Abstract
Deuterium, solvent, and 15N kinetic isotope effects have been used to probe the mechanisms by which flavoproteins oxidize carbon–oxygen and carbon–nitrogen bonds in amines, hydroxy acids, and alcohols. For the amine oxidases d-amino acid oxidase, N-methyltryptophan oxidase, and tryptophan monooxygenase, d-serine, sarcosine, and alanine are slow substrates for which CH bond cleavage is fully rate limiting. Inverse isotope effects for each of 0.992–0.996 are consistent with a common mechanism involving hydride transfer from the uncharged amine. Computational analyses of possible mechanisms support this conclusion. Deuterium and solvent isotope effects with wild-type and mutant variants of the lactate dehydrogenase flavocytochrome b2 show that OH and CH bond cleavage are not concerted, but become so in the Y254F enzyme. This is consistent with a highly asynchronous reaction in which OH bond cleavage precedes hydride transfer. The results of Hammett analyses and solvent and deuterium isotope effects support a similar mechanism for alcohol oxidase.
Keywords: flavoprotein, amine oxidase, kinetic isotope effects, mechanism
Enzymes that contain FMN or FAD are ubiquitous in cellular metabolism due to the chemical versatility of the isoalloxazine ring. One of the most common reactions catalyzed by flavoenzymes is the two-electron oxidation of a metabolite. The net reaction is the transfer of a hydride equivalent to the flavin and thence to an electron acceptor such as oxygen or another redox-active protein. The subset of flavoenzymes which catalyze two-electron oxidation of their substrates include a large number of enzymes which oxidize the bond between carbon and a heteroatom. Their substrates include alcohols, amino acids, hydroxy acids, amines, and nitroalkanes. Despite the chemical and biological importance of these reactions, our present understanding of the catalytic details of these enzymes is far from complete.
To a large extent the uncertainties regarding mechanisms of flavoenzymes are due to the very chemical versatility of the isoalloxazine system. Because the fully oxidized cofactor is readily reduced by either one or two electrons, flavoproteins are capable of participating in reactions involving transfer of a single electron, a hydrogen atom, or a hydride.1 In addition, the oxidized flavin is susceptible to nucleophilic attack at the C(4a) and N(5) positions,1 and several biomimetic systems implicate such flavin adducts as intermediates in oxidation of biological metabolites.2,3 While this range of reactivities is clearly of advantage to biological systems, it complicates the mechanistic analyses of flavoenzymes, in that several viable mechanisms must be considered. In addition, the availability of alternative reactions means that there is always the potential that mechanistic probes such as alternative substrates or mechanism-based inhibitors will utilize pathways not taken by the natural substrates, complicating the mechanistic interpretation of results obtained by such methods. To avoid such complications, we have used a variety of kinetic isotope effects to probe the transition states for carbon–hydrogen bond cleavage in flavoproteins to distinguish among the various mechanistic proposals, since the substitution of a heavier isotope for a lighter one is not expected to alter the mechanism. The present report summarizes our results to date.
Scheme 3.
Amine and amino acid oxidases
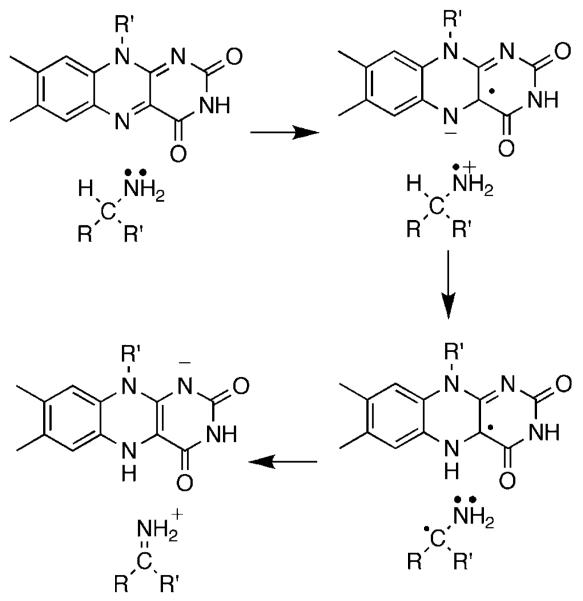
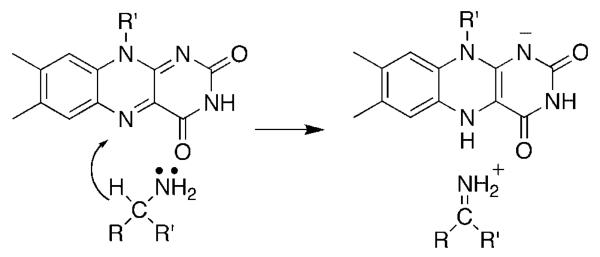
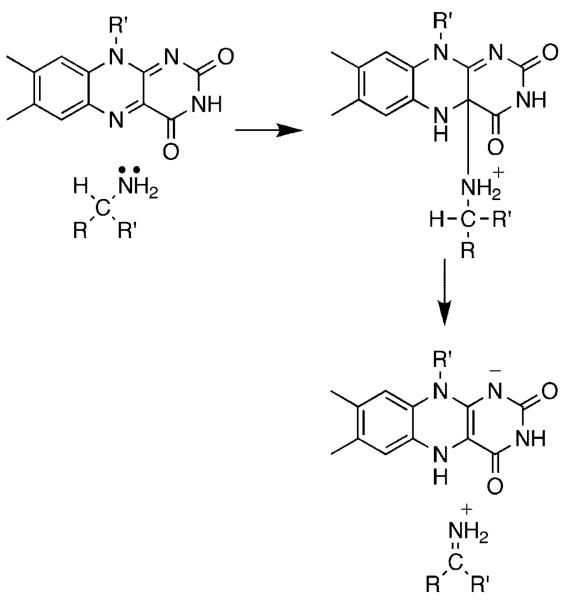
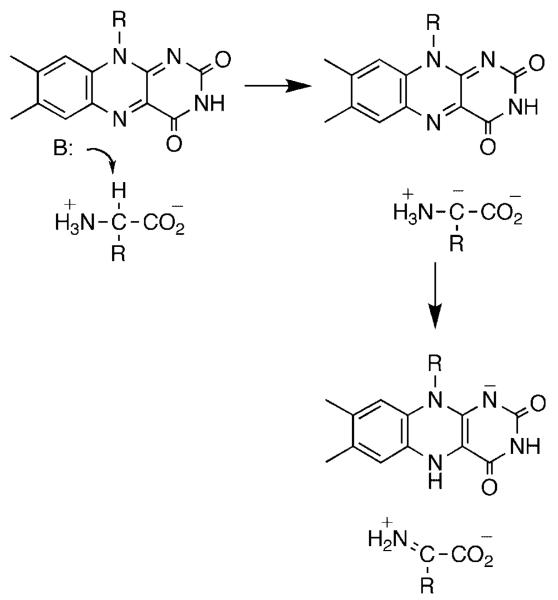
The flavin amine oxidases form two structurally related groups. d-amino acid oxidase (DAAO),4 sarcosine oxidase,5 glycine oxidase,6 and related enzymes have a common fold, differing in the details of substrate orientation and the specific bond which is oxidized. Monoamine oxidase,7 l-amino acid oxidase,8 polyamine oxidase,9 spermine oxidase,10 lysine-specific histone demethylase,11 and related enzymes can be grouped into a second much larger family with a wider range of substrates. The substrate specificities of the two families overlap, in that enzymes that oxidize amino acids, primary amines, and secondary amines are found in both. A remarkable diversity of mechanisms have been proposed for amine oxidation by flavoproteins. These include direct hydride transfer from the neutral amine to the flavin (Scheme 1),12 nucleophilic addition of the amine to the flavin (Scheme 2),13 single electron transfer reactions such as that in Scheme 3,14 and formation of a substrate carbanion by loss of a proton (Scheme 4).15
Scheme 1.
Scheme 2.
Scheme 4.
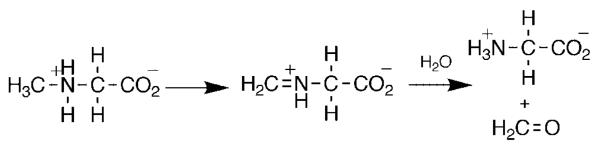
The paradigm for studies of flavoprotein amine oxidases has been DAAO. The enzyme from hog kidney has been the most studied because of ready availability, but DAAO is found throughout the eukaryotes and prokaryotes. The kinetic mechanism of DAAO is summarized in Scheme 5.16,17 The enzyme with oxidized FAD binds the substrate. This is followed by a single kinetic step which yields the reduced flavin and the imino acid product; with most but not all substrates this step is effectively irreversible. The reduced enzyme then reacts with O2 to yield the oxidized flavin and H2O2; in all cases to date this reaction is second order with no evidence for oxygen binding.18 Finally, the product dissociates from the oxidized enzyme and is subsequently hydrolyzed nonenzymatically to the respective keto acid and ammonium. This mechanism is common for all the flavoprotein oxidases, with slight differences in whether the oxidized substrate dissociates before or after flavin oxidation.
Scheme 5.
For many years, the generally accepted mechanism for DAAO was that of Scheme 4, in which the substrate α-hydrogen is removed as a proton to form a carbanion which then transfers electrons to the FAD.19 Based on an analysis of the kinetics with a number of substrates, we identified d-serine as a substrate for which there are no commitments to suppress the intrinsic isotope effect on the step of interest, cleavage of the amino acid carbon-hydrogen bond.20,21 With this substrate, d(kcat/Kser) has a pH-independent value of 4.5, identical to the isotope effect on the limiting rate of flavin reduction measured in a stopped-flow spectrophotomer, and d(kcat/Kser)1.44 equals t(kcat/Kser). Consequently, isotope effects on kcat/Kser report directly on the structure of the transition state for removal of the substrate α-hydrogen. The lack of a solvent isotope effect,20 the timing of proton release in the first turnover,22 and the pH dependence of the kcat/Km values21 are consistent with the active form of the substrate having its amino group uncharged. 15N kinetic isotope effects were used to distinguish between a mechanism involving loss of this atom as a proton (Scheme 4) and a direct hydride transfer (Scheme 1).12 Only in the case of the latter does rehybridization of nitrogen occur in the transition state for CH bond cleavage. The oxidation of d-serine was allowed to proceed till 40–60% completion before isolating the remaining amino acid and analyzing its nitrogen isotopic composition by isotope ratio mass spectrometry.23 The 15(kcat/Kser) value was 1.017 ± 0.007 at pH 7.5, decreasing to 0.9944 ± 0.0025 at pH 10.6. This pH-dependence is due to the 15Keq effect on the amine pKa. At pH 10.6 the amine is almost fully deprotonated, so that the equilibrium effect is not seen and the observed isotope effect equals the 15N effect on the CH bond cleavage step, 0.9944. This value establishes that there is significant rehybridization at nitrogen in the transition state for CH bond cleavage, ruling out the formation of a carbanion and in line for the expectation of the hydride transfer mechanism. It does appear that the rehybridization at the α-carbon may lag that at nitrogen, since the intrinsic alpha secondary tritium isotope effect with glycine is 1.03 ± 0.02.21 However, the intrinsic d(kcat/Kgly) value of 3.6 is much smaller than that for d-serine, suggesting that the transition state with glycine may be earlier than that with d-serine.
The sarcosine oxidase family of amine oxidases catalyzes the oxidative N-demethylation of N-methyl amino acids such as sarcosine (Scheme 6). This family has the same overall three-dimensional structure as DAAO, but binds the amino acid in the reverse orientation.5 Indeed, sarcosine is a substrate for DAAO, but the final products are methylamine and glyoxylate rather than glycine and formaldehyde.24 Despite this similarity, quite different mechanisms have been proposed for the sarcosine oxidase family compared to DAAO, including radical, nucleophilic, and hydride transfer mechanisms.25-27 We selected E. coliN-methyltryptophan oxidase (MTOX) as a representative member of this family for study, and used the slow substrate sarcosine. The kcat and kcat/Km pH profiles for sarcosine are consistent with a requirement that the nitrogen of the substrate be unprotonated and a group in the free enzyme, probably the flavin N(3), be protonated for activity.28 The dkcat and d(kcat/Ksarcosine) values are equal and pH independent and equal to the isotope effect on the rate of flavin reduction by sarcosine. The constant value of 7.2 establishes that there are no commitments with this substrate, so that cleavage of the amino acid CH bond is fully rate limiting for catalysis. There is no solvent isotope effect on the kcat/Km value for sarcosine, establishing that there is no exchangeable proton in flight in the transition state for CH bond cleavage, consistent with a requirement for the anionic form of the substrate.
Scheme 6.
The mechanisms of Schemes 2 and 3 both involve formation of flavin intermediates prior to CH bond cleavage. No intermediates can be detected when the reduction of the flavin by sarcosine is followed in the stopped-flow spectrophotometer, even when the deuterated substrate is used to slow the decay of any species formed prior to CH bond cleavage.28 This suggests that either these mechanisms are incorrect or these intermediates do not accumulate. As a further probe of the mechanism of the sarcosine oxidase family, we measured 15N kinetic isotope effects for the oxidation of sarcosine by MTOX.29 In this case, we isolated both the sarcosine which remained and the glycine which had been formed after the reaction had proceeded till 30–50% completion over the pH range 7.5–9.8. The 15(kcat/K) values determined from either mechanism decrease from 1.0175 at pH 7.5 to 1.0124 at pH 9.8. The 15Keq for sarcosine deprotonation of 1.022629 was used to correct the 15(kcat/K) value to yield the pH-independent 15N kinetic isotope effect for the rate limiting CH bond cleavage step. The values measured independently from sarcosine and glycine are comparable, 0.9952±0.0005 and 0.9940±0.0004, respectively.
The results with MTOX are similar enough to those for DAAO to suggest that both use an identical hydride transfer mechanism, differing only in the relative orientation of the substrate and flavin. To provide a more quantitative basis for interpreting 15N isotope effects for flavin oxidation of secondary amines, the isotope effects for proposed mechanisms were calculated using alloxazine and dimethylamine as a model. Ground-state and transition structures were fully optimized in B3LYP/6-31 + G** calculations using Gaussian 03,30 and tunneling corrections were applied using the one-dimensional Wigner model.31 The calculated isotope effect for direct hydride transfer was 0.993–0.994. In contrast, the nucleophilic mechanism yielded an isotope effect between 1.015 and 1.03, depending upon the details of the model. Thus, the 15N isotope effects rule out the latter mechanism. Direct modeling of a mechanism involving a flavin radical was not possible. Instead, a model involving the transfer of an electron or a hydrogen atom from a secondary amine to ammonia yielded an equilibrium isotope effect of 0.996. Such a value is also consistent with our measured 15N isotope effect. However, the calculations also yield the energetics for the model reactions. The barrier for hydride transfer to a flavin is more than 10 kcal/mol lower than the barriers for mechanisms involving a flavin radical, in agreement with the ~1.7 V difference in the redox potentials of flavins and secondary amines. Overall, the combination of measured 15N isotope effects and computational studies provides strong support for a simple hydride transfer mechanism for the sarcosine oxidase group of enzymes. Thus, all members of this structural family of flavoprotein amine oxidases share a common mechanism.
The most-studied member of the other flavin oxidase family is monoamine oxidase. There have been extensive studies of the mechanism of this enzyme due to its pharmacological importance. The results with mechanism-based inhibitors, deuterium isotope effects, and alternative substrates have been variously interpreted in favor of mechanisms involving nucleophilic addition and radical intermediates for monoamine oxidase and related enzymes.32,33 We recently began to apply 15N kinetic isotope effects to resolve this controversy. To date, we have studied the mechanism of an l-amino acid oxidase, tryptophan monooxygenase from Pseudomonas sativa. The pH dependence of the kcat/K values for amino acid substrates in the wild-type and mutant enzymes had established that the neutral form of the amino group is also required by these enzymes.34,35 While this enzyme prefers amino acids with large hydrophobic side chains,36 these exhibit significant commitments.37 Consequently, we selected the slow substrate alanine for our studies.38 There is no solvent isotope effect on kcat/Kala. The dkcat and d(kcat/Kala) values and the isotope effect on the rate constant for flavin reduction are all equal, with a pH-independent value of 6, establishing that CH bond cleavage is fully rate limiting. Again, no intermediate is seen during flavin reduction, even with deuterated alanine. The 15N isotope effect was measured by analyzing the residual substrate. The enzyme was not stable enough to measure the 15(kcat/Kala) value at high pH, limiting the analysis to pH 8, where a value of 1.0145 was found. Correcting this for the 15Keq value of 1.0223 gave a 15k value of 0.992, again consistent with a hydride transfer mechanism. The results with this enzyme thus suggest that both structural families of flavin amine oxidases utilize hydride transfer mechanisms.
α-Hydroxy acid oxidases and dehydrogenases
In the case of the α-hydroxy acid oxidases, the historical controversy has been whether the α-proton is removed as a proton (Scheme 7, path a) or a hydride (path b).15,39 This was based on the ability of yeast flavocytochrome b2, the most-studied member of this family, to catalyze elimination of bromide and chloride from 3-substituted lactate40 and the similarities of the flavocytochrome b2 and DAAO active sites.4 Flavocytochrome b2 is a dehydrogenase rather than an oxidase. The enzyme contains two domains.41 The flavin domain of about 390 residues is a (ßα)8 barrel containing the FMN cofactor. This domain is homologous to the other members of this flavoprotein family and is capable of converting lactate to pyruvate by itself.42-44 The first 100 residues of the protein contain a heme which accepts electrons from the reduced flavin and transfers them to an acceptor.
Scheme 7.
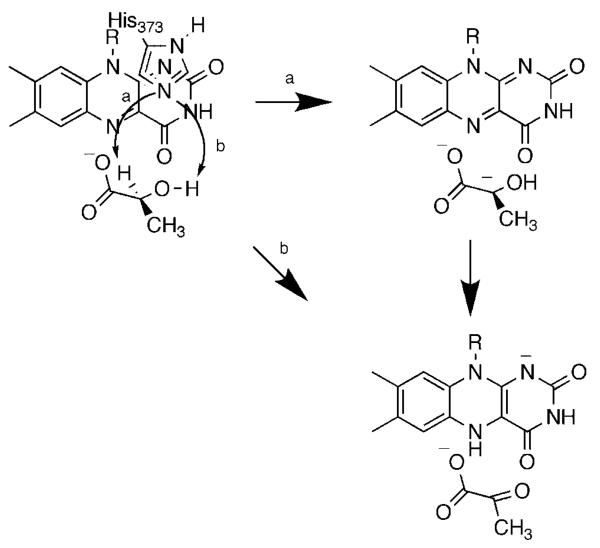
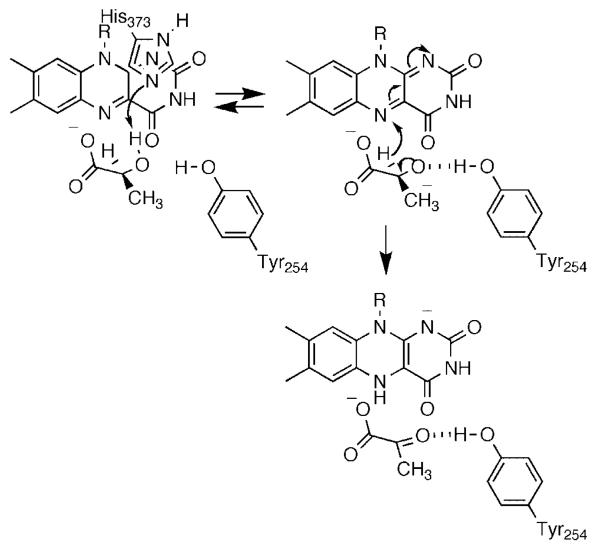
We have been using a combination of isotope effects and mutagenesis to probe the mechanism of lactate oxidation by yeast flavocytochrome b2. Two bonds must be broken in converting lactate to pyruvate, the OH bond and the α-carbon CH bond. We first used solvent and primary deuterium kinetic isotope effects to probe the relative timing of these two bond cleavages with lactate as substrate.45 Because of the limited substrate specificity of the enzyme, these studies were done with the physiological substrate lactate. With the wild-type enzyme, dkcat and d(kcat/Klactate) are both equal to 3.0; this value is significantly less than the isotope effect on the rate of flavin reduction of 5.4, establishing that there are significant commitments with lactate. With the wild-type enzyme d2Okcat equals 1.4 and d2O(kcat/Klactate) equals 0.9. Because we did not have the option of using a slow substrate to eliminate commitments, we utilized active site mutations to slow chemistry. Of the several active site residues that have been subject to site-directed mutagenesis, Asp282 is unusual in not interacting directly with the substrate. Rather, the role of Asp282 is to position the active site base, His373.46 The D282N enzyme shows a 150-fold decrease in the kcat/Klactate value. Both dkcat and d(kcat/Klactate) increase for the mutant enzyme to within error of 5.4; this value is likely the intrinsic isotope effect for the reaction and establishes that there are no commitments for the mutant enzyme. In contrast to the results for the wild-type enzyme, both d2Okcat and d2O(kcat/Klactate) are equal to unity in the mutant enzyme. Thus, the hydroxyl proton is not in flight in the transition state for cleavage of the lactate CH bond in the mutant protein, and presumably also not in the wild-type enzyme. However, these studies did not resolve the order of OH and CH bond cleavage. To further probe the mechanism we analyzed the Y254F enzyme. Tyr254 is within hydrogen bonding distance of the pyruvate carbonyl oxygen in the crystal structure.47 The rate constant for flavin reduction by saturating concentrations of lactate is 40-fold lower in the mutant enzyme. The dkcat and d(kcat/Klactate) values are identical and equal to 5.0. In contrast to the wild-type enzyme, the Y254F enzyme exhibits a significant solvent isotope effect, with d2Okcat and d2O(kcat/Klactate) identical and equal to 1.54. This establishes that CH and OH bond cleavages have become concerted in the mutant enzyme. We explained these results by the mechanism of Scheme 8. In the wild-type enzyme, the OH and CH bond cleavages are highly asynchronous. The lactate hydroxyl proton is abstracted by His373 to form the alkoxide either very early in the reaction or in a pre-equilibrium. The developing charge on the oxygen is stabilized by Tyr254. The hydride is then transferred to the flavin to form pyruvate. Mutagenesis of Tyr254 to phenylalanine destabilizes the alkoxide, delaying its formation until the initiation of hydride transfer from the α-carbon decreases the pKa of the oxygen sufficiently for proton transfer to His373. As a result, the OH and CH bond cleavages are much more synchronous. It is difficult to reconcile the isotope effects for the Y254F enzyme with a mechanism in which OH bond cleavage is subsequent to CH bond cleavage in the wild-type enzyme, such as the carbanion mechanism of Scheme 7. Thus, our results with the Y254F enzyme are more consistent with a highly asynchronous hydride transfer reaction in wild-type flavocytochrome b2 and presumably the other α-hydroxy acid-oxidizing flavoproteins.
Scheme 8.
Alcohol oxidases
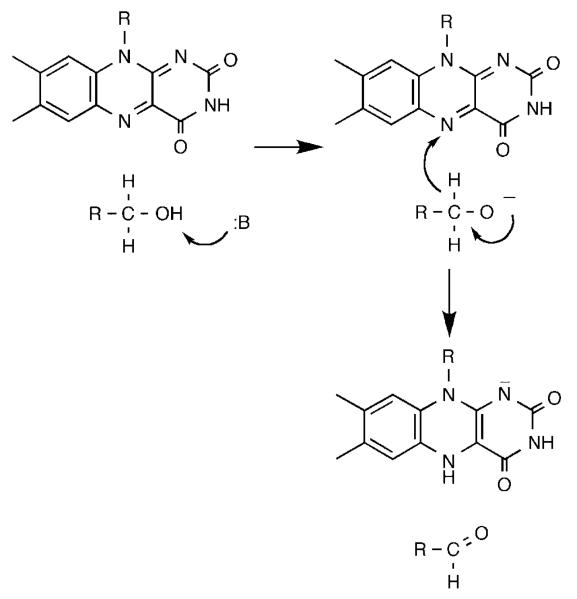
The alcohol-oxidizing flavoproteins make up a separate family of homologous proteins,48 of which glucose oxidase and cholesterol oxidase are the best characterized structurally.49,50 We selected yeast alcohol oxidase for study51 because of its broad substrate specificity. The kcat/Km pH profiles for both methanol and benzyl alcohol show that a single residue with a pKa of 8.3 must be unprotonated for activity. The kcat/Km values for a series of para-substituted benzyl alcohols yield a ρ value of 1.9 and a correlation with σ−, consistent with development of negative charge in the rate limiting transition state. With benzyl alcohol the d2o(kcat/Km) value is quite small, 1.2 ± 0.1 at pH 6.8, while the d2o(kcat/Km) equals 2.0 at high pH. The solvent inventory is linear. This suggests that the removal of the hydroxyl proton by the active site base to form the alkoxide is rate limiting with benzyl alcohols as substrates. In contrast, the kcat/Km values for a series of 2-substituted ethyl alcohols show a ρ value of −1.2 and a correlation with σI. With ethanol, the substrate with the most negative σI value, d(kcat/Km) equals 1.6 at pH 9 and 6.8, while d2o(kcat/Km) is 1.9. In contrast, 2-Br- and 2-Cl–ethanol, with more positive σI values, have d(kcat/Km) values of about 5 and d2o × (kcat/Km) values not significantly different from unity. This suggests that there is a change in the structure of the rate limiting bond cleavage from OH cleavage to CH cleavage as the ability of the substituent to stabilize a negative charge increased. The mechanism in Scheme 9 was proposed to account for these results. As is the case with flavocytochrome b2, the OH and CH bond cleavages are highly asynchronous. A base in the active site, probably a conserved histidine52 removes the hydroxyl proton prior to transfer of a hydride to the flavin. Recent studies of two other members of this flavoprotein family, choline oxidase52 and glucose oxidase,53 have provided additional support for this mechanism.
Scheme 9.
Nitroalkane oxidases
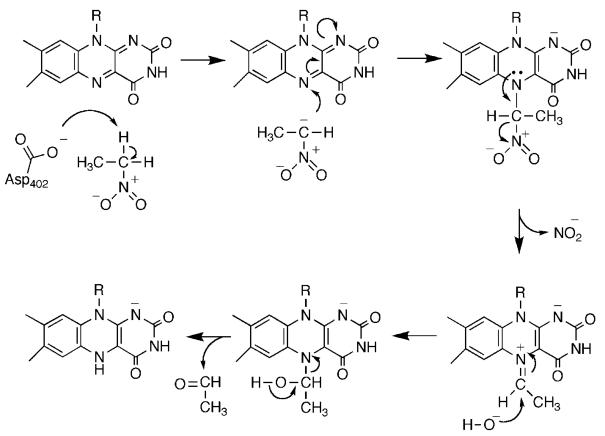
Nitroalkane oxidase (NAO) is a novel flavoprotein which further demonstrates the mechanistic versatility of the isoalloxazine nucleus. NAO catalyzes the oxidation of neutral nitroalkanes to the respective aldehydes or ketones, eliminating nitrite.54 The mechanism of NAO is shown in Scheme 10. Here, catalysis involves the initial formation of a nitroalkane anion by abstraction of an α-proton by Asp402. Nucleophilic attack of the anion on the FAD generates an initial adduct which then eliminates nitrite; attack of hydroxide followed by elimination of the aldehyde would generate the reduced FAD. Critical support for a carbanion intermediate for this enzyme comes from studies of the effects of mutating Asp402, the active site base.55 Neither the D402N nor the D402A enzymes have significant activity with neutral nitroethane. However, nitroalkane anions protonate slowly in solution, so it is possible to use them as substrates. Use of nitroethane anion as substrate restores the activity of D402A NAO, providing strong support for a carbanion intermediate. The kinetic competency of the electrophilic flavin amine cation has been established by trapping it with cyanide,56 while the three-dimensional structure of the enzyme in which this species has been trapped with nitroethane anion has been determined.57
Scheme 10.
Analysis of the substrate specificity of NAO showed that the enzyme prefers linear primary nitroalkanes with four or more carbons.58 Use of deuterated substrates established that CH bond cleavage is fully rate limiting for the reductive half-reaction for nitroethane, with substrate binding becoming increasingly rate limiting with longer chains.59 The initial step in Scheme 10 is the formation of the nitroalkane anion. The nonenzymatic ionization of nitroalkanes has been studied extensively as a model for proton removal from carbon.60-63 Unlike the vast majority of the nonenzymatic counterparts of enzyme-catalyzed reactions, the nonenzymatic reaction occurs at a significant rate at room temperature and pressure. The formation of the anion can readily be followed by trapping it with triiodine.64 We compared the ionization of nitroethane catalyzed by NAO, as reflected in the kcat/Km value, with the reaction catalyzed by acetate. The results are summarized in Table 1. As a catalyst, NAO is 109 times faster than acetate; this is reasonable, especially in that the kcat/Km values for longer chain nitroalkanes are 2–3 orders of magnitude larger than that for nitroethane, while the acetate reaction is unaffected by chain length. The rate increase in the enzyme-catalyzed reaction is due to a decrease in the enthalpy of activation, as is typical for enzymes.66 We also used NAO to address the contribution of quantum mechanical tunneling to the enzymatic rate enhancement. The ability of hydrogen to behave quantum mechanically, transferring from a donor to an acceptor without going completely over the activation barrier, was first discovered in studies of the formation of nitroalkane anions.62 More recently, such behavior has been described in a number of enzyme-catalyzed reactions.67-71 In retrospect, it is not remarkable that hydrogen tunnels in enzyme-catalyzed reactions as well as in solution. A more interesting question is whether enzymes can increase the extent of tunneling, possibly by utilizing enzyme motion to compress the reaction barrier.69 To address this question for NAO, we determined the temperature dependence of the deuterium kinetic isotope effect for the acetate and NAO-catalyzed ionizations of nitroethane. The primary criterion for tunneling is that the kinetic isotope effect on the Arrhenius prefactor A is 1 ± 0.4. Values below this are evidence for moderate tunneling and values above this indicate extensive tunneling.68 AH/AD is not significantly different from one for both the NAO and the acetate-catalyzed reactions (Table 1). The data are consistent with at most a small contribution to tunneling from either catalyst; more importantly, they indicate that the enzyme does not significantly increase the extent of tunneling in this case. To our knowledge, this is the only direct comparison of the contribution of tunneling in which there is an extensive rate enhancement by an enzyme.
Table 1.
Kinetic and thermodynamic parameters for nitroethane anion formation
| Acetate | NAO | |
|---|---|---|
| kH (M−1/s) | 4.14 ± 0.01 × 10−6 | 4800 ± 200 |
| kH/ | 7.8 ± 0.1 | 9.2 ± 0.4 |
| ΔG‡ (kcal/mol) | 24.8 ± 0.2 | 12.5 ± 1.0 |
| ΔH‡ (kcal/mol) | 21.9 ± 0.2 | 3.3 ± 0.4 |
| TΔS‡ (kcal/mol) | 2.9 ± 0.1 | 9.2 ± 0.9 |
| ΔEa (kcal/mol) | 1.38 ± 0.26 | 1.35 ± 0.17 |
| AH/AD | 0.81 ± 0.35 | 0.89 ± 0.25 |
Data from Reference [65].
Conclusion
As described here, the application of deuterium, solvent, and 15N kinetic isotope effects is providing significant insight into the mechanisms by which flavoproteins oxidize the bond between carbon and a heteroatom. The use of mechanism-based inhibitors, alternative substrates, and biomimetic systems has resulted in a wide range of proposals for the different classes of such flavoproteins. In contrast, our results are consistent with a common hydride transfer mechanism for oxidation of amines, amino acids, alcohols, and α-hydroxy acids. With amine substrates, the neutral nitrogen is formed prior to binding the enzyme and subsequent hydride transfer to the flavin. With alcohols and hydroxy acids, enzyme-catalyzed removal of the alcohol proton precedes hydride transfer in a highly asynchronous reaction.
Acknowledgments
Contract/grant sponsor: NIH; contract/grant number: R01 GM58698
Footnotes
Paper published as part of a special issue on ‘Recent Developments in the use of Isotopically Labelled Molecules in Chemistry and Biochemistry’.
REFERENCES
- 1.Muller F. Free flavins: syntheses, chemical and physical properties. In: Muller F, editor. Free Flavins: Syntheses, Chemical and Physical Properties. CRC Press; Boca Raton: 1991. pp. 1–71. [Google Scholar]
- 2.Chan TW, Bruice TC. Biochemistry. 1978;17:4784–4793. doi: 10.1021/bi00615a028. [DOI] [PubMed] [Google Scholar]
- 3.Kim J-M, Hoegy SE, Mariano PS. J Am Chem Soc. 1995;117:100–105. [Google Scholar]
- 4.Mattevi A, Vanoni MA, Todone F, Rizzi M, Teplyakov A, Coda A, Bolognesi M, Curti B. Proc Natl Acad Sci USA. 1996;93:7496–7501. doi: 10.1073/pnas.93.15.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trickey P, Wagner MA, Jorns MS, Mathews FS. Structure. 1999;7:331–345. doi: 10.1016/s0969-2126(99)80043-4. [DOI] [PubMed] [Google Scholar]
- 6.Settembre EC, Dorrestein PC, Park J, Augustine AM, Begley TP, Ealick SE. Biochemistry. 2003;42:2971–2981. doi: 10.1021/bi026916v. [DOI] [PubMed] [Google Scholar]
- 7.Binda C, Newton-Vinson P, Hubalek F, Edmondson DE, Mattevi A. Nat Struct Biol. 2002;9:22–26. doi: 10.1038/nsb732. [DOI] [PubMed] [Google Scholar]
- 8.Pawelek PD, Cheah J, Coulombe R, Macheroux P, Ghisla S, Vrielink A. EMBO J. 2000;19:4204–4215. doi: 10.1093/emboj/19.16.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. Structure. 1999;7:265–276. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 10.Huang Q, Liu Q, Hao Q. J Mol Biol. 2005;348:951–959. doi: 10.1016/j.jmb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Stavropoulos P, Blobel G, Hoelz A. Nat Struct Mol Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- 12.Kurtz KA, Rishavy MA, Cleland WW, Fitzpatrick PF. J Am Chem Soc. 2000;122:12896–12897. [Google Scholar]
- 13.Miller JR, Edmondson DE. Biochemistry. 1999;38:13670–13683. doi: 10.1021/bi990920y. [DOI] [PubMed] [Google Scholar]
- 14.Silverman RB. Acc Chem Res. 1995;28:335–342. [Google Scholar]
- 15.Fitzpatrick PF. Bioorg Chem. 2004;32:125–139. doi: 10.1016/j.bioorg.2003.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Palmer G, Massey V. Mechanisms of flavoprotein catalysis. In: Singer TP, editor. Biological Oxidation. Wiley; New York: 1968. pp. 263–300. [Google Scholar]
- 17.Fitzpatrick PF, Massey V. J Biol Chem. 1982;257:12916–12923. [PubMed] [Google Scholar]
- 18.Massey V. J Biol Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 19.Ghisla S, Massey V. Eur J Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 20.Denu JM, Fitzpatrick PF. Biochemistry. 1994;33:4001–4007. doi: 10.1021/bi00179a029. [DOI] [PubMed] [Google Scholar]
- 21.Denu JM, Fitzpatrick PF. Biochemistry. 1992;31:8207–8215. doi: 10.1021/bi00150a013. [DOI] [PubMed] [Google Scholar]
- 22.Fitzpatrick PF, Massey V. J Biol Chem. 1982;257:9958–9962. [PubMed] [Google Scholar]
- 23.Kurtz K. Studies on the mechanism of D-amino acid oxidase. MS Thesis. 2000 [Google Scholar]
- 24.Naoi M, Yagi K. Biochim Biophys Acta. 1976;438:61–70. doi: 10.1016/0005-2744(76)90223-0. [DOI] [PubMed] [Google Scholar]
- 25.Harris RJ, Meskys R, Sutcliffe MJ, Scrutton NS. Biochemistry. 2000;39:1189–1198. doi: 10.1021/bi991941v. [DOI] [PubMed] [Google Scholar]
- 26.Scrutton NS. Nat Prod Rep. 2004;21:722–730. doi: 10.1039/b306788m. [DOI] [PubMed] [Google Scholar]
- 27.Chen ZW, Zhao G, Martinovic S, Jorns MS, Mathews FS. Biochemistry. 2005;44:15444–15450. doi: 10.1021/bi0515422. [DOI] [PubMed] [Google Scholar]
- 28.Ralph EC, Fitzpatrick PF. Biochemistry. 2005;44:3074–3081. doi: 10.1021/bi047716h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ralph EC, Hirschi JS, Anderson MA, Cleland WW, Singleton DA, Fitzpatrick PF. Biochemistry. 2007;46 doi: 10.1021/bi700482h. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA, editors. Gaussian03. Gaussian, Inc.; Wallingford, CT: [Google Scholar]
- 31.Wigner E. Z Physik Chem B. 1932;19:203–216. [Google Scholar]
- 32.Li M, Binda C, Mattevi A, Edmondson DE. Biochemistry. 2006;45:4775–4784. doi: 10.1021/bi051847g. [DOI] [PubMed] [Google Scholar]
- 33.Rigby SE, Hynson RM, Ramsay RR, Munro AW, Scrutton NS. J Biol Chem. 2005;280:4627–4631. doi: 10.1074/jbc.M410596200. [DOI] [PubMed] [Google Scholar]
- 34.Emanuele JJ, Jr, Fitzpatrick PF. Biochemistry. 1995;34:3716–3723. doi: 10.1021/bi00011a029. [DOI] [PubMed] [Google Scholar]
- 35.Sobrado P, Fitzpatrick P. Biochemistry. 2003;42:13826–13832. doi: 10.1021/bi035299n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emanuele JJ, Jr, Heasley CJ, Fitzpatrick PF. Arch Biochem Biophys. 1995;316:241–248. doi: 10.1006/abbi.1995.1034. [DOI] [PubMed] [Google Scholar]
- 37.Emanuele JJ, Jr, Fitzpatrick PF. Biochemistry. 1995;34:3710–3715. doi: 10.1021/bi00011a028. [DOI] [PubMed] [Google Scholar]
- 38.Ralph EC, Anderson MA, Cleland WW, Fitzpatrick PF. Biochemistry. 2006;45:15844–15852. doi: 10.1021/bi061894o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunane LM, Barton JD, Chen ZW, Le KH, Amar D, Lederer F, Mathews FS. Biochemistry. 2005;44:1521–1531. doi: 10.1021/bi048616e. [DOI] [PubMed] [Google Scholar]
- 40.Urban P, Lederer F. Eur J Biochem. 1984;144:345–351. doi: 10.1111/j.1432-1033.1984.tb08470.x. [DOI] [PubMed] [Google Scholar]
- 41.Xia Z-X, Shamala N, Bethge PH, Lim LW, Bellamy HD, Xuong NH, Lederer F, Mathews FS. Proc Natl Acad Sci USA. 1987;84:2629–2633. doi: 10.1073/pnas.84.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindqvist Y, Brändén C-I, Mathews R, Lederer F. J Biol Chem. 1991;266:3198–3207. [PubMed] [Google Scholar]
- 43.Balme A, Brunt CE, Pallister RL, Chapman SK, Reid GA. Biochem J. 1995;309:601–605. doi: 10.1042/bj3090601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cunane LM, Barton JD, Chen ZW, Welsh FE, Chapman SK, Reid GA, Mathews FS. Biochemistry. 2002;41:4264–4272. [PubMed] [Google Scholar]
- 45.Sobrado P, Daubner SC, Fitzpatrick PF. Biochemistry. 2001;40:994–1001. doi: 10.1021/bi002283d. [DOI] [PubMed] [Google Scholar]
- 46.Gondry M, Lederer F. Biochemistry. 1996;35:8587–8594. doi: 10.1021/bi9529519. [DOI] [PubMed] [Google Scholar]
- 47.Xia Z-X, Mathews FS. J Mol Biol. 1990;212:837–863. doi: 10.1016/0022-2836(90)90240-M. [DOI] [PubMed] [Google Scholar]
- 48.Cavener DR. J Mol Biol. 1992;223:811–814. doi: 10.1016/0022-2836(92)90992-s. [DOI] [PubMed] [Google Scholar]
- 49.Sampson NS, Vrielink A. Acc Chem Res. 2003;36:713–722. doi: 10.1021/ar9800587. [DOI] [PubMed] [Google Scholar]
- 50.Hecht HJ, Kalisz HM, Hendle J, Schmid RD, Schomburg D. J Mol Biol. 1993;229:153–172. doi: 10.1006/jmbi.1993.1015. [DOI] [PubMed] [Google Scholar]
- 51.Menon V, Hsieh C-T, Fitzpatrick PF. Bioorg Chem. 1995;23:42–53. [Google Scholar]
- 52.Ghanem M, Gadda G. Biochemistry. 2005;44:893–904. doi: 10.1021/bi048056j. [DOI] [PubMed] [Google Scholar]
- 53.Brinkley DW, Roth JP. J Am Chem Soc. 2005;127:15720–15721. doi: 10.1021/ja056025l. [DOI] [PubMed] [Google Scholar]
- 54.Fitzpatrick PF, Orville AM, Nagpal A, Valley MP. Arch Biochem Biophys. 2005;433:157–165. doi: 10.1016/j.abb.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 55.Valley MP, Fitzpatrick PF. J Am Chem Soc. 2003;23:8738–8739. doi: 10.1021/ja036045s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valley MP, Tichy SE, Fitzpatrick PF. J Am Chem Soc. 2005;127:2062–2066. doi: 10.1021/ja043542f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagpal A, Valley MP, Fitzpatrick PF, Orville AM. Biochemistry. 2006;45:1138–1150. doi: 10.1021/bi051966w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gadda G, Fitzpatrick PF. Arch Biochem Biophys. 1999;363:309–313. doi: 10.1006/abbi.1998.1081. [DOI] [PubMed] [Google Scholar]
- 59.Gadda G, Choe DY, Fitzpatrick PF. Arch Biochem Biophys. 2000;382:138–144. doi: 10.1006/abbi.2000.2009. [DOI] [PubMed] [Google Scholar]
- 60.Bell RP, Goodall DM. Proc Royal Soc London, Series A. 1966;294:273–297. [Google Scholar]
- 61.Bell RP. Kinetic isotope effects in proton-transfer reactions. In: Bell RP, editor. Kinetic Isotope Effects in Proton-transfer Reactions. Cornell University Press; Ithaca, NY: 1973. pp. 250–296. [Google Scholar]
- 62.Bell RP. The Tunnel Effect in Chemistry. Chapman & Hall; New York: 1980. [Google Scholar]
- 63.Bernasconi CF. Adv Phys Org Chem. 1992;27:119–238. [Google Scholar]
- 64.Thibblin A, Jencks WP. J Am Chem Soc. 1979;101:4963–4973. [Google Scholar]
- 65.Valley MP, Fitzpatrick PF. J Am Chem Soc. 2004;126:6244–6245. doi: 10.1021/ja0484606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolfenden R, Snider MJ. Acc Chem Res. 2001;34:938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 67.Cha Y, Murray CJ, Klinman JP. Science. 1989;243:1325–1330. doi: 10.1126/science.2646716. [DOI] [PubMed] [Google Scholar]
- 68.Kohen A. Prog React Kinet Mech. 2003;28:119–156. [Google Scholar]
- 69.Klinman JP. Pure Appl Chem. 2003;75:601–608. [Google Scholar]
- 70.Roth JP, Wincek R, Nodet G, Edmondson DE, McIntire WS, Klinman JP. J Am Chem Soc. 2004;126:15120–15131. doi: 10.1021/ja047050e. [DOI] [PubMed] [Google Scholar]
- 71.Pavon JA, Fitzpatrick PF. J Am Chem Soc. 2005;127:16414–16415. doi: 10.1021/ja0562651. [DOI] [PMC free article] [PubMed] [Google Scholar]