootstrap replications.
The 13 reference samples of N. micropus and the sample of N. leucodon (outgroup taxon) were combined with 1 representative of each haplotype obtained from the study site. These sequences were examined in a phylogenetic context to ascertain the geographic distribution and evolutionary history of haplotypes obtained from the study site. A neighbor-joining tree was obtained using the software program PAUP* (Swofford 2002) and Tamura and Nei genetic distances (Tamura and Nei 1993) with a minimum evolution model. The Tamura and Nei model was selected because it was developed for use with control region sequences. Parsimony and likelihood methods were evaluated but were not included due to a paucity of informative characters within the data set.
Results
Analysis of 114 d-loop sequences (967 bp) revealed 60 variable sites that defined 42 distinct haplotypes. Haplotypes 1–20 occurred multiple times, with haplotypes 1 and 5 being most frequent (9.6% and 7.0%, respectively). The 22 remaining haplotypes were unique (Table 1). Of these, haplotypes 1, 2, 5, 7, 8, 10, and 17 were present in multiple sampling sites, whereas the remaining haplotypes were restricted to a single locality (Fig. 1, Table 1).
Table 1.
Distribution of individuals possessing unique mitochondrial d-loop sequences (haplotypes) by collecting site. The number of individuals per haplotype is reported for midden sites (1–7) and webs (I–III) and corresponds to sampling localities depicted in Figure 1.
| Midden Site |
Web |
Number of individuals |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | 1 | 2 | 3 | 4 | 5 | 6 | 7 | I | II | III | |
| 1 | 10 | 1 | 11 | ||||||||
| 2 | 1 | 5 | 6 | ||||||||
| 3 | 7 | 7 | |||||||||
| 4 | 2 | 2 | |||||||||
| 5 | 7 | 1 | 8 | ||||||||
| 6 | 3 | 3 | |||||||||
| 7 | 3 | 1 | 1 | 5 | |||||||
| 8 | 1 | 1 | 3 | 5 | |||||||
| 9 | 6 | 6 | |||||||||
| 10 | 3 | 3 | 6 | ||||||||
| 11 | 5 | 5 | |||||||||
| 12 | 4 | 4 | |||||||||
| 13 | 4 | 4 | |||||||||
| 14 | 2 | 2 | |||||||||
| 15 | 2 | 2 | |||||||||
| 16 | 2 | 2 | |||||||||
| 17 | 1 | 3 | 1 | 4 | |||||||
| 18 | 5 | 5 | |||||||||
| 19 | 2 | 2 | |||||||||
| 20 | 2 | 2 | |||||||||
| 21 | 1 | 1 | |||||||||
| 22 | 1 | 1 | |||||||||
| 23 | 1 | 1 | |||||||||
| 24 | 1 | 1 | |||||||||
| 25 | 1 | 1 | |||||||||
| 26 | 1 | 1 | |||||||||
| 27 | 1 | 1 | |||||||||
| 28 | 1 | 1 | |||||||||
| 29 | 1 | 1 | |||||||||
| 30 | 1 | 1 | |||||||||
| 31 | 1 | 1 | |||||||||
| 32 | 1 | 1 | |||||||||
| 33 | 1 | 1 | |||||||||
| 34 | 1 | 1 | |||||||||
| 35 | 1 | 1 | |||||||||
| 36 | 1 | 1 | |||||||||
| 37 | 1 | 1 | |||||||||
| 38 | 1 | 1 | |||||||||
| 39 | 1 | 1 | |||||||||
| 40 | 1 | 1 | |||||||||
| 41 | 1 | 1 | |||||||||
| 42 | 1 | 1 | |||||||||
| Individuals genotyped | 29 | 10 | 11 | 10 | 10 | 9 | 7 | 8 | 10 | 10 | 114 |
Given that 14 pairs of haplotypes differed by a single nucleotide, we were concerned that single nucleotide changes might have been generated in either data collection (PCR and sequencing) or scoring. Polymerase error (2.4 × 10−5 to 8.9 × 10−5; Cariello et al. 1991) is 1 possible source for generation of artifacts in the d-loop nucleotide sequence data. The fact that identical haplotypes were present in multiple individuals suggests that polymerase error would have had to occur at identical sites in different individuals and therefore is an unlikely source for differences among haplotypes. In most instances (92 of the 106 relationship scenarios) haplotypes differed at multiple nucleotide sites, suggesting that differences among haplotypes were valid. Another source of confirmation comes from identical sequences shared by the 12 mother/offspring units included in this study. Because the d-loop is inherited maternally, offspring would be expected to have the same haplotype as their mothers. Additionally, the number of variable sites found within the d-loop region in this study is similar to that reported in other studies of mammals using d-loop sequence data (Matson et al. 2000; Matsuhashi et al. 1999; Mirol et al. 2002).
Overall population haplotype diversity, which represents number and frequency of haplotypes, was 0.974 and within-population haplotype diversity ranged from 0.524 to 0.964 across the 10 collection sites (Table 2). Nucleotide diversity, which reflects frequency of haplotypes and sequence divergence among all haplotypes, was 0.008 and ranged from 0.001 to 0.010 among the 10 collecting sites (Table 2). Percentage of variation attributable to within-group variation was 79.5%, and among-group variation was 20.5%. Genetic differentiation among sites (FST statistic) was 0.205.
Table 2.
Haplotype diversity (h) and nucleotide diversity (π) values estimated for midden sites (1–7) and webs (I–III). Sample sizes for each collection site were adjusted to reflect the exclusion of offspring of the family units. Standard errors for each are shown in parentheses.
| Index |
|||
|---|---|---|---|
| Collection Site | Sample Size | h | π |
| 1 | 18 | 0.941 (0.053) | 0.006 (0.003) |
| 2 | 9 | 0.750 (0.118) | 0.003 (0.002) |
| 3 | 7 | 0.524 (0.134) | 0.003 (0.002) |
| 4 | 10 | 0.600 (0.131) | 0.006 (0.003) |
| 5 | 8 | 0.679 (0.101) | 0.003 (0.002) |
| 6 | 7 | 0.571 (0.090) | 0.001 (0.001) |
| 7 | 5 | 0.700 (0.127) | 0.004 (0.003) |
| I | 8 | 0.964 (0.077) | 0.010 (0.006) |
| II | 10 | 0.911 (0.077) | 0.010 (0.006) |
| III | 10 | 0.933 (0.062) | 0.007 (0.004) |
| Overall | 92 | 0.974 (0.005) | 0.008 (0.004) |
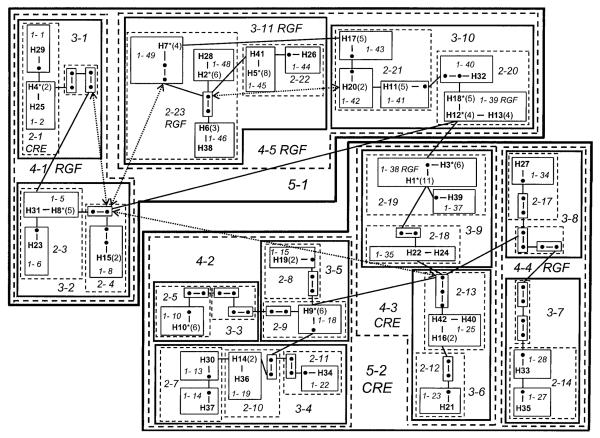
In the nested clade analysis (Fig. 2), 23 mutational steps separated the 2 most divergent haplotypes. The statistical parsimony method of Templeton et al. (1992) predicted that 63 haplotypes were not sampled in this study. Haplotypes that were not sampled were included in order to reconstruct the hypothetical mutational steps linking each haplotype. Within the parsimony network, 4 loops, indicative of ambiguous relationships, were resolved using the logic of Crandall and Templeton (1993). Resolution of these ambiguous relationships allowed the estimation of tip versus interior distance for 3 clades, enabling us to increase the resolution of the nested clade analysis.
Fig. 2.
Nested haplotype network for Neotoma micropus based on an analysis of mitochondrial d-loop sequence data. Haplotypes are represented by the letter “H” followed by the haplotype number. Closed circles represent putative intermediate haplotypes that were not sampled in this study. Nested clades are designated in italics with the nesting level (1 through 5) followed by a dash and the particular nested clade number. CRE = contiguous range expansion, RGF = restricted gene flow with isolation by distance. Sample sizes for haplotypes containing more than 1 individual are in parenthesis next to the haplotype designation. Haplotypes that contain family units (mother and offspring) are indicated with an asterisk. For ease of identification, solid and dashed polygons were used alternatively to delineate between nesting levels. Solid, thin-lined rectangles represent 1-step clades. Dashed, thin-lined rectangles represent 2-step clades. Solid, medium-lined rectangles represent 3-step clades. Dashed, medium-lined polygons represent 4-step clades; and solid, heavy-lined polygons represent 5-step clades. Ambiguous loop connections are indicated in fine dashed arrows.
Using GeoDis (Posada et al. 2000) and the inference key of Templeton et al. (1995) the null hypothesis of no association between genetic and geographic distance was rejected for 19 of the 90 clades. Of these 19 clades, 8 produced inconclusive results, 5 were indicative of restricted gene flow with isolation by distance, and 3 were indicative of contiguous range expansion. For the 3 remaining clades, results were inadequate to distinguish between fragmentation and isolation by distance or contiguous range expansion and long distance colonization (Table 3).
Table 3.
Nested clades with at least 1 significant distance statistic for d-loop haplotypes. Significant distance statistic and final inference (conclusion) are provided. Abbreviations are as follows: Dc= clade distance (estimation of geographic variability represented in each clade), Dn = nested clade distance (description of geographic distribution of a clade in relation to other clades in the same nesting category), I-T = interior to tip distance (average interior distance minus the average tip distance for each nesting level) I = interior clade, T = tip clade, S = significantly small probability, L = significantly large probability, NS = not significant, CD = cannot be determined, and TC = total clade. Abbreviations for Final Inference are as follows: INC = inconclusive, RGF = restricted gene flow, IBD = isolation by distance, CRE = contiguous range expansion, FRA/IBD = geographic sampling scheme inadequate to discriminate between fragmentation and isolation by distance, and CRE/LDC = geographic sampling scheme inadequate to discriminate between contiguous range expansion and long distance colonization.
| Nested Clade | Dc | Dn | I-T Dc | I-T Dn | Chain of Inference |
Final Inference |
|---|---|---|---|---|---|---|
| 1-5 | 1.0245 (IL) | 0.9700 (IL) | CD | CD | 1-2 | INC |
| 1-25 | NS | 3.3178 (TL) | NS | −2.0890 (S) | 1-2-11-17 | INC |
| 1-38 | 0.0000 (TS) | NS | 0.7015 | NS | 1-2-3-4 | RGF and IBD |
| 1-39 | NS | 2.5908 (IL) | NS | NS | 1-2-11-17-4 | RGF and IBD |
| 1-46 | NS | 0.4891 (IS) 1.3205 (TL) |
NS | −0.8314 (S) | 1-2-11-17 | INC |
| 2-1 | 0.0000 (IS) | 0.0466 (IS) | 0.0000-S | −0.0016 (S) | 1-2-11-12 | CRE |
| 2-7 | NS | 5.7228 (TS) | NS | 0.0011 (L) | 1-2-11-17-4-9-10 | FRA/IBD |
| 2-19 | 0.0000 (TS) 0.5098 (IL) |
0.2889 (TL) 0.5001 (IS) |
0.5098 (L) | 0.2112 (L) | 1-2-3-5-6-13-14 | CRE/LDC |
| 2-21 | 0.0000 (IS) | 1.1022 (IS) 4.7966 (IL) |
CD | CD | 1-2 | INC |
| 2-23 | 0.0098 (TS) | 0.5500 (TS) 2.3833 (IL) |
2.6174 (L) | 1.7072 (L) | 1-2-3-4 | RGF and IBD |
| 3-2 | 0.9369 (IS) | 1.3111 (IS) 2.7019 (IL) |
CD | CD | 1-2 | INC |
| 3-4 | NS | 2.0418 (IS) 2.0418 (TS) |
NS | −2.9015 (S) | 1-2-11-17 | INC |
| 3-5 | 0.0000 (TS) 0.0000 (IS) |
8.5846 (TL) 2.8621 (IS) |
NS | −5.7225 (S) | 1-2-11-12-13-14 | CRE/LDC |
| 3-9 | 0.0000 (IS) | 0.4728 (IL) | CD | CD | 1-2 | INC |
| 4-1 | 0.0474 (TS) | 0.7575 (TS) | 1.5669 (L) | 0.8862 (L) | 1-2-3-4 | RGF and IBD |
| 4-3 | 0.4548 (IS) | 1.5622 (IS) | −1.6670 (S) | −1.6025 (S) | 1-2-11-12 | CRE |
| 4-4 | NS | 0.8789 (TS) | NS | 5.4925 (L) | 1-2-11-17-4 | RGF and IBD |
| 5-2 | 2.1351 (IS) | 3.5386 (IS) | −1.6481 (S) | −1.1445 (S) | 1-2-11-12 | CRE |
| TC | 1.8310 (TS) 4.2463 (TL) |
1.9167 (TS) 4.1398 (TL) |
CD | CD | 1-2 | INC |
Fu's test of neutrality (Fu 1996) revealed nonsignificant and positive values (FS) for individuals grouped according to sampling site, with 1 exception. Midden Site I possessed a significant and negative value (FS = −3.815, P = 0.033). When individuals were grouped using a hierarchical nested clade approach, clades 5-1 and 5-2 both possessed negative FS values (−4.949 and −1.650, respectively); however, only clade 5-1 was significant (P = 0.025).
No significant autocorrelation values (GenA1Ex v.5—Peakall and Smouse 2001) were found between spatial distribution and genetic structure. Consequently, we were unable to reject the null hypothesis (no association between genetic and geographic distances) for this test.
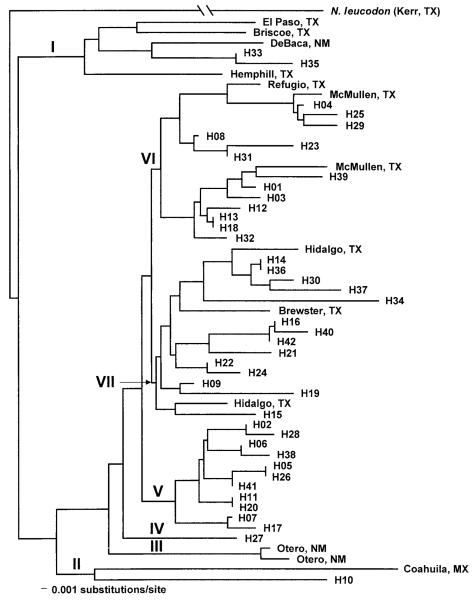
Relationships among haplotypes were examined using a neighbor-joining tree constructed from Tamura and Nei genetic distances (Tamura and Nei 1993). One representative from each of the 42 haplotypes was included in the analysis along with reference and outgroup haplotypes (n = 14). Seven major clades (I–VII) were depicted (Fig. 3). Three clades (I, II, VII) contained haplotypes from the study site and reference haplotypes that were distantly located to the study site (>200 km). One clade (VI) contained haplotypes from the study site and reference haplotypes located <200 km from the study site. Two clades, IV (1 haplotype) and V (11 haplotypes), contained only haplotypes from the study site, and clade (III) contained only reference haplotypes.
Fig. 3.
Neighbor-joining tree constructed with Tamura and Nei genetic distances (Tamura and Nei 1993). Roman numerals are used to depict major clades of haplotypes. Abbreviations are as follows Mexico (MX), New Mexico (NM), and Texas (TX).
Discussion
The woodrat population at Chaparral Wildlife Management Area is characterized by a pattern of low nucleotide diversity and moderate to high haplotype diversity. Nucleotide diversity values were low (0.008 overall and ranged from 0.001 to 0.010 across sites) compared to values reported for populations of species of rodents such as Neotoma fuscipes (0.010 to 0.039—Matocq 2002), Proechimys steerei (0.071—Matocq et al. 2000), Proechimys simonsi (0.118—Matocq et al. 2000), and Lemmus sibiricus (0.018 to 0.028—Ehrich and Stenseth 2001). However, haplotype diversity values were high (0.974 overall and ranged from 0.524 to 0.964 across sites) and similar to those reported for populations of L. sibiricus (0.690 to 0.920—Ehrich and Stenseth 2001) and Clethrionomys glareolus (0.722—Matson et al. 2000). The pattern of low nucleotide diversity and high haplotype diversity indicates that the population is comprised of a high number of closely related haplotypes. The statistical parsimony network corroborated this pattern with 36 of 42 haplotypes being separated by 3 or fewer mutational steps and the remaining 6 haplotypes being separated by 4 or more mutational steps. Additionally, Grant and Bowen (1998) suggested that patterns of low nucleotide diversity and high haplotype diversity are indicative of a low effective population size followed by expansion.
Extensive within-population diversity is confirmed in having 42 haplotypes from 114 individuals collected from an area approximately 4 × 10 km. Other studies on rodents have reported lower values from larger geographical areas. For example, Ehrich and Stenseth (2001) determined 49 haplotypes from 346 individuals (L. sibiricus) collected from sites as far as 600 km from each other and Klaus et al. (2001) reported 29 haplotypes from 142 individuals (Microtus richardsoni) from several populations in adjacent watersheds. Similarly, Matocq et al. (2000) delineated 48 haplotypes from 203 individuals (P. steerei) and 45 haplotypes from 147 individuals (P. simonsi) collected from the Rio Juruá Basin.
The overall FST value (0.205) is indicative of substantial population structure (Wright 1978) among the collection sites. This value is considerably higher than FST values reported from populations of other species of rodents, for example, Clethrionomys gapperi (0.07—Reese et al. 2000) and Spermophilus brunneus (0.167—Galvin et al. 1999).
The 2 historical processes identified by the nested clade analysis as responsible for the observed pattern of genetic diversity were restricted gene flow with isolation by distance and contiguous range expansion. Whereas restricted gene flow seems to play a main role in the genetic structure of clade 5-1, contiguous range expansion seems to be the predominant force in clade 5-2. Based on this, we tentatively assumed that the appropriate interpretation for clade 2-7 was isolation by distance and contiguous range expansion for clades 2-19 and 3-5. Although the nested clade approach is not statistically supported, it is the best available method for estimating historical patterns of genetic diversity at the population level (Knowles and Maddison 2002).
Examination of the distribution of haplotypes across collection sites (Fig. 1) revealed spatial patterns that support the results of the nested clade analysis. First is the presence of “site-specific” haplotypes. Haplotypes are distributed across the 10 collection sites (Fig. 1) and range in frequency from a single site-specific haplotype at Midden Site 7 to 10 haplotypes at Midden Site 1. At 5 sites (middens 1, 2, and 6, and webs I and III) site-specific haplotypes represented the major source of genetic variation. These observations support the premise that gene flow is restricted between collecting sites. Second, 7 haplotypes were shared among collecting sites with 5 sites sharing 2 or more haplotypes. This suggests that a few haplotypes are either widespread in distribution or are of sufficient frequency that they are detectable in multiple collecting sites.
None of 13 reference haplotypes were identical to the 42 haplotype obtained from the study area. However, the neighbor-joining tree indicated that several haplotypes were closely related to haplotypes obtained from the study site. Four of the 7 clades (I, II, VI, and VII) contained both reference and study site samples; however, 3 clades (I, II, and VII) contained haplotypes distributed across a geographic area >200 km. This indicates not only a close relationship among the haplotypes but that these haplotypes have a relatively broad geographic distribution and perhaps a long-standing evolutionary history. Alternatively, haplotypes in clades IV and V appear to be endemic to the study area and indicate a possible restricted distribution or recent evolutionary history. These 2 clades contained 28.6% of the haplotypes represented in the study area. Additional haplotypes, obtained from localities adjacent and distant to the study site, are needed to further address this observation. Clade III contained 2 haplotypes from southeastern New Mexico and does not appear to be associated with any haplotypes from the study site.
The pattern of genetic diversity within this population suggests that gene flow might be restricted among sites separated by as little as 2–5 km. Similarly, Castleberry et al. (2002) using estimates of gene flow, suggested that effective dispersal was limited among subpopulations of Neotoma magister separated by as little as 2–3 km; however, they concluded that isolation by distance was the model best fitting the data. Conversely, Monty et al. (2003) reported genetic differentiation between populations of Neotoma floridana in southern Illinois but found no evidence for correlation between genetic and geographic distance. Other studies (Bowman et al. 2000; Peakall et al. 2003) reported that for some populations the genetic structure at finer scales was homogenized as the geographic scale or the species' ability to disperse increased. For example, genetic structure is lost over an increasing geographic scale; likewise, increased dispersal would interject new genotypes into a previously unoccupied area. However, we have no evidence of an increase in dispersal for N. micropus.
Several processes could result in a pattern of restricted gene flow, including but not limited to, presence of a barrier (geographic, genetic, current, or historical), increased mutation rate at the local level, or inadequate sampling effort. For example, if 2 formerly allopatric populations recently came into contact, insufficient time might have elapsed for the homogenization of genotypes, producing a situation in which only partial or limited gene flow was detected. This scenario is supported by the finding of contiguous range expansion in several of the nested clades. Alternatively, failure to sample at appropriate distances between populations of small sample sizes could result in missing both rare and common haplotypes. Other possibilities such as differential lineage sorting, retention of ancestral haplotypes (Avise 1994), or an interdigitative (complex or infused) pattern of genotypic distributions coupled with temporal expansion and contraction of the distribution of maternal lineages, could be obscuring the processes underlying the genetic diversity found in this population. Additional research, employing different genetic markers and/or analyses, are needed to test these possibilities.
In general, results from the nested clade and genetic distance analyses are compatible, although the geographic scales are different. Both analyses indicated the presence of haplotypes that are restricted or endemic to particular geographic areas, and both identify haplotypes that occupy a broader geographic area. It appears that patterns of restricted gene flow or endemic haplotypes, as well as continual range expansion or broadly distributed haplotypes are applicable in characterizing genetic diversity within a single population or across the geographic range of N. micropus.
The results of Fu's test of neutrality (Fu 1996) revealed nonsignificant values for all but 1 collection site. This indicates that few mutations (rare haplotypes) occur within geographic collection sites. However, when collection sites were disregarded and individuals were grouped based on inclusion into hierarchical nested clades, significant values were recovered. This indicated a greater number of mutations (haplotypes) than expected in the large nested clade (5-1, composite group) than seen in the geographic collection sites. This pattern provides evidence supporting the concept of restricted gene flow within nested clade 5-1. Fu's test (Fu 1996) did not indicate an excess of new haplotypes, and therefore provided no support for contiguous range expansion resulting from population growth in clade 5-2. However, contiguous range expansion might have occurred independently of population growth as a product of migration, redefining of home ranges, response to changing resources, response to local extinction, etc.
This study reports extensive genetic diversity in the examined population of N. micropus. Unfortunately, few studies exist for comparison, although many studies pertaining to phylogenetic relationships and systematics of the genus Neotoma have been conducted (Baker and Mascarello 1969; Birney 1973; Edwards and Bradley 2001, 2002; Edwards et al. 2001; Goldman 1910; Hall and Genoways 1970; Hooper 1960; Koop et al. 1985; Mascarello and Warner 1974; Planz et al. 1996). The finding of contiguous range expansion of populations in this area supports the hypothesis by Fulhorst et al. (2002) that at least some of the genetic variation observed in WWAV on the Chaparral Wildlife Management Area might be due to the genetic interactions of at least 3 populations of N. micropus. Alternatively, Fu's test of neutrality (Fu 1996) supports the presence of restricted gene flow in this area, which could be indicative of a long evolutionary relationship between particular host lineages and viral strains. In order to examine this theory, co-evolutionary studies that compare genetic divergence of the virus strains and their specific hosts are needed. It is possible that the variability observed among the viral strains might be due to a combination of range expansion and restricted gene flow of 3 or more populations of its host in this area.
Acknowledgments
We thank R. E. Strauss, R. Van Den Bussche, and D. A. Ray for valuable help with the data analysis and comments on the manuscript. B. R. Amman, J. G. Brant, B. D. Baxter, D. S. Carroll, N. D. Durish, K. Graham, M. L. Haynie, L. K. Longhofer, R. McAliley, and S. A. Reeder provided valuable help in the field and with comments on the manuscript. M. B. Cajimat, J. E. Comer, S. Gardner, M. Kageyama, C. Milazzo, Jr., J. R. Suchecki, and A. Vestal also provided valuable help in the field. J. J. Harclerode assisted in constructing figures. D. R. Synatzske facilitated the necessary fieldwork. This research was funded by grant DHHSA141435 from the National Institutes of Health and the Texas Tech University Center for Zoonoses and Epidemiology.
Appendix I
Specimens examined
Specimens collected from the Chaparral Wildlife Management Area are listed by locality, TK number (unique reference number), collecting site (M1—M7 for Midden Sites 1—7 or WI-WIII for Web Sites I—III), haplotype number (H1—H42), and GenBank accession number (AY followed by a 6 digit number). Samples are from the United States unless otherwise denoted.
Neotoma micropus
Texas; Dimmit County, Chaparral Wildlife Management Area (TK98008, M1, H39, AY338500; TK98010, M1, H01, AY338501; TK98015, M1, H21, AY338502; TK98016, M1, H22, AY338503; TK98017, M1, H23, AY338504; TK98019, M1, H02, AY338505; TK98022, M1, H24, AY338506; TK98047, M1, H03, AY338507; TK98049, M1, H03, AY338508; TK98050, M1, H01, AY338509; TK98051, M1, H01, AY338510; TK98054, M1, H01, AY338511; TK98055, M1, H01, AY338512; TK98057, M1, H01, AY338513; TK98065, M1, H01, AY338514; TK98066, M1, H01, AY338515; TK98067, M1, H01, AY338516; TK98068, M1, H25, AY338517; TK98070, M1, H04, AY338518; TK98071, M1, H04, AY338519; TK98073, M1, H03, AY338520; TK98074, M1, H03, AY338521; TK98075, M1, H03, AY338522; TK98076, M1, H03, AY338523; TK98077, M1, H03, AY338524; TK98082, M1, H01, AY338525; TK98105, M1, H17, AY338526; TK98106, M1, H26, AY338527; TK98108, M1, H27, AY338528; TK98111, M2, H02, AY338529; TK98112, M2, H02, AY338530; TK98115, M2, H02, AY338531; TK98116, M2, H06, AY338532; TK98121, M2, H02, AY338533; TK98124, M2, H06, AY338534; TK98129, M2, H28, AY338535; TK98131, M2, H02, AY338536; TK98132, M2, H06, AY338537; TK98133, M2, H29, AY338538; TK98143, M3, H07, AY338539; TK98144, M3, H07, AY338540; TK98145, M3, H07, AY338541; TK98146, M3, H05, AY338542; TK98147, M3, H08, AY338543; TK98148, M3, H05, AY338544; TK98149, M3, H05, AY338545; TK98150, M3, H05, AY338546; TK98151, M3, H05, AY338547; TK98152, M3, H05, AY338548; TK98167, M3, H05, AY338549; TK98255, M5, H11, AY338560; TK98256, M5, H11, AY338561; TK98257, M5, H11, AY338562; TK98258, M5, H11, AY338563; TK98259, M5, H11, AY338564; TK98266, M5, H12, AY338565; TK98267, M5, H12, AY338566; TK98269, M5, H12, AY338567; TK98272, M5, H08, AY338568; TK98274, M5, H12, AY338569; TK98361, M6, H13, AY338570; TK98362, M6, H18, AY338571; TK98364, M6, H13, AY338572; TK98368, M6, H18, AY338573; TK98369, M6, H18, AY338574; TK98370, M6, H18, AY338575; TK98371, M6, H13, AY338576; TK98380, M6, H18, AY338577; TK98396, M6, H13, AY338578; TK98417, M7, H08, AY338579; TK98419, M7, H08, AY338580; TK98420, M7, H08, AY338581; TK98422, M7, H31, AY338582; TK98423, M7, H17, AY338583; TK98424, M7, H17, AY338584; TK98432, M7, H17, AY338585; TK100048, WI, H07, AY338587; TK100071, WI, H14, AY338589; TK100102, WI, H05, AY338591; TK100129, WI, H14, AY338593; TK100156, WI, H33, AY338594; TK100157, WI, H34, AY338595; TK100158, WII, H10, AY338596; TK100159, WII, H01, AY338597; TK100189, WII, H17, AY338598; TK100224, WI, H35, AY338599; TK100230, WI, H36, AY338600; TK100349, WII, H38, AY338603; TK100350, WII, H07, AY338604; TK100351, WII, H40, AY338605; TK100352, WII, H15, AY338606; TK100606, WII, H10, AY338611; TK100607, WII, H15, AY338612; TK100608, WII, H10, AY338613). La Salle County, Chaparral Wildlife Management Area (TK98208, M4, H09, AY338550; TK98209, M4, H09, AY338551; TK98210, M4, H09, AY338552; TK98211, M4, H09, AY338553; TK98227, M4, H09, AY338554; TK98238, M4, H10, AY338555; TK98239, M4, H10, AY338556; TK98240, M4, H10, AY338557; TK98241, M4, H09, AY338558; TK98246, M4, H30, AY338559; TK100021, WIII, H32, AY338586; TK100066, WIII, H19, AY338588; TK100082, WIII, H41, AY338590; TK100127, WIII, H42, AY338592; TK100281, WIII, H37, AY338601; TK100282, WIII, H20, AY338602; TK100595, WIII, H20, AY338607; TK100596, WIII, H16, AY338608; TK100597, WIII, H19, AY338609; TK100601, WIII, H16, AY338610).
Reference and outgroup samples:
Neotoma micropus
Mexico: Coahuila; 20 miles S Morelos (sample 1—TK 16501, AY496220). New Mexico; DeBaca County, 16 miles S, 3 miles E Taiban (sample 2—TK31643, AY496223); Otero County, Fort Bliss Military Base (sample 3—TK77270, AY496222; TK77260, AY496221). Texas; Brewster County, Black Gap Wildlife Management Area (sample 4—TK51947, AY496224); Briscoe County, Caprock Canyons State Park (sample 5—TK54620, AY496219); Hemphill County, Gene Howe Wildlife Management Area (sample 6—TK52442, AY496218); Hidalgo County, Las Palomas Wildlife Management Area, Adams Unit (sample 7— TK72536, AY496217); Taormina Unit (sample 8—TK72555, AY496216); Hudspeth County, 5 miles S Sierra Blanca (sample 9—TK4924, AY496214); McMullen County, James E. Daughtrey Wildlife Management Area (sample 10—TK90125, AY496225; TK90112, AY497905); Refugio County, Guadalupe Delta Wildlife Management Area (sample 11—TK49067, AY496215).
Neotoma leucodon
Texas; Kerr County, Kerr Wildlife Management Area (TK49716, AY497904).
Literature Cited
- Anderson DR, Burnham KP, Otis DL. Density estimation of small mammal populations using trapping web and distance sampling methods. Ecology. 1983;64:674–680. [Google Scholar]
- Animal Care and Use Committee Guidelines for the capture, handling, and care of mammals as approved by The American Society of Mammalogists. Journal of Mammalogy. 1998;79:1416–1431. [Google Scholar]
- Avise JC. Molecular markers, natural history, and evolution. Chapman & Hall; New York: 1994. [Google Scholar]
- Baker RJ, Mascarello JT. Karyotypic analyses of the genus Neotoma (Cricetidae, Rodentia) Cytogenetics. 1969;8:187–198. doi: 10.1159/000130061. [DOI] [PubMed] [Google Scholar]
- Birney EC. Systematics of three species of woodrats (genus Neotoma) in central North America. Vol. 58. Miscellaneous Publications, Museum of Natural History, University of Kansas; 1973. pp. 1–173. [Google Scholar]
- Bowman J, Forbes G, Dilworth T. The spatial scale of variability in small mammal populations. Ecography. 2000;23:328–334. [Google Scholar]
- Cariello NF, Swenberg JA, Skopek TR. Fidelity of Thermococcus litoralis DNA polymerase (vent) in PCR determined by denaturing gradient gel electrophoresis. Nucleic Acids Research. 1991;19:4193–4198. doi: 10.1093/nar/19.15.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castleberry SB, King TL, Wood PB, Ford WM. Microsatellite DNA analysis of population structure in Allegheny woodrats (Neotoma magister) Journal of Mammalogy. 2002;83:1058–1070. [Google Scholar]
- Castro-Campillo A, Roberts HR, Schmidly DJ, Bradley RD. Systematic status of Peromyscus boylii ambiguous based on morphologic and molecular data. Journal of Mammalogy. 1999;80:1214–1231. [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Templeton AR. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics. 1993;134:959–969. doi: 10.1093/genetics/134.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CW, Bradley RD. Molecular phylogenetics of the Neotoma floridana species group. Journal of Mammalogy. 2001;82:791–798. [Google Scholar]
- Edwards CW, Bradley RD. Molecular systematics and historical phylobiogeography of the Neotoma mexicana species group. Journal of Mammalogy. 2002;83:20–30. [Google Scholar]
- Edwards CW, Fulhorst CF, Bradley RD. Molecular phylogenetics of the Neotoma albigula species group: further evidence of a paraphyletic assemblage. Journal of Mammalogy. 2001;82:267–279. [Google Scholar]
- Ehrich D, Stenseth NC. Genetic structure of Siberian lemmings (Lemmus sibiricus) in a continuous habitat: large patches rather than isolation by distance. Heredity. 2001;86:716–730. doi: 10.1046/j.1365-2540.2001.00883.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse P, Quattro H. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Applications to human mitochondrial DNA restriction data. Genetics. 1992;136:343–359. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y-X. New statistical test of neutrality for DNA samples from a population. Genetics. 1996;143:557–570. doi: 10.1093/genetics/143.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y-X. Statistical test of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulhorst CF, et al. Isolation and characterization of Whitewater Arroyo virus, a novel North American arenavirus. Virology. 1996;224:114–120. doi: 10.1006/viro.1996.0512. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF, et al. Geographic distribution and genetic diversity of Whitewater Arroyo virus in the southwestern United States. Emerging Infectious Diseases. 2001;7:403–407. doi: 10.3201/eid0703.010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulhorst CF, Milazzo ML, Carroll DS, Charrel RN, Bradley RD. Natural host relationships and genetic diversity of Whitewater Arroyo virus in southern Texas. American Journal of Tropical Medicine and Hygiene. 2002;67:114–118. doi: 10.4269/ajtmh.2002.67.114. [DOI] [PubMed] [Google Scholar]
- Galvin TA, Sherman PW, Yensen E, May B. Population structure of the northern Idaho ground squirrel (Spermophilus brunneus brunneus) Journal of Mammalogy. 1999;80:156–168. [Google Scholar]
- Goldman EA. Revision of the wood rats of the genus Neotoma. North American Fauna. 1910;31:1–124. [Google Scholar]
- Grant WS, Bowen BW. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. Genetics. 1998;89:415–426. [Google Scholar]
- Hall ER, Genoways HH. Taxonomy of the Neotoma albigula-group of woodrats in central Mexico. Journal of Mammalogy. 1970;51:504–516. [Google Scholar]
- Hooper ET. The glans penis in Neotoma (Rodentia) and allied genera. Vol. 618. Occasional Papers, Museum of Zoology, University of Michigan; 1960. pp. 1–20. [Google Scholar]
- Klaus M, Moore RE, Vyse E. Microgeographic variation in allozymes and mitochondrial DNA of Microtus richardsoni, the water vole, in the Beartooth Mountains of Montana and Wyoming, U.S.A. Canadian Journal of Zoology. 2001;79:1286–1295. [Google Scholar]
- Knowles LL, Maddison WP. Statistical phylogeography. Molecular Ecology. 2002;11:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- Koop BF, Baker RJ, Mascarello JT. Cladistic analysis of chromosomal evolution within the genus Neotoma. Vol. 96. Occasional Papers, The Museum, Texas Tech University; 1985. pp. 1–9. [Google Scholar]
- Kosoy MY, et al. Prevalence of antibodies to arenaviruses in rodents from the southern and western United States: evidence for an arenavirus associated with the genus Neotoma. American Journal of Tropical Medicine and Hygiene. 1996;54:570–575. doi: 10.4269/ajtmh.1996.54.570. [DOI] [PubMed] [Google Scholar]
- Mascarello JT, Warner JW. Chromosome variations in the plains woodrat: a pericentric inversion involving constitutive heterochromatin. Experientia. 1974;30:90–91. [Google Scholar]
- Matocq MD. Phylogeographical structure and regional history of the dusky-footed woodrat, Neotoma fuscipes. Molecular Ecology. 2002;11:229–242. doi: 10.1046/j.0962-1083.2001.01430.x. [DOI] [PubMed] [Google Scholar]
- Matocq MD, Patton JL, da Silva MNF. Population genetic structure of two ecologically distinct Amazonian spiny rats: separating history and current ecology. Evolution. 2000;54:1423–1432. doi: 10.1111/j.0014-3820.2000.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Matson CW, Rodgers BE, Chesser RK, Baker RJ. Genetic diversity of Clethrionomys glareolus populations from highly contaminated sites in the Chornobyl region, Ukraine. Environmental Toxicology and Chemistry. 2000;19:2130–2135. [Google Scholar]
- Matsuhashi T, Masuda R, Mano T, Yoshida MC. Microevolution of the mitochondrial DNA control region in the Japanese brown bear (Ursus arctos) population. Molecular Biology and Evolution. 1999;16:676–684. doi: 10.1093/oxfordjournals.molbev.a026150. [DOI] [PubMed] [Google Scholar]
- Mirol PM, Peral Garcia P, Vega-Pla JL, Dulout FN. Phylogenetic relationships of Argentinean Creole horses and other South American and Spanish breeds inferred from mitochondrial DNA sequences. Animal Genetics. 2002;33:356–363. doi: 10.1046/j.1365-2052.2002.00884.x. [DOI] [PubMed] [Google Scholar]
- Monty A-M, Heist EJ, Wagle ER, Emerson RE, Nicholson EH, Feldhamer GA. Genetic variation and population assessment of eastern woodrats in southern Illinois. Southeastern Naturalist. 2003;2:243–260. [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Peakall R, Rubial M, Lindenmayer DB. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution. 2003;57:1182–1195. doi: 10.1111/j.0014-3820.2003.tb00327.x. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. Population genetic software for teaching and research. Australian National University; Canberra, Australia: 2001. GenA1Ex v.5: Genetic analysis in Excel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G, Gissi C, De Chirico A, Saccone C. Nucleotide substitution rate of mammalian mitochondrial genomes. Journal of Molecular Evolution. 1999;48:427–434. doi: 10.1007/pl00006487. [DOI] [PubMed] [Google Scholar]
- Planz JV, Zimmerman EG, Spradling TA, Akins DR. Molecular phylogeny of the Neotoma floridana species group. Journal of Mammalogy. 1996;77:519–535. [Google Scholar]
- Posada D, Crandall KA. Intraspecific gene genealogies: trees grafting into networks. Trends in Ecology and Evolution. 2001;16:37–45. doi: 10.1016/s0169-5347(00)02026-7. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA, Templeton AR. GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Molecular Ecology. 2000;9:487–488. doi: 10.1046/j.1365-294x.2000.00887.x. [DOI] [PubMed] [Google Scholar]
- Reese CL, Waters JM, Pagels JF, Brown BL. Genetic structuring of relict populations of Gapper's red-backed vole. Journal of Mammalogy. 2000;82:289–301. [Google Scholar]
- Rooney AP, Honeycutt RL, Derr JN. Historical population size change of bowhead whales inferred from DNA sequence polymorphism data. Evolution. 2001;55:1678–1685. doi: 10.1111/j.0014-3820.2001.tb00687.x. [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin v.2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva; Geneva, Switzerland: 2000. [Google Scholar]
- Suchecki JR, Ruthven DC, III, Fulhorst CF, Bradley RD. Natural history of the southern plains woodrat (Neotoma micropus) Texas Journal of Science. 2004;56:131–140. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (* and other methods), Version 4.0b10. Sinauer Associates, Inc., Publishers; Sunderland, Massachusetts: 2002. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Templeton AR, Boerwinkle E, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basis theory and an analysis of alcohol dehydrogenase activity in Drosophila. Genetics. 1987;117:343–351. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Routman E, Phillips CA. Separating population structure from population history: a cladistic analysis of the geographic distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambyostoma tigrinum. Genetics. 1995;140:767–782. doi: 10.1093/genetics/140.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II: methods for discrete population genetic data. Sinauer Associates, Inc., Publishers; Sunderland, Massachusetts: 1996. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetic structure of populations. Annals of Eugenetics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]
- Wright S. Evolution and the genetics of populations. Vol. 4. University of Chicago Press; Chicago, Illinois: 1978. [Google Scholar]