Abstract
Annexin 1 is an anti-inflammatory protein that plays a key role in innate immunity by modulating the activation of several types of cells, including neutrophils. Here we have developed a cleavage assay using tagged annexin 1 and observed marked activity in the membrane fraction of activated neutrophils. A combination of inhibitors, transfected cells, and proteomic analyses allowed us to identify proteinase 3 as the main enzyme responsible for this cleavage in the N terminus region of the protein, at least in the context of neutrophil activation. Because annexin 1 is an important endogenous anti-inflammatory mediator, blocking its cleavage by proteinase 3 would augment its homeostatic pro-resolving actions and could represent an opportunity for innovative anti-inflammatory drug discovery.
Annexin 1 (AnxA1)5 belongs to the superfamily of annexin proteins characterized by the presence of two principal domains, the N-terminal, which is believed to confer the particular biological effects of each annexin, and the large C-terminal-containing core (1). Studies over the past 20 years have shown that AnxA1 acts as an endogenous down-regulator of blood neutrophils (PMN). AnxA1 released by these cells inhibits in a time-dependent fashion their interaction with activated endothelial cells (2, 3). Similarly, treatment of animals with exogenous AnxA1 reduces PMN adhesion and emigration to inflamed vessels, thereby affecting leukocyte recruitment to the site of inflammation (4, 5). The possibility that the actions of AnxA1 could be modulated in the microenvironment of an activated PMN has been scantly investigated. Association studies detected intact (37 kDa) and cleaved (33 kDa) AnxA1 proteins concomitant to elastase presence (6, 7). AnxA1 cleavage also occurs following human PMN adhesion (2), a process that activates protein externalization (8). More recently, the proteolytic action of human recombinant neutrophil elastase (HNE) on AnxA1 catabolism has been described (9).
Proteinase 3 (PR3) is another serine protease expressed in neutrophil organelles, including secretory vesicles (10). Distinct regulatory roles have been ascribed to PR3, given its ability to modulate neutrophil cell differentiation (11, 12), cytokine bioactivity (such as tumor necrosis factor α (TNF-α), interleukin 1β, transforming growth factor β, and interleukin 8) and receptor (such as interleukin 2) synthesis (13). In view of the wider distribution of PR3 in PMN membranes and organelles relative to HNE, mainly localized in azurophilic granules (14, 15), we tested the hypothesis that PR3 could cleave AnxA1 in the PMN microenvironment. Here, we have devised a novel AnxA1 cleavage assay to compare (i) the effects of HNE and PR3 and (ii) the actions of enzyme inhibitors. The ultimate aim was to assess their contribution to AnxA1 cleavage in the context of cell activation.
EXPERIMENTAL PROCEDURES
Cells and Culture Conditions
The epithelial cell line HEK-293T (Amersham Biosciences) was maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% (v/v) fetal bovine serum, 2 mm l-glutamine, 1% (v/v) non-essential amino acids, 1 μg/ml Geneticin, and 50 μg/ml G418. The human mastocytic cell line (HMC1 or human mast cell clone 1) and clones overexpressing human PR3, inactive mutant PR3 (S203A), or HNE were cultured as previously described (16).
PMN Activation and Fractionation
PMN of healthy individuals were isolated by Hystopaque sedimentation as reported (2). In activation assays PMN (107 cells/ml) were incubated with 10 ng/ml TNF-α (Sigma) for 30 min at 37 °C. For preparation of whole cell lysates (WL), PMN were pelleted and resuspended (3 × 107/ml) in lysis buffer (200 mm NaCl, 20 mm Tris-HCl, pH 8.0, 1% Triton X-100) for 15 min. To obtain cytosolic (CYT) and membrane (MEM) fractions, cells (3 × 107/ml) were resuspended in 20 mm Tris-HCl, pH 7.5, centrifuged briefly for 2 min at 300 × g, transferred to a new microcentrifuge tube, and centrifuged again for 45 min at 800 × g. The resultant supernatant (cytosolic fraction, CYT) was collected and the remaining pellet resuspended in 20 mm Tris-HCl, 1% Triton X-100, pH 7.5, at 30 × 106 cells/ml for 15 min (membrane fraction, MEM). Membrane-bound AnxA1 was recovered by washing 3 × 106 pelleted cells twice with 50 μl of 1 mm ethylene-diaminetetraacetic acid (EDTA). When purified PMN (107 cells/ml) were prepared for Western blot analysis only, a mixture of protease and phosphatase inhibitors was added to the lysis buffers. Extracts for use in the cleavage assay (see below) were prepared in the absence of inhibitors.
Construction of MF-AnxA1 Expression Vector and Cell Transfection
Total RNA from 10−15 × 106 HL-60 cells was extracted using the RNeasy® extraction kit (Qiagen) and reverse transcribed using avian myeloblastosis virus reverse transcriptase (Promega) according to the manufacturers' instructions. AnxA1 with c-Myc tagged on the N terminus and FLAG tagged on the C terminus (MF-AnxA1) was generated using the primers reported in the supplemental information. PCR products were cloned into BamHI and XhoI sites of pcDNA3.1 vector (Invitrogen). MF-AnxA1 (2 μg) was transfected into HEK-293T cells using 6 μl of FuGENE 6 transfection reagent (Roche Applied Science) and 92 μl of OptiMEM™ (Invitrogen) culture medium for each well of a 6-well culture plate (containing 5 × 105 cells). After 24 h, cells from each well were lysed in 500 μl of lysis buffer and stored at −20 °C until further use. HMC1 cell lines were transfected with MF-AnxA1 by nucleofection (Amaxa GmbH). Briefly, 106 cells were nucleofected using 2 μg of plasmid DNA in supplemented Nucleofector Solution V. T-030 program was selected, and cells were incubated in a humidified incubator (37 °C, 5% CO2) for 24 h prior to collection.
AnxA1 Cleavage Assay
MF-AnxA1-transfected HEK-293T cell lysates were immunoprecipitated (IP) with anti-c-Myc-(Santa Cruz Biotechnology) or anti-FLAG (Sigma) -conjugated agarose beads overnight at 4 °C (20 μl of beads/reaction). The beads were washed three times with 200 mm NaCl, 20 mm Tris-HCl, pH 8.0, and stored on ice until further use. For the analysis of cleavage activity of cell lysates, extracts from PMN (104–106) or HMC1 cells (3 × 106) were diluted into a reaction volume 100 μl of Hanks' buffered salt solution with Ca2+/Mg2+ and added to the c-Myc IP containing intact MF-AnxA1. The reaction was incubated at 37 °C for 30 min, and thereafter a mixture of protease inhibitors (see supplemental information) was added to prevent further proteolysis. The reaction mixture was then IP with anti-FLAG-conjugated agarose beads overnight at 4 °C, while the remaining pellet was boiled in 20 μl of 6× Laemmli buffer for 5 min. The anti-FLAG beads were subsequently washed and boiled (95 °C, 5 min) and samples subjected to 12% SDS-PAGE. Where purified PR3 (Elastin Products, Owensville, MO) or HNE (Elastin Products) were added to MF-AnxA1, this was done in 100 μl of Hanks' buffered salt solution with Ca2+/Mg2+. Phenylmethylsulfonyl fluoride (PMSF) (1 mm) or eglin C (0.1, 1 mm) was added to MEM fractions for 15 min at 37 °C prior to addition to the cleavage assay.
Western Blot Analysis
Samples boiled in 6× Laemmli buffer were subjected to standard SDS-polyacrylamide gel electrophoresis (12%) and electrophoretically blotted onto polyvinylidene diflouride membranes (Millipore, Watford, UK). Membranes were incubated with primary antibodies in Tris-buffered saline solution containing Tween 20 and 5% (w/v) nonfat dry milk at room temperature for 1 h. Membranes were washed for 20 min with the solution changed at 5-min intervals and secondary antibodies and proteins detected using the enhanced chemiluminescence (ECL) detection kit and visualized on Hyperfilm (Amersham Biosciences). The primary antibodies used were rabbit polyclonal anti-AnxA1 (Zymed Laboratories Inc., Cambridge, UK), mouse monoclonal anti-c-Myc (Santa Cruz), and mouse monoclonal anti-FLAG M2 (Sigma). Secondary antibodies used (polyclonal goat anti-rabbit and goat anti-mouse) were conjugated to horseradish peroxidase (Dako, Cambridge, UK).
MALDI-TOF Analysis of MF-AnxA1 N Terminus Fragments
MF-AnxA1 was incubated with PMN membrane fractions according to the cleavage assay. The c-Myc IP was washed with 200 mm NaCl, 20 mm Tris-HCl, pH 8, and then eluted by competition with 50 μl (1 mg/ml) of c-Myc peptide (Sigma). The sample was subjected to MALDI-TOF analysis (Mass Spectrometry Facility School of Pharmacy, University of London, UK), run in positive mode (Applied Biosystems Voyager DEPRO MALDI-TOF).
Confocal Microscopy
Resting or stimulated PMN (10 ng/ml TNF-α, 30 min at 37 °C) were adhered onto poly-l-lysine pre-coated coverslips and fixed in 3% paraformaldehyde prior to permeabilization with either ice-cold methanol or streptolysin-O, as described (17). The primary and secondary antibodies were as follows: mouse monoclonal anti-human PR3 (2 μg/ml; Sanquin) followed by ALEXA-488 anti-mouse IgG (6.7 μg/ml; Molecular Probes); mouse monoclonal anti-HNE (25 μg/ml; Bio-genesis) followed by ALEXA-488 anti-mouse IgG; mouse rabbit polyclonal anti-human AnxA1 (1:50 dilution; Zymed Laboratories) followed by ALEXA-555 anti-rabbit IgG (13.3 μg/ml; Molecular Probes). Slides were mounted using Permafluor mounting medium (Dako) and analyzed by confocal microscopy with a Zeiss LSM-5 confocal scanning laser microscope as previously described (17).
RESULTS
Localization and Cleavage of Endogenous AnxA1 in Human PMN
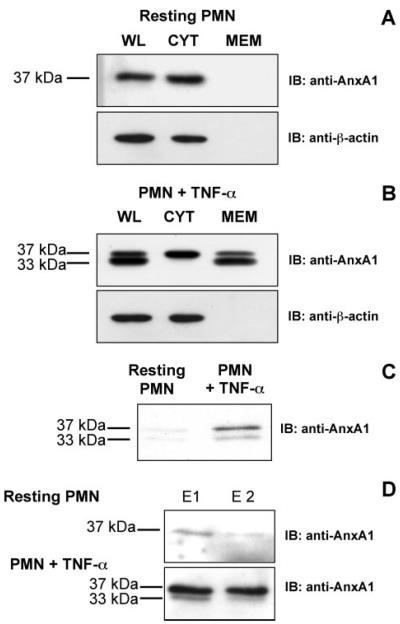
PMN activation or adhesion induces AnxA1 externalization onto the membrane and simultaneous release in the extracellular milieu (2). Consistent with these findings, in resting PMN AnxA1 was localized predominantly in the cytosol (Fig. 1A, CYT). Upon PMN activation with TNF-α, the protein was markedly mobilized onto the membrane fraction (Fig. 1B, MEM) and secreted into the culture medium (Fig. 1C). Furthermore, the 37-kDa intact AnxA1 form was detected in the whole lysate of resting cells while it was clipped into the 33-kDa form upon treatment with TNF-α (Fig. 1, A and B, WL).
FIGURE 1. Profile of AnxA1 protein expression in human PMN.
Resting PMN (A) or PMN stimulated with 10 ng/ml TNF-α (B) were processed for whole lysate (WL), cytosolic (CYT), and membrane (MEM) fractions and immunoblotted with anti-AnxA1 or anti-actin antibody (upper and lower panels, respectively). C, conditioned medium from resting or TNF-α-stimulated PMN was collected and immunoblotted for AnxA1. D, intact and activated PMN (upper and lower panels, respectively) were washed consecutively twice (E1, E2) with 1 mm EDTA to elute membrane-bound AnxA1.
To determine the cellular compartment where the protein is clipped, we isolated the CYT and MEM fractions; stimulation with TNF-α mobilized the protein onto the membrane, where it was present as a clipped form (Fig. 1B, 33 kDa). Importantly, elution of membrane-bound AnxA1 with two consecutive washes of EDTA (E1 and E2) resulted in both intact and clipped AnxA1 being recovered from stimulated PMN, compared with control unstimulated cells where only modest levels of the intact form were detected (Fig. 1D). Together, these results indicate AnxA1 to be present in the cytosol predominantly as an N-terminal-intact 37-kDa species. In response to cell stimulation, the protein translocates to the membrane compartment, where it is cleaved and released as an N-terminal-clipped 33-kDa species.
AnxA1 Cleavage Assay
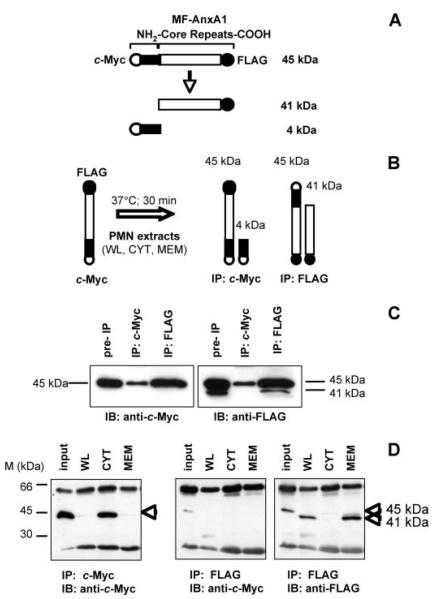
A number of studies over the past ten years have suggested that AnxA1 is cleaved at the N terminus during inflammatory conditions (6, 7). To isolate and identify the enzyme(s) responsible for this cleavage we developed an “AnxA1 cleavage assay.” To discern endogenous from exogenous AnxA1, a double-tagged protein derivative with a c-Myc tag on the N terminus and a FLAG tag on the C terminus was cloned (MF-AnxA1; see schematic in Fig. 2A). This construct was transfected into HEK-293T cells and further purified from the cell lysates by IP. MF-AnxA1 was then used as an in vitro substrate to identify PMN cellular fractions containing “cleavage activity” (Fig. 2B). At the end of the reaction, the “clipped forms” were isolated by IP for the relative tags (c-Myc for the N terminus and FLAG for the C terminus). As shown in Fig. 2C, transfection of MF-AnxA1 into HEK-293T cells and analysis of the whole cell lysate (pre-IP) for the c-Myc or FLAG tags yielded a single band of ~45 kDa (37 kDa of the full-length protein plus the c-Myc and FLAG tags) in the c-Myc blot and a double band of 45 and 41 kDa in the FLAG blot, corresponding to the intact (37-kDa) and clipped (33-kDa) forms of AnxA1, respectively. IP of cell lysates with c-Myc-conjugated agarose beads and immunoblotting with anti-c-Myc or anti-FLAG detected a single N terminus-intact form (Fig. 2C). Conversely, IP with FLAG-conjugated agarose beads and immunoblotting with anti-c-Myc or anti-FLAG showed a single and a double band, respectively (Fig. 2C). This demonstrates that IP of the FLAG tag recovers both intact and clipped MF-AnxA1, whereas IP of the c-Myc tag enables isolation of the intact form only. We next incubated MF-AnxA1 (purified from HEK-293 cell lysates) with distinct PMN extracts. As shown in Fig. 2D, addition of non-stimulated PMN WL resulted in loss of c-Myc immunoreactivity. Collection and IP of the reaction buffer with anti-FLAG beads, followed by immunoblot analysis with anti-c-Myc (Fig. 2D, middle panel) or anti-FLAG (Fig. 2D, right panel) detected a single 41-kDa FLAG-positive but c-Myc-negative band. This corresponds to the clipped 33-kDa AnxA1 fragment, suggesting that cleavage occurs at the N terminus. Addition of the PMN CYT fraction had minimal cleavage activity, as the intact MF-AnxA1 was still detected. Importantly, and in line with the profile of endogenous AnxA1, the majority of PMN cleavage activity measured in the WL appears to be derived from the MEM fraction. Here, c-Myc immunoreactivity was absent (Fig. 2D, IP with c-Myc), while the N terminus-cleaved 41-kDa portion was present in the wash (Fig. 2D, IP with FLAG). The validation of the cleavage assay was achieved by testing commercial preparations of purified HNE and PR3. Supplemental Fig. S1, A and B, shows that either enzyme was able to produce a concentration-dependent disappearance of c-Myc immunoreactivity within the 30-min reaction time, but PR3 appeared to be more efficient when concentrations were compared.
FIGURE 2. AnxA1-specific proteolytic activity resides in the membrane fraction of human PMN.
A, schematic representation of MF-AnxA1 and fragments resulting from proteolysis. B, depiction of the AnxA1 cleavage assay. PMN fractions (WL, CYT, MEM) were incubated with MF-AnxA1 for 30 min at 37 °C (left). Intact MF-AnxA1 could be recovered from the reaction mixture by analysis of the c-Myc IP (right), while N terminus-cleaved MF-AnxA1 could be recovered by IP for FLAG (far right). C, HEK-293T cells were transfected with MF-AnxA1. Cell lysates were IP for c-Myc or FLAG epitopes and immunoblotted with anti-c-Myc (left panel) or anti-FLAG (right panel). D, resting PMN-derived cell fractions were incubated with MF-AnxA1 (input) and assessed for AnxA1-specific proteolytic activity using the cleavage assay described above. Intact MF-AnxA1 was obtained by recovering the c-Myc IP and immunoblotting for c-Myc (left panel). N-terminal-cleaved MF-AnxA1 fragments were obtained by IP for FLAG and immunoblotting with anti-c-Myc (middle panel) or anti-FLAG (right panel) antibodies. Data are representative of five distinct preparations.
MALDI-TOF Analysis of theNTerminus-clipped MF-AnxA1
To identify the fragments and potential cleavage sites resulting from proteolytic activity within the PMN membrane fraction, N terminus fragments that remained bound to the c-Myc beads were analyzed. Here, MALDI-TOF analysis revealed three major peptides of masses 2436, 3900, and 5186Da(Table 1 and supplemental Fig. S2). Comparison of the mass of these peptides with the known AnxA1 N terminus sequence suggested the presence of major cleavage sites occurring at Ala11, Val22, Val36.
TABLE 1. N terminus-located MF-AnxA1 fragments cleaved by the PMN membrane fraction.
MF-AnxA1 was eluted from c-Myc-conjugated agarose beads and subjected to MALDI-TOF analysis. Three predominant peptide fragments were recovered.
| Fragment | N terminus sequence | Residues |
|---|---|---|
| Da | ||
| 2436 | EQKLISEEDL MAMVSEFLKQA | 1–11 |
| 3900 | EQKLISEEDL MAMVSEFLKQAWFIENEEQEYV | 1–22 |
| 5186 |
EQKLISEEDL MAMVSEFLKQAWFIENEEQEYV QTVKSSKGGPGSAV |
1–36 |
Cleavage of AnxA1 by PR3
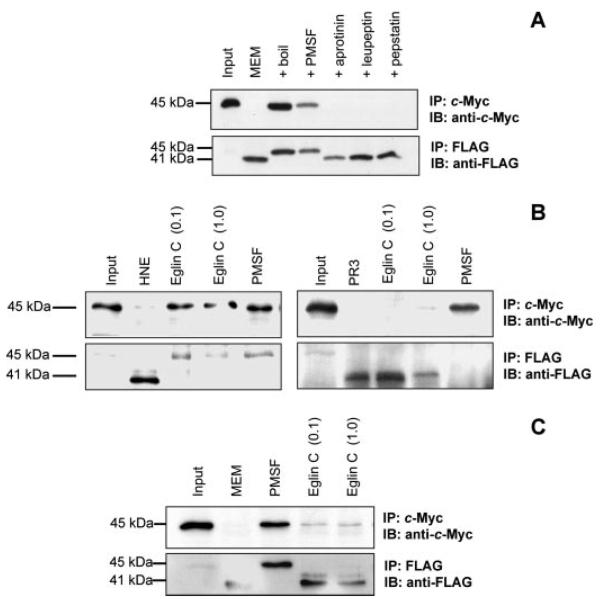
Next, we examined the contribution of serine proteases to AnxA1 cleavage. The ability of the PMN MEM fraction to cleave MF-AnxA1 was specific because addition of the denatured fraction (boiled at 95 °C for 5 min) did not produce any effect (Fig. 3A). Moreover, MEM fraction activity was blocked by preincubation with PMSF, a pan-inhibitor of serine proteases, but not by other protease inhibitors, including aprotinin, leupeptin, and pepstatin. Thus, PMSF, at a concentration active in blocking the cleavage effect of purified HNE and PR3 (Fig. 3B), reverted the cleavage of the N-terminal tag by MEM fraction, also apparent by the recovery of the intact 45-kDa species (Fig. 3, A and C).
FIGURE 3. Abrogation of AnxA1 proteolysis by a serine-type protease inhibitor.
A, MF-AnxA1 (input) was incubated with resting PMN-derived membrane (MEM) fractions in the absence or presence of phenylmethylsulfonylfluoride (PMSF, 1 mm) or heat-inactivated samples (boil). B, MF-AnxA1 (input) was incubated with 50 milliunits of HNE (left panel) or 10 milliunits of PR3 (right panel), treated with/without 1 mm PMSF or 0.1–1 mm eglin C. C, as in panel A, PMN-derived membrane (MEM) fractions were incubated with 1 mm PMSF or 0.1–1 mm eglin C. In all cases, intact MF-AnxA1 was recovered and immunoblotted with anti-c-Myc antibody (upper panels) and resultant N-terminal-cleaved proteolytic fragments obtained by IP and immunoblotting with anti-FLAG antibody (lower panels). Blots are representative from at least three distinct experiments.
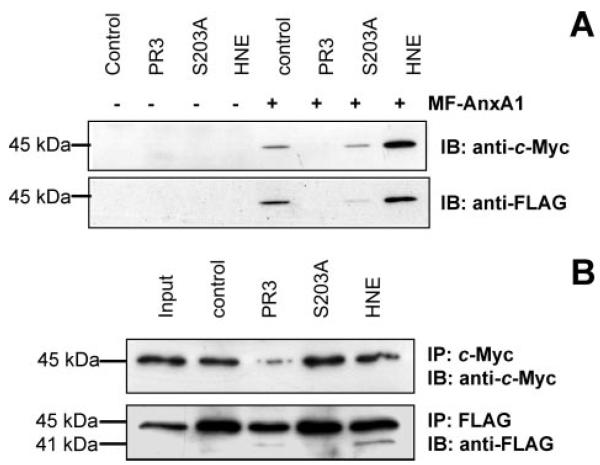
A hint toward the identification of the catabolic enzyme present in MEM fractions was obtained by the ineffectiveness of eglin C; this inhibitor, active in blocking HNE, but not PR3 actions (Fig. 3B), did not significantly affect the endogenous MEM activity (Fig. 3C). To further demonstrate the specificity of PR3 cleavage, we used HMC1 cells stably transfected with active PR3, the inactive mutant S203A, or HNE (16). Transfection with the MF-AnxA1 construct led to equivalent expression of the protein, which was, however, markedly degraded on its N terminus only in PR3/HMC1 cells, as detected following IP with c-Myc (Fig. 4A). Overexpression of MF-AnxA1 in cells transfected with PR3 resulted in the loss of both the c-Myc (Fig. 4A, top panel) and FLAG tags (bottom panel). Absence of the 41-kDa fragment likely results from the dynamic effect of constitutively active PR3 with excess substrate degradation and possibly release of small fragments into the cell supernatant. Finally, these data were validated in the in vitro assay whereby addition of cell lysates from untransfected HMC1 cells (control) or S203A mutant did not reduce immunoreactivity for intact 45-kDa MF-AnxA1, whereas PR3/HMC1 and HNE/ HMC1 cell extracts clipped, although to a different extent, the c-Myc N terminus portion of the substrate (Fig. 4B, top panel). Analysis of FLAG immunoreactivity showed recovery of the 41-kDa clipped fragment (Fig. 4B, bottom panel). These results highlight the difference between the in vivo and in vitro settings, where only PR3 cleaves AnxA1 in intact cells. Therefore, colocalization, rather than substrate specificity, dictates the bone fide AnxA1 catabolic enzyme. To verify this point in the context of PMN biology, we next conducted confocal microscopy analyses chasing AnxA1, PR3, and HNE.
FIGURE 4. N-terminal proteolysis of MF-AnxA1 by PR3 in HMC1 cells.
A, HMC1 cells (control, PR3, PR3 mutant S203A, and HNE) were transfected with MF-AnxA1. After 24 h cell lysates were immunoblotted for c-Myc (upper panel) and FLAG (lower panel) with anti-c-Myc and anti-FLAG antibodies, respectively. B, extracts prepared from control, PR3, PR3 mutant S203A, and HNE cells were processed and incubated with MF-AnxA1 (input). Intact MF-AnxA1 (upper panel) and N-terminal-cleaved fragments (lower panel) were obtained as described above. Blots are representative from at least three distinct experiments.
Endogenous AnxA1 and PR3 Localization in Resting and Stimulated PMN
The localization of endogenous AnxA1 and PR3 (and HNE, for comparative purposes) was monitored in resting and stimulated PMN. Cell treatment with streptolysin-O, which leads to controlled plasma membrane permeabilization allowing access only to extra-granular proteins in the PMN, revealed a high degree of overlay between the staining obtained for AnxA1 and PR3 in resting PMN (Fig. 5A). This is likely to reflect membrane co-localization and, of importance, it was augmented after cell incubation with TNF-α (Fig. 5B). Subjecting the cells to total permeabilization procedure (methanol treatment) confirmed a high degree of co-localization between the two stainings. In contrast, when cells were prepared either with streptolysin-O or methanol treatment, minimal or no co-localization between AnxA1 and HNE staining was detected (supplemental Fig. S3).
FIGURE 5. Localization of endogenous AnxA1 and PR3 in resting and activated PMN.
PMN were incubated in the absence (A) or presence (B) of 10 ng/ml TNF-α for 30 min at 37 °C before being subjected to treatment with streptolysin-O (SLO) or methanol (metOH) as described under “Experimental Procedures.” Staining with anti-AnxA1 or anti-PR3 antibodies is shown, including the overlay panels on the right-hand side, showing a high degree of co-localization between the two proteins in either PMN state. Images are representative of four or five analyses.
DISCUSSION
In this study we searched for the AnxA1 cleavage activity present in activated PMN. Analyses of AnxA1 localization in both resting and activated PMN, in relation to the different protein species (intact and cleaved), identified a link between AnxA1 N terminus cleavage and the simultaneous process of cell activation. This link was centered on PR3.
At least three pools of AnxA1 are known to exist in leukocytes: a cytosolic pool, a membrane-bound pool sensitive to elution by di-cation chelators such as EDTA, and a membrane pool resistant to EDTA washes (18, 19). In resting PMN, the majority of AnxA1 is localized to the gelatinase granules (8, 20) and to a much lesser extent on the cell membrane (2). Consistent with these studies, we found that while resting PMN contained only intact AnxA1 mainly localized within the cell cytosol, as a result of PMN activation the 37-kDa species was both externalized and processed into a 33-kDa fragment. The latter event is likely explained by mobilization of sub-cellular organelles that act as a store for proteases that only become membrane-expressed upon translocation during cell activation. Interestingly, analysis of the conditioned medium revealed both intact and clipped forms of the protein to be present irrespective of the treatment group, indicating the inherent ability for AnxA1 to be cleaved at the level of the cell membrane once mobilized in this microenvironment. Together these results indicate that a protease, localized to the membrane fraction of the PMN, may be responsible for the cleavage of the intact AnxA1 to form the characteristic 37/33-kDa doublet.
To track down the protease(s), we developed a cleavage assay where a double-tagged recombinant AnxA1 was used as substrate. With this tool we were able to recapitulate the cleavage of AnxA1 by the PMN, demonstrating that this occurs in presence of the membrane and not the cytosolic fraction. PMN contain an array of bactericidal proteins within their cytoplasmic organelles, including the serine proteases HNE and PR3. We restricted our attention to these proteases, because of the clear-cut actions of pan-inhibitors whereby only PMSF produced remarkable inhibition of the cleavage activity in the MEM fractions. In addition, when we analyzed by MALDI-TOF the products obtained from the cleavage assay, we found three peptide fragments corresponding to cleavage sites occuring at Ala11, Val22, VAl36. Cleavage of these small aliphatic residues is consistent with PR3 site of action (21, 22); in addition, loss of the N terminus at Val36 in bronchoalveolar lavage fluid of cystic fibrosis patients has previously been reported (7). As the clipped fragments that we have examined remain attached to the c-Myc epitope, we could detect smaller species that might be successively cleaved and lost from the N terminus. It is possible therefore that in the context of AnxA1 catabolism, and in the microenvironment of an adherent PMN, fragments corresponding to residues 1–11 and 1–22 are more unstable than the longer 1–36 one, as loss of the first 36 amino acids is consistent with the formation of the 33-kDa N terminus-cleaved fragment. It is interesting to note that loss of residues 1–22 might correspond to the ~43-kDa MF-AnxA1 fragment detected in some of our assays.
Descriptive work has reported HNE ability to cleave AnxA1 in vitro (9, 23) and possibly also in vivo (6, 7, 24). Indeed, addition of HNE to MF-AnxA1 generated the expected cleavage products. However, here we also tested the hypothesis that PR3 could be operative in the context of PMN activation, given that a pool of this serine protease is present in the plasma membrane as well as in the membrane of secretory vesicles (10), the earliest and most easily mobilized organelles upon PMN activation (14).
Addition of recombinant PR3 or membrane fraction of cells stably transfected with PR3 caused AnxA1 cleavage with a pattern similar to that observed in PMN. This held true also in vivo when MF-AnxA1 was transfected in HMC1 cells expressing a constitutively active or cleavage-inactive mutant PR3. Here we observed a complete loss of the N terminus tag in cells overexpressing PR3 and no effects in cells expressing the inactive mutant. HNE-transfected HMC1 cells did not display MF-AnxA1 degradation, possibly because of a different localization of the two proteins. This conclusion is reminiscent of our previous study with p21, where a similar distinction between in vitro and in vivo effects of HNE and PR3 on p21 degradation could be unveiled (16). In addition, our results were consistent with the experiments where enzyme inhibitors were added: as mentioned, the pan-serine protease inhibitor PMSF blocked the cleavage activity of PMN MEM fraction (as well as of commercial preparations of HNE and PR3). In contrast, eglin C was not active on the endogenous enzymatic activity whereas, in line with published data (25), it could inhibit the effect of HNE, but not human PR3.
Finally, the conclusion that PR3 might be the most important, though perhaps not exclusive (26), catabolic player for AnxA1 in the context of PMN activation, reached by a combination of molecular, biochemical, and enzymatic approaches, was corroborated by the confocal analyses of resting and activated PMN. In line with the described distribution of PR3 (27), a diffuse presence of anti-PR3 immunostaining was observed in partially permeabilized PMN, attained with streptolysin-O treatment protocol (17). The degree of overlay between the AnxA1 and PR3 immunostaining was remarkable and, again, in line with the discrete yet multiple pools of this anti-inflammatory protein in this leukocyte (5, 8, 20, 28, 29) it was, if possible, even higher after PMN stimulation with TNF-α. The absence of such degree of co-localization between AnxA1 and HNE was strident and not affected by the PMN activation protocol. Indeed, elastase abounds in the azurophilic granules that are thought to be required only when the PMN reaches the site of infection, i.e. after extravasation and the ensuing full activation (14). In addition, HNE is much more soluble than PR3, which tends to be sequestered at the cell membrane, hence secreted in the inflammatory milieu.
A high proportion of PMN expressing membrane PR3 has been described in patients with anti-neutrophil cytosolic protein-associated vasculitis as well as in those affected by rheumatoid arthritis (27, 30). These data suggest that membrane PR3 could have a pro-inflammatory effect and be considered a risk factor for developing chronic inflammatory disease (31). The data presented here indicate a novel cellular event underlying the pro-inflammatory phenotype of PR3, that is, removal of the endogenous counter-regulatory inhibitory properties of AnxA1. Analyses in PMN taken from patients suffering vasculitis would provide support to this hypothesis. Experimental data in gut inflammation have described marked AnxA1 secretion and cleavage associated with PMN tissue infiltration (32).
To conclude, we interpret these data by saying that PR3 is predominantly responsible for the AnxA1 cleavage activity in PMN membrane fraction; once the PMN is activated, the substrate (AnxA1) translocates to the membrane (8), becoming available for the enzyme (PR3). Clearly, this novel biochemical mechanism would act as a brake in the inhibitory/homeostatic functions of AnxA1 and must be visualized in the temporal and spatial context of the activated PMN. Future studies will address whether a phosphorylation event is required in the PMN for AnxA1 exportation and subsequent cleavage, as reported in other cells (33-35). Collectively, these findings may direct the design of inhibitors aimed at neutralizing the cleavage activity of PR3 toward endogenous AnxA1 and hence suitable for development as novel anti-inflammatory therapeutics.
Supplementary Material
Footnotes
This work was supported in part by the Research Advisory Board of Barts and the London (Ph.D. studentship to L. V.), the British Heart Foundation (Grant 06/153/22042), the Medical Research Council (to F. D. A.), the Arthritis Research Campaign UK (to M. P.), and Wellcome Trust (to R. J. F.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and supplemental references and experimental procedures.
The abbreviations used are: AnxA1, annexin 1; HNE, human recombinant neutrophil elastase; PMN, polymorphonuclear; PR3, proteinase 3; TNF, tumor necrosis factor; HEK, human embryonic kidney; WL, whole lysate; CYT, cytosolic; MEM, membrane; IP, immunoprecipitate or immunoprecipitation; PMSF, phenylmethylsulfonyl fluoride; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; HMC1, human mast cell clone 1.
REFERENCES
- 1.Gerke V, Moss SE. Physiol. Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 2.Perretti M, Croxtall JD, Wheller SK, Goulding NJ, Hannon R, Flower RJ. Nat. Med. 1996;2:1259–1262. doi: 10.1038/nm1196-1259. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee BE, Yona S, Rosignoli G, Young RE, Nourshargh S, Flower RJ, Perretti M. J. Leukocyte Biol. 2005;78:639–646. doi: 10.1189/jlb.0405206. [DOI] [PubMed] [Google Scholar]
- 4.Lim LH, Solito E, Russo-Marie F, Flower RJ, Perretti M. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14535–14539. doi: 10.1073/pnas.95.24.14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perretti M, Flower RJ. J. Leukocyte Biol. 2004;76:25–29. doi: 10.1189/jlb.1103552. [DOI] [PubMed] [Google Scholar]
- 6.Smith SF, Tetley TD, Guz A, Flower RJ. Environ. Health Perspect. 1990;85:135–144. doi: 10.1289/ehp.85-1568329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsao FH, Meyer KC, Chen X, Rosenthal NS, Hu J. Am. J. Respir. Cell Mol. Biol. 1998;18:120–128. doi: 10.1165/ajrcmb.18.1.2808. [DOI] [PubMed] [Google Scholar]
- 8.Perretti M, Christian H, Wheller SK, Aiello I, Mugridge KG, Morris JF, Flower RJ, Goulding NJ. Cell Biol. Int. 2000;24:163–174. doi: 10.1006/cbir.1999.0468. [DOI] [PubMed] [Google Scholar]
- 9.Rescher U, Goebeler V, Wilbers A, Gerke V. Biochim. Biophys. Acta. 2006;1763:1320–1324. doi: 10.1016/j.bbamcr.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 10.Witko-Sarsat V, Cramer EM, Hieblot C, Guichard J, Nusbaum P, Lopez S, Lesavre P, Halbwachs-Mecarelli L. Blood. 1999;94:2487–2496. [PubMed] [Google Scholar]
- 11.Bories D, Raynal MC, Solomon DH, Darzynkiewicz Z, Cayre YE. Cell. 1989;59:959–968. doi: 10.1016/0092-8674(89)90752-6. [DOI] [PubMed] [Google Scholar]
- 12.Dublet B, Ruello A, Pederzoli M, Hajjar E, Courbebaisse M, Canteloup S, Reuter N, Witko-Sarsat V. J. Biol. Chem. 2005;280:30242–30253. doi: 10.1074/jbc.M414609200. [DOI] [PubMed] [Google Scholar]
- 13.van der Geld YM, Limburg PC, Kallenberg CG. J. Leukocyte Biol. 2001;69:177–190. [PubMed] [Google Scholar]
- 14.Borregaard N, Cowland JB. Blood. 1997;89:3503–3521. [PubMed] [Google Scholar]
- 15.Witko-Sarsat V, Rieu P, Descamps-Latscha B, Lesavre P, Halbwachs-Mecarelli L. Lab. Investig. 2000;80:617–653. doi: 10.1038/labinvest.3780067. [DOI] [PubMed] [Google Scholar]
- 16.Witko-Sarsat V, Canteloup S, Durant S, Desdouets C, Chabernaud R, Lemarchand P, Descamps-Latscha B. J. Biol. Chem. 2002;277:47338–47347. doi: 10.1074/jbc.M202789200. [DOI] [PubMed] [Google Scholar]
- 17.Pederzoli M, Kantari C, Gausson V, Moriceau S, Witko-Sarsat V. J. Immunol. 2005;174:6381–6390. doi: 10.4049/jimmunol.174.10.6381. [DOI] [PubMed] [Google Scholar]
- 18.Flower RJ, Rothwell NJ. Trends Pharmacol Sci. 1994;15:71–76. doi: 10.1016/0165-6147(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 19.Peers SH, Smillie F, Elderfield AJ, Flower RJ. Br. J. Pharmacol. 1993;108:66–72. doi: 10.1111/j.1476-5381.1993.tb13441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lominadze G, Powell DW, Luerman GC, Link AJ, Ward RA, McLeish KR. Mol. Cell. Proteomics. 2005;4:1503–1521. doi: 10.1074/mcp.M500143-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Hajjar E, Korkmaz B, Gauthier F, Brandsdal BO, Witko-Sarsat V, Reuter N. J. Med. Chem. 2006;49:1248–1260. doi: 10.1021/jm051018t. [DOI] [PubMed] [Google Scholar]
- 22.Korkmaz B, Hajjar E, Kalupov T, Reuter N, Brillard-Bourdet M, Moreau T, Juliano L, Gauthier F. J. Biol. Chem. 2007;282:1989–1997. doi: 10.1074/jbc.M608700200. [DOI] [PubMed] [Google Scholar]
- 23.Huang KS, McGray P, Mattaliano RJ, Burne C, Chow EP, Sinclair LK, Pepinsky RB. J. Biol. Chem. 1987;262:7639–7645. [PubMed] [Google Scholar]
- 24.Vishwanatha JK, Davis RG, Rubinstein I, Floreani A. Clin. Cancer Res. 1998;4:2559–2564. [PubMed] [Google Scholar]
- 25.Wiesner O, Litwiller RD, Hummel AM, Viss MA, McDonald CJ, Jenne DE, Fass DN, Specks U. FEBS Lett. 2005;579:5305–5312. doi: 10.1016/j.febslet.2005.08.056. [DOI] [PubMed] [Google Scholar]
- 26.Movitz C, Sjolin C, Dahlgren C. Biochim. Biophys. Acta. 1999;1416:101–108. doi: 10.1016/s0005-2736(98)00212-0. [DOI] [PubMed] [Google Scholar]
- 27.Witko-Sarsat V, Lesavre P, Lopez S, Bessou G, Hieblot C, Prum B, Noel LH, Guillevin L, Ravaud P, Sermet-Gaudelus I, Timsit J, Grunfeld JP, Halbwachs-Mecarelli L. J. Am. Soc. Nephrol. 1999;10:1224–1233. doi: 10.1681/ASN.V1061224. [DOI] [PubMed] [Google Scholar]
- 28.Francis JW, Balazovich KJ, Smolen JE, Margolis DI, Boxer LA. J. Clin. Investig. 1992;90:537–544. doi: 10.1172/JCI115892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosales JL, Ernst JD. J. Immunol. 1997;159:6195–6202. [PubMed] [Google Scholar]
- 30.Schreiber A, Busjahn A, Luft FC, Kettritz R. J. Am. Soc. Nephrol. 2003;14:68–75. doi: 10.1097/01.asn.0000040751.83734.d1. [DOI] [PubMed] [Google Scholar]
- 31.Harper L, Savage CO. J. Pathol. 2000;190:349–359. doi: 10.1002/(SICI)1096-9896(200002)190:3<349::AID-PATH524>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 32.Vergnolle N, Coméra C, Buéno L. Eur. J. Biochem. 1995;232:603–610. doi: 10.1111/j.1432-1033.1995.tb20850.x. [DOI] [PubMed] [Google Scholar]
- 33.Solito E, Christian HC, Festa M, Mulla A, Tierney T, Flower RJ, Buckingham JC. FASEB J. 2006;20:1498–1500. doi: 10.1096/fj.05-5319fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulla A, Christian HC, Solito E, Mendoza N, Morris JF, Buckingham JC. Clin. Endocrinol. (Oxf.) 2004;60:107–119. doi: 10.1111/j.1365-2265.2004.01936.x. [DOI] [PubMed] [Google Scholar]
- 35.Dorovkov MV, Ryazanov AG. J. Biol. Chem. 2004;279:50643–50646. doi: 10.1074/jbc.C400441200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.