Abstract
Mixed Lineage Kinase 3 (MLK3) is a mitogen-activated protein kinase kinase kinase (MAP3K) that activates multiple mitogen activated protein kinase (MAPK) pathways in response to growth factors, stresses and the pro-inflammatory cytokine, tumor necrosis factor (TNF). MLK3 is required for optimal activation of stress activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) signaling by TNF, however, the mechanism by which MLK3 is recruited and activated by the TNF receptor remains poorly understood. Here we report that both TNF and Interleukin-1β (IL-1β) stimulation rapidly activate MLK3 kinase activity. We observed that TNF stimulates an interaction between MLK3 and TNF receptor associated factor (TRAF) 2 and IL-1β stimulates an interaction between MLK3 and TRAF6. RNA interference (RNAi) of traf2 or traf6 dramatically impairs MLK3 activation by TNF indicating that TRAF2 and TRAF6 are critically required for MLK3 activation. We show that TNF also stimulates ubiquitination of MLK3 and MLK3 can be conjugated with lysine 48 (K48)- and lysine 63 (K63)-linked polyubiquitin chains. Our results suggest that K48-linked ubiquitination directs MLK3 for proteosomal degradation while K63-linked ubiquitination is important for MLK3 kinase activity. These results reveal a novel mechanism for MLK3 activation by the proinflammatory cytokines TNF and IL-1β.
Keywords: Mixed Lineage Kinase 3 (MLK3), mitogen activated protein kinase (MAPK), tumor necrosis factor (TNF), interleukin 1 β (IL-1β), TNF receptor associated factor (TRAF), ubiquitination
1. Introduction
MAPK signaling pathways function to transduce extracellular signals into a wide range of cellular responses. MAPK signaling pathways consist of a protein kinase cascade, where an activated MAP3K phosphorylates and activates a MAPK kinase (MAP2K), which in turn, phosphorylates and activates a MAPK [1]. MAPKs translocate to the nucleus to activate transcription factors and regulate genes involved cellular processes such as proliferation, survival, differentiation and apoptosis [1]. The three most well characterized MAPKs are extracellular signal-regulated kinase (ERK), SAPK/JNK and p38. ERK is predominantly activated by growth factors, whereas SAPK/JNK and p38 MAPK are primarily activated by environmental stresses and pro-inflammatory cytokines [1, 2].
MLK3 is a member of a family of MAP3Ks. Upon activation, MLKs directly phosphorylate and activate the MAP2Ks, MKK4/SEK1 and MKK3/6 [3, 4]. Activated MKK4/SEK1 and MKK3/6 directly phosphorylate and activate SAPK/JNK and p38, respectively [1]. MLK3 is activated by epidermal growth factor (EGF), T cell receptor costimulation, TNF, sorbitol, ceramide and nerve growth factor deprivation in neuronal cells [5-9]. RNAi studies showed that MLK3 promotes microtubule instability, is required for cell proliferation in colon epithelial and lung fibroblast cells, and is required for activation of ERK and SAPK/JNK by TNF and EGF [8, 10]. MLK3 has also recently been found to limit Rho GTPase activity [11].
Activation of MLK3 occurs through autophosphorylation on amino acid residues threonine 277 (Thr277) and serine 281 (Ser281) within the MLK3 kinase domain [9]. Autoinhibition of MLK3 kinase activity is mediated by an interaction between its SH3 domain and proline 495 (Pro495) in the C-terminus [12]. MLK3 is required for optimal TNF activation of the SAPK/JNK pathway, however, the proteins required for MLK3 activation in TNF signaling have not been identified [8, 13].
In inflammatory responses, TNF binds the TNF receptor (TNFR) to activate several signal transduction pathways including SAPK/JNK [14]. TRAFs are adaptor proteins recruited to the cytoplasmic tails of receptors in the TNFR and IL-1 receptor/Toll-like receptor (IL-1R/TLR) superfamilies [15]. Different TRAFs have different targets to facilitate the activation of multiple downstream effectors. The signaling pathways activated by TRAFs result in many different responses including cell survival, proliferation, apoptosis and differentiation [15]. TRAF proteins contain a TRAF domain at the C-terminus, which is important for interactions with upstream regulators and for mediating TRAF homo or hetero-oligomerization [16]. All TRAFs, except for TRAF1, also contain an N-terminal RING finger and several zinc finger motifs, which function in downstream signaling [17].
Multiple studies have demonstrated a requirement for TRAF2 in TNF-stimulated SAPK/JNK activation and TRAF2 has also been shown to activate MAP3Ks such as MEKK1 and apoptosis-stimulated kinase 1 (ASK1) [18-23]. TRAF6 binds receptors in the IL-1R/TLR superfamily and cells deficient in TRAF6 fail to respond to lipopolysaccharide (LPS), IL-1 and IL-8 [24, 25]. IL-1- and IL-8-induced TRAF6 activation leads to AP-1 activation via the SAPK/JNK pathway [26]. TRAF6 is ubiquitinated with K63-linked polyubiquitin chains via the same E2 complex (Ubc13/Uev1A) as TRAF2 resulting in activation of the MAP3K, TGFβ activated kinase 1 (TAK1), and the IκB kinase complex [26, 27]. The TRAF6 RING domain is required for the activation of TAK1 or SAPK/JNK by TRAF6 [28]. Collectively these data indicate that TRAF2 and TRAF6 play critical roles in the activation of MAP3Ks in cytokine signaling.
In this study, we wished to determine if the activation of MLK3 by TNF and IL-1β occurs through a TRAF-dependent mechanism.We show that MLK3 interacts with TRAF2 and TRAF6 in response to TNF and IL-1β, respectively.Depletion of TRAF2 or TRAF6 with RNAi impaired MLK3 activation by TNF indicating that TRAF2 and TRAF6 are required for TNF-stimulated MLK3 activation. We show that MLK3 ubiquitination occurs rapidly in response to TNF and IL-1-β and that MLK3 can be conjugated with K63- and K48-linked polyubiquitin chains. In addition, our results suggest that K48-linked ubiquitination directs MLK3 for proteosomal degradation while K63-linked ubiquitination is important for MLK3 kinase activity.
2. Materials and methods
2.1 Cell culture and reagents
Human embryonic kidney 293 (HEK293), human epithelial ovarian cancer (SKOV3) and human colon cancer cells (HT29) were obtained from the American Type Culture Collection. Cells were grown in a humidified atmosphere with 5% CO2 at 37°C. HEK293, SKOV3 and HT29 cells were cultured in Dulbecco’s Modified Eagle’s Medium (Cellgro) supplemented with 10% fetal bovine serum (Hyclone). All culture media was supplemented with 25 βg/ml streptomycin and 25 I.U. penicillin (Cellgro).
Human recombinant TNF and IL-1β were from Biosource. TNF and IL-1β were diluted in fresh culture media to 50 ng/ml and 20 ng/ml respectively for treatment of cells. MG132 (Peptide International) was used at a final concentration in the media of 10 μM.
2.2. Expression plasmids
Expression constructs for glutathione-S-transferase (GST) fusion proteins of human TRAF2 used in this study were pEBG-GST-WT-TRAF2, pEBG-GST-ΔTRAF-TRAF2, pEBG-GST-ΔRING-TRAF2. Other constructs used in the study were pRK5-FLAG-MLK3 and pCMV-Myc-TRAF6. pRK5-HA-WT-Ubiquitin, pRK5-HA-K48-Ubiquitin, pRK5-HA-K48R-Ubiquitin and pRK5-HA-K63-Ubiquitin constructs were provided by Dr. T. Dawson [29]. pRK5-HA-K63R-Ubiquitin was prepared using site-directed mutagenesis with the QuikChange II Site Directed Mutagenesis Kit (Stratagene). The DNA sequence of the mutant was verified by sequencing at the University of Michigan DNA sequencing core facility.
2.3. Transfections
Transient transfections of DNA plasmids were preformed using Lipofectamine (Invitrogen) reagent according to manufacturer’s protocol. Transfections of siRNA oligos were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. For transfections, cells were seeded in either 6 or 10 cm dishes and transfected when cells reached 60 to 70% confluency.
2.4. Immunoblot analysis
Cell extracts were separated by 15% SDS-PAGE. Proteins were transferred to polyvinylidene fluoride membrane and Western blotting was performed with the following antibodies from Santa Cruz Biotechnologies: MLK3 (sc-536), GST (sc-459), β-Actin (sc-47778), TRAF2 (sc-876), TRAF6 (sc-7221), HA (sc-57592) and Myc (sc-40). Western blotting was also performed with Ubiquitin antibody (550944) from BD Pharmingen and FLAG antibody (200472-21) from Stratagene. The phosphorylation-specific MLK3 antibody (28115) was from Cell Signaling Technology. The secondary antibodies were Immun-Star goat anti-mouse horseradish peroxidase conjugate (170-5047) and Immun-Star goat anti-rabbit horseradish peroxidase conjugate (170-5046) from Bio-Rad. Quantitation of signal intensity of Western blot analysis was performed using Image J software (National Institutes of Health).
2.5. Immunoprecipitation
Immunoprecipitation of endogenous or overexpressed proteins in HEK293 or SKOV3 cells was performed as previously described [21]. N-ethylmaleimide was added to the cell lysis buffer to a final concentration of 200 μM to inhibit deubiquitin enzymes where necessary.
2.6. RNAi
TRAF2 and TRAF6 siRNA oligos used in this study were as previously described [30]. Nonspecific siRNA oligos were from Dharmacon.
3. Results
3.1. TNF and IL-1β activate MLK3 kinase activity
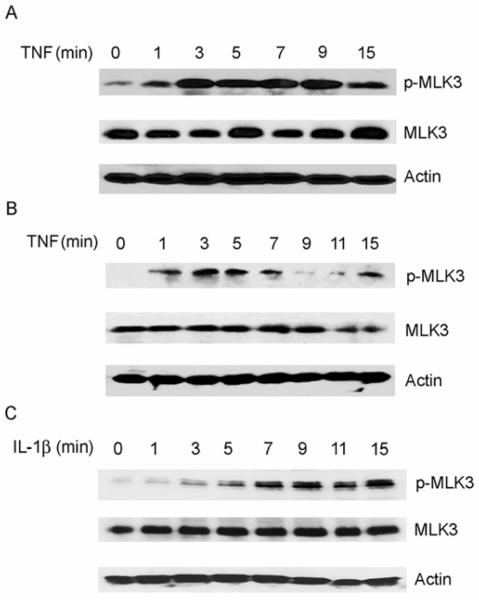
TNF activates the TNFR1 receptor and this leads to activation of SAPK/JNK and p38 MAPK signaling pathways [31]. To analyze the timing of the activation of endogenous MLK3 by TNF, SKOV3 and HT29 cells were treated with 50 ng/ml of human recombinant TNF for time periods ranging from 0 to 15 min. Cellular extracts were analyzed by Western blotting with an antibody that specifically recognizes phosphorylated residues (Thr 277/Ser 281) in the activation loop of the MLK3 kinase domain. Phosphorylation of these residues is required for MLK3 kinase activity. We observed that SKOV3 cells had a detectable basal level of endogenous active MLK3 that rapidly increased in response to TNF treatment after 1-3 min (Fig. 1A). Similarily, in HT29 cells, a rapid increase in the level of active MLK3 was observed after TNF stimulation for 1 min (Fig. 1B). Thus, endogenous MLK3 is activated in response to TNF treatment. These results are consistent with previous findings that TNF stimulates MLK3 activity in Jurkat T cells [6]. Interestingly, we observed that IL-1β treatment of SKOV3 cells also caused an increase in MLK3 kinase activity that occurred at 3 min, gradually increased through 7 min, and remained at the highest level between 7 and 15 min (Fig.1C). These data indicate that treatment of colon or ovarian cancer cells with TNF or IL-1β stimulates a rapid increase in endogenous MLK3 kinase activity.
Fig. 1. TNF and IL-1β stimulate endogenous MLK3 kinase activity.
A) SKOV3 cells were treated with 50 ng/ml TNF for indicated time periods and whole cell lysates were subjected to SDS-PAGE and Western blotting with a phospho-specific-MLK3 (p-MLK3), β-Actin and total MLK3 antibodies. B) HT-29 cells were treated with 50 ng/ml TNF for the indicated time periods and cell extracts were analyzed by Western blotting as described inA). C) SKOV3 cells were treated with IL-1β for the indicated time periods and cell extracts were analyzed by Western blotting as described in A).
3.2. MLK3 interacts with TRAF2 and TRAF6
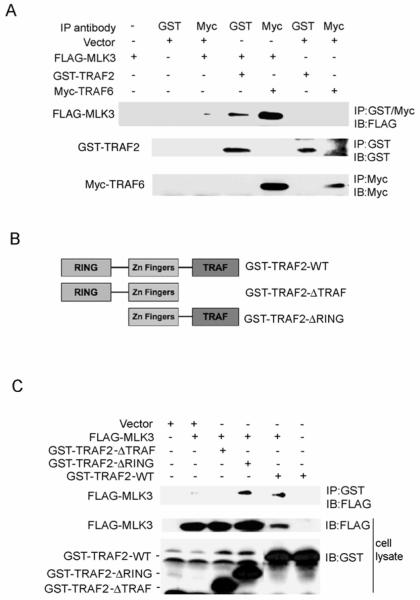
Previous reports indicated that both TRAF2 and MLK3 are required for TNF stimulated-SAPK/JNK activation [8, 13, 19]. We hypothesized that TRAF2 or TRAF6 could function to recruit MLK3 to the TNFR upon TNF stimulation, thereby facilitating MLK3 activation. To investigate this possibility, we first tested if TRAF2 or TRAF6 associated with MLK3 in cells. HEK293 cells were transiently co-transfected with FLAG-MLK3 and either GST-TRAF2 or Myc-TRAF6. GST-TRAF2 and Myc-TRAF6 were immunoprecipitated with anti-GST and anti-Myc antibodies, respectively. Immunoprecipitated complexes were subjected to SDS-PAGE and Western blot analysis with anti-FLAG antibody to detect co-immunoprecipitated FLAG-MLK3. We observed that FLAG-MLK3 specifically co-immunoprecipitated with GST-TRAF2 and Myc-TRAF6 (Fig. 2A). These results demonstrate that MLK3 associates with TRAF2 and with TRAF6 in cells.
Fig. 2. MLK3 interacts with TRAF2 and TRAF6.
A) HEK293 cells were transiently transfected with FLAG-MLK3 and either GST-TRAF2 or MYC-TRAF6. GST-TRAF2 and Myc-TRAF6 were immunoprecipitated from cell lysates with anti-GST and anti-Myc antibodies, respectively, and immunoprecipitates were subjected to SDS-PAGE and Western blotting with anti-FLAG antibody. Cell extracts were also immunoblotted with anti-GST, anti-Myc and anti-FLAG antibodies. B) Diagram indicating the TRAF2 truncation mutants analyzed in experiments in C). C) HEK293 cells were transiently transfected with FLAG-MLK3 and GST-TRAF2 truncation mutant expression constructs. GST-TRAF2 truncation mutants were immunoprecipitated from cell lysates and immunoprecipitates were subjected to SDS-PAGE and Western blotting with anti-FLAG antibody. Whole cell lysates were also analyzed by Western blotting with anti-GST and anti-FLAG antibodies.
The TRAF domain of the TRAF proteins is required for interactions with downstream targets such as NIK, GCK, and MEKK1 [21, 32, 33]. Next, we wished to determine the region of TRAF2 that is necessary for the interaction with MLK3. HEK293 cells were transiently co-transfected with FLAG-MLK3 and GST-TRAF2 (wildtype and truncation mutants) into HEK293 cells. The TRAF2 truncation mutants consisted of TRAF2 lacking the C-terminal TRAF domain (GST-TRAF2-ΔTRAF) and TRAF2 lacking the N-terminal RING domain (GST-TRAF2-ΔRING) (Fig. 2B). FLAG-MLK3 co-immunoprecipitated with GST-TRAF2-ΔRING and with wildtype GST-TRAF2 (GST-TRAF2-WT) but failed to co-immunoprecipitate with GST-TRAF2-ΔTRAF (Fig. 2C). These data indicate that the TRAF domain of TRAF2 is required for the TRAF2-MLK3 interaction.
3.3. TNF and IL-1β induce association of MLK3 and TRAFs
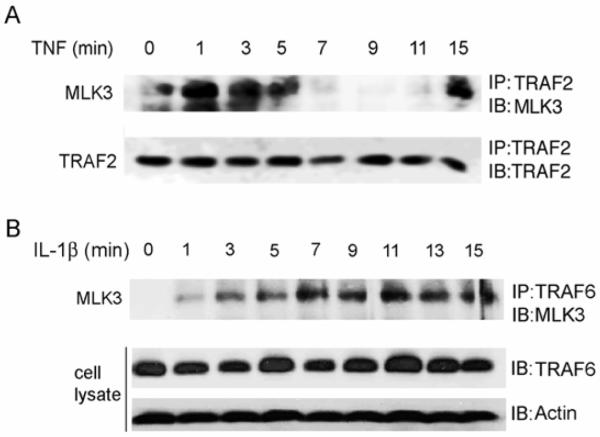
To determine if TNF stimulates an association between endogenous MLK3 and TRAF2, we treated SKOV3 cells with TNF and immunoprecipitated TRAF2 from cell lysates with anti-TRAF2 antibody. In the absence of TNF, little endogenous MLK3 co-immunoprecipitated with TRAF2 (Fig. 3A). However, when cells were stimulated for 1-5 min with TNF, a substantial increase in the association between MLK3 and TRAF2 was observed (Fig. 3A). This association was biphasic and decreased substantially at 7-11 min and then reappeared at 15 min. Interestingly, the MLK3-TRAF2 interaction occurred before the TNF-stimulated increase in MLK3 kinase activity (refer to Fig. 1A and Fig. 1B). Thus, it is plausible that recruitment of MLK3 to the TNFR1 could occur through its association with TRAF2, which subsequently facilitates MLK3 activation.
Fig. 3. TNF and IL-1β treatment of cells induces an association between MLK3 and TRAF proteins.
A) SKOV3 cells were treated with 50 ng/ml of TNF for indicated time periods. TRAF2 was immunoprecipitated from cell lysates with anti-TRAF2 antibody and immunoprecipitates were subjected to SDS-PAGE and Western blotting with anti-MLK3 and anti-TRAF2 antibodies. B) SKOV3 cells were treated with 20 ng/ml of IL-1β for indicated time periods. TRAF6 was immunoprecipitated from lysates with anti-TRAF6 antibody and immunoprecipitates were subjected to SDS-PAGE and Western blotting with anti-MLK3 antibody. Whole cell lysates were also analyzed by Western blotting with anti-TRAF6 and anti-Actin antibodies.
IL-1β binds receptors in the IL-1R/Toll-like receptor (TLR) superfamily. TRAF6 is required for IL-1β signaling and lack of TRAF6 leads to a marked decrease in signaling through these receptors [24]. We postulated that MLK3 is recruited to the IL-1β receptor through its interaction with TRAF6. In order to determine whether IL-1β stimulates an endogenous association between MLK3 and TRAF6 we treated SKOV3 cells with IL-1β and immunoprecipitated TRAF6 from lysates with anti-TRAF6 antibody. In the absence of IL-1β treatment, no endogenous MLK3 co-immunoprecipitated with TRAF6 (Fig. 3B). However, when cells were stimulated with IL-1β, a rapid increase in the association of MLK3 with TRAF6 was observed starting at 1 min (Fig. 3B). The IL-1β-stimulated association between MLK3 and TRAF6 occurred before the observed increase in IL-1β stimulated MLK3 kinase activity (see Fig. 1C) suggesting that the interaction with TRAF6 is required for the activation of MLK3 by IL-1β. Possibly, MLK3 is recruited to the IL-1β receptor via its interaction with TRAF6 and this interaction facilitates MLK3 activation.
3.4. Silencing either traf2 or traf6 impairs TNF activation of MLK3
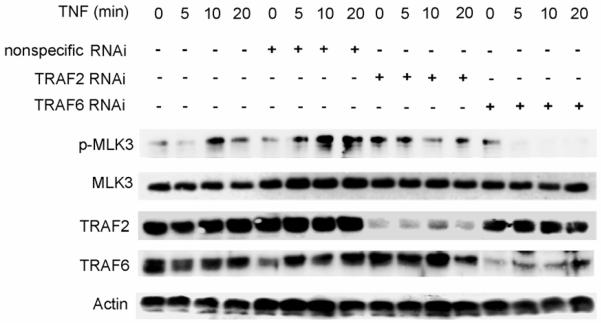
Next, we wished to determine if TRAF2 or TRAF6 is required for activation of MLK3 by TNF. Cells were depleted of TRAF2 or TRAF6 with small interfering RNA (siRNA) and TNF-stimulated MLK3 activity was analyzed. In SKOV3 cells that were untransfected or transfected with the non-specific siRNA oligos, we observed that TNF stimulation resulted in a substantial increase in MLK3 kinase activity after 10 min (Fig. 4). This TNF-stimulated increase in MLK3 kinase activity was significantly reduced in SKOV3 cells lacking TRAF2 and dramatically reduced in cells lacking TRAF6 (Fig. 4). These results suggest that TRAF6, and to a lesser extent TRAF2, is required for activation of MLK3 by TNF.
Fig. 4. Silencing traf2 or traf6 impairs TNF activation of MLK3.
HEK 293 cells were transfected with non-specific, TRAF2 or TRAF6 siRNA oligos.Cells were then treated with vehicle or TNF for the indicated time periods. Cell lysates were prepared and subjected to SDS PAGE and Western blotting with anti-phospho-MLK3, anti-MLK3, anti-Actin, anti-TRAF2, and anti-TRAF6 antibodies.
3.5. TNF stimulates ubiquitination of MLK3
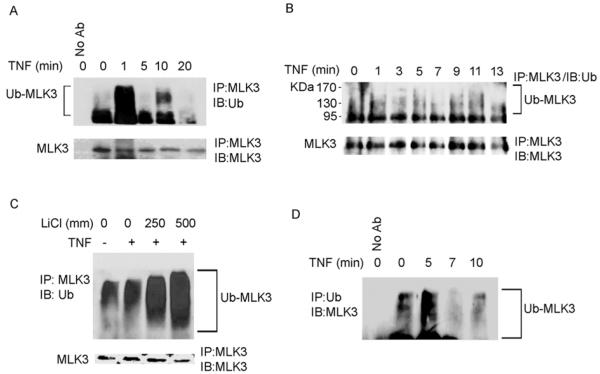
Most TRAF proteins contain a RING domain that facilitates ubiquitination of targets with K63-linked polyubiquitin chains [34]. K63 linked polyubiquitination targets proteins for fates other than proteosomal degradation [26]. Interestingly, activation of the SAPK/JNK pathway by TNF is dependent on TRAF2 K63-linked ubiquitination [35]. We postulated that MLK3 could be ubiquitinated in response to TNF stimulation and this may also be an important component of the mechanism of regulation of MLK3 kinase activity. To determine if TNF stimulation induces MLK3 ubiquitination, HEK293 and SKOV3 cells were treated with TNF and cell lysates were subjected to immunoprecipitation with anti-MLK3 antibody. Immunoprecipitates were probed with anti-ubiquitin antibody to detect ubiquitinated MLK3. Fig. 5A shows that ubiquitinated MLK3 was detected within 1 min after TNF stimulation in HEK293 cells. In SKOV3 cells, TNF treatment for 1 min stimulated an increase in the level of ubiquitinated MLK3 (Fig. 5B). When HEK293 cells were treated with TNF for 5 min and MLK3 immunoprecipitates were washed with a lysis buffer containing 250 mM or 500 mM LiCl to disrupt non-specific interactions, a high level of ubiquitinated MLK3 was still observed (Fig. 5C). Also, when HEK293 cells were treated with TNF and ubiquitinated proteins were immunoprecipitated from cell lysates with anti-ubiquitin antibody, and then immunoblotted with anti-FLAG antibody to detect ubiquitinated FLAG-MLK3, a similar ladder of bands was observed indicating that MLK3 is directly conjugated with ubiquitin (Fig. 5D). To our knowledge, this is the first report that MLK3 is ubiquitinated in response to TNF stimulation.
Fig. 5. TNF stimulates ubiquitination of MLK3.
(A) HEK293 and (B) SKOV3 cells were treated with 50 ng/ml TNF for time periods indicated. MLK3 was immunoprecipitated from cell lysates with anti-MLK3 antibody (or no antibody control) and immunoprecipitates were subjected to SDS-PAGE and immunoblotted with anti-Ubiquitin (Ub) and anti-MLK3 antibodies. (C) HEK293 cells were treated with 50 ng/ml of TNF for 5 min and MLK3 was immunoprecipitated from cell lysates with anti-MLK3 antibody. Immunoprecipitates were washed with different concentrations of LiCl and subjected to SDS-PAGE and Western blotting with anti-Ubiquitin and anti-MLK3 antibodies. (D) HEK293 cells were treated with 50 ng/ml of TNF for time periods as indicated. Ubiquitinated proteins were immunoprecipitated with anti-Ubiquitin antibody. Control immunoprecipitation was performed with no antibody. Immunoprecipitates were subjected to SDS-PAGE and Western blotting with anti-MLK3 antibody.
3.6 MLK3 is conjugated with K63- and K48- linked ubiquitin chains
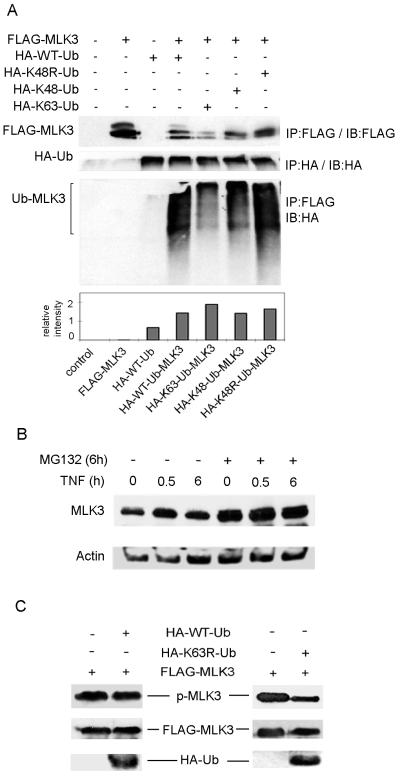
The lysine residues at positions 48 and 63 of ubiquitin provide sites for isopeptide linkages of other ubiquitin molecules. K48-linked polyubiquitin chains target proteins for proteosomal degradation while K63-linked ubiquitin chains serve non-proteolytic functions such as localization and activation [27]. To determine the linkage between the polyubiquitin chains that are conjugated on MLK3, HA-tagged Ubiquitin with K48 mutated to arginine (HA-K48R), with all lysines except K48 mutated to arginine (HA-K48), and all lysines except K63 mutated to arginine (HA-K63), were transiently overexpressed together with FLAG-MLK3 in HEK293 cells (Fig 6A). Cell lysates were prepared and FLAG-MLK3 was immunoprecipitated with anti-FLAG antibody.FLAG immunoprecipitates were immunoblotted with anti-HA antibody to detect ubiquitinated FLAG-MLK3. Ubiquitinated FLAG-MLK3 was observed in the immunoprecipitates from cells expressing HA-wildtype (WT), HA-K48R, HA-K48 or HA-K63 Ubiquitin (Fig 6A). These results indicate that MLK3 can be conjugated with K63- or K48-linked ubiquitin chains.
Fig. 6. MLK3 is conjugated with K48- and K63-linked ubiquitin chains.
(A) HA-K48R-Ub, HA-K48-Ub, HA-K63-Ub or HA-WT-Ub expression plasmids were co-expressed with FLAG-MLK3 in HEK293 cells. FLAG-MLK3 was immunoprecipitated with anti-FLAG antibody. Immunoprecipitates were subjected to SDS-PAGE and Western blotting with anti-HA antibody to detect ubiquitinated FLAG-MLK3 and with anti-FLAG antibody to detect total FLAG-MLK3 protein. The levels of ubiquitinated FLAG-MLK3 from the Western blot were quantitated and normalized to the level of FLAG-MLK3 expression as indicated in lower panel. To verify expression, HA-ubiquitin proteins were immunoprecipitated from cell lysates with anti-HA antibody and Western blotting was performed with anti-HA antibody. (B) HEK293 cells were treated with 10 μM MG132 for 6 h and treated with vehicle or TNF (50 ng/ml) for indicated time periods. Whole cell extracts were prepared by lysing cells in 6X SDS sample buffer. Cell extracts were subjected to SDS PAGE and immunoblotting with anti-MLK3 and anti-Actin antibodies. (C) HEK293 cells were transiently transfected with FLAG-MLK3 and HA-WT-Ub or HA-K63R-Ub expression plasmids. Cell extracts were subjected to SDS PAGE and immunoblotting with anti-phospho-MLK3, anti-FLAG and anti-HA antibodies.
Since MLK3 is conjugated with K48-linked ubiquitin chains we wished to investigate if MLK3 protein undergoes proteosomal degradation. HEK293 cells were treated with MG132, a proteosome inhibitor, for 6 h and stimulated with vehicle or TNF for 0, 0.5 or 6 h (Fig. 6B). We observed that inhibition of proteosome activity resulted in an increase in the total amount of endogenous MLK3 protein in cells treated with vehicle or TNF (Fig. 6B). These results indicate that MLK3 could be degraded via a ubiquitin-proteosome pathway and that K48-linked ubiquitination of MLK3 may function to target MLK3 for proteosomal degradation.
To investigate the role of K63-linked ubiquitination in the activation MLK3 kinase activity, the levels of active MLK3 were analyzed in HEK293 cells co-expressing MLK3 and a ubiquitin mutant that disrupts K63-linked polyubiquitin chain formation (K63R-Ub). Overexpression of K63R-Ub with MLK3 resulted in a substantial decrease in the level of active MLK3 in TNF-treated cells in comparison to cells expressing WT-Ub, indicating that K63-linked ubiquitination is indeed important for MLK3 kinase activity (Fig. 6C).
4. Discussion
Both MLK3 and TRAF2 are required for TNF-induced SAPK/JNK activation. Our data suggest that MLK3 is recruited to the TNFR via binding to TRAF2 and/or TRAF6, and this interaction is necessary for MLK3 activation by cytokines. We found that MLK3 associates with both TRAF2 and TRAF6, and MLK3 specifically interacts with the TRAF domain of TRAF2. This finding is similar to that observed with other MAP3Ks such as ASK1, which binds the TRAF domain of TRAF2 to facilitate SAPK/JNK activation [22, 23]. The TNF-induced interaction between MLK3 and TRAF2 occurred within 1-3 min whereas activation of MLK3 did not occur until 3 min after TNF stimulation suggesting that the association between TRAF2 and MLK3 could be a prerequisite for MLK3 activation. Indeed, silencing of traf2 or traf6 significantly impaired TNF-stimulated MLK3 activation indicating a requirement for TRAF2 and TRAF6 in MLK3 activation.
TRAF6 interaction with members of the TNFR and IL-1R families is mediated by the death domain containing adaptor protein MyD88, which recruits IRAK, then TRAF6 associates with IRAK to elicit signaling by IL-1β and LPS [36]. IRAK phosphorylation results in release of the IRAK1-TRAF6 complex into the cytoplasm where they activate NF-κB and SAPK/JNK pathways. Similar to TRAF2, TRAF6 activates both NF-κB and SAPK/JNK signaling [24]. We observed an IL-1β-dependent increase in the association between TRAF6 and MLK3 that occurred before the observed activation of MLK3 by IL-1β, suggesting that this association is required for MLK3 activation by IL-1β. These results suggest that MLK3 is recruited to the IL-1 receptor through its association with TRAF6. Hence, in addition to its critical role in TNF signaling, MLK3 may also have an important role in IL-1β signaling.
We observed that MLK3 was conjugated with K48-linked polyubiquitin chains which primarily functions to target proteins for proteosomal degradation. Since inhibition of proteosome activity elevates total MLK3 protein levels we suggest that K48-linked ubiquitination of MLK3 may function to target MLK3 for proteosomal degradation by the ubiquitin-proteosome pathway.
Our results indicate that MLK3 is rapidly ubiquitinated in response to TNF in both HEK293 and SKOV3 cells. This ubiquitination, that occurred within 20 min, had little effect on total MLK3 protein levels and is likely not K48-linked ubiquitination. However, it occurs at times consistent with MLK3-TRAF2 association and MLK3 activation. Therefore, it could be K63-linked ubiquitination which we have shown to be important for MLK3 kinase activity.
TRAF2 and TRAF6 are ubiquitin ligases that mediate K63-linked polyubiquitination in TNFR and IL-1R/TLR signaling pathways [35, 37]. Furthermore, TNF activation of the SAPK/JNK pathway is dependent on TRAF2 K63-linked ubiquitination [35]. TRAF2 catalyzes K63-linked polyubiquitination of receptor interacting protein (RIP) which is required for TNF-induced activation of NF-κB [38]. In addition, TRAF6 K63-linked ubiquitination is critically required for activation of the MAP3K, TAK1 [26]. Possibly, in a similar fashion, TRAF2 and/or TRAF6 signaling promotes TNF-dependent, K63-linked polyubiquitination of MLK3 which facilitates its activation.Alternatively, TRAF2 or TRAF6 may recruit MLK3 to the intracellular domains of cytokine receptors where it is ubiquitinated by an E3 ligase, which remains to be identified.
5. Conclusion
Our findings indicate that upon TNF or IL-1 receptor ligation, TRAF2 and TRAF6 interact with MLK3 and are required for TNF-stimulated activation of MLK3. TNF stimulation promotes MLK3 ubiquitination and MLK3 can be ubiquitinated with K48- or K63-linked chains. We observed that K63-linked ubiquitination is required for optimal MLK3 kinase activity and K48-linked ubiquitination is important for proteosomal degradation of MLK3. Taken together, our results suggest that the interaction with TRAF proteins and ubiquitination are both integral components in the regulation of MLK3 by the pro-inflammatory cytokines, TNF and IL-1β.
Acknowledgements
We gratefully acknowledge Dr. J. Kyriakis for providing the GST-TRAF2 constructs, Dr. D. Leaman for the TRAF6 construct and Dr. T. Dawson for the Ubiquitin constructs.This work was supported by from a National Institutes of Health grant 1 R15 CA132006-01 (to D.N.C).
Glossary
Abbreviations
- MLK3
Mixed Lineage Kinase 3
- MAPK
mitogen activated protein kinase
- MAP3K
MAP kinase kinase kinase
- RNAi
RNA interference
- NF-κB
nuclear factor κB
- JNK
c-Jun N-terminal kinase
- SAPK
stress activated protein kinase
- MAP2K
MAPK kinase
- EGF
epidermal growth factor
- TNF
tumor necrosis factor
- TNFR
TNF receptor
- TRAF
tumor necrosis factor receptor associated factor
- IL-1
interleukin 1
- siRNA
short interfering RNA
- ERK
extracellular signal-regulated kinase
- TGF-β
transforming growth factor-β
- TAK1
TGF-β activated kinase 1
- ASK1
apoptosis-stimulated kinase 1
- IL-1R/TLR
IL-1 receptor/Toll-like receptor
- HEK293
human embryonic kidney 293
- LPS
lipopolysaccharide
- RIP
receptor interacting protein
- GST
glutathione-S-transferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kyriakis JM, Avruch J. Physiol. Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- [2].Raman M, Chen W, Cobb MH. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- [3].Rana A, Gallo K, Godowski P, Hirai S, Ohno S, Zon L, Kyriakis JM, Avruch J. J. Biol. Chem. 1996;271:19025–19028. doi: 10.1074/jbc.271.32.19025. [DOI] [PubMed] [Google Scholar]
- [4].Tibbles LA, Ing YL, Kiefer F, Chan J, Iscove N, Woodgett JR, Lassam NJ. Embo J. 1996;15:7026–7035. [PMC free article] [PubMed] [Google Scholar]
- [5].Hehner SP, Hofmann TG, Ushmorov A, Dienz O, Wing-Lan Leung I, Lassam N, Scheidereit C, Droge W, Schmitz ML. Mol. Cell. Biol. 2000;20:2556–2568. doi: 10.1128/mcb.20.7.2556-2568.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sathyanarayana P, Barthwal MK, Kundu CN, Lane ME, Bergmann A, Tzivion G, Rana A. Mol. Cell. 2002;10:1527–1533. doi: 10.1016/s1097-2765(02)00734-7. [DOI] [PubMed] [Google Scholar]
- [7].Sathyanarayana P, Barthwal MK, Lane ME, Acevedo SF, Skoulakis EM, Bergmann A, Rana A. Biochim. Biophys. Acta. 2003;1640:77–84. doi: 10.1016/s0167-4889(03)00022-3. [DOI] [PubMed] [Google Scholar]
- [8].Chadee DN, Kyriakis JM. Nat. Cell. Biol. 2004;6:770–776. doi: 10.1038/ncb1152. [DOI] [PubMed] [Google Scholar]
- [9].Gallo KA, Johnson GL. Nat. Rev. Mol. Cell. Biol. 2002;3:663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- [10].Swenson KI, Winkler KE, Means AR. Mol. Biol. Cell. 2003;14:156–172. doi: 10.1091/mbc.E02-02-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Swenson-Fields KI, Sandquist JC, Rossol-Allison J, Blat IC, Wennerberg K, Burridge K, Means AR. Mol. Cell. 2008;32:43–56. doi: 10.1016/j.molcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang H, Gallo KA. J. Biol. Chem. 2001;276:45598–45603. doi: 10.1074/jbc.M107176200. [DOI] [PubMed] [Google Scholar]
- [13].Brancho D, Ventura JJ, Jaeschke A, Doran B, Flavell RA, Davis RJ. Mol. Cell. Biol. 2005;25:3670–3681. doi: 10.1128/MCB.25.9.3670-3681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Barbin G, Roisin MP, Zalc B. Neurochem. Res. 2001;26:107–112. doi: 10.1023/a:1011086426652. [DOI] [PubMed] [Google Scholar]
- [15].Bradley JR, Pober JS. Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- [16].Takeuchi M, Rothe M, Goeddel DV. J. Biol. Chem. 1996;271:19935–19942. doi: 10.1074/jbc.271.33.19935. [DOI] [PubMed] [Google Scholar]
- [17].Rothe M, Sarma V, Dixit VM, Goeddel DV. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- [18].Lee SY, Reichlin A, Santana A, Sokol KA, Nussenzweig MC, Choi Y. Immunity. 1997;7:703–713. doi: 10.1016/s1074-7613(00)80390-8. [DOI] [PubMed] [Google Scholar]
- [19].Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa JL, Ferrick D, Hum B, Iscove N, Ohashi P, Rothe M, Goeddel DV, Mak TW. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- [20].Reinhard C, Shamoon B, Shyamala V, Williams LT. Embo J. 1997;16:1080–1092. doi: 10.1093/emboj/16.5.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chadee DN, Yuasa T, Kyriakis JM. Mol. Cell. Biol. 2002;22:737–749. doi: 10.1128/MCB.22.3.737-749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, Miyazono K, Ichijo H. Mol. Cell. 1998;2:389–395. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- [23].Liu H, Nishitoh H, Ichijo H, Kyriakis JM. Mol. Cell. Biol. 2000;20:2198–2208. doi: 10.1128/mcb.20.6.2198-2208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, van der Heiden A, Itie A, Wakeham A, Khoo W, Sasaki T, Cao Z, Penninger JM, Paige CJ, Lacey DL, Dunstan CR, Boyle WJ, Goeddel DV, Mak TW. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Naito A, Azuma S, Tanaka S, Miyazaki T, Takaki S, Takatsu K, Nakao K, Nakamura K, Katsuki M, Yamamoto T, Inoue J. Genes Cells. 1999;4:353–362. doi: 10.1046/j.1365-2443.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- [26].Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- [27].Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- [28].Kobayashi N, Kadono Y, Naito A, Matsumoto K, Yamamoto T, Tanaka S, Inoue J. Embo J. 2001;20:1271–1280. doi: 10.1093/emboj/20.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM. J. Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ea CK, Sun L, Inoue J, Chen ZJ. Proc. Natl. Acad. Sci. U S A. 2004;101:15318–15323. doi: 10.1073/pnas.0404132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Aggarwal BB. Nat. Rev. Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- [32].Yuasa T, Ohno S, Kehrl JH, Kyriakis JM. J. Biol. Chem. 1998;273:22681–22692. doi: 10.1074/jbc.273.35.22681. [DOI] [PubMed] [Google Scholar]
- [33].Malinin NL, Boldin MP, Kovalenko AV, Wallach D. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- [34].Pineda G, Ea CK, Chen ZJ. Adv. Exp. Med. Biol. 2007;597:80–92. doi: 10.1007/978-0-387-70630-6_7. [DOI] [PubMed] [Google Scholar]
- [35].Shi CS, Kehrl JH. J. Biol. Chem. 2003;278:15429–15434. doi: 10.1074/jbc.M211796200. [DOI] [PubMed] [Google Scholar]
- [36].Cao Z, Henzel WJ, Gao X. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- [37].Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. J. Biol. Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee TH, Shank J, Cusson N, Kelliher MA. J. Biol. Chem. 2004;279:33185–33191. doi: 10.1074/jbc.M404206200. [DOI] [PubMed] [Google Scholar]