Abstract
The rapidly increasing incidence of melanoma, coupled with its highly aggressive metastatic nature, is of urgent concern. In order to design rational therapies, it is of critical importance to identify the genetic determinants that drive melanoma formation and progression. To date, signaling cascades emanating from the EGF receptor, c-MET and other receptors are known to be altered in melanoma. Important mutations in signaling molecules, such as BRAF and N-RAS, have been identified. In this review, some of the major genetic alterations and signaling pathways involved in melanoma will be discussed. Given the great deal of genetic heterogeneity observed in melanoma, it is likely that many more genetic determinants exist. Through the use of powerful genomic technologies, it is now possible to identify these additional genetic alterations in melanoma. A critical step in this analysis will be culling bystanders from functionally important drivers, as this will highlight genetic elements that will be promising therapeutic targets. Such technologies and the important points to consider in understanding the genetics of melanoma will be reviewed.
Keywords: array-comparative genomic hybridization, BRAF, CDKN2A, genomics, MAPK, metastasis, N-RAS, PI3K, receptor tyrosine kinase
Clinical presentation of melanoma
Melanoma is the most aggressive form of skin cancer, surpassing most solid tumors in terms of propensity to metastasize. Based on the National Cancer Institute estimates, the lifetime risk of a person in the USA developing melanoma of the skin is an astounding 1 in 55 and, in the Western world, cases of melanoma have doubled in the last 20 years [1,2]. Meanwhile, the risk of invasive melanoma in the USA has increased almost tenfold in the last 50 years [3]. So, while the incidence of many cancers has decreased or is leveling off, the occurrence of melanoma is still on the rise. Since the risk of developing melanoma may be correlated with UV exposure given an individual’s genetic background, and as sun exposure trends change worldwide, the personal and economic toll on society from melanoma is likely to increase. This will be exacerbated by the fact that there are few therapeutic options for metastatic melanoma, a lethal end-stage progression of the disease. Therefore, it will be of critical importance for the scientific and healthcare community to develop diagnostic and therapeutic strategies to manage and treat melanoma. This will require a much more comprehensive understanding of the genetic basis of melanoma than currently exists.
While a description of melanoma has been noted as far back as the 5th Century BC, much remains to be elucidated about the causal events in melanoma pathogenesis. Cutaneous melanoma arises from uncontrolled proliferation of melanocytes, melanin-producing cells located in the basal layer of the epidermis [4]. By distributing pigment from melanosomes to keratinocytes present in the skin, melanocytes serve a protective role against UV radiation for the skin. Melanoma can also occur in the eye (ocular), meninges and digestive tract. Clinically, four subtypes exist. Lentigo maligna is associated with chronic sun-exposed areas of the body, such as the face. Acral lentiginous melanoma is found on non-sun-exposed regions, such as the palms, nail beds and soles of the feet. Nodular melanoma is a raised nodule that may or may not have an associated superficial spreading component. The most common subtype – particularly among young adults in the USA – is superficial spreading melanoma [1], which generally occurs on areas of the body with intermittent sun exposure, such as the trunk and proximal extremities.
For chronic sun-exposure sites, lentigo maligna may be the most common subtype [5]. In an analysis of a military population in Texas (USA) assumed to have high sun exposure, most patients presented with lentigo maligna [6]. This has clinical relevancy, since sun exposure might vary from population to population, thereby altering the predominant melanoma subtype.
Melanoma progression can begin with the development of either dysplastic or benign nevi (common acquired or congenital) [4]. These can then progress to the radial growth phase, in which the growth expands laterally but remains localized to the epidermis. At this phase, cells are still dependent on growth factors, and are not anchorage independent or tumorigenic. Progression to the vertical growth phase is hallmarked by invasion into the dermis, subcutaneous tissue and upper epidermis. In the vertical growth phase, cells are no longer growth factor dependent, are anchorage independent and presage distal metastasis.
Clinical staging of melanoma progresses from an in situ growth to one increasing in thickness and vertical invasion, to regional lymph-node spread and, finally, to distal metastasis. Vertical invasion may be representative of the degree of progression and is often measured by the Breslow thickness, a measure of the thickness of the tumor from the upper layer of the epidermis to the innermost depth of invasion [7]. The Breslow thickness remains the most significant predictor of metastasis and prognosis. However, although most thin melanomas do not metastasize, a portion do, suggesting that there is a high-risk subgroup of melanomas among the generally low-risk thin melanomas [8]. This highlights the urgent need to develop prognostic molecular biomarkers that can complement gold-standard clinicopathological parameters in stratifying patients with a high risk of metastasis.
Genetics of melanoma
To date, some of the genetic events underlying the development of melanoma have been characterized. These include alterations in receptor tyrosine kinase (RTK) function and the emanating signaling cascades or in intracellular regulatory molecules. These changes have been identified through genomic structure and sequence analysis, some of which are summarized below.
CDKN2A
The complexion of an individual can suggest propensity for melanoma development. Light complexion, inability to tan or presentation of atypical moles or freckles correlates with increased risk for melanoma [9,10]. However, familial history of melanoma is also a strong predictor of melanoma development. A meta-analysis showed that first-degree relation to a person with melanoma increased one’s risk 2.24-fold [11,12]. Such relationships led to the identification of 9p21 as an important locus in melanoma, with loss of heterozygosity or mutation at 9p21 occurring in melanomaprone families [13,14]. Within this region are the CDKN2A and CDKN2B genes. Interestingly, CDKN2A encodes for two tumor-suppressor proteins, INK4A (p16INK4A) and ARF (p14ARF in humans and p19ARF in mice) [15].
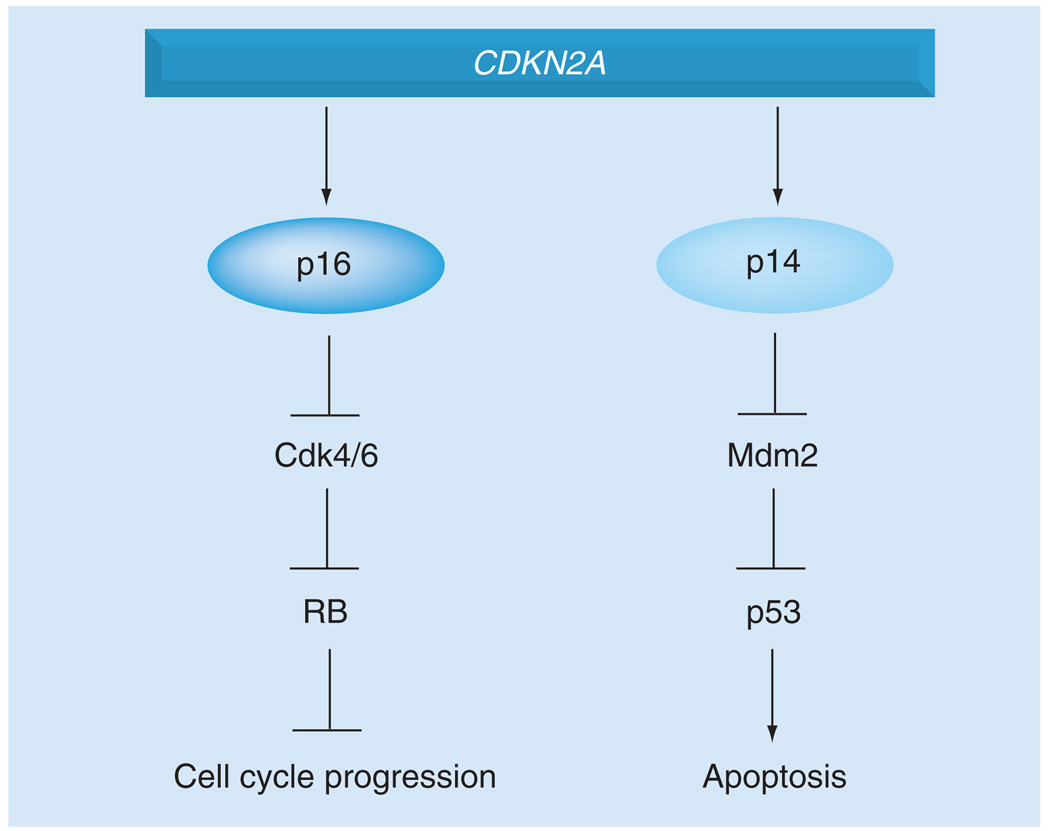
INK4A (p16INK4a) is a cyclin-dependent kinase inhibitor that activates the tumor-suppressor gene retinoblastoma (RB) via negative regulation of Cdk4/6 (Figure 1). This ultimately leads to progression through the G1/S transition. Tellingly, 25–40% of familial melanomas have mutations in the INK4A coding region [16,17]. Albeit rarely, alterations in CDK4 and RB have also been associated with melanoma in humans [18–22].
Figure 1. CDKN2A encodes two important regulators of melanoma.
The CDKN2A gene encodes both p16INK4A and p14ARF, which control the RB and p53 pathways, respectively. Germline alterations in CDKN2A are common events in familial melanoma. RB: Retinoblastoma.
Alternative reading frame is a negative regulator of the tumor suppressor p53, a functional relationship that explains the infrequent direct p53 mutation in human melanoma in contrast to other solid tumors [4,23–25]. Genetic evidence in mice also supports this phenomenon. In Tyr-HRASV12 transgenic mice with cutaneous melanoma sustaining loss of heterozygosity of Trp53, Arf remains wild type; in Tyr-HRASV12 transgenic mice on Ink4a/Arf−/− background, p53 remains wild type [26,27].
Receptor tyrosine kinases
Receptor tyrosine kinases are widely dysregulated in various cancers. In melanoma, alterations in the EGF receptor (EGFR), Met RTK (c-MET) and Kit receptor tyrosine kinase (c-KIT) result in changes to the associated signaling cascades (Figure 2).
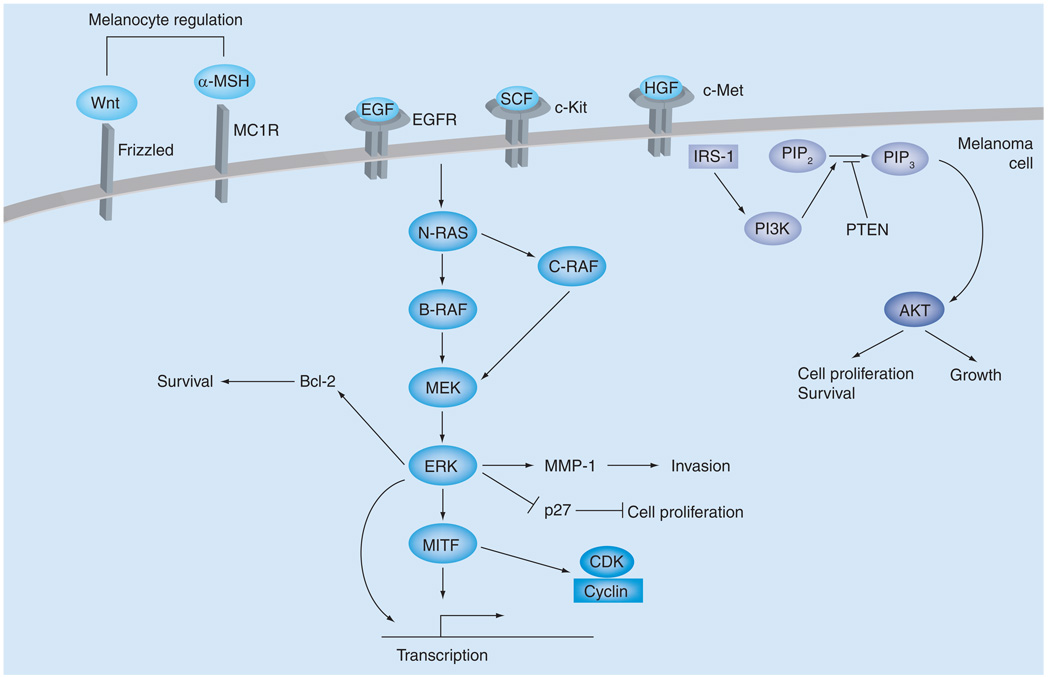
Figure 2. Signaling pathways involved in melanoma.
Various receptors can activate the ERK and PI3K pathways, both important signaling cascades in melanoma that control cell proliferation and survival. In addition, signaling pathways important for melanocyte regulation may be co-opted in tumors.
EGFR: Endothelial growth factor receptor; MMP: Matrix metalloproteinase.
The EGFR can be activated by EGF, TGF-α, amphiregulin and heparin-binding EGF. Such binding can activate the MAPK and PI3K signaling cascades. EGFR is located on chromosome 7 and copy number gains of chromosome 7 have been observed in the later stages of melanoma [28–30]. However, no focal amplifications have been detected in melanoma, making assignment of EGFR as the target of such genomic changes difficult. Although EGFR mutation has not been reported, it should be noted that systematic resequencing in a large number of melanoma samples has not been conducted. In the tetracycline-regulated H-RASV12-inducible mouse model of melanoma, transcriptome analysis revealed an EGFR signaling loop driven by RAS (from rat sarcoma virus) via the regulation of EGF family ligands [31]. When the inducible RAS allele is shut off in an explanted tumor, a sustained EGFR signal promotes survival through PI3K activation of AKT. Consistent with this, loss of EGFR inhibits RAS-driven tumorigenicity of melanoma cells [32–34]. This suggests that EGFR may be an alternate target for therapeutic intervention in RAS-driven melanoma.
The c-MET receptor is activated by the binding of its ligand, HGF [35]. The c-MET receptor is known to be expressed in melanocytes. Located at 7q33, copy number gains of this locus are considered to be a late-stage event in melanoma [28,36]. As with the EGFR, no focal amplifications or mutations of the c-Met gene have been identified in melanoma [15]. However, additional data suggest that c-Met is a critical regulator of melanoma progression. For example, c-Met expression is upregulated in metastatic melanoma [37]. This may be accounted for by the fact that while MET activation is predominantly paracrine, in melanoma, autocrine activation of MET has been described [38]. Interestingly, expression of c-Met in melanocytes drives metastasis rather than the initial steps of melanoma development [15]. Furthermore, increased c-Met expression or activity promotes invasiveness and, in explant models, correlates with metastasis [39]. In genetically engineered HGF-mouse models, continuous expression of HGF drives a c-MET autocrine loop that promotes cutaneous and metastatic melanoma [40–42]. Interestingly, Ink4a/Arf deficiency enhanced melanoma development in HGF mice exposed to UVB [42]. In addition, c-Met has been identified as a target of the microphthalmia-associated transcription factor (discussed later), which is itself amplified in a subset of melanoma [43,44].
c-KIT is a RTK that binds to the stem-cell factor ligand. c-KIT activation can stimulate various signaling pathways, including RAS/ERK and PI3K. Oddly, c-KIT does not represent a typical RTK in melanoma, since immunohistochemical analysis has indicated a frequent loss of c-KIT with melanoma progression [45–47]. Furthermore, metastatic melanoma cells expressing c-Kit have a greater responsiveness to stem cell factor-mediated apoptosis [48]. However, a prosurvival and proliferation role for c-KIT has also been suggested. In mice or humans, loss or inhibition of c-KIT, respectively, results in loss of melanocytes [49,50]. c-KIT activation also promotes proliferation of cells with a melanocytic lineage [51]. Additionally, a study of human metastatic melanomas showed three out of 153 samples to have high c-KIT expression as detected by immunohistochemistry. Through high-resolution amplicon melting and DNA sequencing, it was found that a L576P mutation was present in three samples, with a loss of the normal allele [52,53]. The L576P mutation, found in exon 11, maps to the 5´ juxtamembrane domain, the region in which most KIT activating mutations are found [54,55]. The L576P mutation has also been found in other tumors (e.g., gastrointestinal stromal cell tumors) [55].
More recently, through copy number analysis and sequencing, a study found that 21% of mucosal, 11% of acral and 28% of chronic sun-damaged skin had alterations in KIT [56]. Recently, a screen for mutations in melanoma subtypes suggested that 23% of acral, 15% of mucosal and only 2% of cutaneous melanomas have KIT mutations [57]. This suggests that KIT status might define a subpopulation of melanomas. Interestingly, imatinib can inhibit KIT and has been effective in patients with gastrointestinal stromal tumors with KIT mutations [58–61]. Indeed, in a patient with rectal acral melanoma harboring an exon duplication in the juxtamembrane domain of KIT, imatinib was effective in reducing tumor volume by half in 4 weeks, with prolonged survival for 4 months [62]. In another case, a patient presenting with mucosal melanoma responded to imatinib and exhibited regression of anal nodules for up to 6 months [63]. These case reports emphasize the existence of genetically distinct melanoma subtypes and also the ability of genetic alterations to stratify patients into therapeutic subgroups.
Signaling cascades
Two major signaling cascades emanate from the aforementioned RTKs, the RAS/RAF/MEK/ERK and the PI3K pathways (Figure 2). Growth-factor independence is a requisite for cancer cells, and dysregulation of receptor signaling in cancers can occur at various levels, from alterations at the receptor to changes in the intracellular signaling cascades [64]. While a growth factor may offer a degree of signaling stimulation, robust activation of signaling can arise from the activation of individual signaling molecules. Disruption in these signaling pathways may result in aberrant cell proliferation and/or apoptosis, and eventual tumor development.
The RAS/RAF/MEK/ERK pathway is activated by various receptors, including c-KIT, FGF receptor and c-MET. The RAS family of small G proteins is made up of N-RAS, H-RAS and K-RAS. Mutations in N-RAS, the most common in melanoma, are found in 33% of primary and 26% of metastatic tumors, and are correlated with sun exposure and nodular lesions [65,66]. N-RAS mutations are rarely found in dysplastic nevi, one of the potential starting points for melanoma [65,67,68]. H-RAS point mutations and genomic 11p amplification have only been identified in Spitz nevi, a benign lesion that does not progress to melanoma [69]. It should be noted, however, that some atypical Spitz nevi have been shown to metastasize to regional lymph nodes but whether H-RAS mutations correlate with metastasis is uncertain and, furthermore, the distinction of atypical Spitz nevi from melanoma is controversial [70,71]. No mutations of K-RAS have been described in melanoma. These data suggest that the RAS family members have distinct roles in melanoma. Consistent with this, while activated H-RAS expression on an Ink4a, Arf or p53 mutation background promotes nonmetastatic melanoma in mice [26,27,72], N-RAS expression in Ink4a/Arf deficiency promotes metastasis of cutaneous melanoma with high penetrance and short latency [73].
The RAF family consists of ARAF, BRAF and CRAF. In a systematic screen for mutations, BRAF mutations were identified in a variety of tumor cell lines, with the highest occurrence in melanoma. Sequencing revealed up to 67% of melanoma tumors samples had a mutation in BRAF. An amino acid substitution (V600E) was, by far, the most common mutation both in cell lines and tumor samples [74]. Alterations in BRAF may be an early somatic event (see later) as germline mutations in BRAF are not commonly found in familial melanoma [75–77].
BRAF can regulate various aspects of cell survival. Activated BRAF promotes IκB degradation, while inhibition of BRAF sensitizes cells to apoptosis [78]. BRAF can also regulate cell growth by regulating p27kip1 levels [79]. The majority of BRAF mutations occur in defined areas of biochemical function, with a single phosphomimetic substitution of V600E in the kinase activation domain being the most common [80]. Mutations in BRAF are found most frequently in melanomas at sites with intermittent UV exposure and only occur in approximately 11% of lentigo maligna melanomas arising from chronically sun-exposed areas [81–83]. BRAF mutation is associated with germline variants of the MCR1 gene, which promotes melanin production [83,84]. The melanocortin receptor 1 (MC1R) is the receptor for α-melanocyte-stimulating hormone, which is stimulated by UV radiation, perhaps suggesting a degree of interplay between BRAF and UV exposure [85]. In this regard, it is interesting that BRAF mutation is most prevalent in the melanomas associated with intermittent sun exposure [81]. MC1R itself is also interesting, since MC1R variants are associated with fair skin, red hair and freckles; physical attributes that have been associated with melanoma risk [10,86,87].
The role of BRAF in melanoma is complicated by the fact that BRAF mutations are also found in benign and dysplastic nevi [88–91]. These nevi often remain growth-arrested for their lifetime and rarely progress to melanoma. This may suggest that BRAFV600E promotes a checkpoint for malignant transformation. In fact, congenital nevi are positive for the senescence marker senescence-associated β-galactosidase and for p16INK4A [92]. Furthermore, while BRAFV600E cannot transform human melanocytes, it can transform murine melanocytes that are p16INK4A deficient [44,93] and can induce INK4A, senescence-associated β-galactosidase and cell cycle arrest in normal cells [15]. The BRAF effect, therefore, is an example of oncogene-induced senescence – a method by which premalignant lesions are inhibited from progressing [94]. Interestingly, INK4A expression is not present in 100% of BRAF oncogene-induced senescence cases, suggesting that another pathway aside from INK4A may exist [92]. BRAF-dependent oncogene-induced senescence may rely on signaling through IGF binding protein (IGFBP7) and other genes. A genome-wide shRNA screen for factors necessary for BRAFV600E-mediated senescence identified IGFBP7. IGFBP7 was required for BRAFV600E-induced apoptosis and senescence in melanocytes, and media containing IGFBP7 induced apoptosis in BRAF mutant melanoma cells lines. In vivo, recombinant IGFBP7 administration suppressed BRAFV600E tumors and in humans, benign nevi with BRAFV600E had high levels of IGFBP7, while BRAFV600E melanomas did not [95].
Additional evidence shows that BRAF is not sufficient for transformation of melanocytes. In zebra fish, BRAF activation forms benign nevi, but requires p53 deficiency to progress to full melanoma [79]. Additionally, while N-RAS expression in telomerase reverse transcriptase (TERT)-immortalized RB–p53 mutant human melanocytes produced invasive melanoma, BRAFV600E expression only produced junctional nevi, or a nevi between the dermis and epidermis [96]. Again, this demonstrates that BRAF requires the cooperation of other determinants to drive melanoma progression.
It is of note that BRAFV600E and N-RAS mutations are mutually exclusive in melanoma, and in fact, the occurrence of each mutation may be specific to certain subtypes of melanoma [74,97,98]. For example, BRAF mutations are associated with melanomas arising on body sites of intermittent sun exposure, while N-RAS mutations may be more common in sites of chronic sun damage [99]. This suggests that there are innate differences in the roles that BRAF and N-RAS may play in melanoma and this may be reflected in shifts in signaling. In BRAF mutant cells, BRAF is required for ERK signaling. However, in the context of N-RAS mutation, melanoma cells signal to ERK through CRAF after a concomitant alteration in cAMP signaling to CRAF (Figure 2) [100]. Furthermore, with supervised clustering, the gene-expression profile of primary melanomas with BRAF mutations was shown to be distinct from those with N-RAS mutations, although it must be noted that others have not found such differences in cell lines [101,102]. Interestingly, even the specific BRAF mutation may determine signaling. In non-V600E BRAF mutant melanoma cells, CRAF inhibition induced apoptosis, suggesting that, in these specific cells, CRAF is of consequence [103].
The ERK signaling pathway can regulate various molecules important for tumorigenesis (Figure 2). In wild-type BRAF or NRAS cells, ERK activation is quite low compared with mutant cells and can control proteins involved in extracellular adherence, cell motility and angiogenesis [102]. In melanoma cells, ERK can inhibit the cell cycle regulator p27kip1 [104]. ERK can also alter in vitro invasion capability by regulating the production of matrix metalloproteinase -1 [105]. However, ERK can also regulate survival and senescence. In a microarray screen, Bcl-2 was found to be regulated by microphthalmia-associated transcription factor (MITF) in melanocytes and melanoma cells [106]. Therefore, a close examination of the phenotypic outcome of ERK activation in melanoma should be carried out [107].
The PI3K pathway is responsive to various extracellular signals, including those from integrins, extracellular matrix proteins, HGF and insulin-like growth factors. Upon receptor stimulation, PI3K generates the lipid second messenger, phosphoinositol-3,4,5 triphosphate (PIP3). PIP3 triggers the phosphorylation and activation of the serine/threonine kinase, AKT (Figure 2). Interestingly, patient survival is inversely related to phospho-AKT levels [108]. There are three AKT isoforms and each may have a distinct role in cancers, including melanoma. Of the three, DNA copy number gain of AKT3 is observed in melanoma, and 40–60% of sporadic tumors may have AKT3 activation [109]. Additionally, AKT3 correlates well with melanoma progression and loss of AKT3-triggered apoptosis [109]. On the other hand, AKT1 activation inhibits the migration and invasion of various cell lines, including a melanoma cell line [44,110,111]. Although AKT may have a role in melanoma, the extent to which AKT is a critical regulator of melanoma is yet to be determined.
By far, the majority of genetic alterations that do occur in the PI3K pathway in melanoma occur in the phosphatase and tensin homologue deleted on chromosome 10 (PTEN) tumor suppressor. PTEN is a bona fide tumor suppressor. By dephosphorylating the D3 position of PIP3, PTEN negatively regulates the PI3K pathway. Loss of heterozygosity of 10q, where the PTEN gene is located, has been observed in various cancers, including melanoma [112,113]. Loss of an allele or expression changes in PTEN occur in 20 and 40% of melanoma tumors, respectively, and somatic point mutations and homozygous deletions are rarely observed [98,114–116]. Expression of PTEN can inhibit AKT, promote apoptosis and inhibit growth, tumorigenicity and metastatic potential of PTEN-deficient melanoma cells [117–120]. Somatic or germline mutations of PTEN promote tumor phenotypes in the mouse, including melanoma [121–125].
Phosphatase and tensin homologue deleted on chromosome 10 itself can be regulated by NEDD4-1-mediated polyubiquitination [126]. Interestingly, PTEN inactivation through NEDD4-1 enhances the efficiency of transformation of mouse fibroblasts with RAS expression and p53 deficiency [127].
The importance of both the ERK and PI3K signaling in tumor induction is also conserved in melanoma. In transgenic TP-Ras mice and in 3D melanoma cultures, inhibition of both pathways is required for suppression of cell growth [128,129]. While mutations in PTEN and N-RAS are generally discordant in melanoma, BRAF and PTEN mutations are found concurrently in 20% of melanomas [83,130].
Genomic approach to melanoma
While various signaling molecules have been identified in melanoma, the genomic heterogeneity of melanoma suggests that many more genetic drivers remain to be discovered. The genetic heterogeneity is illustrated by array-comparative genomic hybridization (aCGH) analysis, which shows that different subtypes of melanoma have different copy number alteration profiles [83]. Furthermore, upon sequencing, it was found that BRAF mutations are not distributed equally but instead are predominantly found in melanomas without sun damage, rather than in chronic sun-exposed melanomas [83]. Such distinctions between different melanoma types may, in fact, indicate the predominance of a signaling pathway in a particular subgroup. A critical extension of this observation is that a melanoma subgroup defined by a molecular signature may respond favorably to a therapy specifically directed against that signature. Therefore, understanding the alterations present in melanoma will be critical for the development and application of therapies. These alterations will include changes in sequence (i.e., BRAF), copy number gains and losses (i.e., CDKN2A), epigenetic changes, alterations in non-coding miRNAs and changes in protein-coding RNA (Figure 3). This will define the melanoma atlas – the genomic, epigenomic and proteomic changes that define melanoma. Identification of such changes can be accomplished through the use of various technologies, including sequencing efforts, comparative genomic hybridizations, chromatin immmunoprecipitation plus microarray technology (ChIP–Chip) and various microarrays.
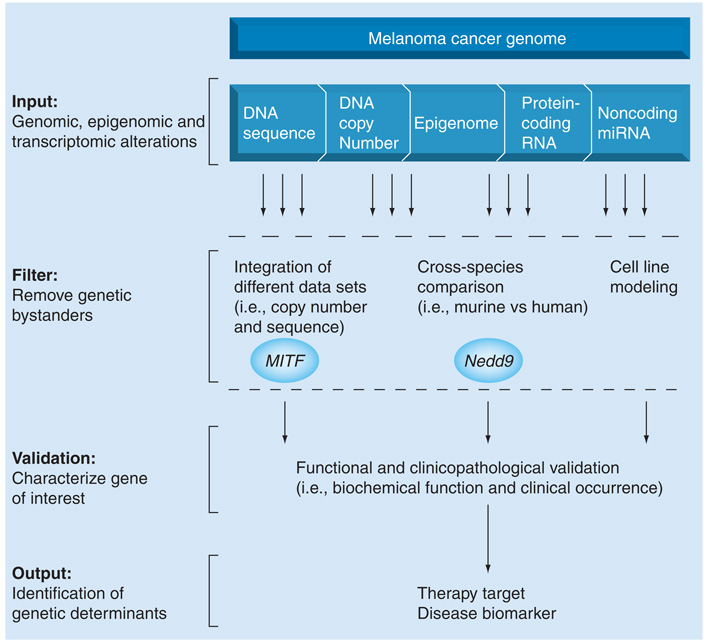
Figure 3. Filtering of genomic alterations identifies genetic determinants of melanoma.
The melanoma cancer genome is composed of genomic, epigenomic and transcriptomic alterations found in melanoma. Bystanders may be culled from true genetic drivers of disease through integration of different data sets, cross-species comparison for evolutionarily conserved alterations and analysis in cell lines. These filtering methods have been used successfully to identify MITF and Nedd9 as important regulators of melanoma. All genes of interest must undergo extensive functional and clinicopathological validation in order to identify genetic determinants of melanoma that may serve as therapeutic targets or disease biomarkers.
Sequence and copy number analyses have provided insight into the melanoma genetic profile. As discussed previously, sequencing efforts identified BRAF and KIT mutations as critical alterations in melanoma. As sequencing becomes less expensive, allowing for the sequencing of a greater number of tumor samples and a greater portion of the genome, it is expected that the identification of mutations of known and novel genes important in melanoma will increase dramatically. For copy number analysis, a powerful tool has been the use of array-based high-resolution comparative genomic hybridization (aCGH). Through comparison of samples (i.e., normal vs tumor), aCGH can identify regions of copy number gains or losses, which may contain genes that regulate tumor formation, development or progression. In an analysis of metastatic tumor samples grouped by unsupervized methods and matched against clinical outcome, a particular group correlated with poor prognosis; rather than being defined by metastatic site, this group was defined by similar patterns of chromosomal alterations [15]. This suggests that analysis of genomic alterations has biological validity. Another analysis demonstrates how aCGH might stratify melanoma patients based on risk. Over 400 copy number alterations were identified in metastatic lesions upon comparison of melanocytic lesions, primary and metastatic melanomas, melanoma cell lines and melanocytic nevi [15]. This comprised previously described events, including gain of 1q, 6p, 7 and loss of 6q, 9p and 10. The number of copy number alterations, which is, in metastatic melanoma, reflective of genomic heterogeneity, was found to be significantly greater than that in primary melanomas. Using such comparisons, genes that may represent useful prognostic biomarkers for the clinical stratification of patients or serve as therapeutic targets might be identified.
As a result of such alterations (i.e., sequence and copy number), overall gene expression may be altered. While many gene-expression profiles have been based on cell line comparisons, more recent work has concentrated on primary human tumor samples. Such analyses have identified gene-expression signatures relevant to melanoma prognosis. Unsupervised hierarchieal clustering of 83 primary tumor samples identified 254 genes that defined the expression profile of patients with 4-year metastasis-free survival [131]. This data set showed differential expression between mutant and wild-type BRAF samples, suggesting that gene-expression analysis of primary tumor samples will give insight into the distinct genetic signatures of melanoma subtypes [101].
A specific example of the value of analyzing genetic aberrations is the identification of MITF as an important factor in melanoma progression. MITF amplification was detected in single nucleotide polymorphism analysis of a collection of cancer cell lines. In determining lineage-specific alterations, gain of 3p14–3p13 was identified in melanoma. Integration of the 3p amplified melanoma cell lines with expression data identified MITF as the gene showing the greatest change in expression and copy number gain in the 3p region [44]. Subsequent analysis showed MITF to be amplified in 10% of primary cutaneous and 15–20% of metastatic melanoma. Furthermore, MITF amplification was inversely related to 5-year survival [44]. However, such data cannot discriminate between driver and bystander effects; for that, functional studies are required. In TERT-immortalized melanocytes with BRAFV600E and RB and p53 deficiency, MITF expression drove transformation capability (e.g., soft agar colony formation). Loss of MITF also inhibited growth and survival in 3p13–14 amplified cell lines [44]. These in vitro studies indicate the existence of genetic subsets in which MITF functions, suggesting that additional molecular subclasses of melanoma may exist.
It must be noted that the role of MITF is not entirely clear-cut. In mouse melanocytes with BRAFV600E, MITF expression reduced cell proliferation, while re-expression of MITF in vivo reduced tumor formation [132]. Follow-up by the same group showed that while short-term inhibition of ERK increased MITF levels in melanoma cell lines, longer term inhibition increased MITF levels [133]. It is possible that while some melanomas have MITF expression, some may not [134].
In cancer, tumor cells often demonstrate characteristics and signaling pathways of the developmental lineage of the tumor cell, and MITF is a perfect example of a lineage-specific gene in melanoma. MITF is a central regulator of normal melanocyte differentiation and survival. Indeed, disruption of the MITF gene results in graying of the coat color of mice and abnormal pigmentation in humans [135–137]. Tumors may use these pathways for their own benefit. In fact, BRAF may promote the selection of variants with amplified MITF. Therefore, MITF may promote the survival of both normal and neoplastic melanocytic cells. Other melanocyte signaling may also have a role in tumorigenesis, including c-KIT, Wnt and MC1R [84–86,138].
Epigenetic changes, such as DNA methylation and histone modification, are regulatory mechanisms in melanoma. Through the use of 5-Aza-2´-deoxycytidine to inhibit DNA methylation, it was found that expression of 0.8% of genes on a microarray were altered in melanoma cells lines compared with untreated controls [139]. Further analysis found that HOXB13 and SYK expression increased in melanocytes with 5-Aza-2´-deoxycytidinetreatment. Overexpression of either gene reduced cell proliferation and decreased tumor size in a xenograft model [139]. Other genes, including the estrogen receptor α, have differential methylation status in primary and metastatic melanoma [140]. Stratification of patients for treatment will require disease biomarkers, and analysis of DNA methylation state might be a usable biomarker. It has been demonstrated that the levels of methylated PTEN and estrogen receptor α DNA in the serum is increased in metastatic patients [140,141]. Furthermore, histone deacetylase inhibitors have inhibited growth of melanoma cells in vitro and in xenografts, suggesting that epigenetic changes are of importance in tumors [142]. Currently, epigenetic alterations can be analyzed through the use of various methodologies, including ChIP–Chip, restriction landmark genome scanning and modification of histones. As the technologies improve, it is likely that a greater number of epigenetic changes in melanoma will be identified.
miRNAs are noncoding RNAs that can regulate translation and stability of protein-coding RNAs. In some solid tumors, miRNAs have been found to target tumor suppressors and oncogenes [143]. Interestingly, melanoma can be segregated from other tumors types by patterns of miRNA expression [144]. Furthermore, miRNA copy numbers are altered in melanoma cell lines [145]. While not conclusive, such data suggest that miRNAs in melanoma may be a fruitful line of research.
As future genomic analyses will generate a wealth of data, the ability to cull bystanders from true genetic drivers of melanoma will be of critical importance. This may be carried out through integration of different data sets (i.e., copy number and sequencing), identification of evolutionarily conserved alterations (i.e., cross-species analysis) and cell-based analysis (Figure 3). In all cases, genes of interest must be validated through biological assays that determine the functional and clinicopathological relevance of an alteration. A greater understanding of the role of a genetic alteration in tumor development can be garnered, using model systems (i.e., mouse), genetic manipulation (i.e., gain-of-function and loss-of-function), functional assays (i.e., migration) and investigation of biochemical function (i.e., signaling). This must be placed in the context of the clinicopathological characteristics of the gene of interest, such as prevalence in tumor samples, occurrence in different histological or molecular subtypes and responsiveness of a tumor with the alteration to specific therapies. Together, such analyses will address three necessary questions: is the gene genetically targeted for deregulation? Does this alteration have a functional consequence? And, if so, what is the clinical relevance of such alterations?
Since tumor promoting alterations can occur at various levels, integrating data sets may allow for the identification of genes important in melanoma. For example, sequencing identified c-KIT activation in mucosal and acral melanoma, while aCGH revealed the amplification of the genomic region [56]. A gene altered by multiple mechanisms may represent a critical player in tumorigenesis, and must be altered by cancer. The Cancer Genome Atlas has begun to catalog alterations in glioblastoma multiforme. A similar effort on a melanoma cancer genome project will undoubtedly transform this field.
Since genes that are drivers for tumor development are likely to be conserved evolutionarily, comparing murine genomic profiles against human profiles can act as a filter to identify critical genomic events. This methodology of cross-species comparison was used successfully to identify Nedd9 as a novel melanoma gene. Using a genetically engineered mouse model, metastatic tumors independent of inducible H-RAS status were isolated [146]. aCGH analysis of these tumor lines compared with nonmetastatic parental cells identified a recurrent focal amplification associated with metastatic progression. A syntenic amplification, observed in 36% of metastatic tumors, is found at 6p24–25 in humans. While the amplification of this region in humans quite wide, making gene identification difficult, comparison with the mouse amplification narrowed the region of interest. Further triangulation against expression data demonstrated Nedd9 as is the most likely gene candidate. Not only did cross-species analysis filter human genetic data, it also provided the logical framework for further in vitro characterization. Focal adhesion kinase was shown to be an important mediator of Nedd9 function and important in in vitro invasion and attachment to matrigel. These in vitro assays are reflective of characteristics (i.e., cell motility) that are important for a tumor cell to attain in order to metastasize in vivo. Therefore, the metastatic phenotype of the amplification (e.g., Nedd9) led to directed in vitro tests of invasion.
It must be noted that the degree of genomic aberrations observed in melanoma in humans is much greater than that seen in mice, suggesting that while cross-species comparison may help to filter out genomic noise, inherent differences between species may limit the completeness of the investigation. However, engineering genomic instability onto a mouse model can provide DNA double-strand breaks and consequent copy number aberrations and translocation. The use of Atm−/−, Trp53-deficient mice with telomere dysfunction generated lymphomas with greater number of chromosomal aberrations than seen in lymphomas with intact telomeres [147]. Many of the copy number alterations that emerged in the murine genome were syntenic to human T-cell acute lymphoblastic leukemia/lymphoma. Even more interestingly, most of the minimal common regions from the murine model overlapped with minimal common regions from aCGH data sets of various other human tumor types, including melanoma. This suggests that murine models designed with genetic instability may provide a more humanized model with which to filter genomic noise in the human cancer genomes and, thereby, identify significant genomic aberrations.
Upon gene identification, the use of cell-based assays has provided insight into the role of different genes in melanoma. Commonly, gain-of-function (e.g., overexpression) and loss-of-function (e.g., siRNA) studies can shed light on the biological consequence occurring from the genetic alterations. Knowledge of a gene’s role in a biological context (i.e., cell migration) and in signaling will be useful in deciding how a therapy might affect a tumor (i.e., limit invasiveness). However, cell-based analysis will be limited by the genetic background of any cell line and artifacts arising from time in culture. Even though 3D culture systems and xenograft models attempt to mimic the tumor in vivo environment, neither can fully recapitulate in vivo human tumors. Therefore, concurrently, extensive clinicopathological analysis must be performed in order to establish the relevance of any alteration in human cancers. That said, cell line-based experiments can provide an important starting point for mechanistic and phenotypic analyses.
Expert commentary
With the rapid advances of high throughput genomic technologies, particularly the emerging next-generation sequencing technology, researchers will soon be able to define the complete somatic architecture of human melanoma through multidimensional genomic analyses of melanoma specimens. Such an atlas will enable not only identification of genetic events associated with melanoma but also comprehensive data for molecular subclassification that can enhance the clinicians’ ability to stratify and personalize treatment of melanoma patients.
Five-year view
Over the next 5 years, there will be great advances in the understanding of the genetic and epigenetic changes underlying melanoma genesis and progression. As technologies that allow high-throughput analysis of the genome develop at break-neck speed, the rate at which genetic alterations associated with melanoma are discovered will only increase. Efforts such as The Cancer Genome Atlas, will propagate the integration of different types of genomic aberrations found in melanoma. In parallel, methodologies to sift through the data and discover ‘true’ drivers of disease will become of critical importance. Interestingly, as the cost of genome sequencing drops, it may also become common to sequence each individual’s tumor. However, given the gross amount of aberrations found, the clinical relevancy of such data will have to be rigorously established.
Currently, the prognosis for patients with metastatic melanoma is very grim. Research over the next few years will shed light on the genetic determinants of metastasis, thereby allowing the therapeutic targeting of tumor cells that are about to, or have, metastasized. As the cases of melanoma increase worldwide, and since sun exposure is a cooperative factor in melanoma, research into the relationship between the molecular underpinnings of melanoma and UV radiation will be of critical importance. Through such research, the next 5 years may bring forth promising treatments for melanoma.
Key issues
The incidence of melanoma has increased in the last 50 years. As patterns of sun exposure change, and given the potential link between UV radiation and melanoma, understanding the determinants of melanoma will be of critical importance.
While localized melanoma is surgically excisable, therapies for metastatic melanoma have poor clinical efficacy. This has resulted in a grave prognosis for patients with metastatic melanoma.
Genetic alterations in CDKN2A, PTEN, BRAF and N-RAS are common in melanoma.
Various signaling pathways have been shown to be involved in melanoma, including the MAPK and PI3K pathways.
Alterations in c-KIT, c-MET and EGF receptor copy numbers have also been observed in melanoma.
Melanoma shows a high degree of heterogeneity, suggesting the existence of patient subgroups defined by specific genetic alterations. Understanding the genetic and molecular underpinnings of these subgroups will be necessary for the development of therapies.
Analyses of genomic changes in melanoma (i.e., sequence, copy number, genomic structure and epigenetic) have identified key regulators of melanoma. More are likely to be identified.
As a testament to the power of genomic analysis to identify genetic determinants in a disease, Nedd9 and MITF were identified as critical regulators of melanoma. However, such work requires extensive biological validation.
Of critical importance will be the culling of bystanders from genetic drivers of melanoma. This can be achieved through the integration of different data sets, cross-species analysis and cell line-based assays.
Functional and clinicopathological validation of any gene or genetic element of interest must be performed rigorously to identify the critical regulators of melanoma initiation and progression.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Papia Ghosh, Dana-Farber Cancer Institute, Department of Medical Oncology, 44 Binney Street, Boston, MA 02215, USA, Tel.: +1 617 258 8614, Fax: +1 617 582 8169, papia_ghosh@dfci.harvard.edu.
Lynda Chin, Dana-Farber Cancer Institute, Department of Medical Oncology, 44 Binney Street, Boston, MA 02215, USA, Tel.: +1 617 632 6472, Fax: +1 617 582 8169, lynda_chin@dfci.harvard.edu.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445(7130):851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 2.Ries LAG, Melbert D, Krapcho M, Stinchcomb DG, editors. National Cancer Institute. SEER Cancer Statistics Review, 1975–2005. Bethesda, MD, USA: National Cancer Institute; 2008. [Google Scholar]
- 3.Rigel DS. Cutaneous ultraviolet exposure and its relationship to the development of skin cancer. J. Am. Acad. Dermatol. 2008;58(5 Suppl 2):S129–S132. doi: 10.1016/j.jaad.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 4.Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat. Rev. Cancer. 2003;3(8):559–570. doi: 10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- 5.Swetter SM, Boldrick JC, Jung SY, Egbert BM, Harvell JD. Increasing incidence of lentigo maligna melanoma subtypes: northern California and national trends 1990–2000. J. Invest. Dermatol. 2005;125(4):685–691. doi: 10.1111/j.0022-202X.2005.23852.x. [DOI] [PubMed] [Google Scholar]
- 6.Forman SB, Ferringer TC, Peckham SJ, et al. Is superficial spreading melanoma still the most common form of malignant melanoma? J. Am. Acad. Dermatol. 2008;58(6):1013–1020. doi: 10.1016/j.jaad.2007.10.650. [DOI] [PubMed] [Google Scholar]
- 7.Balch CM, Soong SJ, Gershenwald JE, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J. Clin. Oncol. 2001;19:3622–3634. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- 8.Gimotty PA, Van Belle P, Elder DE, et al. Biologic and prognostic significance of dermal Ki67 expression, mitoses, and tumorigenicity in thin invasive cutaneous melanoma. J. Clin. Oncol. 2005;23(31):8048–8056. doi: 10.1200/JCO.2005.02.0735. [DOI] [PubMed] [Google Scholar]
- 9.Gilchrest BA, Eller MS, Geller AC, Yaar M. The pathogenesis of melanoma induced by ultraviolet radiation. N. Engl. J. Med. 1999;340(17):1341–1348. doi: 10.1056/NEJM199904293401707. [DOI] [PubMed] [Google Scholar]
- 10.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur. J. Cancer. 2005;41(1):28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Ford D, Bliss JM, Swerdlow AJ, et al. The International Melanoma Analysis Group (IMAGE). Risk of cutaneous melanoma associated with a family history of the disease. Int. J. Cancer. 1995;62(4):377–381. doi: 10.1002/ijc.2910620403. [DOI] [PubMed] [Google Scholar]
- 12.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur. J. Cancer. 2005;41(14):2040–2059. doi: 10.1016/j.ejca.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 13.Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat. Genet. 1994;8(1):15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 14.Kamb A, Shattuck-Eidens D, Eeles R, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat. Genet. 1994;8(1):23–26. doi: 10.1038/ng0994-22.• Identification of CDKN2A as the gene deleted at 9p21
- 15.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20(16):2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 16.Aitken J, Welch J, Duffy D, et al. CDKN2A variants in a population-based sample of Queensland families with melanoma. J. Natl Cancer Inst. 1999;91(5):446–452. doi: 10.1093/jnci/91.5.446. [DOI] [PubMed] [Google Scholar]
- 17.Tsao H, Zhang X, Kwitkiwski K, et al. Low prevalence of germline CDKN2A and CDK4 mutations in patients with early-onset melanoma. Arch. Dermatol. 2000;136(9):1118–1122. doi: 10.1001/archderm.136.9.1118. [DOI] [PubMed] [Google Scholar]
- 18.Wolfel T, Hauer M, Schneider J, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269(5228):1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 19.Zuo L, Weger J, Yang Q, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat. Genet. 1996;12(1):97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 20.Molven A, Grimstvedt MB, Steine SJ, et al. A large Norwegian family with inherited malignant melanoma, multiple atypical nevi, and CDK4 mutation. Genes Chromosomes Cancer. 2005;44(1):10–18. doi: 10.1002/gcc.20202. [DOI] [PubMed] [Google Scholar]
- 21.Draper GJ, Sanders BM, Kingston JE. Second primary neoplasms in patients with retinoblastoma. Br. J. Cancer. 1986;53(5):661–671. doi: 10.1038/bjc.1986.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fletcher O, Easton D, Anderson K, et al. Lifetime risks of common cancers among retinoblastoma survivors. J. Natl Cancer Inst. 2004;96(5):357–363. doi: 10.1093/jnci/djh058. [DOI] [PubMed] [Google Scholar]
- 23.Albino AP, Vidal MJ, McNutt NS, et al. Mutation and expression of the p53 gene in human malignant melanoma. Melanoma Res. 1994;4(1):35–45. doi: 10.1097/00008390-199402000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Castresana JS, Rubio MP, Vazquez JJ, et al. Lack of allelic deletion and point mutation as mechanisms of p53 activation in human malignant melanoma. Int. J. Cancer. 1993;55(4):562–565. doi: 10.1002/ijc.2910550407. [DOI] [PubMed] [Google Scholar]
- 25.Lubbe J, Reichel M, Burg G, Kleihues P. Absence of p53 gene mutations in cutaneous melanoma. J. Invest. Dermatol. 1994;102(5):819–821. doi: 10.1111/1523-1747.ep12381544. [DOI] [PubMed] [Google Scholar]
- 26.Bardeesy N, Bastian BC, Hezel A, et al. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol. Cell. Biol. 2001;21(6):2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin L, Pomerantz J, Polsky D, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11(21):2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58(10):2170–2175. [PubMed] [Google Scholar]
- 29.Koprowski H, Herlyn M, Balaban G, et al. Expression of the receptor for epidermal growth factor correlates with increased dosage of chromosome 7 in malignant melanoma. Somat. Cell Mol. Genet. 1985;11(3):297–302. doi: 10.1007/BF01534687. [DOI] [PubMed] [Google Scholar]
- 30.Udart M, Utikal J, Krahn GM, Peter RU. Chromosome 7 aneusomy. A marker for metastatic melanoma? Expression of the epidermal growth factor receptor gene and chromosome 7 aneusomy in nevi, primary malignant melanomas and metastases. Neoplasia. 2001;3(3):245–254. doi: 10.1038/sj.neo/7900156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bardeesy N, Kim M, Xu J, et al. Role of epidermal growth factor receptor signaling in RAS-driven melanoma. Mol. Cell. Biol. 2005;25(10):4176–4188. doi: 10.1128/MCB.25.10.4176-4188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dlugosz AA, Hansen L, Cheng C, et al. Targeted disruption of the epidermal growth factor receptor impairs growth of squamous papillomas expressing the υ-ras(Ha) oncogene but does not block in vitro keratinocyte responses to oncogenic ras. Cancer Res. 1997;57(15):3180–3188. [PubMed] [Google Scholar]
- 33.Gangarosa LM, Sizemore N, Graves-Deal R, et al. A raf-independent epidermal growth factor receptor autocrine loop is necessary for Ras transformation of rat intestinal epithelial cells. J. Biol. Chem. 1997;272(30):18926–18931. doi: 10.1074/jbc.272.30.18926. [DOI] [PubMed] [Google Scholar]
- 34.Sibilia M, Fleischmann A, Behrens A, et al. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell. 2000;102(2):211–220. doi: 10.1016/s0092-8674(00)00026-x. [DOI] [PubMed] [Google Scholar]
- 35.Bottaro DP, Rubin JS, Faletto DL. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251:802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 36.Wiltshire RN, Duray P, Bittner ML, et al. Direct visualization of the clonal progression of primary cutaneous melanoma: application of tissue microdissection and comparative genomic hybridization. Cancer Res. 1995;55:3954–3957. [PubMed] [Google Scholar]
- 37.Natali PG, Nicotra MR, Di Renzo MF, et al. Expression of the c-Met/HGF receptor in human melanocytic neoplasms: demonstration of the relationship to malignant melanoma tumour progression. Br. J. Cancer. 1993;68(4):746–750. doi: 10.1038/bjc.1993.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vande Woude GF, Jeffers M, Cortner J, et al. Met-HGF/SF: tumorigenesis, invasion and metastasis. Ciba Found. Symp. 1997;212:119–130. doi: 10.1002/9780470515457.ch8. [DOI] [PubMed] [Google Scholar]
- 39.Rusciano D, Lorenzoni P, Burger MM. Expression of constitutively activated hepatocyte growth factor/scatter factor receptor (c-met) in B16 melanoma cells selected for enhanced liver colonization. Oncogene. 1995;11(10):1979–1987. [PubMed] [Google Scholar]
- 40.Otsuka T, Takayama H, Sharp R, et al. c-Met autocrine activation induces development of malignant melanoma and acquisition of the metastatic phenotype. Cancer Res. 1998;58(22):5157–5167. [PubMed] [Google Scholar]
- 41.Noonan FP, Recio JA, Takayama H, et al. Neonatal sunburn and melanoma in mice. Nature. 2001;413(6853):271–272. doi: 10.1038/35095108. [DOI] [PubMed] [Google Scholar]
- 42.Recio JA, Noonan FP, Takayama H, et al. Ink4a/arf deficiency promotes ultraviolet radiation-induced melanomagenesis. Cancer Res. 2002;62(22):6724–6730. [PubMed] [Google Scholar]
- 43.McGill GG, Haq R, Nishimura EK, Fisher DE. c-Met expression is regulated by Mitf in the melanocyte lineage. J. Biol. Chem. 2006;281(15):10365–10373. doi: 10.1074/jbc.M513094200. [DOI] [PubMed] [Google Scholar]
- 44.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664.•• Through single nucleotide polymorphism and gene-expression analyses, MITF is identified as an example of a lineage addiction oncogene in melanoma.
- 45.Montone KT, van Belle P, Elenitsas R, Elder DE. Proto-oncogene c-kit expression in malignant melanoma: protein loss with tumor progression. Mod. Pathol. 1997;10(9):939–944. [PubMed] [Google Scholar]
- 46.Shen SS, Zhang PS, Eton O, Prieto VG. Analysis of protein tyrosine kinase expression in melanocytic lesions by tissue array. J. Cutan. Pathol. 2003;30(9):539–547. doi: 10.1034/j.1600-0560.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 47.Isabel Zhu Y, Fitzpatrick JE. Expression of c-kit (CD117) in Spitz nevus and malignant melanoma. J. Cutan. Pathol. 2006;33(1):33–37. doi: 10.1111/j.0303-6987.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 48.Huang S, Luca M, Gutman M, et al. Enforced c-KIT expression renders highly metastatic human melanoma cells susceptible to stem cell factor-induced apoptosis and inhibits their tumorigenic and metastatic potential. Oncogene. 1996;13:2339–2347. [PubMed] [Google Scholar]
- 49.Yoshida H, Kunisada T, Kusakabe M, Nishikawa S, Nishikawa SI. Distinct stages of melanocyte differentiation revealed by anlaysis of nonuniform pigmentation patterns. Development. 1996;122(4):1207–1214. doi: 10.1242/dev.122.4.1207. [DOI] [PubMed] [Google Scholar]
- 50.Grichnik JM, Burch JA, Burchette J, Shea CR. The SCF/KIT pathway plays a critical role in the control of normal human melanocyte homeostasis. J. Invest. Dermatol. 1998;111(2):233–238. doi: 10.1046/j.1523-1747.1998.00272.x. [DOI] [PubMed] [Google Scholar]
- 51.Sviderskaya EV, Wakeling WF, Bennett DC. A cloned, immortal line of murine melanoblasts inducible to differentiate to melanocytes. Development. 1995;121(5):1547–1557. doi: 10.1242/dev.121.5.1547. [DOI] [PubMed] [Google Scholar]
- 52.Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum. Pathol. 2005;36(5):486–493. doi: 10.1016/j.humpath.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 53.Willmore-Payne C, Holden JA, Hirschowitz S, Layfield LJ. BRAF and c-kit gene copy number in mutation-positive malignant melanoma. Hum. Pathol. 2006;37(5):520–527. doi: 10.1016/j.humpath.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Nakahara M, Isozaki K, Hirota S, et al. A novel gain-of-function mutation of c-kit gene in gastrointestinal stromal tumors. Gastroenterology. 1998;115(5):1090–1095. doi: 10.1016/s0016-5085(98)70079-4. [DOI] [PubMed] [Google Scholar]
- 55.Fukuda R, Hamamoto N, Uchida Y, et al. Gastrointestinal stromal tumor with a novel mutation of KIT proto-oncogene. Intern. Med. 2001;40(4):301–303. doi: 10.2169/internalmedicine.40.301. [DOI] [PubMed] [Google Scholar]
- 56.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J. Clin. Oncol. 2006;24(26):4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 57.Beadling C, Jacobson-Dunlop E, Hodi FS, et al. KIT gene mutations and copy number in melanoma subtypes. Clin. Cancer Res. 2008;14(21):6821–6828. doi: 10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- 58.van Oosterom AT, Judson IR, Verweij J, et al. Update of Phase I study of imatinib (STI571) in advanced soft tissue sarcomas and gastrointestinal stromal tumors: a report of the EORTC Soft Tissue and Bone Sarcoma Group. Eur. J. Cancer. 2002;38 Suppl. 5:S83–S87. doi: 10.1016/s0959-8049(02)80608-6. [DOI] [PubMed] [Google Scholar]
- 59.van Oosterom AT, Judson I, Verweij J, et al. Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: a Phase I study. Lancet. 2001;358(9291):1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- 60.Verweij J, Casali PG, Zalcberg J, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet. 2004;364(9440):1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 61.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 2002;347(7):472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 62.Hodi FS, Friedlander P, Corless CL, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J. Clin. Oncol. 2008;26(12):2046–2051. doi: 10.1200/JCO.2007.14.0707.• Case study of a patient with acral melanoma and KIT mutation responding to imatinib.
- 63.Lutzky J, Bauer J, Bastian BC. Dose-dependent, complete response to imatinib of a metastatic mucosal melanoma with a K642E KIT mutation. Pigment Cell Melanoma Res. 2008;21(4):492–493. doi: 10.1111/j.1755-148X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 64.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 65.Jafari M, Papp T, Kirchner S, et al. Analysis of ras mutations in human melanocytic lesions: activation of the ras gene seems to be associated with the nodular type of human malignant melanoma. J. Cancer Res. Clin. Oncol. 1995;121(1):23–30. doi: 10.1007/BF01202725. [DOI] [PubMed] [Google Scholar]
- 66.van Elsas A, Zerp SF, van der Flier S, et al. Relevance of ultraviolet-induced N-ras oncogene point mutations in development of primary human cutaneous melanoma. Am. J. Pathol. 1996;149(3):883–893. [PMC free article] [PubMed] [Google Scholar]
- 67.Papp T, Pemsel H, Zimmermann R, et al. Mutational analysis of the N-ras, p53, p16INK4a, CDK4, and MC1Rgenes in human congenital melanocytic naevi. J. Med. Genet. 1999;36:610–614. [PMC free article] [PubMed] [Google Scholar]
- 68.Albino AP, Nanus DM, Mentle IR, et al. Analysis of ras oncogenes in malignant melanoma and precursor lesions: correlation of point mutations with differentiation phenotype. Oncogene. 1989;4:1363–1374. [PubMed] [Google Scholar]
- 69.Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am. J. Pathol. 2000;157:967–972. doi: 10.1016/S0002-9440(10)64609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barnhill RL. The Spitzoid lesion: rethinking Spitz tumors, atypical variants, ‘Spitzoid melanoma’ and risk assessment. Mod. Pathol. 2006;19 Suppl. 2:S21–S33. doi: 10.1038/modpathol.3800519. [DOI] [PubMed] [Google Scholar]
- 71.Smith KJ, Barrett TL, Skelton HG, 3rd, Lupton GP, Graham JH. Spindle cell and epithelioid cell nevi with atypia and metastasis (malignant Spitz nevus) Am. J. Surg. Pathol. 1989;13(11):931–939. doi: 10.1097/00000478-198911000-00003. [DOI] [PubMed] [Google Scholar]
- 72.Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin L. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene. 2003;22(32):5055–5059. doi: 10.1038/sj.onc.1206809. [DOI] [PubMed] [Google Scholar]
- 73.Ackermann J, Frutschi M, Kaloulis K, et al. Metastasizing melanoma formation caused by expression of activated N-RasQ61Kon an INK4a-deficient background. Cancer Res. 2005;65(10):4005–4011. doi: 10.1158/0008-5472.CAN-04-2970. [DOI] [PubMed] [Google Scholar]
- 74.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766.•• Demonstrates that sequencing identifies BRAF mutation as a common event in malignant melanoma.
- 75.Lang J, Boxer M, MacKie R. Absence of exon 15 BRAF germline mutations in familial melanoma. Hum. Mutat. 2003;21(3):327–330. doi: 10.1002/humu.10188. [DOI] [PubMed] [Google Scholar]
- 76.Casula M, Colombino M, Satta MP, et al. BRAF gene is somatically mutated but does not make a major contribution to malignant melanoma susceptibility: the Italian Melanoma Intergroup Study. J. Clin. Oncol. 2004;22(2):286–292. doi: 10.1200/JCO.2004.07.112. [DOI] [PubMed] [Google Scholar]
- 77.Laud K, Kannengiesser C, Avril MF, et al. BRAF as a melanoma susceptibility candidate gene? Cancer Res. 2003;63(12):3061–3065. [PubMed] [Google Scholar]
- 78.Liu J, Suresh Kumar KG, Yu D, et al. Oncogenic BRAF regulates β-Trcp expression and NF-κB activity in human melanoma cells. Oncogene. 2007;26(13):1954–1958. doi: 10.1038/sj.onc.1209994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhatt KV, Hu R, Spofford LS, Aplin AE. Mutant B-RAF signaling and cyclin D1 regulate Cks1/S-phase kinase-associated protein 2-mediated degradation of p27Kip1 in human melanoma cells. Oncogene. 2007;26(7):1056–1066. doi: 10.1038/sj.onc.1209861. [DOI] [PubMed] [Google Scholar]
- 80.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6(4):313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 81.Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J. Natl Cancer Inst. 2003;95(24):1878–1890. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
- 82.Edwards RH, Ward MR, Wu H, et al. Absence of BRAF mutations in UV-protected mucosal melanomas. J. Med. Genet. 2004;41(4):270–272. doi: 10.1136/jmg.2003.016667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005;353(20):2135–2147. doi: 10.1056/NEJMoa050092.•• Copy number aberrations profiles were distinct in melanoma histological subtypes.
- 84.Landi MT, Bauer J, Pfeiffer RM, et al. MC1R germline variants confer risk for BRAF-mutant melanoma. Science. 2006;313(5786):521–522. doi: 10.1126/science.1127515. [DOI] [PubMed] [Google Scholar]
- 85.Kennedy C, ter Huurne J, Berkhout M, et al. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J. Invest. Dermatol. 2001;117(2):294–300. doi: 10.1046/j.0022-202x.2001.01421.x. [DOI] [PubMed] [Google Scholar]
- 86.Duffy DL, Box NF, Chen W, et al. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum. Mol. Genet. 2004;13(4):447–461. doi: 10.1093/hmg/ddh043. [DOI] [PubMed] [Google Scholar]
- 87.Raimondi S, Sera F, Gandini S, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int. J. Cancer. 2008;122(12):2753–2760. doi: 10.1002/ijc.23396. [DOI] [PubMed] [Google Scholar]
- 88.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat. Genet. 2003;33(1):19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 89.Kumar R, Angelini S, Snellman E, Hemminki K. BRAF mutations are common somatic events in melanocytic nevi. J. Invest. Dermatol. 2004;122(2):342–348. doi: 10.1046/j.0022-202X.2004.22225.x. [DOI] [PubMed] [Google Scholar]
- 90.Saldanha G, Purnell D, Fletcher A, et al. High BRAF mutation frequency does not characterize all melanocytic tumor types. Int. J. Cancer. 2004;111(5):705–710. doi: 10.1002/ijc.20325. [DOI] [PubMed] [Google Scholar]
- 91.Yazdi AS, Palmedo G, Flaig MJ, et al. Mutations of the BRAF gene in benign and malignant melanocytic lesions. J. Invest. Dermatol. 2003;121(5):1160–1162. doi: 10.1046/j.1523-1747.2003.12559.x. [DOI] [PubMed] [Google Scholar]
- 92.Michaloglou C, Vredeveld LC, Soengas MS, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 93.Wellbrock C, Ogilvie L, Hedley D, et al. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004;64(7):2338–2342. doi: 10.1158/0008-5472.can-03-3433. [DOI] [PubMed] [Google Scholar]
- 94.Sharpless NE, DePinho RA. Cancer: crime and punishment. Nature. 2005;436(7051):636–637. doi: 10.1038/436636a. [DOI] [PubMed] [Google Scholar]
- 95.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132(3):363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chudnovsky Y, Adams AE, Robbins PB, Lin Q, Khavari PA. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nat. Genet. 2005;37(7):745–749. doi: 10.1038/ng1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Edlundh-Rose E, Egyhazi S, Omholt K, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006;16(6):471–478. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 98.Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J. Invest. Dermatol. 2006;126(1):154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 99.Jiveskog S, Ragnarsson-Olding B, Platz A, Ringborg U. N-ras mutations are common in melanomas from sun-exposed skin of humans but rare in mucosal membranes or unexposed skin. J. Invest. Dermatol. 1998;111(5):757–761. doi: 10.1046/j.1523-1747.1998.00376.x. [DOI] [PubMed] [Google Scholar]
- 100.Dumaz N, Hayward R, Martin J, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66(19):9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 101.Kannengiesser C, Spatz A, Michiels S, et al. Gene expression signature associated with BRAF mutations in human primary cutaneous melanomas. Mol. Oncol. 2008:425–430. doi: 10.1016/j.molonc.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shields JM, Thomas NE, Cregger M, et al. Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma. Cancer Res. 2007;67(4):1502–1512. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- 103.Smalley KS, Xiao M, Villanueva J, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2008;28(1):85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kortylewski M, Heinrich PC, Kauffmann ME, et al. Mitogen-activated protein kinases control p27/Kip1 expression and growth of human melanoma cells. Biochem J. 2001;357(Pt 1):297–303. doi: 10.1042/0264-6021:3570297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huntington JT, Shields JM, Der CJ, et al. Overexpression of collagenase 1 (MMP-1) is mediated by the ERK pathway in invasive melanoma cells: role of BRAF mutation and fibroblast growth factor signaling. J. Biol. Chem. 2004;279(32):33168–33176. doi: 10.1074/jbc.M405102200. [DOI] [PubMed] [Google Scholar]
- 106.McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109(6):707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 107.Kwong L, Chin L, Wagner SN. Growth factors and oncogenes as targets in melanoma: lost in translation? Adv. Dermatol. 2007;23:99–129. doi: 10.1016/j.yadr.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: a clinicopathologic study of 292 cases. J. Clin. Oncol. 2005;23(7):1473–1482. doi: 10.1200/JCO.2005.07.168. [DOI] [PubMed] [Google Scholar]
- 109.Stahl JM, Sharma A, Cheung M, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64(19):7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 110.Ross P, Hall L, Haff LA. Quantitative approach to single-nucleotide polymorphism analysis using MALDI-TOF mass spectrometry. Biotechniques. 2000;29(3):620–626. 628–629. doi: 10.2144/00293rr05. [DOI] [PubMed] [Google Scholar]
- 111.Yoeli-Lerner M, Yiu GK, Rabinovitz I, et al. Atk blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol. Cell. 2005;20:539–550. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Bastian BC. Understanding the progression of melanocytic neoplasia using genomic analysis: from fields to cancer. Oncogene. 2003;22(20):3081–3086. doi: 10.1038/sj.onc.1206463. [DOI] [PubMed] [Google Scholar]
- Wu H, Goel V, Haluska FG. PTEN signaling pahtways in melanoma. Oncogene. 2003;22:3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 114.Pollock PM, Walker GJ, Glendening JM, et al. PTEN inactivation is rare in melanoma tumours but occurs frequently in melanoma cell lines. Melanoma Res. 2002;12(6):565–575. doi: 10.1097/00008390-200212000-00006. [DOI] [PubMed] [Google Scholar]
- 115.Mikhail M, Velazquez E, Shapiro R, et al. PTEN expression in melanoma: relationship with patient survival, Bcl-2 expression, and proliferation. Clin. Cancer Res. 2005;11(14):5153–5157. doi: 10.1158/1078-0432.CCR-05-0397. [DOI] [PubMed] [Google Scholar]
- 116.Slipicevic A, Holm R, Nguyen MT, et al. Expression of activated Akt and PTEN in malignant melanomas: relationship with clinical outcome. Am. J. Clin. Pathol. 2005;124(4):528–536. doi: 10.1309/YT58WWMTA6YR1PRV. [DOI] [PubMed] [Google Scholar]
- 117.Robertson GP, Furnari FB, Miele ME, et al. In vitro loss of heterozygosity targets the PTEN/MMAC1 gene in melanoma. Proc. Natl Acad. Sci. USA. 1998;95(16):9418–9423. doi: 10.1073/pnas.95.16.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stewart AL, Mhashilkar AM, Yang XH, et al. PI3 kinase blockade by Ad-PTEN inhibits invasion and induces apoptosis in RGP and metastatic melanoma cells. Mol. Med. 2002;8(8):451–461. [PMC free article] [PubMed] [Google Scholar]
- 119.Stahl JM, Cheung M, Sharma A, et al. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Res. 2003;63(11):2881–2890. [PubMed] [Google Scholar]
- 120.Robertson GP. Functional and therapeutic significance of Akt deregulation in malignant melanoma. Cancer Metastasis Rev. 2005;24(2):273–285. doi: 10.1007/s10555-005-1577-9. [DOI] [PubMed] [Google Scholar]
- 121.You MJ, Castrillon DH, Bastian BC, et al. Genetic analysis of Pten and Ink4a/Arf interactions in the suppression of tumorigenesis in mice. Proc. Natl Acad. Sci. USA. 2002;99:1455–1460. doi: 10.1073/pnas.022632099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 1998;19(4):348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 123.Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 124.Podsypanina K, Ellenson LH, Nemes A, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl Acad. Sci. USA. 1999;96(4):1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ma X, Ziel-van der Made AC, Autar B, et al. Targeted biallelic inactivation of Pten in the mouse prostate leads to prostate cancer accompanied by increased epithelial cell proliferation but not by reduced apoptosis. Cancer Res. 2005;65(13):5730–5739. doi: 10.1158/0008-5472.CAN-04-4519. [DOI] [PubMed] [Google Scholar]
- 126.Trotman LC, Wang X, Alimonti A, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128(1):141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang X, Trotman LC, Koppie T, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128(1):129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bedogni B, Welford SM, Kwan AC, et al. Inhibition of phosphatidylinositol-3-kinase and mitogen-activated protein kinase kinase 1/2 prevents melanoma development and promotes melanoma regression in the transgenic TPRas mouse model. Mol. Cancer Ther. 2006;5(12):3071–3077. doi: 10.1158/1535-7163.MCT-06-0269. [DOI] [PubMed] [Google Scholar]
- 129.Smalley KS, Haass NK, Brafford PA, et al. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol. Cancer Ther. 2006;5(5):1136–1144. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 130.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J. Invest. Dermatol. 2004;122(2):337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J. Natl Cancer Inst. 2006;98(7):472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 132.Wellbrock C, Marais R. Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation. J. Cell. Biol. 2005;170(5):703–708. doi: 10.1083/jcb.200505059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wellbrock C, Rana S, Paterson H, et al. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS ONE. 2008;3(7):e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer. 2006;6(8):593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 135.Hemesath TJ, Steingrimsson E, McGill G, et al. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8(22):2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 136.Steingrimsson E, Moore KJ, Lamoreux ML, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 1994;8(3):256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- 137.Price ER, Fisher DE. Sensorineural deafness and pigmentation genes: melanocytes and the Mitf transcriptional network. Neuron. 2001;30(1):15–18. doi: 10.1016/s0896-6273(01)00259-8. [DOI] [PubMed] [Google Scholar]
- 138.Weeraratna AT, Jiang Y, Hostetter G, et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1(3):279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 139.Muthusamy V, Duraisamy S, Bradbury CM, et al. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res. 2006;66(23):11187–11193. doi: 10.1158/0008-5472.CAN-06-1274. [DOI] [PubMed] [Google Scholar]
- 140.Mori T, Martinez SR, O’Day SJ, et al. Estrogen receptor-α methylation predicts melanoma progression. Cancer Res. 2006;66(13):6692–6698. doi: 10.1158/0008-5472.CAN-06-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mirmohammadsadegh A, Marini A, Nambiar S, et al. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006;66(13):6546–6552. doi: 10.1158/0008-5472.CAN-06-0384. [DOI] [PubMed] [Google Scholar]
- 142.Kato Y, Salumbides BC, Wang X-F. Antitumor effect of the histone deacetylase inhibitor LAQ824 in combination with 13-cis-retinoic acid in human malignant melanoma. Mol. Cancer Ther. 2007;6:70–81. doi: 10.1158/1535-7163.MCT-06-0125. [DOI] [PubMed] [Google Scholar]
- 143.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 145.Zhang L, Huang J, Yang N, et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad. Sci. USA. 2006;103(24):9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim M, Gans JD, Nogueira C, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125(7):1269–1281. doi: 10.1016/j.cell.2006.06.008.•• Cross-species analysis filters genomic data and identifies Nedd9 as an important regulator of melanoma.
- 147.Maser RS, Choudhury B, Campbell PJ, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447(7147):966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]