Abstract
In humans, cystathionine β-synthase (CBS) is a hemeprotein, which catalyzes a pyridoxal phosphate (PLP)-dependent condensation reaction. Changes in the heme environment are communicated to the active site, which is ~20 Å away. In this study, we have examined the role of H67 and R266, which are in the second coordination sphere of the heme ligands, H65 and C52 respectively, in modulating the heme's electronic properties and in transmitting information between the heme and active sites. While the H67A mutation is comparable to wild-type CBS, interesting differences are revealed by mutations at the R266 site. The pathogenic mutant, R266K, is moderately PLP-responsive while the R266M mutation shows dramatic differences in the ferrous state. The electrostatic interaction between C52 and R266 is critical for stabilizing the ferrous heme and its disruption leads to the facile formation of a 424 nm (C-424) absorbing ferrous species, which is inactive, compared to the active 449 nm ferrous species for wild-type CBS. Resonance Raman studies on the R266M mutant reveal that the kinetics of C52 rebinding after Fe-CO photolysis are comparable to that of wild-type CBS. EXAFS studies on C-424 CBS are consistent with the presence of two axial N/O low Z scatters with only one being a rigid unit of a histidine residue while the other could be a solvent molecule, an oxygen atom from the peptide backbone or a side chain nitrogen. The redox potential for the heme in full-length CBS is −350 ± 4 mV and is substantially lower than the value of −287 ± 2 mV determined for truncated CBS. A redox-regulated ligand change has the potential to serve as an allosteric on/off switch in human CBS and the second sphere ligand, R266, plays an important role in this transition.
Introduction
Cystathionine β-synthase (CBS) plays a key role in mammalian sulfur metabolism by catalyzing the condensation of serine and homocysteine to give cystathionine [1, 2]. CBS belongs to the fold type II family of the pyridoxal 5′-phosphate (PLP)-dependent enzymes. However, in addition to PLP, needed to catalyze a canonical β-replacement reaction, mammalian CBS harbors a second cofactor, a b type heme with unusual electronic properties [3, 4] and whose function remains to be elucidated. Mutations in CBS cause homocystinuria, an inborn error of metabolism that affects four major organ systems including the cardiovascular, ocular, skeletal, and central nervous system [5]. More than 130 pathogenic mutations have been reported in CBS, and are dispersed across the coding sequence [6].
Recombinant human CBS is isolated as a mixture of homooligomers with a subunit molecular mass of 63 kDa. The oligomeric state ranges from a dimer to a 16-mer [7, 8]. CBS is a modular protein in which the N-terminal stretch of ~70 amino acids constitutes the heme binding domain. The heme domain is followed by a core domain in which PLP is covalently linked via a Schiff base in the active site and leads into the C-terminal regulatory domain to which the allosteric effector, S-adenosylmethionine (AdoMet) binds [9]. AdoMet binding results in an ~2 to 3-fold increase in the specific activity of the purified enzyme [10]. While yeast and protozoan CBSs are devoid of heme [11, 12], the latter cofactor is predicted, based on the presence of the N-terminal heme domain extension and the presence of conserved heme ligands, to be present in a range of eukaryotes [4]. C-terminal truncation, which results in deletion of the regulatory domain, is accompanied by a change in the oligomeric state from mixed oligomers to the dimer [10, 13]. The truncated enzyme is highly active exhibiting an ~4-fold higher kcat and a 2- to 3-fold higher Vmax than the full-length enzyme but is unresponsive to further activation by AdoMet [13-15]. These observations indicate that the C-terminal domain exerts intrasteric inhibition, which is relieved upon binding of AdoMet [14].
The heme iron in human CBS is six-coordinate in both the ferric and ferrous states and exhibits unusual electronic properties [4]. Ferric heme is inert towards all ligands that have been tested except mercuric chloride, which coordinates to the cysteinate ligand in both the ferric and ferrous states (4). In contrast, ferrous heme binds gaseous molecules like CO and NO [16, 17] in addition to cyanide and isonitriles [16, 18]. Binding of exogenous ligands to the heme is correlated with loss of CBS activity suggesting a regulatory role for the heme [16, 17, 19]. The reaction of ferrous CBS with oxygen leads directly to the formation of ferric CBS with concomitant formation of superoxide without detectable formation of an intermediate species, consistent with an outer sphere electron transfer to oxygen [20].
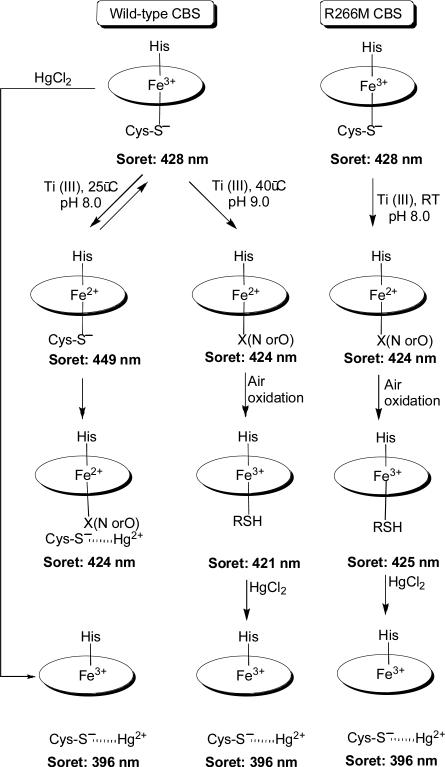
The heme in CBS has been proposed to be a redox sensor based on the ~1.7-fold decrease in enzyme activity associated with conversion of the ferric CBS to the ferrous state under in vitro assay conditions [21, 22]. Upon reduction, the Soret maximum of CBS undergoes a red shift from 428 nm associated with the ferric state, to 449 nm in the ferrous state. More recently, it has been demonstrated that the activity of the 449 nm-containing ferrous CBS is similar to that of the 428 nm ferric form, and the decrease in activity reported previously is associated with a slow time-dependent conversion of the 449 nm species to a 424 nm species (hereafter referred to as the C-424 species for the CBS form absorbing at 424 nm) [23]. Formation of the C-424 species is favored under nonphysiological conditions, i.e., by alkaline pH and high temperatures, and its formation appears to be irreversible [24]. Thus, the C-424 species is formed by heating ferrous CBS (at 40°C or higher at pH 9.0) or by reducing ferric CBS under similar conditions. In either case, the C-424 product is inactive and in addition to displaying a Soret maximum at 424 nm has α and β bands at 560 and 530 nm respectively [24]. Various spectroscopic studies have suggested that the C-424 species is ligated by two neutral ligands [24]. Together, this new information on C-424 suggests that the reduced activity associated with CBS under in vitro assay conditions (pH 8.6 and 37°C for 30 min), could be due to partial conversion of the 449 nm ferrous species to C-424. The reported reversibility of this phenomenon by ferricyanide [21] would be explained by conversion of the 449 nm ferrous CBS to the ferric state at the start of the assay.
Changes in the heme environment are transmitted to the PLP site as evidenced by a change in the chemical shift and line width of the 31P NMR signal associated with the phosphorus atom in PLP in the ferric versus ferrous states [25]. Furthermore, as described above, binding of exogenous ligands to ferrous heme is associated with loss of CBS activity [16, 17, 19], providing additional evidence for communication between the two cofactor-binding sites. Based on the crystal structure of truncated CBS lacking the AdoMet-binding regulatory domain [22, 26], we have postulated that one route for communication between the heme and PLP domains may involve the heme ligand C52, and the second sphere ligand, R266, which is at one end of α-helix 8. The other end of this helix projects into the PLP binding site [27]. Alternatively, the signal transduction between the heme and PLP domains could involve ligand switching as seen in the CO-sensing transcription factor, CooA [28] and transcriptional regulator RcoM-2 [29]. In CBS, the heme ligand, H65, is the first in a tandem repeat of three histidines. In principle, switching between histidines could induce a conformational change that is subsequently transmitted to the active site. Based on the crystal structure, H67 appears to be better positioned than H66 to serve as an alternate heme ligand in the event of ligand switching [22, 26].
Interestingly, R266 is the locus of a couple of pathogenic missense mutations, R266K and R266G. The R266K (c.797G→A) mutant was first identified in the Norwegian population [30] and its prevalence amongst Norwegian homocystinuric patients is quite high at ~35%. Patients harboring this mutation are characterized as being pyridoxine-responsive, i.e., they show clinical improvement when administered therapeutic doses of the PLP precursor, pyridoxine. The second pathogenic mutant, R266G (c.796A→G), was identified in a Japanese patient [31] who exhibited a severe homocystinuric phenotype and was unresponsive to pyridoxine supplementation.
In this study, we have tested the hypothesis that the second sphere ligands, R266 and/or H67, are involved in relaying information about the heme environment to the active site of CBS. To achieve this goal we have characterized various CBS mutants including R266K, R266G, R266M, and H67A. Two of the mutations used in this study, R266K and R266G mimic patient mutations. The other two mutations, R266M and H67A were generated either to introduce an approximately isosteric substitution at position 266 but lacking the positive charge associated with the native side chain (R266M) or to assess the influence of a second sphere residue, which interacts with distal heme ligand, H65. We have also tested the relevance of the C-424 species to redox regulation of human CBS activity by studying second sphere mutants that promote its facile formation under milder conditions. Extended X-ray absorption fine structure spectroscopy (EXAFS) studies on the C-424 species further confirm its ligation by two neutral ligands. While our results suggest that H67 is not involved in communication between the heme and PLP domains, retention of the salt-bridge between C52 and R266 is required for CBS activity. This study also reveals the importance of second sphere ligands for modulating the spectroscopic and redox properties of the heme in full-length CBS.
Methods and Materials
Chemicals
[14C]-Serine (158 mCi/mmol) was purchased from Amersham. L-Serine, ampicillin, pyridoxal 5’-phosphate, pyridoxine, AdoMet, and DL-homocysteine were purchased from Sigma.
Preparation of CBS Expression Constructs
The CBS mutants were generated using the Quick Change Kit from Stratagene. The template for wild-type human CBS was the pGEX4T1/hCBS plasmid in which CBS is fused in frame with glutathionine S-transferase (GST) [32]. The set of primers that were used are as follows:
H67A forward, 5′- CCGAGTCCCCACATCACGCCACTGCCCCGGC-3′;
R266K forward, 5′-ATCACGGGCATTGCCAAAAAGCTGAAGGAGAAG-3′;
R266G forward, 5′-ATCACGGGCATTGCCGGCAAGCTGAAGGAGAAG-3′;
R266M forward, 5′-CGGGCATTGCCATGAAGCTGAAGG-3′;
The reverse primers were complementary to the forward primer sequence.
The R266M mutant was also expressed in the truncated form (CBS△C143) by introducing the mutation using the expression vector pGEXCBSN as template [32]. The mutations were confirmed by nucleotide sequencing at the Genomics Core facility at the University of Nebraska Lincoln.
Expression and Purification of CBS
E. coli cells containing the desired CBS-expression plasmid, were grown and harvested as described previously [21]. The cell pellet was then resuspended in 200 ml of 50 mM Tris, pH 8.0, containing one protease inhibitor cocktail tablet (Roche) and 35 mg of lysozyme and stirred for 1.5 h at 4°C. The cells were lysed by sonication and the supernatant was obtained by centrifuging at 12,000 × g for 40 min. The supernatant was loaded onto a 5 × 4 cm glutathione sepharose column pre-equilibrated with phosphate buffered saline (PBS). The column was washed with 700 ml of PBS and the GST-fused CBS was eluted using 10 mM glutathione in 50 mM Tris, pH 8.0. This was followed by thrombin digestion at 4°C to remove the GST tag following which the protein was loaded onto a 5 × 7.5 cm MonoQ column (131 POROS HQ/H,) equilibrated with buffer A (50 mM Tris-HCl, pH 8.0, 50 mM KCl). The protein was loaded at a flow-rate of 3 ml/min. The column was then washed with 100 ml of buffer A at 10 ml/min. The protein was eluted with a 500 ml gradient ranging from 50 to 450 mM KCl in 50 mM Tris-HCl, pH 8.0 at the same flow rate. The fractions of interest were pooled, concentrated and stored at –80°C.
UV-Visible Absorption Spectroscopy
Spectra of reduced CBS were recorded at room temperature in an anaerobic chamber (Vacuum Atmospheres Co., Hawthorne, CA) with an atmosphere of 95:5 nitrogen:hydrogen and <1 ppm O2. CBS samples were prepared in anaerobic 50 mM Tris buffer, pH 8.4 and reduced with 1 mM titanium citrate. Potassium ferricyanide (final concentration 1mM) was added to reoxidize the samples, when needed.
The electronic absorption spectra of CBS were also obtained in 50 mM potassium phosphate buffer, pH 7.4, which had been degassed for 15 min using nitrogen. An anaerobic solution of CBS was injected into the anaerobic buffer and 1 mM titanium citrate was added when needed to reduce the enzyme. The spectra were monitored for 60 min at 37°C.
CO and HgCl2 Binding Experiments
For CO binding experiments, anaerobic 50 mM Tris buffer, pH 8.4 placed in a sealed cuvette was firstly saturated with CO and then R266M CBS (2.8 μM) was added followed by addition of 1 mM titanium citrate. For the HgCl2 binding experiments, R266M was first reduced with 1 mM titanium citrate to form 424 nm. This was followed by reoxidation with 1 mM potassium ferricyanide. The reoxdized sample (425 nm) was treated with 400 μM HgCl2 (final concentration) and chelation by mercury of the cysteine ligand to heme was monitored by UV-visible spectroscopy.
Aerobic Enzyme Assay
Specific activities of proteins were determined by using the fixed-time radiolabeled assay as described previously [21]. The reaction mixture contained 50 mM Tris pH 8.6, 30 mM [14C]-serine and 30 mM DL-homocysteine, 0.25 mM PLP and 0.38 mM AdoMet. The reaction was initiated by addition of CBS and quenched following incubation for 30 min at 37°C with 200 μl of 10 % TCA. Product formation was analyzed as described [21]. The activity of CBS was also assayed under anaerobic conditions using 50 mM potassium phosphate buffer, pH 7.4. Here the reaction was performed in the chamber, quenched with 10% TCA and then samples were brought outside the chamber to assay as described previously [21].
Spectroelectrochemistry of Full-Length Wild-type CBS and the R266K Mutant
Potentiometric measurements were performed under a nitrogen atmosphere in a Belle Technology glove box with <1 ppm oxygen using a two-electrode single compartment spectroelectrochemical cell containing a gold foil working electrode and a silver chloride reference electrode and the data were analyzed as described previously [20].
Resonance Raman Spectroscopy of R266M CBS
Studies on the R266M mutant were conducted using a variant in which the regulatory domain had been truncated (R266M-△C-143). Our previous studies on CBS have also been conducted on the truncated variant containing the wild-type sequence [33].
To study the ligand rebinding kinetics the samples were prepared as follows. A Sephadex G25M PD10 desalting column (5 × 1.5 cm) equilibrated with the desired buffer, was used to desalt CBS and the protein was collected and concentrated at 4°C using a 30 kDa molecular weight cut-off Centricon centrifuge filter. The buffers used were 100 mM phosphate at pH 7.6, 100 mM Tris at pH 8.5 or 50 mM glycine-NaOH at pH 10.5. The enzyme solution was prepared by purging truncated ferric CBS (~30 μM in heme) with argon gas and then adding an excess of sodium dithionite. To prepare the CO bound form of CBS, the ferrous CBS solution was purged with CO for 2 h, followed by addition of an excess of sodium dithionite. Time-resolved resonance Raman Spectroscopy and measurement of relative Raman cross sections of the ν4 band (hCBS-5c scale factor = 0.4, ferrous hCBS scale factor = 0.8) were done using a 527 nm pump pulse to photolyse the CO and a 435 nm probe pulse according to the method described previously [33, 34].
EXAFS Study of the Ferrous C-424 State of Wild-type CBS
Concentrated CBS samples (~1 mM) in 50 mM Tris buffer, pH 9.0, with 30% glycerol were loaded in an anaerobic chamber into Lucite samples cells of 70 μl with a removable fourth wall of Kapton tape and flash frozen in liquid nitrogen. The samples were reduced with 1 mM titanium citrate in an anaerobic chamber prior to loading in the sample cells. For XAS collection, the samples were kept at 10 K-12 K in an Oxford liquid helium flow cryostat. Fe K-edge XAS was collected at SSRL on beamline 9-3 at 3 GeV with currents between 80 and 100 mA with a Si[220] double crystal monochromator and a Rh-coated mirror upstream of the monochromator with a 10 KeV energy cutoff to reject harmonics. Data were collected in the fluorescence mode using a high-count-rate Canberra 30-element Ge array detector with maximum count rates held below 120 kHz per channel. Fe K-edge spectra were collected using 5 eV steps in the pre-edge (6900–7100 eV), 0.2 eV steps from 7100-7140 and 0.05 Å−1 steps in the EXAFS region to a k of 18 Å−1. Data collection was integrated for 1s in the pre-edge, 1.5 s in edge regions, and 1.5 to 25 s (k2-weighted of 2.6 to 8 Å−1 and k3−weighted 8 to 18 Å−1) in the EXAFS region for a total scan time of roughly 45 min. A 6 μ path length Mn filter was placed in front of the detector to reduce the elastic scatter peak and Soller slits were used to reduce the Mn Kβ fluorescence. Each sample was scanned multiple times to give a total of 109 counts total in the windowed Fe Kα peak at k = 18 Å−1 giving a total of 5 scans for one sample, 7 scans for a duplicate sample and 8 scans for a third sample. Comparison of the first and last scans for each sample showed no detectable change in the XANES spectrum, demonstrating that there was no x-ray induced radiation damage.
X-ray energies were calibrated by simultaneous measurement of a Fe foil absorption spectrum and assigning the first inflection point of the absorption edge to 7111.2 eV. Data from each detector channel were inspected for glitches before inclusion in the final average. Pre-edge subtraction and normalization to McMaster values were carried out using the program MBACK [35]. The data was converted to k space via the equation:
where E0 = 7112 eV. Data was fit, using Excurv98, to the EXAFS equation:
where Ns is the number of scatterers with atom type s at a distance of Ras. Aas(k) is the effective back-scattering amplitude function, σ2as is the Debye-Waller factor and φas(k) is the phase shift of the photoelectron wave traveling between the potentials of the absorbing and scattering atoms. Fourier transforms of the EXAFS data were calculated using k3-weighted data from k = 2.0-18 Å−1. Multiple scattering paths where calculated in the Excurv98 program. Raw data were k3-weighted and the individual fits were preformed allowing Ras of the first shell atoms to vary. Distances of the outer shell carbons in both the heme and histidine residues were set and linked to the distance of the first shell nitrogen as a rigid body, while Debye-Waller factors were held constant at values suggested by Bunker et al [36].
Results
Purification of CBS Mutants
In this study, we have purified and characterized the patient mutations R266K and R266G and a third site-specific mutation at this position, R266M. In addition, a residue neighboring the distal H65 heme ligand, H67 (Fig. 1) was mutated to alanine. We were unable to characterize the R266G patient mutation due to the instability of the protein and extensive degradation of this mutant protein was observed during thrombin treatment. To circumvent this problem, the R266G mutant was partially characterized with the GST tag. However, the GST-R266G fusion protein did not exhibit any detectable activity unlike the GST-tagged wild-type CBS, which exhibits some, albeit reduced activity [37]. All other mutants used in this study were purified to ~95% purity (not shown).
Figure 1.
Structure of the heme in CBS and location of other second coordination sphere residues. The ligands to heme are shown in stick representation. The second coordination sphere residues that were mutated in this study are shown in ball and stick representation.
Kinetic Properties of R266K, R266M and H67A Mutants of CBS
When assayed in the absence of exogenous PLP and AdoMet, R266K CBS exhibited a specific activity of 115 ± 7 μmol h−1 mg protein−1 in the standard assay (Table 1). Addition of PLP resulted in a modest increase in the specific activity to 154 ± 19 μmol h−1 mg protein−1. The specific activity of the R266K mutant increased to 343 ± 21 μmol h−1 mg protein−1 in the presence of AdoMet, which is comparable to the value observed with wild-type enzyme. Together, PLP and AdoMet, resulted in a 3-fold increase in the Vmax of the R266K mutant protein. The R266M variant showed a significantly lower basal activity (41 ± 1.4 μmol h−1 mg protein−1). Upon addition of PLP, the specific activity increased ~2-fold to 72.0 ± 5.0 μmol h−1 mg protein−1 and the addition of AdoMet further enhanced the activity (234 ± 22 μmol h−1 mg protein−1). Thus, the R266M variant exhibited an ~6-fold increase in activity when both PLP and AdoMet were added to the reaction mixture.
Table 1.
Comparison of kinetic parameters for wild-type CBS and various second sphere heme-pocket mutants
| aSpecific Activity | wild-type | H67A | R266K | R266M |
|---|---|---|---|---|
| +PLP, +AdoMet | 316 ± 10 | 316 ± 17 | 343 ± 21 | 234 ± 22 |
| −PLP, −AdoMet | 220 ± 20 | 139 ± 12 | 115 ± 7 | 41 ± 1.4 |
| KMSer, mM | 2.0 ± 0.3 | 5.4 ± 1.6 | 4.4 ± 1.7 | 5.5 ± 0.5 |
| KMHcy, mM | 5.0 ± 0.9 | 3.4 ± 1.0 | 1.3 ± 0.3 | 1.6 ± 0.3 |
Specific activity is expressed in units of μmol h−1 mg protein−1.
The H67A mutation had a very minor effect on CBS activity. In the absence of PLP and AdoMet, the specific activity was 139 ± 12 μmol h−1 mg protein−1, which increased ~2.3 fold upon addition of PLP and AdoMet. The resulting specific activity (316 ±17 μmol h−1 mg protein−1) is indistinguishable from that obtained for wild-type CBS (Table 1). The KM values for serine were slightly elevated for all three mutants compared to wild-type CBS. In contrast, the KM for homocysteine for the H67A mutant was comparable to that for wild-type CBS and slightly lower for the R266K and R266M mutants (Table 1).
The activities of the R266 mutants in the presence of mercuric chloride and CO were compared to that of wild-type CBS. Both R266K and R266M exhibited very low activity in the ferrous-CO and the mercuric chloride-chelated state, which was similar to the behavior observed with wild-type CBS (Table 2).
Table 2.
Comparison of specific activities of wild-type CBS and the R266 mutantsa
| wild-type | R266K | R266M | |
|---|---|---|---|
| − HgCl2 | 352 ± 27 | 343 ± 21 | 234 ± 22 |
| 300 μM HgCl2 | 6.1 ± 1.6 | bND | bND |
| CO bound-CBS | 33.8 ± 4.2 | 5.2 ± 1.0 | 8.2 ± 3.5 |
Assays were performed in 50 mM Tris-HCl, pH 8.6 in the presence of PLP and AdoMet and specific activity is reported in units ofμmol h−1 mg protein−1.
Not detected.
R266M Forms the C-424 Species upon Reduction
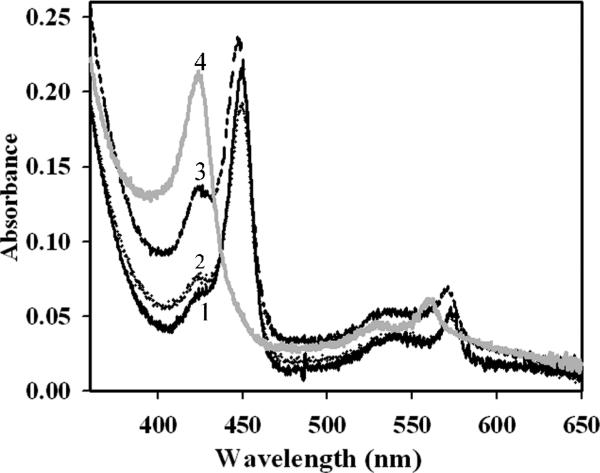
In the oxidized state, all the mutants exhibit absorption spectra with a Soret peak at 428 nm, which is similar to that of wild-type CBS (data not shown). Upon reduction of the H67A and R266K mutants, the Soret peak shifted to 449 nm and the α and β bands sharpened at 570 and 540 nm respectively, as seen with wild-type CBS (Fig. 2 and Scheme I). In both wild-type CBS and in the H67A mutant, a shoulder is observed at 424 nm, which is more prominent in the spectrum of the R266K mutant. In contrast, reduction of the R266M mutant resulted in the Soret peak shifting over to 424 nm (with α and β bands at 559 and 529 nm respectively).
Figure 2.
Comparison of the UV-visible absorption spectra of wild-type CBS and other second coordination sphere mutants in the reduced state. The spectra were recorded in 50 mM Tris-HCl, pH 8.4 at room temperature. Wild-type, 2.2 μM (solid black line, 1) and H67A, 1.5 μM (dotted line, 2) shows the reduced Soret peak at 449 nm, whereas R266K, 2.2 μM (medium dashed line, 3) shows a major Soret peak at 449 nm with a shoulder at 424 nm. Reduction of R266M CBS, 2.2 μM (solid gray line, 4) leads directly to the formation of 424 nm.
Scheme I.
To further characterize the ferrous R266M mutant, binding of CO and the thiol chelator mercuric chloride, were studied. Binding of CO to ferrous R266M results in a shift in the Soret peak from 424 nm to 421 nm (Fig. 3A). Binding of CO to wild-type CBS results in a large blue shift from 449 nm to 422 nm [17], consistent with the displacement of the thiolate ligand. Addition of mercuric chloride to wild-type ferrous CBS results in a shift in the Soret peak from 449 nm (α and β bands at 571 and 540 nm) to 424 nm (α and β bands at 559 and 529 nm), consistent with the formation of a six-coordinate low spin ferrous species in which the identity of the sixth ligand is not known [19, 38]. Addition of up to 400 μM mercuric chloride did not elicit a detectable change in the spectrum of the ferrous R266M mutant (not shown). Air oxidation or ferricyanide treatment of ferrous R226M CBS, resulted in a minor shift in the Soret peak to 425 nm and broadening of the α/β absorption band. Treatment of this species with mercuric chloride, resulted in a shift in the Soret peak to 396 nm (Fig. 3B), suggesting formation of a five-coordinate species as seen with wild-type ferric CBS [38].
Figure 3.
Electronic absorption characteristics of R266M CBS A. Comparison of the absorption spectra of 2.5 μM ferric R266M CBS (solid line) with a Soret peak at 428 nm and ferrous-CO bound R266M (dashed line) with a Soret peak at 421 nm (dashed line). B. Mercuric chloride binding to the reoxidized R266M. Ferrous R266M (2.3 μM) with a Soret peak at 424 nm (dashed line) is air-oxidized to yield a spectrum with a Soret peak at 425 nm (solid line) and broadened α/β bands at 550 nm. Binding of mercuric chloride to this reoxidized species yields a spectrum with a Soret peak at 396 nm (dotted line).
Electronic Absorption Spectroscopy and CBS Activity at Physiolgical pH
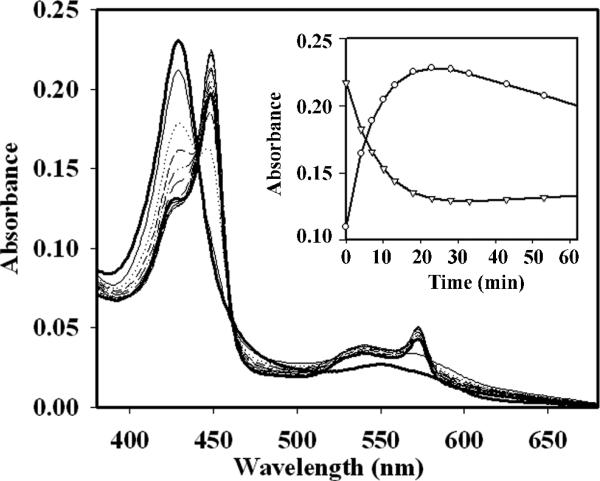
Since the pH optimum for CBS activity under in vitro assay conditions is 8.6, previous spectroscopic investigations on this enzyme have been conducted at this relatively alkaline pH. In comparison to pH 8.6, reduction of ferric CBS at pH 7.4 with titanium citrate was slow (Fig. 4), which is consistent with the redox potential of titanium citrate being higher at lower pH [39]. The gradual increase in 449 nm is accompanied by a corresponding decrease in the 428 nm absorbance (Fig. 4 inset). Next, the activity of wild-type CBS in the ferric and ferrous states was determined in 50 mM potassium phosphate, pH 7.4 at 37°C in the anaerobic chamber. Under these conditions, the activity of ferric CBS was 207 ± 12 μmol h−1 mg protein−1 and the activity of ferrous CBS was 155 ± 11 μmol h−1 mg protein−1. The modestly lower activity of ferrous CBS can be ascribed in part to the conversion of the active 449 nm-absorbing ferrous CBS to the inactive C-424 state over the 30 min time course of the assay.
Figure 4.
Spectrum of wild-type CBS (2.5 μM in 50 mM KPi, pH 7.4 at 37°C) reduced with 1 mM titanium citrate. The solid dark line with a Soret peak at 428 nm represents the spectrum of oxidized wild-type CBS. The inset shows the time-dependent changes in absorption at 449 nm (circles) and 428 nm (triangle).
Redox Potential Determination for full-length wild-type CBS and the R266K mutant
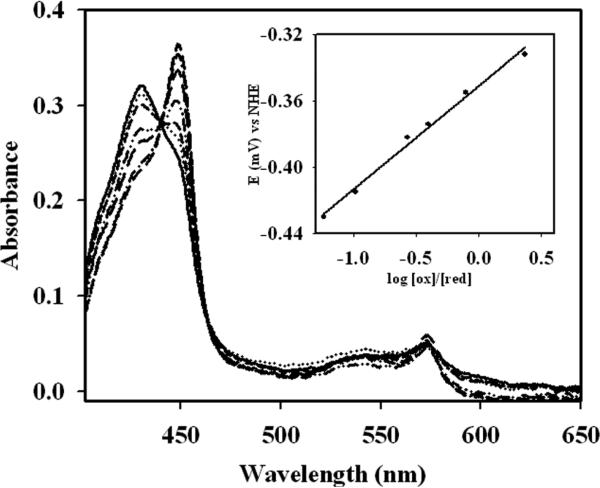
The redox potential of the heme in wild-type full-length CBS was determined to be −350 ± 4 mV (slope = 73.3 ± 7.5) in the reductive direction and −349 ± 2 mV (slope = 65.0 ± 3.8 mV) in the oxidative direction (Fig. 5). The slopes are consistent with a one-electron reduction. The presence of AdoMet (100 μM) had no effect on the redox potential and a value of −359 ± 5 mV (slope = 68.7 ± 8.1) was obtained. Mutation of the second sphere residue, R266, to lysine had no apparent effect on the heme redox potential and a value of −366 ± 8 mV was obtained. Since the R266M mutant converted irreversibly to the C-424 species upon reduction, its redox potential could not be measured.
Figure 5.
Redox potential of full-length wild-type CBS. The UV-visible spectrum of CBS was monitored at 428 nm and 449 nm for the ferric and ferrous states in wild-type human CBS. The potentiometric titration was performed in the presence of 20 μM benzyl viologen, 20 μM methyl viologen, 5 μM wild-type CBS with incremental 1 μM additions of sodium dithionite. The spectra are shown after subtraction of the dye mediator. Inset shows the redox potential of wild-type CBS.
Resonance Raman Studies on R266M CBS
Resonance Raman studies were performed on the truncated R266M mutant to evaluate the influence of the electrostatic interaction between R266 and the heme ligand, C52, on the kinetics of C52 rebinding after Fe-CO photolysis. The C52 rebinding rate depends in part upon the thiol protonation state, and an alteration to the pH dependence of C52 rebinding after Fe-CO photolysis would be evidence that the pKa of the dissociated C52 (for a thiolate in aqueous solution, the pKa is ~8.8 [40]) was changed due to the loss of the salt bridge interaction in the R266M mutation. The intensity of the ν4 band of CBS at three pH values were fitted with a first order exponential to yield the rate constants listed in Table 3. Since the intensity of the ν4 band in ferrous CBS was low under the conditions of this experiment compared to 5-coordinate CBS and for CBS-CO, the rate constant for formation of ferrous CBS could not be measured directly. Instead, the rate constant for ferrous CBS was calculated from the other difference between CBS (5c) decay and CBS-CO formation according to the following equation.
A pH dependence in the kinetics of cysteine rebinding was not observed in the R266M mutant. Rather, the rate constants were similar to the values determined previously for truncated wild-type CBS, which are also pH independent [34]. This indicates that the pKa for the dissociated C52 is shifted from its value in aqueous solution and is not simply due to salt bridge formation with R266. Other factors, such as the hydrophobic heme pocket are probably responsible for the absence of a pH dependence in the C52 rebinding kinetics.
Table 3.
Rate constants (s–1) determined by time-resolved Raman spectroscopy for the formation (or disappearance) of CBS species following photolysis of CBS-CO.
| pH | CBS-5c | CBS-CO | Fe(II)-hCBS (calc) | |
|---|---|---|---|---|
| R266Ma | 7.6 | 1.8(0.3) × 104 | 1.07(0.16) × 104 | 7.4(2.5) × 103 |
| 8.5 | 2.0(0.3) × 104 | 1.18(0.17) × 104 | 8.2(2.4) × 103 | |
| 10.5 | 2.4(0.3) × 104 | 1.33(0.12) × 104 | 1.05(0.23) × 104 | |
| WTb | 7.6 | 1.8(0.3) × 104 | 1.32(0.16) × 104 | 4.6(1.4) × 103 |
| 8.6 | 2.2(0.7) × 104 | 1.8(0.4) × 104 | 4.0(2.2) × 103 | |
| 10.5 | 2.1 (0.3) × 104 | 1.49(0.16) × 104 | 6.4(1.6) × 103 |
This work.
[34]
EXAFS Spectroscopy on the C-424 species of CBS
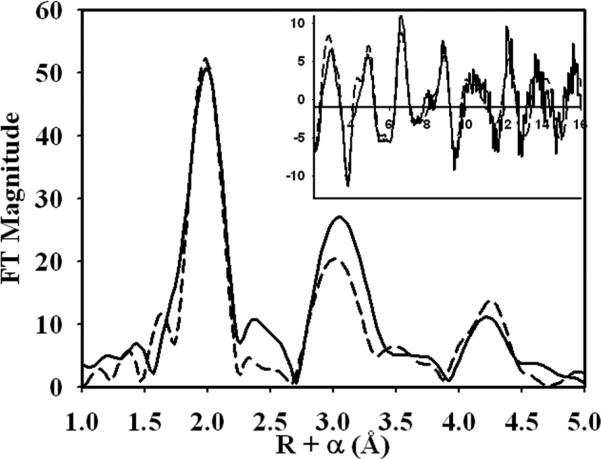
Fourier transforms (FT) of the EXAFS region of the XAS spectrum of the wild-type C-424 species show a large narrow peak at around 2.0 Å (phase shifted) suggesting a large coordination number consisting solely of ligands roughly 2.0 Å from the iron center (Fig. 6), accompanied by a large second shell peak at approximately 3.0 Å consistent with previous studies on heme compounds [41]. Fitting of the EXAFS spectra was preformed by first modeling a heme moiety using previously defined distances from crystallographic data [22], and then comparing fits with axially bound sulfur, nitrogens and histidine residues. The fit index for the fit with an axial sulfur and an axial histidine while reasonable as compared to the other fits, suffers from the drawback that the amplitude in the FT is far too intense at the 2.3 Å distance seen for Fe-S (Supplementary Fig. 2).
Figure 6.
Fourier transforms of the EXAFS spectra of C-424 state of wild-type CBS. The inset shows the EXAFS spectra of the C-424 species. In both figures, the experimental spectra are depicted by solid lines. The theoretical FT for the heme iron with an axial O/N low Z scatterer and a histidine residue is shown in broken line.
The best fit for the EXAFS spectra of C-424 CBS was obtained with two axial N/O low Z scatters with one being modeled as a rigid unit of a histidine residue (Fig. 6, inset). This fit suggests that the coordination environment of the C-424 species is one of a heme with an axial histidine ligand and another low Z scatterer without the signature multiple scattering of histidine. The alternate fit with two histidines still has an N component at 1.86 Å, which is too low for distances seen for either a heme or histidine Fe-N distance (Supplementary Table 1). When one of the nitrogens is left as a single O/N scatter as in the alternate fit, the distance of 1.86 Å while unreasonable for a Fe-histidine distance is acceptable for a solvent molecule or an oxygen atom from the peptide backbone.
Discussion
The binding of exogenous ligands to the heme in human CBS is communicated over a significant distance to the catalytic site and results in allosteric inhibition. Although traditionally viewed as a property of oligomeric proteins exhibiting cooperative behavior, allostery is now recognized to also be a property of monomeric proteins [42]. NMR spectroscopy and statistical clustering analysis approaches have been useful for identifying amino acid networks that are important for allosteric signal transmission [43, 44]. In this study, we have investigated the hypothesis that a second sphere ligand interaction plays a role in communicating changes from the heme to the PLP site in human CBS.
While the heme in the ferric state appears to be relatively inert to ligand exchange, perturbations to the heme environment by binding of exogenous ligands e.g., CO and NO to the ferrous state lead to inhibition of CBS activity [45]. Furthermore, mutation of C52 or chelation of the thiolate ligand with mercuric chloride also adversely affects CBS activity [37, 46]. A considerable distance, ~20 Å, separates the active site PLP from the heme. Based on the crystal structure [22, 47], we have proposed that communication between the heme and PLP domains could involve the R266 residue, which resides at the tip of α-helix 8 and is within hydrogen bonding distance to the axial ligand, C52. Thus disruption of this interaction between C52 and R266 would affect CBS activity. Interestingly, two patient mutations at R266, R266K and R266G, have been reported. In this study, we have examined the effects of both pathogenic mutations as well as the site-direct mutation, R266M, on the transmission of information on the heme electronic state to the PLP site.
R266K CBS has been previously reported to exhibit ~2-3 fold lower activity than the wild-type enzyme and addition of exogenous PLP did not increase its activity [48]. In our hands, a small but significant increase in the activity of R266K CBS was observed in the presence of exogenous PLP (Table 1) and may explain the pyridoxine-responsiveness in patients harboring this mutation [30]. The R266G mutant is unstable and exhibited no activity under in vitro assay conditions, properties that correlate well with the severe homocystinuric phenotype of patients harboring this mutation [31]. The R266M mutant generated in this study, like the pathogenic R266G mutation, lacks a positive charge at this position but unlike R266G CBS, is stable. The R266M mutant exhibits an ~5.5-fold lower basal activity than wild-type CBS, which increases ~2-fold upon addition of PLP. These results indicate the potential importance of the salt-bridge between C52 and R266 in modulating PLP content and therefore, CBS activity in the ferric state.
Treatment of the R266K and R266M mutants with HgCl2 inhibits enzyme activity (Table 2), suggesting that the salt bridge between C52 and R266 is not the only determinant of communication between the heme and PLP sites. However, a potential caveat of this assay for evaluating communication between the two cofactors is that HgCl2 chelates all free thiols in the protein, and the observed inhibition could result from conformational changes induced by thiol chelation at sites distal to the heme. Full-length human CBS has 11 thiols. To circumvent this limitation, we have examined the effect of CO-binding to ferrous heme in the R266K and R266M mutants. The activity of the ferrous R266K and R266M mutants decreased to near background levels in the presence of CO, similar to wild-type CBS. While these results appear to argue against R266 playing a primary role in signal transduction between the cofactor domains, a closer inspection of the spectroscopic features of the mutant proteins suggests an alternative explanation.
The R266K mutation represents a conservative substitution in which the positive charge on the side chain is preserved and is predicted to stabilize the sulfur-ligated ferrous heme by charge delocalization. In fact, the spectroscopic features of ferrous R266K are indistinguishable from that of wild-type CBS. The presence of the Soret peak with a maximum at 449 nm indicates the presence of a cysteinate ligand (Scheme I). Binding of CO to ferrous R266K is expected to disrupt the salt bridge as in wild-type CBS, a change that is propagated to the active site. In contrast, reduction of the R266M mutant, leads to the facile formation of the C-424 species, which is slowly generated from the ferrous R266K mutant and from wild-type ferrous CBS at unphysiologically high pH and high temperature [24]. Hence, R266 plays a key role in stabilizing the cysteinate-ligated ferrous state of CBS. The C-424 species formed upon reduction of the R266M mutant is inactive and while binding of CO shifts the Soret peak to 421 nm, it can have no further effect on inhibiting CBS activity.
The identity of the ligand that replaces C52 in the C-424 species is not known but is predicted to be a neutral ligand [24] and candidates include water, a protonated thiol, a backbone amide oxygen and an imidazole side chain. Treatment of the C-424 species in the R266M mutant and in wild-type CBS with HgCl2, did not elicit any changes in the heme spectrum, consistent with the absence of a thiol ligand to the heme. It has been noted previously that the crystal structure of the truncated CBS does not reveal any obvious ligand candidates within 10 Å distance of the heme. Nevertheless, H232, K267, K271 and P2 have been suggested as possible ligands in the C-424 species [49]. Our EXAFS analysis does not support the presence of a bis-histidine or thiol ligation and suggests instead that a solvent molecule, an oxygen atom from a peptide backbone or a lysine might replace the cysteinate ligand.
Exposure of C-424 CBS to air results in a very small shift in the Soret maximum to 421 nm and 425 nm in wild-type and R266M CBS respectively, and a broadening of the α,β bands consistent with conversion to ferric heme (Scheme 1). The source of the differences in the Soret band positions seen upon reoxidation of the C-424 species in R266M versus wild type CBS are not known. The reoxidized form of the enzyme like the C-424 state, also exhibits no activity. Treatment of this species with HgCl2 results in the Soret maximum shifting to 396 nm, suggesting that the heme switches back to the cysteinate ligand upon air oxidation of C-424. The absence of activity in air oxidized CBS derived from the C-424 state together with the small differences in the positions of the Soret maxima between the reoxidized enzyme (425 nm) versus ferric enzyme as isolated (428 nm), indicates that a conformational change accompanying formation of the reduced C-424 species traps the enzyme in an inactive state, which is not reversed by simply switching back to the cysteinate ligand. These results raise the possibility that an allosteric ligand or a protein partner might exist in the cell that can modulate a ligand-switch induced conformational change in CBS and play a role in regulating its activity.
However, since the inactivating ligand switching in CBS is only observed in the ferrous state, its relevance to regulation in vivo raises the obvious question of the reduction potential of the heme iron in the full-length protein. We have recently reported that the redox potential for the ferric/ferrous couple in truncated dimeric CBS is −287 ± 2 mV [50]. We have now determined the redox potential of the heme in full-length CBS, which is significantly lower at −350 ± 4 mV. Other heme centers that have a thiolate ligand have redox potentials in a similar range and include cytochrome P450 (−360 to −170 mV) [51], nitric oxide synthase (−347 to −239 mV) and Rhodospirillum rubrum CooA (−320 mV) [52] For a low-spin heme with cysteinate ligand, a low reduction potential is expected. However, the 60 mV difference in the potential between the truncated dimer and full-length CBS is surprising and suggests the possibility that the regulatory domain of CBS interacts with the heme domain, thus modulating its redox potential. Another possibility is that the change in the oligomeric state of the enzyme from a mixture of higher-order oligomers in full-length CBS to a dimer in the truncated form, influences the heme environment. The redox potential of the R266K mutant is similar to that of wild-type CBS and consistent with the preservation of the salt bridge between C52 and K266, which would be important in delocalizing the charge and stabilizing a sulfur-ligated ferrous heme. It is not known if a redox partner in the cell can reduce the heme and therefore make CBS susceptible to heme-based allosteric redox inhibition.
In CBS, H67 is a conserved residue in the heme-binding domain that is close to the heme ligand, H65. This led us to consider that H67 may be involved in ligand switching during conversion of ferric CBS to the ferrous state and thus provide an alternative pathway for signal transduction. However, since the kinetic and spectroscopic properties of H67A are very similar to those of wild type CBS, the hypothesis that H67 is involved in the ligand switching is not supported. Thus, a second sphere ligand interaction with the distal heme ligand, H65, has minimal effects as compared to the prominent effects of the R266M mutation, which perturbs the proximal C52 ligand.
We have determined the fate of 449 nm ferrous species under more physiological pH conditions (i.e., 50 mM potassium phosphate, pH 7.4 at 37°C) and find that the activities of ferric and ferrous CBS (determined over a 30 min time period) are similar. In contrast, earlier studies [21, 49] reported an ~1.7-1.9-fold lower activity for ferrous versus ferric CBS (in 100 mM Tris buffer, pH 8.6), which can be explained by the higher 424:449 nm ratio of CBS, which in turn is explained by the higher pH of the assay conditions. The instability of the 449 nm-absorbing ferrous CBS and its conversion to the C-424 state may be associated with the unfavorable electronics of having a thiolate ligand to ferrous heme. In other hemeproteins in which thiolate ligands are seen, reduction is accompanied by ligand switching in which the thiolate group is replaced by a neutral ligand [29, 53]. The cellular relevance of ferrous CBS is not known. Its formation is a prerequisite for binding CO or NO, gaseous signaling molecules which CBS has been shown to bind under in vitro conditions, leading to its inhibition [45, 54].
In conclusion, a redox-regulated ligand change rather than the oxidation state of the heme iron per se has the potential to serve as an allosteric on/off switch for CBS. The 449 nm-reduced state of CBS is stabilized by the interaction between the second sphere ligand R266, and the heme ligand, C52. In the absence of this stabilizing interaction, heme reduction leads directly to the formation of the C-424 species, as seen with the R266M mutant (Scheme 1). Although the C-424 CBS species can be reoxidized, it does not regain activity. The identity of the ligand that replaces C52 in the C-424 state is not known although EXAFS studies rule out a neutral thiol ligand. Insights into the ligand switch and the conformational change that it appears to elicit, will have to await structural characterization of CBS in this state.
Supplementary Material
Abbreviations Used
- CBS
cystathionine β-sythase
- PLP
pyridoxal 5′-phosphate
- AdoMet
S-adenosylmethionine
- EXAFS
extended X-ray absorption fine structure
- PBS
phosphate-buffered saline
Abbreviations
- CBS
cystathionine β-synthase
- PLP
pyridoxal 5′-phosphate
- TCA
trichloroacetic acid
- XAS
X-ray absorption spectroscopy
- GST
glutathione S-transferase
- FT
fourier transform
Footnotes
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (HL58984 to R.B., GM33576 to T.G.S. GM061068 to D.B. and GM38047 to J. P-H).
References
- 1.Miles EW, Kraus JP. J Biol Chem. 2004;279:29871–29874. doi: 10.1074/jbc.R400005200. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee R, Zou CG. Arch Biochem Biophys. 2005;433:144–156. doi: 10.1016/j.abb.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 3.Kery V, Bukovska G, Kraus JP. J. Biol. Chem. 1994;269:25283–25288. [PubMed] [Google Scholar]
- 4.Singh S, Madzelan P, Banerjee R. Nat. Prod. Rep. 2007;24:631–639. doi: 10.1039/b604182p. [DOI] [PubMed] [Google Scholar]
- 5.Mudd SH, Levy HL, Kraus JP. In: The Online Metabolic and Molecular Bases of Inherited Disease. Scriver CR, Beaudet AL, Sly WS, Valle D, Vogelstein B, Kinzler KW, Childs B, editors. The McGraw-Hill Companies; New York: 2001. pp. 2007–2056. [Google Scholar]
- 6.Kraus JP, Janosik M, Kozich V, Mandell R, Shih V, Sperandeo MP, Sebastio G, de Franchis R, Andria G, Kluijtmans LA, Blom H, Boers GH, Gordon RB, Kamoun P, Tsai MY, Kruger WD, Koch HG, Ohura T, Gaustadnes M. Hum Mutat. 1999;13:362–375. doi: 10.1002/(SICI)1098-1004(1999)13:5<362::AID-HUMU4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Frank N, Kery V, Maclean KN, Kraus JP. Biochemistry. 2006;45:11021–11029. doi: 10.1021/bi060737m. [DOI] [PubMed] [Google Scholar]
- 8.Sen S, Banerjee R. Biochemistry. 2007;46:4110–4116. doi: 10.1021/bi602617f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taoka S, Widjaja L, Banerjee R. Biochemistry. 1999;38:13155–13161. doi: 10.1021/bi990865t. [DOI] [PubMed] [Google Scholar]
- 11.Jhee KH, McPhie P, Miles EW. J Biol Chem. 2000;275:11541–11544. doi: 10.1074/jbc.c000056200. [DOI] [PubMed] [Google Scholar]
- 12.Nozaki T, Shigeta Y, Saito-Nakano Y, Imada M, Kruger WD. J Biol Chem. 2001;276:6516–6523. doi: 10.1074/jbc.M009774200. [DOI] [PubMed] [Google Scholar]
- 13.Kery V, Poneleit L, Kraus JP. Arch Biochem Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 14.Evande R, Boers GHJ, Blom HJ, Banerjee R. Biochemistry. 2002;41:11832–11837. doi: 10.1021/bi026248d. [DOI] [PubMed] [Google Scholar]
- 15.Shan X, Dunbrack RL, Jr., Christopher SA, Kruger WD. Hum Mol Genet. 2001;10:635–643. doi: 10.1093/hmg/10.6.635. [DOI] [PubMed] [Google Scholar]
- 16.Taoka S, West M, Banerjee R. Biochemistry. 1999;38:2738–2744. doi: 10.1021/bi9826052. [DOI] [PubMed] [Google Scholar]
- 17.Taoka S, Banerjee R. J. Inorg. Bioc. 2001;87:245–251. doi: 10.1016/s0162-0134(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 18.Vadon-Le Goff S, Delaforge M, Boucher JL, Janosik M, Kraus JP, Mansuy D. Biochem Biophys Res Commun. 2001;283:487–492. doi: 10.1006/bbrc.2001.4807. [DOI] [PubMed] [Google Scholar]
- 19.Taoka S, Green EL, Loehr TM, Banerjee R. J. Inorg. Bioc. 2001;87:253–259. doi: 10.1016/s0162-0134(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 20.Carballal S, Madzelan P, Zinola CF, Grana M, Radi R, Banerjee R, Alvarez B. Biochemistry. 2008;47:3194–3201. doi: 10.1021/bi700912k. [DOI] [PubMed] [Google Scholar]
- 21.Taoka S, Ohja S, Shan X, Kruger WD, Banerjee R. J Biol Chem. 1998;273:25179–25184. doi: 10.1074/jbc.273.39.25179. [DOI] [PubMed] [Google Scholar]
- 22.Taoka S, Lepore BW, Kabil O, Ojha S, Ringe D, Banerjee R. Biochemistry. 2002;41:10454–10461. doi: 10.1021/bi026052d. [DOI] [PubMed] [Google Scholar]
- 23.Cherney MM, Pazicni S, Frank N, Marvin KA, Kraus JP, Burstyn JN. Biochemistry. 2007;46:13199–13210. doi: 10.1021/bi701159y. [DOI] [PubMed] [Google Scholar]
- 24.Pazicni S, Cherney MM, Lukat-Rodgers GS, Oliveriusova J, Rodgers KR, Kraus JP, Burstyn JN. Biochemistry. 2005;44:16785–16795. doi: 10.1021/bi051305z. [DOI] [PubMed] [Google Scholar]
- 25.Kabil Ö, Taoka S, LoBrutto R, Shoemaker R, Banerjee R. J. Biol. Chem. 2001;276:19350–19355. doi: 10.1074/jbc.M100029200. [DOI] [PubMed] [Google Scholar]
- 26.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. EMBO J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evande R, Ojha S, Banerjee R. Arch Biochem Biophys. 2004;427:188–196. doi: 10.1016/j.abb.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Aono S, Ohkubo K, Matsuo T, Nakajima H. J. Biol. Chem. 1998;273:25757–25764. doi: 10.1074/jbc.273.40.25757. [DOI] [PubMed] [Google Scholar]
- 29.Marvin KA, Kerby RL, Youn H, Roberts GP, Burstyn JN. Biochemistry. 2008;47:9016–9028. doi: 10.1021/bi800486x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim CE, Gallagher PM, Guttormsen AB, Refsum H, Ueland PM, Ose L, Folling I, Whitehead AS, Tsai MY, Kruger WD. Hum Mol Genet. 1997;6:2213–2221. doi: 10.1093/hmg/6.13.2213. [DOI] [PubMed] [Google Scholar]
- 31.Katsushima F, Oliveriusova J, Sakamoto O, Ohura T, Kondo Y, Iinuma K, Kraus E, Stouracova R, Kraus JP. Mol Genet Metab. 2006;87:323–328. doi: 10.1016/j.ymgme.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Shan X, Kruger WD. Nat Genet. 1998;19:91–93. doi: 10.1038/ng0598-91. [DOI] [PubMed] [Google Scholar]
- 33.Green EL, Taoka S, Banerjee R, Loehr TM. Biochemistry. 2001;40:459–463. doi: 10.1021/bi0010874. [DOI] [PubMed] [Google Scholar]
- 34.Puranik M, Weeks CL, Lahaye D, Kabil O, Taoka S, Nielsen SB, Groves JT, Banerjee R, Spiro TG. J Biol Chem. 2006;281:13433–13438. doi: 10.1074/jbc.M600246200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weng TC, Waldo GS, Penner-Hahn JE. J Synchrotron Radiat. 2005;12:506–510. doi: 10.1107/S0909049504034193. [DOI] [PubMed] [Google Scholar]
- 36.Dimakis N BG. Physical review B. 2002;65:201103–201106. [Google Scholar]
- 37.Ojha S, Wu J, LoBrutto R, Banerjee R. Biochemistry. 2002;41:4649–4654. doi: 10.1021/bi011827o. [DOI] [PubMed] [Google Scholar]
- 38.Ojha S, Hwang J, Kabil O, Penner-Hahn JE, Banerjee R. Biochemistry. 2000;39:10542–10547. doi: 10.1021/bi000831h. [DOI] [PubMed] [Google Scholar]
- 39.Zehnder AJB, Wuhrman K. Science. 1976;194:1165–1166. doi: 10.1126/science.793008. [DOI] [PubMed] [Google Scholar]
- 40.Kortemme T, Creighton TE. J Mol Biol. 1995;253:799–812. doi: 10.1006/jmbi.1995.0592. [DOI] [PubMed] [Google Scholar]
- 41.Paola D'Angelo DL, della Longa Stefano, Benfatto Maurizio, Louis Hazemann Jean, Feis Alessandro, Smulevich Giulietta, Ilari Andrea, Bonamore Alessandra, Boffi Alberto. Biophys J. 2004;86:3882–3892. doi: 10.1529/biophysj.103.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodey NM, Benkovic SJ. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 43.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Proc Natl Acad Sci U S A. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suel GM, Lockless SW, Wall MA, Ranganathan R. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 45.Taoka S, Banerjee R. J Inorg Biochem. 2001;87:245–251. doi: 10.1016/s0162-0134(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 46.Taoka S, Green EL, Loehr TM, Banerjee R. J Inorg Biochem. 2001;87:253–259. doi: 10.1016/s0162-0134(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 47.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. EMBO J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Wang L, Fazlieva R, Kruger WD. Hum Mutat. 2006;27:474–482. doi: 10.1002/humu.20320. [DOI] [PubMed] [Google Scholar]
- 49.Cherney MM, Pazicni S, Frank N, Marvin KA, Kraus JP, Burstyn JN. Biochemistry. 2007;46:13199–13210. doi: 10.1021/bi701159y. [DOI] [PubMed] [Google Scholar]
- 50.Carballal S, Madzelan P, Zinola CF, Grana M, Radi R, Banerjee R, Alvarez B. Biochemistry. 2008;47:3194–3201. doi: 10.1021/bi700912k. [DOI] [PubMed] [Google Scholar]
- 51.Huang YY, Hara T, Sligar S, Coon MJ, Kimura T. Biochemistry. 1986;25:1390–1394. doi: 10.1021/bi00354a030. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima H, Honma Y, Tawara T, Kato T, Park SY, Miyatake H, Shiro Y, Aono S. J Biol Chem. 2001;276:7055–7061. doi: 10.1074/jbc.M003972200. [DOI] [PubMed] [Google Scholar]
- 53.Reynolds MF, Parks RB, Burstyn JN, Shelver D, Thorsteinsson MV, Kerby RL, Roberts GP, Vogel KM, Spiro TG. Biochemistry. 2000;39:388–396. doi: 10.1021/bi991378g. [DOI] [PubMed] [Google Scholar]
- 54.Taoka S, West M, Banerjee R. Biochemistry. 1999;38:7406. doi: 10.1021/bi995077i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.