SUMMARY
Ferrochelatase (protoheme ferrolyase, E.C. 4.99.1.1) is the terminal enzyme in heme biosynthesis and catalyzes the insertion of ferrous iron into protoporphyrin IX to form protoheme IX (heme). Within the past two years x-ray crystallographic data obtained with human ferrochelatase have clearly shown that significant structural changes occur during catalysis that are predicted to facilitate metal insertion and product release. One unanswered question about ferrochelatase involves defining the mechanism whereby some metals, such as divalent Fe, Co, Ni and Zn, can be used by the enzyme in vitro to produce the corresponding metalloporphyrins while other metals, such as divalent Mn, Hg, Cd or Pb, are inhibitors of the enzyme. Through the use of high resolution x-ray crystallography along with characterization of metal species via their anomalous diffraction, the identity and position of Hg, Cd, Ni, or Mn in the center of enzyme-bound porphyrin macrocycle was determined. When Pb, Hg, Cd or Ni was present in the macrocycle the conserved π helix was in the extended, partially unwound “product release” state. Interestingly, in the structure of ferrochelatase with Mn-porphyrin bound the π helix is not extended or unwound and is in the “substrate-bound” conformation. These findings show that at least in the cases of Mn, Pb, Cd and Hg, metal “inhibition” of ferrochelatase is not due to the inability of the enzyme to insert the metal into the macrocycle or by binding to a second metal binding site as has been previously proposed. Rather, inhibition occurs after metal insertion and results from poor or diminished product release. Possible explanations for the lack of product release are proposed herein.
Keywords: Heme synthesis, Ferrochelatase, Protoporphyrin IX, X-ray crystallography, metal inhibition
INTRODUCTION
The terminal step of heme biosynthesis, the insertion of ferrous iron into protoporphyrin IX to form protoheme IX (heme) is catalyzed by the enzyme ferrochelatase (protoheme ferrolyase, E.C. 4.99.1.1). In eucaryotes and Gram negative bacteria this enzyme is membrane associated while in Gram positive bacteria it is soluble1. The tertiary structure of the enzyme appears to be highly conserved although the amino acid sequence is less than 10% conserved between mammals and bacteria. Additional differences of note are that the ferrochelatases from higher, nonplant eucaryotes are mitochondrial membrane-associated homodimers and possess one [2Fe-2S] cluster per subunit while the cytoplasmic membrane-associated bacterial enzymes are monomeric. A few bacterial enzymes possess a [2Fe-2S] cluster,2,3 although the position within the primary sequence of the cysteine ligands for some varies considerably from what is found in eucaryotic, cluster-containing ferrochelatases.3
Deficiency of ferrochelatase in humans is the basis of the inherited disease erythropoietic protoporphyria (EPP).4,5 A mutation in one or both alleles may occur, but for most heterozygous EPP patients a variant in the promoter region of one allele results in diminished ferrochelatase protein being produced. This variant is frequently associated with a normal ferrochelatase allele and by itself does not result in clinical EPP.6,7,8
Among ferrochelatases the two best characterized are those from the bacterium Bacillus subtilis and human. For both of these enzymes, multiple crystal structures are available, and also kinetic and mutational analyses have been performed.1,9 A complete catalytic cycle involves acquisition of substrates, desolvation of ferrous iron, deprotonation of the two pyrrole protons, macrocycle distortion to allow metallation, and finally planarization and product release.1,9,10 Currently there is general agreement about several aspects of the overall catalytic mechanism, but there remain a few unresolved issues including the identification of the route for substrate metal entry into the active site, identification of the residue(s) involved in metal insertion, and an explanation for the metal specificity observed in previous steady-state kinetic studies.
What is known is that both human and the B. subtilis ferrochelatase utilize Fe, Ni, and Zn, while Pb, Hg, Cd, and Mn serve as competitive inhibitors in vitro and in vivo.11 In addition, species dependent specificity has been observed wherein the B. subtilis enzyme can incorporate Cu but not Co, while the opposite has been observed for human ferrochelatase.1 Various mechanisms have been proposed to explain the different metal specificities.12,13,14 One point in common to the proposed models for metal specificity is that they involve a selection mechanism that occurs prior to the chelation reaction. It has been suggested that metal specificity results from the nature of active site residues or from variations in macrocycle distortion.9,12,13,14 However, these hypotheses have rested heavily on steady-state kinetics and structural information that is not completely unambiguous. It is important to point out that more recent investigations into the transient kinetics of human ferrochelatase revealed that substrate binding (metal and porphyrin) and chelation occurs rapidly in contrast to product release.15
The model suggesting that metal specificity depended upon the amount of macrocycle distortion has become less attractive after structural studies with human ferrochelatase demonstrated that there is only a 10 to 12° porphyrin macrocycle distortion16 rather than a previously proposed ~ 30° distortion. Theoretical modeling predictions for macrocycle metallation are consistent with the minor distortion17,18,19 that was observed and together these observations weaken the argument that macrocycle distortion is a significant modulator of metal selectivity. Another model suggests that differences in the active site residues may provide the mechanism for metal selectivity.14,20 For example, an obvious difference in active site residues is observed at the top of the active site pocket. At this position a methionine residue (M76) is found in the human enzyme, while a tyrosine residue (Y13) is found in B. subtilis ferrochelatase.
In order to address the question of metal specificity and insertion, our laboratory has worked to capture structural “snapshots” of the human enzyme during catalysis. Starting with the assumption that in the presence of an inhibitory metal it may be possible to capture a metal-inhibited transition state, we set about trying to trap such a structure hoping that it might reveal the specific site of pre-chelation metal binding. In this work we employed the porphyrin substrates protoporphyrin and deuteroporphyrin in conjunction with the previously identified inhibitory metals Mn, Cd, and Hg as well as an alternative substrate Ni to investigate the mechanism of metal inhibition. Herein we present structural data clearly demonstrating that human ferrochelatase is catalytically competent to insert all of these “inhibitory” metals into the porphyrin macrocycle. Moreover, these data show that the adequacy of a metal to serve as an effective substrate, as opposed to being inhibitory, depends upon the ability of the metallated porphyrin to “escape” from the active site pocket post-metallation. These observations are inconsistent with current models and therefore alter the way in which metal substrate specificity should be viewed for human ferrochelatase and potentially all chelatase reactions.
RESULTS
Structure of the wild-type human ferrochelatase with “inhibitory metals”
Our strategy for capturing a physiologically relevant porphyrin- and metal-bound ferrochelatase structure was to put the enzyme under normal turnover conditions and then attempt to stop the reaction by addition of an inhibitory metal such as Hg, Cd or Mn. Since the current hypotheses suggest that metal selection occurs prior to the chelation reaction, it was our hope that this approach would produce a crystallizable metal and/or porphyrin bound ferrochelatase that would represent a pre-chelation intermediate and provide new insight into the mechanism of porphyrin deprotonation as well as the initial site of substrate metal binding. Such a structure would also address questions involving porphyrin distortion, the mechanism of porphyrin deprotonation, and metal specificity.
Earlier published attempts to capture pre-metallation reaction intermediates using Pb as the inhibitory metal were inconclusive because the Pb atom was found to partially occupy positions on both sides of the porphyrin plane.21 Such observations have been made in crystal structures of small inorganic Pb-metallated porphyrins22,23 so it was not clear if the Pb had in fact been inserted into the macrocycle or if it represented a pre-metallation intermediate. To address this issue we chose to examine the structures of Hg (FC-HG) and Cd (FC-CD) inhibited ferrochelatase. Hg is of particular interest because it has an ionic radius that is similar to that of Pb but structures of Hg-porphyrin compounds have been reported that demonstrate that the Hg atom can occupy a single position on one side of the porphyrin plane.24 We were able to obtain diffraction quality crystals of the wild-type human ferrochelatase enzyme following incubation of the enzyme with protoporphyrin and Hg. The crystals obtained were of substantial size (0.3mm × 0.8 mm × 0.8mm) and diffracted to better than 2.0 Å (Table 1).
Table 1.
Data collection and refinement statistics.
| Data | FC-HGa | FC-CDb | MN-DEUTc | NI-DEUTd |
| Space Group | P212121 | P212121 | P212121 | P212121 |
| Wavelength | 0.98 | 0.98 | 0.98 | 0.98 |
| Resolution Range (Ǻ) | 50.0-1.6 | 50.0-1.8 | 50.0-2.0 | 50.0-2.2 |
| Outer Shell | 1.66-1.6 | 1.86-1.8 | 2.07-2.0 | 2.49-2.2 |
| Unique Observations | 224,750 | 82,065 | 116,112 | 64,956 |
| Completeness (%) | 99.9(99.6)e | 99.9(99.7) | 99.8(98.4) | 99.6(96.8) |
| Rsym (%)f | 0.08(0.24) | 0.05(0.19) | 0.07(0.22) | 0.06(0.26) |
| I/σ | 34.2(4.5) | 37.5(6.2) | 32.8(5.6) | 23.2(3.0) |
| Model | FC-HG | FC-CD | MN-DEUT | NI-DEUT |
| Unit Cell (a, b, c) in Ǻ | 86,93,109 | 86,93,109 | 88,94,109 | 86,93,110 |
| Protein Atoms | 5,985g | 5,848 | 5,839 | 5,790 |
| Solvent Atoms | 799 | 393 | 534 | 336 |
| Resolution Limits (Ǻ) | 50.0-1.6 | 50.0-1.8 | 50.0-2.0 | 50.0-2.2 |
| Rcryst (%) | 20.4 | 21.3 | 16.9 | 21.5 |
| Rfree (%) | 24.2 | 25.1 | 20.1 | 24.9 |
| rmsd bonds (Ǻ) | 0.006 | 0.009 | 0.006 | 0.008 |
| rmsd angles (°) | 1.23 | 1.31 | 1.24 | 1.29 |
| average B factor (Ǻ2) | 24.7 | 27.3 | 19.7 | 36.6 |
Data for crystals obtained with wild-type human ferrochelatase treated with protoporphyrin IX and either Hga or Cdb.
Data for crystals obtained with the F110A variant of human ferrochelatase treated with deuteroporphyrin either Mnc or Nid.
Numbers in parentheses denote values for the outermost resolution shell.
Rsym = ∑hkl[∑I(|Ihkl,I - <Ihkl>|)]/∑hkl,I<Ihkl>, where Ihkl is the intensity of an individual measurement of the reflection with indices hkl and <Ihkl> is the mean intensity of that reflection.
Number includes atoms for residues in alternate conformations. Coordinates for the FC-HG, FC-CD, MN-DEUT, and NI-DEUT data have been deposited in the Protein Data Bank under the PDBID codes 3HCN, 3HCO, 3HCP, and 3HCR respectively.
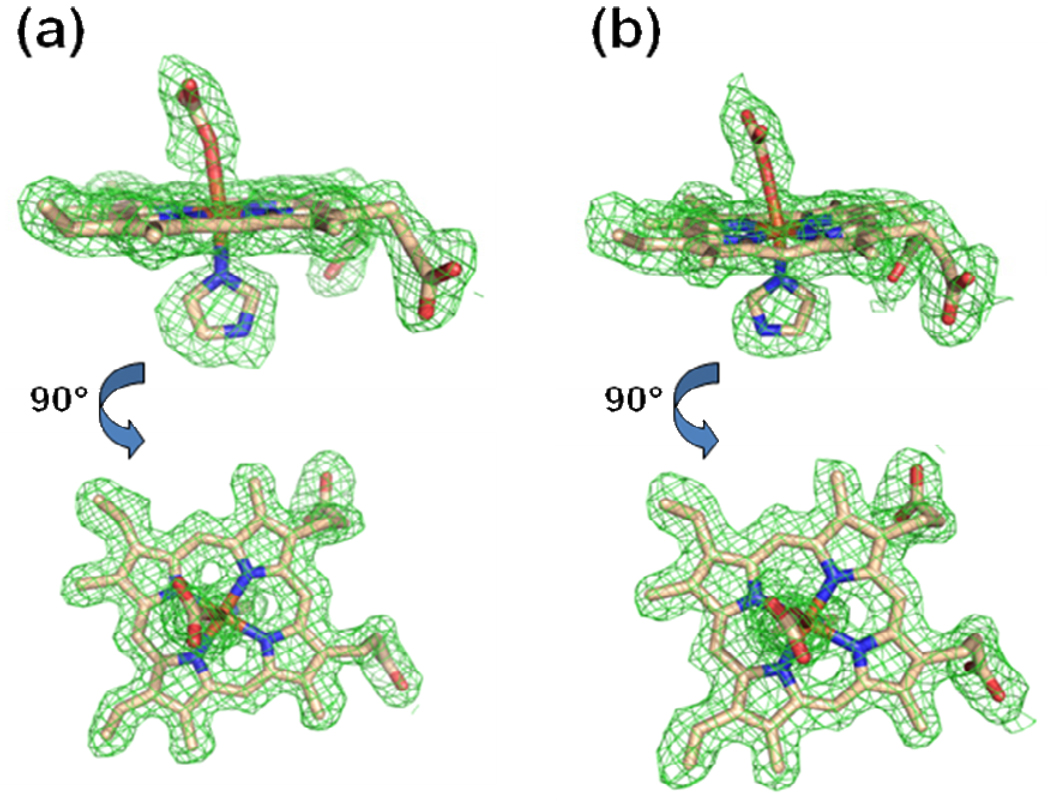
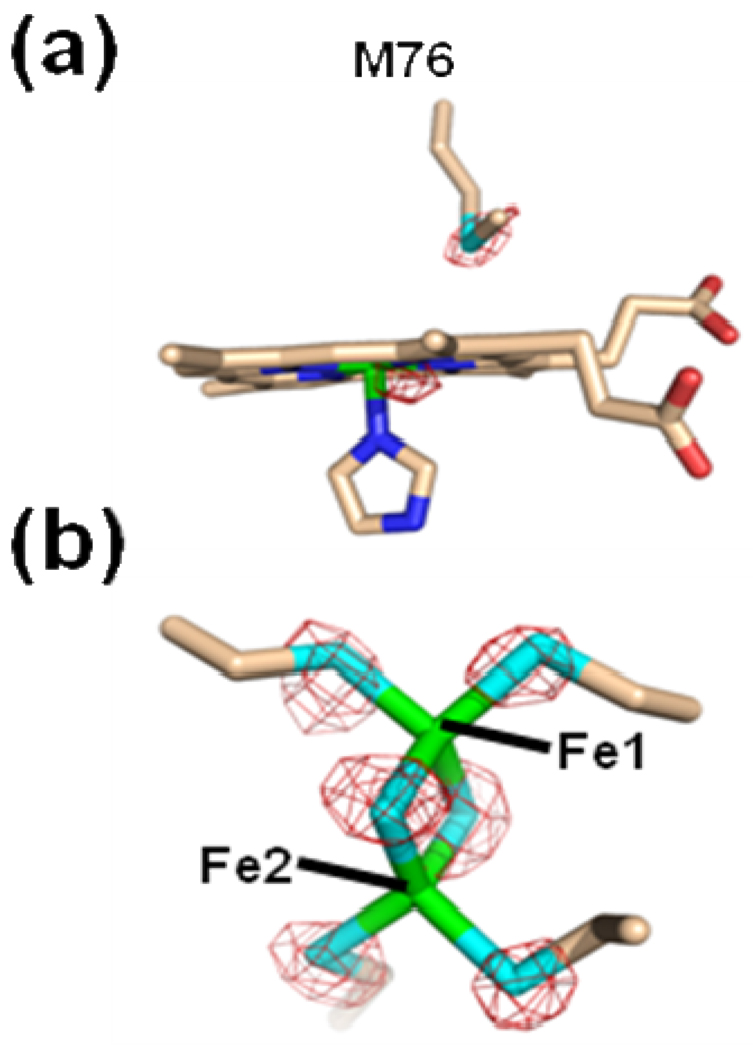
It was somewhat of a surprise that iterative rounds of model building and refinement revealed a structure that contained a metallated protoporphyrin in both subunits. The composite omit map and model for the metallated protoporphyrin IX for both the Hg and Cd data sets are shown in Figure 1 and reveal very clean density for a six coordinate metallated protoporphyrin. We have modeled a metallated porphyrin in both active sites with an imidazole ligand on the H263 side of the porphyrin plane. It is likely that this free imidazole originates from the affinity column elution buffer. The metallated porphyrin in the A monomer for both the FC-HG and FC-CD data were modeled with a bicarbonate ligand on the M76 side of the porphyrin plane. In both cases (FC-HG and FC-CD) the metallated porphyrin in the B monomer was best modeled with bis-imidazole ligation (Supplemental Figure 1).
Figure 1.
Model and electron density revealing a metallated protoporphyrin IX bound within the active site of the A monomer for the wild-type human ferrochelatase enzyme treated with protoporphyrin IX and either Hg (Panel a) or Cd (Panel b). The model is shown in stick format and the 2Fo-Fc composite omit map (green cage) was generated using the simulated annealing protocol with 7% of the model omitted per cycle. The lower image in both panels is the same data shown in the top image rotated by 90°. Carbon, nitrogen, oxygen, and iron atoms are colored tan, blue, red, and orange respectively and the composite omit map is contoured at 1 σ.
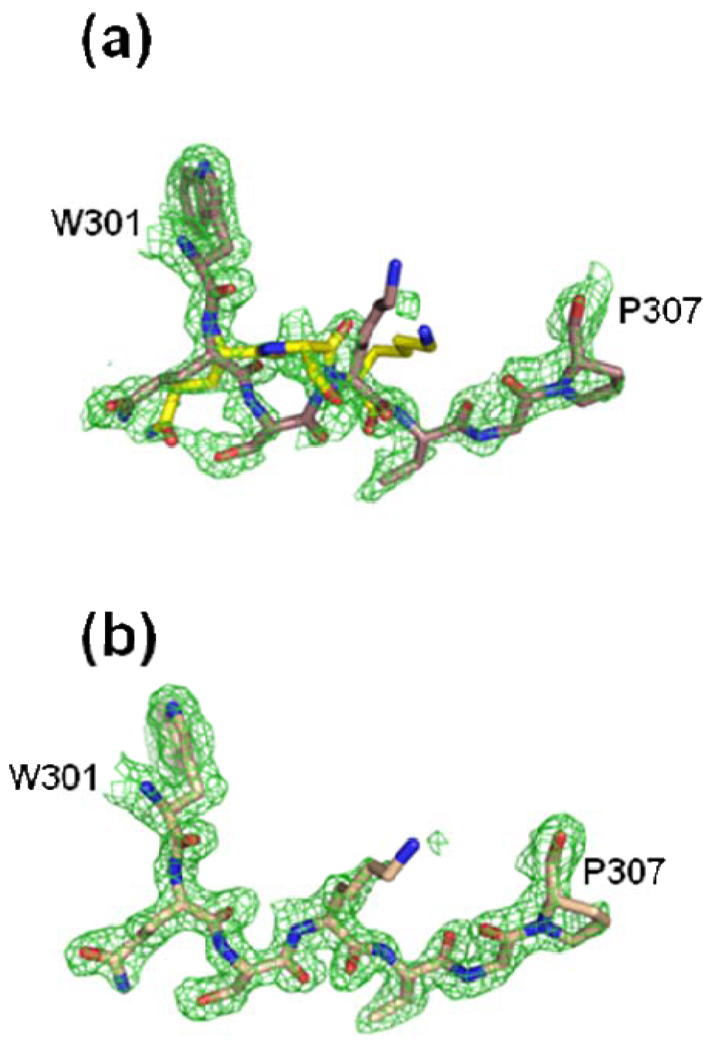
The Hg data are of high enough resolution (refined to 1.6 Å) to show that atomic details within the active site structures of the two monomers (A and B) are not equivalent. Of particular interest was the position of several residues in a structurally conserved loop (residues 301–314). The conformation of these residues was clearly different between monomers A and B (Figure 2, Panels a and b). Specifically, these alternate conformations are due to the different spatial arrangement of the peptide backbone and side chains for residue Q302 through residue K304 in the A monomer as compared to the B monomer for both the Cd and the Hg data sets.
Figure 2.
Alternate conformations observed in the conserved loop region of human ferrochelatase consisting of residues 301–307 for monomer A (Panel a). The electron density for the same residues in monomer B (Panel b) was much cleaner and indicated a single conformation. In both panels the green cage represents a 2Fo-Fc composite omit map that was generated using the simulated annealing protocol with 7% of the model omitted per cycle and contoured at one sigma. Atom coloring is the same as that shown in Figure 1 except that the carbon atoms are colored yellow for the alternate conformations of residues 302, 303, and 304.
Identification of bound metals in ferrochelatase subunits
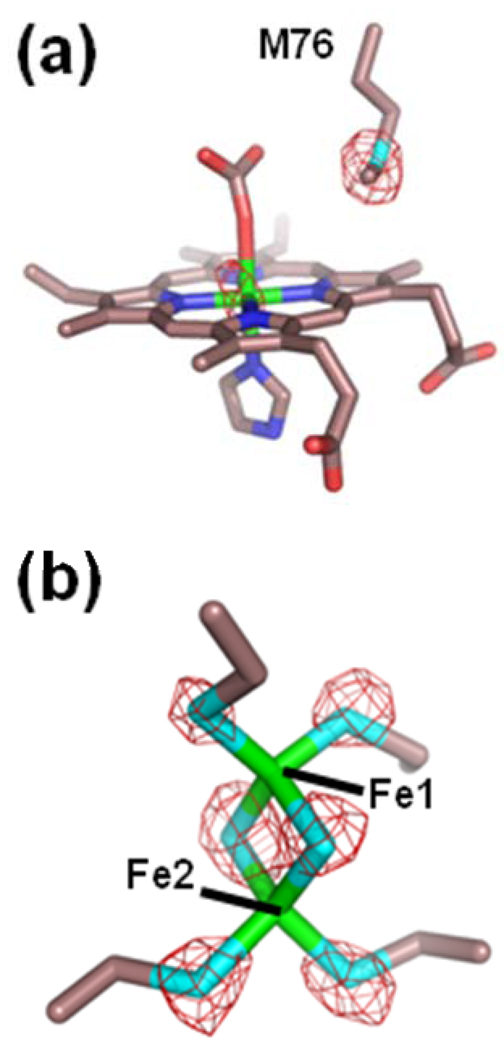
In addition to the differences seen in the electron density for the metallated porphyrin ligands and the conformations of the 302–304 loop in the A monomer, there are also significant peaks observed in the difference map between the two subunits when the metal was modeled as iron. Of specific interest was the observation of a 6.0 σ peak in the Fourier difference map for the A monomer in the FC-HG data. While the electron density appeared to be well modeled by the iron atom in the B monomer, an iron atom does not have sufficient electron density to fully account for the observed data in subunit A. Given the manner in which the enzyme was prepared, only iron or mercury could account for the electron density arising from a metal atom. A reasonable conclusion would be that a Hg atom is present in this site in the A subunit. This would account for the additional density as well as the other differences seen in the active site of monomer A.
However, to address this question experimentally, we took advantage of the fact that the anomalous signal (Δf’’) for Hg is 14 electrons at an X-ray wavelength of 1.90 Å while the anomalous signals for Fe and a number of other metals of biological significance such as Mn, Co, Ni, Cu, and Zn are all less than 2 electrons at this wavelength. Each subunit of human ferrochelatase contains a [2Fe-2S] cluster whose constituents, and their respective anomalous signals, can be used as built-in controls for comparison to any Hg anomalous signal observed. Specifically, at the longer wavelength (1.9 Å), the iron anomalous will essentially disappear and the sulfur and Hg anomalous signals will become stronger.
The data from the anomalous scattering experiments are presented in Figure 3. Peaks for all the sulfur atoms were also determined in the model allowing for further refinement and accurate placement of these residues. The data presented in Figure 3 show an anomalous signal at the 1.9 Å wavelength for the metal ion in the porphyrin of monomer A. This signal is similar to that observed for the ordered sulfur atoms in the model and is not attributable to iron since under these conditions no signal is seen for the well ordered iron atoms in the [2Fe-2S] cluster of either monomer (See Figure 3, Panel b). An identical anomalous signal was obtained for anomalous data collected with the FC-CD crystals (Δf’’ of ~8 electrons for Cd at a wavelength of 1.9 Å). These data strongly suggest that the metal seen in the porphyrin in subunit A is due to Hg (or Cd for the FC-CD data set) although the degree of occupancy cannot be accurately determined in these experiments.
Figure 3.
Model and anomalous data for the HG-PROTO crystals (Table 1) of wild-type human ferrochelatase for subunit A. The electron density (red cage) represents anomalous signal for data collected at a wavelength of 1.9 Å and is contoured at 6 σ. The model is represented in stick format with carbon, oxygen, nitrogen, sulfur, and iron colored yellow, red, blue, cyan, and green, respectively.
The significance of these observations is two-fold. First, they show that both enzyme subunits of the ferrochelatase dimer are functionally active. The observation that only iron is found in the B monomer and only the A monomer has a heavy metal present was unexpected and interesting. Such data might suggest an enzyme mechanism that may be cooperative in nature, something that has not been observed to date. Second, the data clearly demonstrate that human ferrochelatase is capable of catalyzing the insertion of an “inhibitory” metal ion in this case Hg, Cd, or Pb21 into a porphyrin macrocycle.
Deuteroporphyrin binding and Mn “inhibited” ferrochelatase structures
The current hypotheses surrounding metal selectivity in the ferrochelatase mechanism are built on in vitro investigations of the enzyme kinetics in the presence and absence of inhibitory metals.11,12,25,26,27,28 It is important to note that many kinetic investigations utilize deuteroporphyrin instead of protoporphyrin IX. In fact, ferrochelatase utilizes a variety of IX isomer porphyrins1 and deuteroporphyrin is popular as an alternative substrate in vitro since it yields faster turnover numbers, although with slightly lower affinity.28 In contrast to protoporphyrin IX, deuteroporphyrin lacks the vinyl groups at the 2 and 4 ring positions and, therefore, it is possible to structurally discriminate enzyme-bound (metallated) deuteroporphyrin from any possible heme (protoheme) carryover from the enzyme preparation.
For structural studies with the “inhibitor” metal Mn (MN-DEUT) and the “substrate” metal Ni (NI-DEUT), we were successful in obtaining crystals and solving structures of both the previously characterized F110A variant and wild-type ferrochelatase (Table I). In all cases, the protein was incubated with either Mn or Ni and deuteroporphyrin IX prior to crystallization. For a given metal (either Mn or Ni), the models obtained with either the wild-type enzyme or the F110A variant were identical. Therefore, for brevity we have presented only the higher resolution data sets for the current discussion.
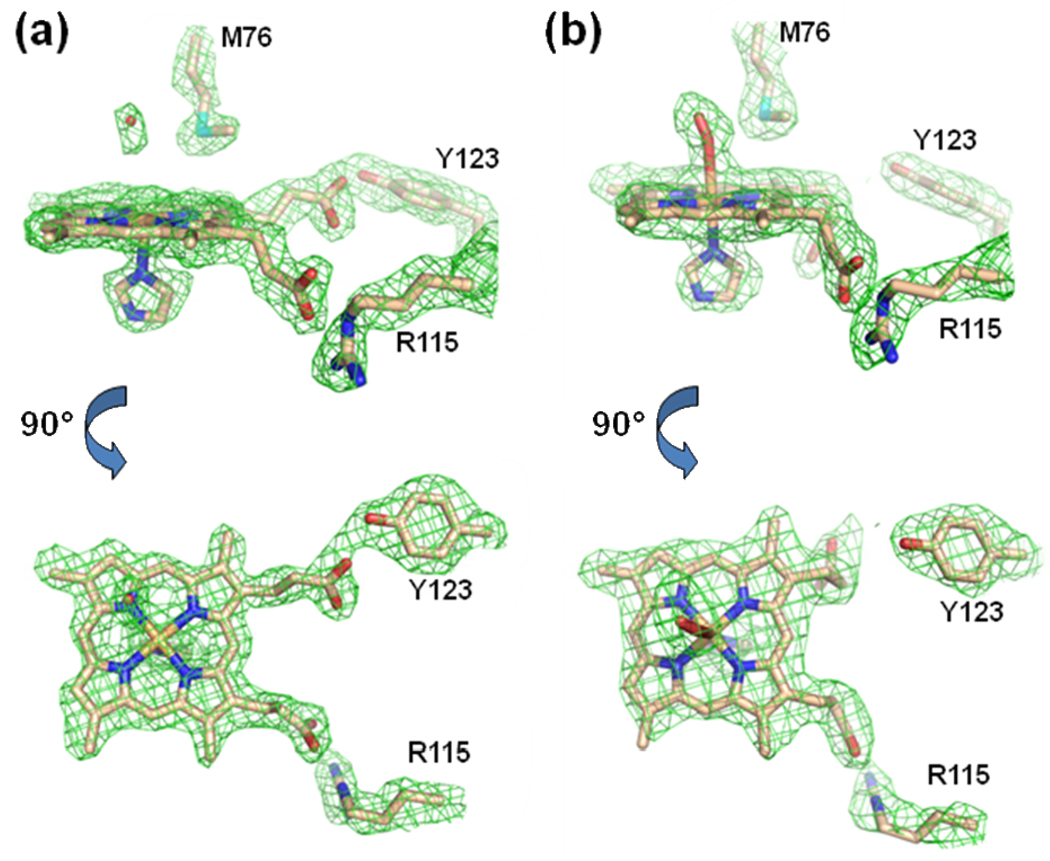
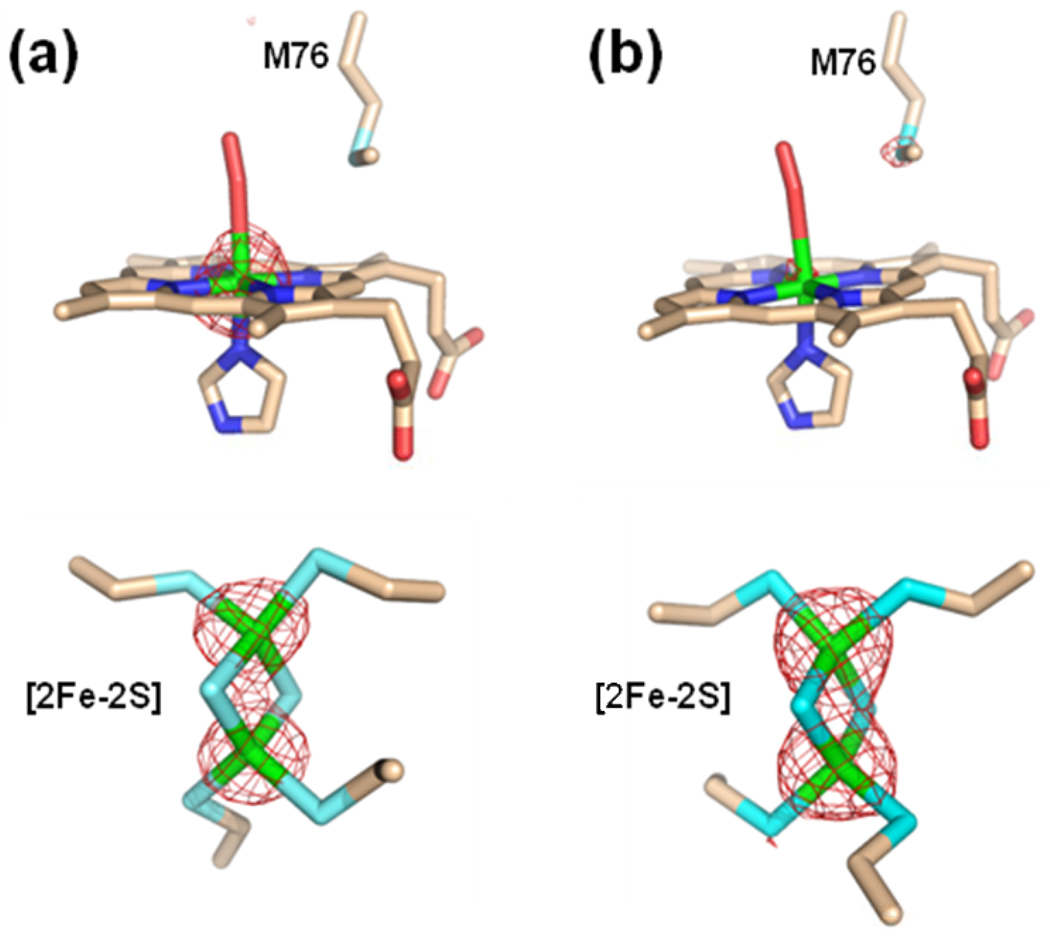
Several interesting observations are apparent from these data. First, as with Hg and Cd-containing structures discussed above, metallated porphyrins are found in both the A and B subunits (Figure 4). Second, in contrast to what was previously observed in ferrochelatase with bound metallated porphyrins,21 but similar to the published non-metallated protoporphyrin IX-bound structure of human ferrochelatase,16 the MN-DEUT data revealed electron density for the porphyrin propionates of the metallated deuteroporphyrin juxtaposed to the side chain of R115 (via propionate 7) and the side chain of Y123 (via propionate 6) (Figure 4, Panel a). However, the spatial orientation of the deuteroporphyrin macrocycle in the MN-DEUT structure is not as deep in the active site cleft in comparison to what was observed with free (non-metallated) protoporphyrin IX. The binding mode of protoporphyrin IX has been observed to be slightly deeper in the active site allowing for the propionate at position 6 to also interact with the side chain of S130.16 For the MN-DEUT data, this propionate forms a single hydrogen bond with the side chain of Y123 (Figure 4, Panel a). While both the MN-DEUT and NI-DEUT data sets revealed a metallated porphyrin with the propionate at position 7 forming a salt bridge with the side chain of R115, the position of the propionate at position 6 was distinctly different (Figure 4, compare Panel a with Panel b). This observation was somewhat surprising because, at the present time, all of the structures reported for human ferrochelatase that contain a bound metallated porphyrin have shown that the propionate at position 6 is bent back towards the center of the porphyrin ring rather than interacting with Y123 and S130. The data presented in Figure 4 reveals that for deuteroporphyrin, this conformational adjustment is only observed for the substrate metal, nickel. It is also of note that the metallated porphyrin in the MN-DEUT structure is five coordinate and has been modeled with the ligand as an imidazole molecule (Figure 4, Panel A). In contrast, the metallated porphyrin for the NI-DEUT data has a ligand on the M76 side of the active site pocket whose density is best modeled as molecular oxygen (Figure 4, Panel b).
Figure 4.
Model and 2Fo-Fc composite omit map generated with the simulated annealing protocol (7% of the model was omitted per cycle) for the MN-DEUT (Panel a) and the NI-DEUT (Panel b) crystals of human ferrochelatase (Table 1). For both Panel A and Panel B the lower image is a 90° rotation of the data presented in the upper image. The electron density is contoured a 1 σ and represented by the green cage. Atoms are shown in the stick configuration and carbon, nitrogen, oxygen, and iron atoms are colored tan, blue, red, and orange respectively.
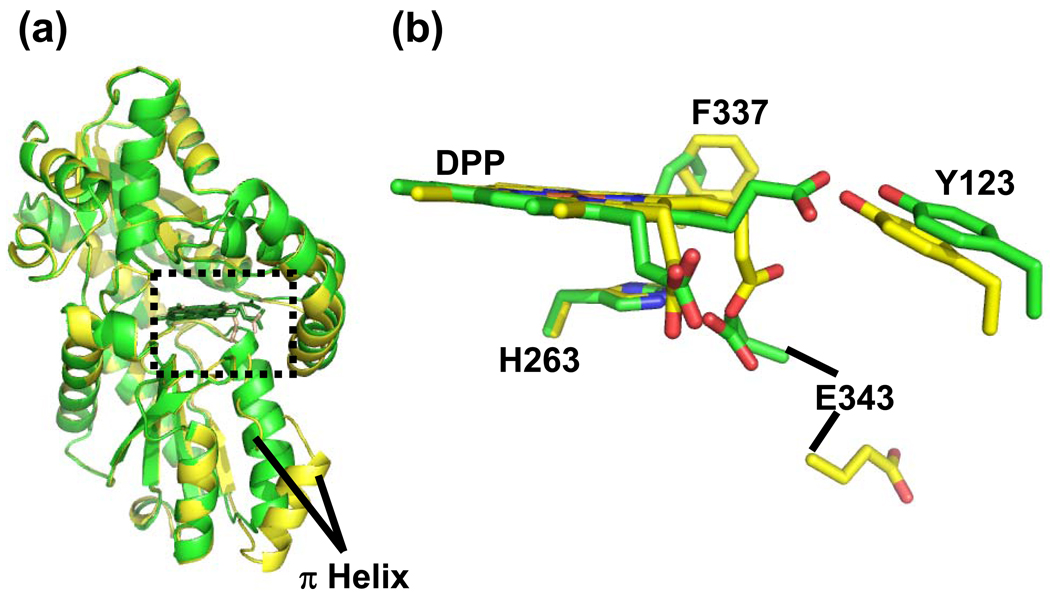
Further comparison of the MN-DEUT and NI-DEUT data sets reveal another significant and somewhat surprising observation. Despite the presence of a metallated porphyrin in the active site of the MN-DEUT data, the structurally conserved π helix is not unwound and the overall structure of the enzyme is in a significantly more “closed” conformation (Figure 5 panel a). In all of the metallated protoporphyrin structures of human ferrochelatase reported to date, the π helix has been observed in an “open” conformation referred to as the “release” state (Supplemental Figure 2). The structural differences between the MN-DEUT and NI-DEUT models reported here are shown in the overlays in Figure 5. In summary, Figure 5 highlights the overall differences in the models and highlights the relative orientations of the metallated porphyrins, the π helix, and key active site residues. In addition to the unwinding and extension of the π helix, the propionate at position is positioned back towards the center of the porphyrin macrocycle and we see some reorientation of the side chain for the conserved residue F337. These data clearly show that the NI-DEUT structure is distinctly different from the MN-DEUT model despite the only variation in enzyme preparation being the addition of either Ni or Mn.
Figure 5.
Comparison of the MN-DEUT and NI-DEUT models. Panel a; Cartoon representation of secondary structure showing an overlay of the MN-DEUT (Green) and NI-DEUT (Yellow) models. The structurally conserved π helix is labeled and the active site is highlighted by a dashed box. Panel b; Model in stick format for the metallated deuteroporphyrin and the relative positions of some strictly conserved residues in both data sets. Carbon atoms in the MN-DEUT model are shown in green while the carbon atoms in the NI-DEUT model are shown in yellow. Oxygen, nitrogen, and metal (currently modeled as iron) atoms are colored red, blue and orange, respectively.
Identification of Mn and Ni in metalloporphyrin-containing structures
In order to determine whether or not the structural differences described above and shown in Figure 4 and Figure 5 are due to Mn or Ni incorporation we investigated the identity of the metal using anomalous scattering in a fashion similar to what is detailed above for Hg and Cd. Similar to Hg, Mn also has a significant anomalous signal at wavelengths beyond the iron edge. Specifically, this signal will get progressively stronger up to the K-edge for Mn which is at an X-ray wavelength of 1.89 Å. To this end, we collected data at 1.8 Å in order to address whether or not Mn was the metal incorporated into the porphyrin. This data is shown in Figure 6 and reveals an anomalous signal consistent with Mn being bound in the active sites of both the A and B subunits. This is in contrast to the data we obtained with the Hg and Cd protoporphyrin treated wild-type enzyme discussed above where the anomalous signal was only observed in the A monomer. Similarly, the electron density in the composite omit maps for residues 302–304 was very clean in both monomers indicating a well defined structure for this loop region in both monomers of the MN-DEUT model.
Figure 6.
Model and anomalous signal for subunit A the MN-DEUT crystals (Table 1). The electron density (purple cage) represents anomalous signal for data collected at a wavelength of 1.8 Å and is contoured at 6 σ. The model is represented in stick format with carbon, oxygen, nitrogen, sulfur, and the metal (modeled as iron) colored tan, red, blue, cyan, and orange, respectively.
The anomalous signal data are also very clear with respect to the identity of the metal incorporated into the deuteroporphyrin for the NI-DEUT data. These data are presented in Figure 7 and indicate that the majority of the metal incorporated into the deuteroporphyrin for the NI-DEUT data set is nickel. Specifically, the intensity of the anomalous signal for the Ni atom at the center of the deuteroporphyrin is greatest just prior to the nickel edge (Figure 7, Panel a). At longer wavelengths, beyond the nickel edge, the intensity for any Ni atoms decreases significantly while the intensity for any iron atoms, such as those in the [2Fe-2S] cluster, become more intense as the X-ray energy gets closer to the iron edge (Figure 7, Panel B).
Figure 7.
Model and anomalous signal for the NI-DEUT crystals for data collected at an X-ray energy of 1.45 Å(Panel a) and 1.70 Å (Panel b). For both panels the upper image shows the model and anomalous signal around the metallated deuteroporphyrin and the lower panel show the model and anomalous signal around the [2Fe-2S] cluster. In both cases the anomalous map is contoured at 8 σ and represented by the red cage. The model is shown in stick format and carbon, nitrogen, oxygen, sulfur, and iron are colored tan, blue, red, cyan, and green respectively.
DISCUSSION
Metal chelatases are evolutionarily old enzymes that are necessary for producing cofactors with broad usage and incredible catalytic flexibility. Perhaps the most ancient of the chelatases that we know about currently is the small CbiX cobalt chelatase of Archaeoglobus fulgidus.29 This protein contains only one of the usual two domains found in class II chelatases such as ferrochelatase. Nature has taken this domain via gene fusion and mutation and crafted chelatases with increasing size and specificity. Interestingly, this evolutionary journey somehow resulted in the production of enzymes that in vivo do not make metallation mistakes. Part of the observed specificity can be attributed to compartmentalization or specific metal chaperones, such that the chelatase is only exposed to the correct metal, part to basic chemistry, and part to enzyme design.
At the inception of the current work our intent was to define the intramolecular spatial elements or residues responsible for substrate metal approach and chelation. This question is not easily addressed since the purified enzyme in solution in vitro will have global access to substrate iron, including the opening to the active site pocket that in vivo is believed to be membrane-embedded for eucaryotes and therefore prohibits iron access via the active site “mouth”. In addition, ferrous iron may bind at random sites on proteins so distinguishing the actual iron entry path unambiguously may be difficult to determine in vitro. In fact, the serendipitous binding of metals to proteins is well documented and was a necessity for early crystallographers whom relied on chance metal binding in order to perform Multiple Isomorphous Replacement (MIR). At the time, this technique was the only approach available in order to solve the phase problem and find a starting point for building an atomic model.
Side chains that are good Lewis bases such glutamic acid, aspartic acid, cysteine, and histidine are the most common protein ligands for the heavier transition metals. However, the atomic interactions that result in metal binding to these side chains are identical for the more common first row transition metals. These side chains are commonly found as ligands to metals in enzyme cofactors, with histidine, cysteine, and tyrosine having well defined interactions with heme iron. Tyrosine has been identified as a heme ligand in a eukaryotic heme sensor30 and as a ligand in a bacterial heme transport proteins.31 In both cases, the evolution of tyrosine as a ligand to the heme iron has some functional significance as both proteins must efficiently bind heme but only transiently. Both of these functions (heme sensor or heme transporter) are similar to ferrochelatase in that the heme must not be bound too tightly by the protein. For all ferrochelatases, proper enzyme function requires the quick removal of the product and any interaction that stabilizes the bound heme state would result in a bottleneck in the heme biosynthetic pathway resulting in accumulation of protoporphyrin. Employment of H263 as a metal donor, as has been suggested by others, rather than as the pyrrole proton acceptor, would result in the formation of a metallated porphyrin with the imidazole of H263 as the axial ligand to the metal. Removal of this highly stable product would require a mechanism to disrupt the metal-imidazole ligand bond to allow displacement of the heme product. Evidence for such a mechanism does not exist.
In order to address metal specificity and chelation, our approach was to employ inhibitory metals which we assumed would compete with substrate iron, bind to the enzyme, and result in an unchelated metal and free porphyrin bound to the enzyme. In such a situation it would be anticipated that the crystal structure of this complex would clearly identify the site from which the metal was to be inserted. Surprisingly, we found that the “inhibitory” metals Hg, Cd, and Mn all serve as substrates for the enzyme in that they are inserted into the porphyrin macrocycle. The crystallographic data further indicate that the inhibition of ferrochelatase by these metals occurs because the metallated product is poorly released from the enzyme and is not due to a selection mechanism that occurs prior to chelation.
Thus, by serendipity, we have addressed the question of the basis for ferrochelatase metal specificity. It is clear that neither the amount of macrocycle ring distortion nor the specific active site residues play a significant pre-chelation role in selectivity. In addition, the suggestion that a second metal binding site exists and serves a regulatory function12 is not supported by the current structural work. Our data, however, do not rule out the role that active site residues may play in subtle, pre-chelation interactions that affect the rate of metal insertion or the order in which competing metals might be chelated. Such issues would be anticipated based upon the specific electronic properties of the metal involved and could be addressed by looking at transient state kinetics and specifically the chelation rate versus release rate with different metals. Kinetic models that employ competing metals must now account for altered rates of product release for the different metallated porphyrins. At the present time, all that can be said is that properties of the metal that have yet to be precisely identified must account for the persistent product binding that results in the apparent inhibition. This is especially true since the porphyrin macrocycle into which the various metals are inserted is identical in all situations studied herein. The precise mechanism of the inhibition could be due to a number of possibilities including subtle differences in the release pathway, binding mode of the metallated porphyrin product (as seen in the MN-DEUT data set), or slower/less efficient triggering of the conformational changes that are required for product release.
Considering the structural information presented above, the remaining question is, why do some metallated porphyrins fail to exit the active site effectively? Given the differences found between Mn and other inhibitory metals the answer to this question is clearly metal specific. For Mn, the structural data show that the active site mouth is in the closed position even after metallation has occurred and both Y123 and R115 are interacting with the propionates as has been observed for the physiological substrate.16 Moreover, the π helix is neither unwound nor extended. Clearly this “closed” structure is not static and some release must occur since Mn inhibition is competitive and not irreversible. However, since the crystal lattice simply represents capture of one of several possible thermodynamic minima (typically the most populated), our data suggest that when Mn is incorporated into the deuteroporphyrin, the closed conformation is simply more stable. This by no means indicates that a certain population of the release conformation does not exist. All one can conclude is that the most populated conformation has simply been trapped in the lattice during crystal growth. Such a suggestion is consistent with the degree of protein flexibility that we have observed and previously detailed.16,21
The ferrochelatase structure with bound Mn-porphyrin raises something of a conundrum since we had proposed previously that metallation per se triggers the π helix extension.21 Yet, this does not appear to be the case for Mn. There are, however, two hints from the structural data that provide clues. One finds that unlike all other human ferrochelatase structures with metallated porphyrins where the metal has axial ligands on both sides of the ring (i.e. six coordinate metals), when Mn has been incorporated into the macrocycle, there is no ligand on the M76 side indicating that the Mn is five coordinate. Additionally, the porphyrin 6 propionate remains in the porphyrin “substrate-bound” conformation rather than being flipped back towards the center of the macrocycle as is observed in other metallated porphyrin-containing structures. Whether these features are independent or dependent upon one another, the observations provide additional evidence of the importance of the spatial location of the macrocycle propionate and adds to the decades old data related to the preference for IX isomer porphyrins.32
Unlike Mn, the metals Pb, Hg, and Cd all are chelated and result in porphyrin propionate 6 movement and π helix extension. However, the simple fact that the metallated porphyrin is still captured in the active site during crystallization suggests that these products also release much more slowly. One distinction is that Pb, Hg, and Cd are significantly larger in size than Fe and other “substrate” metals. For Pb, the largest metal of the three, density maps are consistent with a metal that does not sit in the plane of the macrocycle which may prevent easy exit. Clearly, new structures with metals such as Ru and Os may provide additional clues. An interesting observation is that the altered conformation of segment 302–304 exists only in the “inhibited” subunit. These data suggest that this region of the active site is more sensitive to the properties of the incoming metal cation.
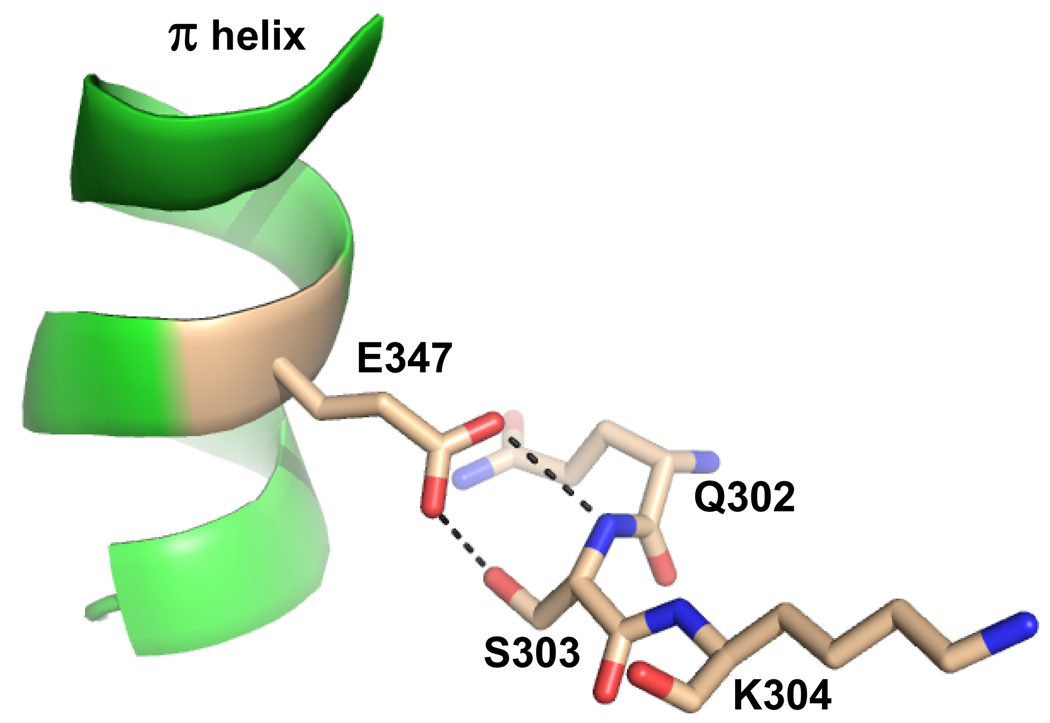
One possible explanation for the slow rate of product release in the Hg and Cd inhibited forms may be in the interactions between conserved residues found in both the π helix and the 302–304 loop. For example, in the wound form the strictly conserved residue E347, in the π helix, forms a hydrogen bond with the side chains of Q302 and S303 (Figure 8). These interactions provide stabilizing interactions for the π helix either in the free or porphyrin bound form. If the 302–304 loop is sensitive to the incoming metal and the hydrogen bonding between this loop and the π helix is disrupted, then this could be enough to destabilize the π helix. When comparing the conformations of residues 302–304 in the A and B monomers, it is clear that one of the alternate conformations for these residues in A, which has a mixture of Hg and iron or Cd and iron, is distinct from what is found in B, which contains iron (Figure 2, Panel A). The alternate conformation for residue S303, where the side chain is rotated 180° out of the active site pocket, results in interactions between Q302 with H341 and H263. The observed reorientation of H263 away from the inserted metal in the macrocycle along with kinetics data suggest this residue is protonated and would, therefore, most likely be hydrogen bonded with the oxygen atom of the Q302 side chain. Q302 is also oriented to hydrogen bond with H341. These interactions would stabilize the enzyme in a post chelation state, which would result in slow product release.
Figure 8.
Cartoon and ball-and-stick representation showing the spatial relationship and hydrogen bonding between the structurally conserved π helix and residues 302–304 in an active site loop. The coordinates are taken from the human model with protoporphyrin IX bound (PDB ID 2HRE). Hydrogen bonds are represented by the dashed lines. The π helix is colored green and nitrogen, carbon, and oxygen atoms are colored blue, tan, and red respectively.
The current data also raise another question. Specifically, why is it possible to obtain crystals of wild-type human ferrochelatase with protoheme bound in one subunit and Hg-protoporphyrin in the second subunit, but not wild-type enzyme with protoheme bound in both subunits? The answer is undoubtedly tied to the fact that crystallographic data represent solution states that are thermodynamically trapped in the crystal lattice. In our case, there are no crystal contacts in the π helix region so the crystal lattice can accommodate both the closed and the release conformations. Clearly, both of these conformational states are thermodynamically close in free energy.
In vivo the issue of enzyme-based metal selectivity is moot because specific metal chaperones33 will insure that only ferrous iron is delivered to the enzyme. Moreover, the release and clearing of the product heme may very well be accomplished through additional chaperones or heme transport proteins. In vitro, the absence of such transporters and the presence of additional metal or heme ligands such as imidazole, bicarbonate, dithiothreitol, β-mercaptoethanol, or oxygen may also facilitate or interfere with metal delivery as well as product release. This conclusion is clearly supported by both biochemical and structural observations.12,21
The current data are consistent with a model for human ferrochelatase where coordinated rearrangement of active site residues both at the top and bottom of the pocket result in unwinding of the π helix upon metallation. Residues 302 – 304, in a conserved metal sensitive loop of the protein, most likely stabilize the π helix in the resting or porphyrin bound state (See Figure 8). Upon metal insertion these residues are found in an altered location which results in a destabilized π helix. In the presence of a large inhibitory metal this region is found in a conformation which allows it to interact with key active site residues locking them into a post-chelation form, thus acting as an electrostatic sensor. This, though, is not the complete story as an alternative metal, similar in size and electrostatics to iron is inserted, but does not result in the unwinding of the π helix. Since the main difference in the Mn inhibited form is the absence of a sixth ligand at the top of the active site pocket and an altered orientation for the side chain of the strictly conserved F337 at the back of the pocket, it seems likely that movement of active site residues at the top of the pocket must play a role in further destabilizing the π helix causing it to unwind, possibly via F337.
Previous studies have demonstrated the importance of the active site hydrogen bonding network in catalysis,34 and these new structures suggest a role for this network in product release. Investigation with additional inhibitory metals as well as other active site variants should clarify the specific role residues at the top of the pocket play in product release and winding or unwinding of the structurally conserved π helix.
METHODS
Expression, purification and crystallization
Wild-type and F110A variant ferrochelatases were expressed and purified as previously described.35,36 Prior to affinity purification using Talon™ (Clontech, Palo Alto, California) metal affinity resin, approximately 10 µmole porphyrin, either deuteroporphyrin IX or protoporphyrin IX (Frontier Scientific, Logan, Utah) and metal salt (HgCl2, CdCl2, MnCl2 or NiCl2) were added to crude extracts. Following purification wild-type and variant enzymes were concentrated to between 500 and 800 µM prior to crystallization. In all cases, crystals were grown in mother liquor composed of 0.05M ammonium acetate, 0.05M Bis-Tris, pH 6.5 and 35% (v/v) pentaerythritol ethoxylate (15/4 EO/OH).21
Data collection, structure determination, and refinement
Data were collected at SER-CAT using the ID line. Single anomalous dispersion experiments were performed at X-ray wavelengths of 1.45 Å, 1.7 Å, 1.8 Å, and 1.9 Å in order to optimize the anomalous signal for Ni, Fe, Mn, and Hg respectively. A helium cone was used at the X-ray wavelengths of 1.8 Å and 1.9 Å in order to minimize air absorption of the diffracted X-rays. Frames were processed using HKL2000.37 Phases were obtained by using the poly alanine form of either the existing wild-type (PDB ID 2QD4) or the F110A (PDB ID 1HRK) model. Molecular replacement and subsequent rounds of model building and refinement were performed using the programs COOT38 and CNS.39
Supplementary Material
Acknowledgments
This work was support by NIH grant DK 32303 to H.A.D and by NSF grant MCB 0835432 to W.N.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dailey HA, Dailey TA. Ferrochelatase. In: Kadish KM, Smith KM, Guilard R, editors. The Porphyrin Handbook. Vol. 12. San Diego: Academic Press; 2003. pp. 93–122. [Google Scholar]
- 2.Dailey TA, Dailey HA. Identification of [2Fe-2S] clusters in microbial ferrochelatases. J Bacteriol. 2002;184:2460–2464. doi: 10.1128/JB.184.9.2460-2464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepherd M, Dailey TA, Dailey HA. A new class of [2Fe-2S]-cluster-containing protoporphyrin (IX) ferrochelatases. Biochem J. 2006;397:47–52. doi: 10.1042/BJ20051967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner DA, Didier JM, Frasier F, Christensen SR, Evans GA, Dailey HA. A molecular defect in human protoporphyria. Am J Hum Genet. 1992;50:1203–1210. [PMC free article] [PubMed] [Google Scholar]
- 5.Lamoril J, Boulechfar S, de Verneuil H, Grandchamp B, Nordmann Y, Deybach JC. Human erythropoietic protoporphyria: two point mutations in the ferrochelatase gene. Biochem Biophys Res Commun. 1991;181:594–599. doi: 10.1016/0006-291x(91)91231-z. [DOI] [PubMed] [Google Scholar]
- 6.Gouya L, Puy H, Lamoril J, Da Silva V, Grandchamp B, Nordmann Y, Deybach JC. Inheritance in erythropoietic protoporphyria: a common wild-type ferrochelatase allelic variant with low expression accounts for clinical manifestation. Blood. 1999;93:2105–2110. [PubMed] [Google Scholar]
- 7.Gouya L, Puy H, Robreau AM, Bourgeois M, Lamoril J, Da Silva V, Grandchamp B, Deybach JC. The penetrance of dominant erythropoietic protoporphyria is modulated by expression of wildtype FECH. Nat Genet. 2002;30:27–28. doi: 10.1038/ng809. [DOI] [PubMed] [Google Scholar]
- 8.Gouya L, Puy H, Robreau AM, Lyoumi S, Lamoril J, Da Silva V, Grandchamp B, Deybach JC. Modulation of penetrance by the wild-type allele in dominantly inherited erythropoietic protoporphyria and acute hepatic porphyrias. Hum Genet. 2004;114:256–262. doi: 10.1007/s00439-003-1059-5. [DOI] [PubMed] [Google Scholar]
- 9.Al-Karadaghi S, Franco R, Hansson M, Shelnutt JA, Isaya G, Ferreira GC. Chelatases: distort to select? Trends Biochem Sci. 2006;31:135–142. doi: 10.1016/j.tibs.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanzilotta WN, Dailey HA. Human Ferrochelatase. In: Messerschmidt A, editor. Handbook of Metalloproteins. UK: John Wiley & Sons, Ltd; 2007. [Google Scholar]
- 11.Dailey HA. Metal inhibition of ferrochelatase. Ann N Y Acad Sci. 1987;514:81–86. doi: 10.1111/j.1749-6632.1987.tb48763.x. [DOI] [PubMed] [Google Scholar]
- 12.Hunter GA, Sampson MP, Ferreira GC. Metal ion substrate inhibition of ferrochelatase. J Biol Chem. 2008;283:23685–23691. doi: 10.1074/jbc.M803372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlberg T, Hansson MD, Yengo RK, Johansson R, Thorvaldsen HO, Ferreira GC, Hansson M, Al-Karadaghi S. Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues. J Mol Biol. 2008;378:1074–1083. doi: 10.1016/j.jmb.2008.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sellers VM, Wu CK, Dailey TA, Dailey HA. Human ferrochelatase: characterization of substrate-iron binding and proton-abstracting residues. Biochemistry. 2001;40:9821–9827. doi: 10.1021/bi010012c. [DOI] [PubMed] [Google Scholar]
- 15.Hoggins M, Dailey HA, Hunter CN, Reid JD. Direct measurement of metal ion chelation in the active site of human ferrochelatase. Biochemistry. 2007;46:8121–8127. doi: 10.1021/bi602418e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medlock A, Swartz L, Dailey TA, Dailey HA, Lanzilotta WN. Substrate interactions with human ferrochelatase. Proc Natl Acad Sci U S A. 2007;104:1789–1793. doi: 10.1073/pnas.0606144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Y, Ryde U. The structure of sitting-atop complexes of metalloporphyrins studied by theoretical methods. J Inorg Biochem. 2004;98:878–895. doi: 10.1016/j.jinorgbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Ryde U. Reaction mechanism of porphyrin metallation studied by theoretical methods. Chemistry. 2005;11:1549–1564. doi: 10.1002/chem.200400298. [DOI] [PubMed] [Google Scholar]
- 19.Sigfridsson E, Ryde U. The importance of porphyrin distortions for the ferrochelatase reaction. J Biol Inorg Chem. 2003;8:273–282. doi: 10.1007/s00775-002-0413-8. [DOI] [PubMed] [Google Scholar]
- 20.Hansson M. Doctoral Disseration. Lund University; 2009. Structural and physicochemical studies on metal and porphyrin binding by the Bacillus subtilis ferrochelatase. [Google Scholar]
- 21.Medlock AE, Dailey TA, Ross TA, Dailey HA, Lanzilotta WN. A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase. J Mol Biol. 2007;373:1006–1016. doi: 10.1016/j.jmb.2007.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barkigia KM, Fajer J, Adler AD. Crystal and Molecular Structure of (5,10,15,20-Tetra-N-propylporphinato)lead(II): A "Roof" Porphyrin. Inorg Chem. 1980;19:2057–2061. [Google Scholar]
- 23.Plater JM, Aiken S, Gelbrich T, Hursthouse MB, Bourhill G. Structures of Pb(II) porphyrins. Polyhedron. 2001;20:3219–3224. [Google Scholar]
- 24.Wang MC, Sue LS, Liau BC, Ko BT, Elango S, Chen JH. Mercury complexes of meso-tetra-(p-cyanophenyl)porphyrin and N-methylporphyrin: meso-tetra(p-cyanophenyl)porphyrinatomercury(II) and chloro(N-methyl-meso- tetraphenylporphyrinato)mercury(II) Inorg Chem. 2001;40:6064–6068. doi: 10.1021/ic010275v. [DOI] [PubMed] [Google Scholar]
- 25.Dailey HA, Fleming JE. Bovine ferrochelatase. Kinetic analysis of inhibition by N-methylprotoporphyrin, manganese, and heme. J Biol Chem. 1983;258:11453–11459. [PubMed] [Google Scholar]
- 26.Gaertner RR, Hollebone BR. The in vitro inhibition of hepatic ferrochelatase by divalent lead and other soft metal ions. Can J Biochem Cell Biol. 1983;61:214–222. doi: 10.1139/o83-030. [DOI] [PubMed] [Google Scholar]
- 27.Leung YK, Ho JW. Purification and properties of ferrochelatase from Chironomidae larvae. Mol Cell Biochem. 2001;220:161–167. doi: 10.1023/a:1010976829463. [DOI] [PubMed] [Google Scholar]
- 28.Dailey HA, Jones CS, Karr SW. Interaction of free porphyrins and metalloporphyrins with mouse ferrochelatase. A model for the active site of ferrochelatase. Biochim Biophys Acta. 1989;999:7–11. doi: 10.1016/0167-4838(89)90021-6. [DOI] [PubMed] [Google Scholar]
- 29.Brindley AA, Raux E, Leech HK, Schubert HL, Warren MJ. A story of chelatase evolution: identification and characterization of a small 13–15-kDa "ancestral" cobaltochelatase (CbiXS) in the archaea. J Biol Chem. 2003;278:22388–22395. doi: 10.1074/jbc.M302468200. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh K, Thompson AM, Goldbeck RA, Shi X, Whitman S, Oh E, Zhiwu Z, Vulpe C, Holman TR. Spectroscopic and biochemical characterization of heme binding to yeast Dap1p and mouse PGRMC1p. Biochemistry. 2005;44:16729–16736. doi: 10.1021/bi0511585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho WW, Li H, Eakanunkul S, Tong Y, Wilks A, Guo M, Poulos TL. Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins. J Biol Chem. 2007;282:35796–35802. doi: 10.1074/jbc.M706761200. [DOI] [PubMed] [Google Scholar]
- 32.Honeybourne CL, Jackson JT, Jones OT. The interaction of mitochondrial ferrochelatase with a range of porphyrin substrates. FEBS Lett. 1979;98:207–210. doi: 10.1016/0014-5793(79)80185-4. [DOI] [PubMed] [Google Scholar]
- 33.Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, Trede NS, Barut BA, Zhou Y, Minet E, Donovan A, Brownlie A, Balzan R, Weiss MJ, Peters LL, Kaplan J, Zon LI, Paw BH. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 34.Dailey HA, Wu CK, Horanyi P, Medlock AE, Najahi-Missaoui W, Burden AE, Dailey TA, Rose J. Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis. Biochemistry. 2007;46:7973–7979. doi: 10.1021/bi700151f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burden AE, Wu C, Dailey TA, Busch JL, Dhawan IK, Rose JP, Wang B, Dailey HA. Human ferrochelatase: crystallization, characterization of the [2Fe-2S] cluster and determination that the enzyme is a homodimer. Biochim Biophys Acta. 1999;1435:191–197. doi: 10.1016/s0167-4838(99)00196-x. [DOI] [PubMed] [Google Scholar]
- 36.Mayer MR, Dailey TA, Baucom CM, Supernak JL, Grady MC, Hawk HE, Dailey HA. Expression of human proteins at the Southeast Collaboratory for Structural Genomics. J Struct Funct Genomics. 2004;5:159–165. doi: 10.1023/B:JSFG.0000029202.77832.34. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica, Section D: Biological Crystallography. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.