Abstract
A highly conserved 35 nucleotide RNA stem-loop, the dimerization initiation site (DIS), in the 5′ untranslated region (UTR) of the human immunodeficiency virus type I (HIV-1) genome has been identified as the sequence primarily responsible for initiation of viral genome dimerization. The DIS initiates viral genome dimerization through a loop-loop ‘kissing’ interaction and is converted from an intermediate ‘kissing’ to a more thermodynamically stable extended duplex dimer in a conformational rearrangement that is chaperoned by the HIV-1 nucleocapsid protein (NCp7). Here we describe fluorescence methods designed to probe local RNA dynamics and structural transitions associated with the DIS dimer formation and its NCp7 chaperoned structural conversion. These methods take advantage of the exquisite sensitivity of the quantum yield of the fluorescent nucleotide base analog, 2-aminopurine (2-AP), to its immediate structural and dynamic environment. The 2-AP fluorescence methods described allow a detailed kinetic and thermodynamic examination of this type of RNA-RNA interaction, as well as an analysis of the molecular mechanism of NCp7 chaperone activity.
Keywords: HIV-1, dimerization initiation site (DIS), viral genome dimerization, RNA-RNA ‘kissing’ interaction, folding kinetics, equilibrium binding, 2-aminopurine fluorescence
Introduction
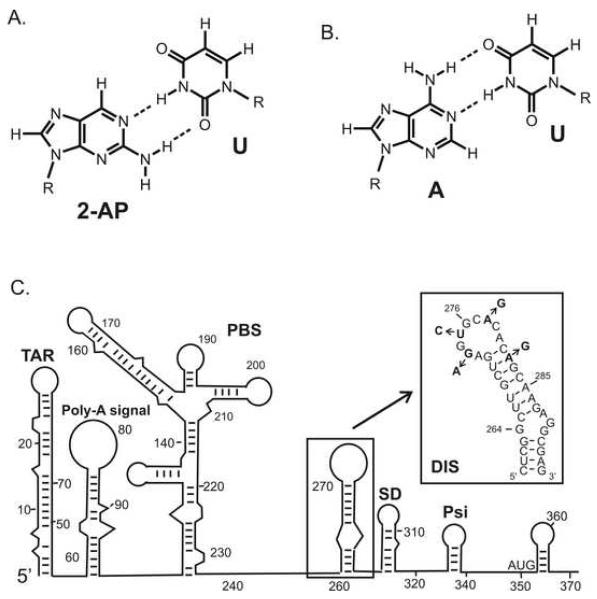
Fluorescent nucleotide analogs have found widespread utility as probes for detecting the folding, structural dynamics and molecular interactions of nucleic acids (1, 2). The sensitivity of the quantum yield (ΦF) of 2-aminopurine (2-AP) to its local environment (3), as well as its commercial availability as a phosphoramidite, has made this nucleotide analog a particularly popular fluorescent probe for monitoring conformational changes in nucleic acid structure and dynamics. 2-AP fluorescence is highly quenched when it is stacked with other nucleotide bases, but can increase as much as 100-fold when fully exposed to solvent. In general, however, the dynamic range of fluorescence quenching observed for 2-AP when inserted within oligonucleotides has been found to be about an order of magnitude smaller due to the fact that 2-AP fluorescence is efficiently quenched in these sequences even in ‘solvent exposed’ regions of the structure. In this respect, it should be noted that all bases have been found to exhibit similar extents of static as well as dynamic quenching of 2-AP, while base pairing and hydrogen bonding are not considered significant modulators of 2-AP fluorescence (4, 5). The sensitivity of 2-AP’s fluorescence quantum yield to its microenvironment allows nucleic acid binding to be detected through direct contact, as well as indirectly through distal conformational changes in oligonucleotides that may accompany macromolecular interactions or chemistry. In addition to its advantage as a highly sensitive fluorescence probe of its local environment, 2-AP has been found generally to not perturb native RNA and DNA structures and cognate interactions even when inserted in close proximity to binding interfaces (1, 2). This is because 2-AP is similar in constitution to adenine (6-aminopurine) and can form well stacked base pairs with either uridine (RNA) or thymine (DNA) in the context of standard A- and B-form helices (Fig. 1a, b).
Fig. 1.
The fluorescent nucleotide base analog 2-aminopurine (2-AP) is a generally non-perturbing substitution within nucleic acid sequences owing to its close structural similarity to adenosine and its ability to form well stacked, wobble pairs with uracil or thymidine. (A) Structure and base pairing of 2-AP with Uracil (2-AP:U). (B) Standard Watson Crick pairing of adenosine with uracil (A:U). (C) Secondary structure rendering of the 5′-leader sequence of the HIV-1 genomic RNA, with the DIS stem-loop boxed. The RNA sequence and secondary structure of the DIS stem-loop found in the HIV-1 subtype-A isolate. Positions in the sequences which differ for the subtype-B isolate of HIV-1 are indicated by arrows and substituted nucleotides.
2-AP fluorescent detected methods have been applied in a number of studies to characterize RNA dynamics (6-9) and conformational changes associated with RNA folding (10-14), catalysis (12) and protein and small molecule binding to RNA (15-23). For example, the utility of 2-AP as a fluorescent probe has been demonstrated in analysis of the interaction of Rev peptides and small molecule inhibitors with the HIV-1 rev responsive element (RRE) (17, 24), aminoglycoside and Tat peptide binding to the HIV-1 trans activator region (TAR) (19), small molecule binding to an RNA tetraloop (25), aminoglycoside binding to prokaryotic and eukaryotic ribosomal RNA sequences (26), and hammerhead ribozyme inhibition (27). In our laboratory, 2-AP fluorescence detection methods have been developed to probe RNA structural transitions associated with the HIV-1 dimerization initiation site (DIS) dimer formation and NCp7 mediated structural conversion of the DIS dimer from a ‘kissing’ to an extended duplex isoform (28-30). The DIS sequence is located in the 5′ untranslated region (UTR) and forms a highly conserved 35-nucleotide stem-loop (Fig. 1c), which is primarily responsible for dimerization of genomic RNA. Its loop contains a palindromic hexanucleotide sequence flanked by highly conserved 5′ and 3′ purines and can form a homodimer through a loop-loop ‘kissing’ interaction. Furthermore, HIV-1 nucleocapsid protein (NCp7) catalyzes conversion of the DIS stem-loop from a kinetic intermediate ‘kissing’ to a more thermodynamically stable extended duplex mediated dimer linkage in a RNA structural rearrangement suggested to be associated with maturation of the budded viral particle. Proper packaging of genomic RNA as a homodimer appears to be functionally relevant to a number of aspects of retroviral replication, including recombination and reverse transcription, translation of gag, and selective encapsidation (31-33). Here we describe in detail the 2-AP fluorescence detection methods we have developed for determining equilibrium and kinetic binding constants of the DIS dimer formation, for distinguishing thermal melting transitions of the DIS dimer, for characterizing local structural conformations of unpaired dynamic bases, and for determining in real time, kinetic rates of NCp7 chaperoned DIS structural maturation.
2. RNA Chemical Synthesis and Sequence Design
2.1 Synthesis of 2-AP labeled and unlabeled DIS stem-loops
RNA oligonucleotides, containing 2-aminopurine 2′-O-methyl riboside and 2-aminopurine riboside, are typically synthesized at a 1 μM scale on either an Applied Biosystems 390 synthesizer (Perkin-Elmer, Forest City, CA) or a MerMade6 DNA/RNA oligonucleotide synthesizer (BioAutomation, Plano, TX) using standard TOM-protected phosphoramidite chemistry. Synthesis is carried out following protocols established by Glen Research (Sterling, VA) and standard script file settings by BioAutomation. Each phosphoramidite is incorporated with three consecutive six minute coupling steps using 0.25 M ethylthiotetrazole (ETT) in acetonitrile employed as the activator. One micromole scale 500 Å standard RNA CPG columns are purchased from BioAutomation. Nucleoside phosphoramidites, including 2-aminopurine riboside, are purchased from Glen Research. Cleavage and deprotection of RNA oligonucleotides is carried out following standard procedures. Briefly, methylamine (MA) solution (1 volume 40% aqueous methylamine plus 1 volume of 33% ethanolic methylamine) is added to the column resin and left overnight at room temperature, or for at least 6 hours at 35 °C. Oligonucleotides are then dried with N2 gas. Removal of the 2′ silyl protecting group is achieved by adding 1 mL of tetrabutylammonium fluoride (TBAF) and leaving it for 6 hours at 35 °C or overnight at room temperature. The 2′ deprotection reaction is quenched by addition of 1ml of ddH2O and the oligonucleotides desalted using an Amicon Centriplus concentrator (Millipore, Bedford, MA).
Unlabeled RNA oligonucleotides are prepared by in vitro T7 polymerase run-off transcription with synthetic DNA templates according to the method of Milligan and Uhlenbeck (34, 35). All RNA oligonucleotides are purified using preparative-scale denaturing polyacrylamide gel electrophoresis (PAGE), recovered by electrophoretic elution using an Elutrap eluter (Scheiler & Schuell, Keene, NH) and then desalted and exchanged into standard buffer using an Amicon Centriplus concentrator. If necessary, further buffer exchange is carried out using a Pierce Microdialysis system (Pierce Instruments, Rockford, IL). Standard buffer conditions for all experiments are 1 mM sodium cacodylate [pH=6.5] and 25 mM NaCl. Sodium cacodylate, which is a temperature insensitive pH buffer, was used in all experiments since it allows Mg+2 to be added to the solutions without causing precipitation, as might be observed with phosphate buffers. All RNA oligonucleotides are stored in aliquots at −20 °C and were ‘snap cooled’ prior to use in each experimental series by first heating the RNA containing buffer solution to 90 °C for 2 minutes and then quickly cooling the samples on dry ice. RNA concentrations are determined by measuring the absorbance at 260 nm using extinction coefficients determined by the web based program, OligoAnalyzer (IDT, Coralville, IA), http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/. NCp7 protein is expressed in E. coli and purified as previously described (36).
2.2 Altering the DIS stem-loop to create heterodimeric complexes
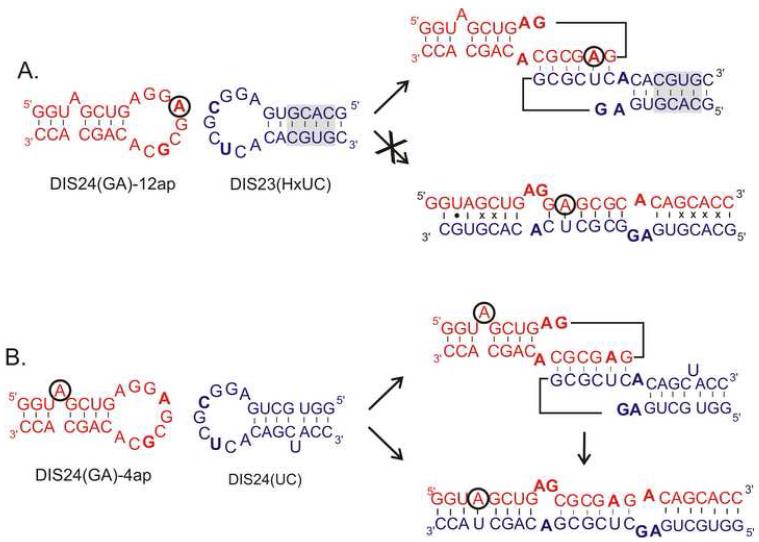
The DIS loop contains a self associating palindromic sequence (Fig. 1c), which is found most often to be either GUGCAC (subtype A) or GCGCGC (subtype B) (37). Since the native DIS stem-loops form dimers to various extents and affinities depending on RNA concentrations and experimental conditions, heterodimeric forming DIS stem-loops were first designed to allow a finer control over the monomer versus dimer state of the DIS stem-loops, and thereby facilitate quantitative analysis of the binding reactions. To eliminate the propensity of the wild-type DIS stem-loops to form homodimers and instead direct formation of a heterodimeric complex, two unique DIS stem-loops (Fig. 2) have been designed through point mutations in the palindromic sequence [U275 to A275 and A278 to G278 to form the DIS(GA) stem-loop and U275 to C275 and A278 to U278 to form the complementary DIS(UC) stem-loop]. These sequences form heterocomplexes through complementary hexamer loop sequences; while, homodimers are energetically disfavored due to the mismatches created in the homodimeric loop-loop interactions. Experiments with the modified DIS stem-loops show that they exhibit wild-type ‘kissing’ dimer properties, while predominately forming hetero- rather than homodimer complexes (28, 29) In addition to creating matched heterodimeric DIS stem-loop pairs, both truncated DIS(24) constructs which lack the DIS stem bulge and full length DIS stem-loop DIS(35) constructs, this approach has also been used to alter DIS loop sequences to create heterodimeric versions of larger fragments of the HIV-1 genomic RNA, including the packaging sequence element and the full 5′-UTR (30).
Fig. 2.
RNA sequence and secondary structure of an example set of DIS stem-loop constructs designed for the fluorescence experiments. In these DIS constructs, mutations have been made in the hexanucleotide sequence [U275 • A275 and C278 • G278] to form the DIS(GA) stem-loop and U275 • C275 and C278 • U278 to form the complementary DIS(UC) stem-loop] to eliminate the palindrome and promote heterodimer formation. Nucleotide positions in the DIS(GA) stem-loop that were substituted with the fluorescent probe 2-AP are circled. (A) The exchanged stem helix of DIS23(HxUC) is shadowed and disfavors conversion of the ‘kissing’ dimer to an extended duplex owing to the mispairing which result in this designed system. Using these constructs, the ‘kissing’ interaction is measured as a function of the quenching of the 2-AP probe in the DIS loop. (B) Schematic of the intermolecular base pair interactions of the DIS24(GA)-4ap with DIS24(UC) construct which form the basis for the DIS loop-loop ‘kissing’ complex structural conversion to an extended duplex dimer. Using these constructs, the structural conversion to an extended duplex dimer is measured as a function of the quenching of the 2-AP probe that results from base pairing and stacking during the stem strand exchanges.
2.3 Designing 2-AP labeled DIS stem-loops to distinguish the DIS dimer isoforms
Native polyacrylamide gel electrophoresis (PAGE) or gel filtration is typically used to visualize nucleic acid complexes and in the case of the DIS dimerization can be used to readily distinguish monomer from dimer states. However, since the DIS dimer isoforms share similar conformations and are the same molecular weight, these two dimers can not be readily distinguished using these standard methods which differentiate macromolecules and macromolecular complexes based on size and/or molecular shape. The two dimer isoforms can be distinguished based on differences in the Mg+2 dependence on native gel stability of the DIS ‘kissing’ and extended duplex dimers (38). While both ‘kissing’ and extended duplex dimers are observed as stable complexes on native gels that contain Mg+2 in the gel buffers, only the extended duplex is observed as a stable complex on native gels run without Mg+2. In practice, however, the use of native gels to distinguish dimer isoforms is limiting in that binding or NCp7 chaperone experiments either must be carried out in buffers that do not contain Mg+2, or other divalent cations, or after the reaction is complete, the Mg+2 must be removed from the buffer solution before the dimer state can be assessed.
To address the technical challenge of distinguishing the DIS dimer states and allow real time detection of RNA-RNA equilibrium binding, DIS stem-loops have been designed with 2-AP inserted into single positions in the sequence to provide unambiguous fluorescence probes for the formation of each of the two structural conformations of the DIS dimer in solution: ‘kissing’ versus extended duplex dimers. Distinguishing these two states fluorescently also enables the analysis of the structural rearrangement of the DIS ‘kissing’ dimer catalyzed by the HIV-1 NCp7 protein. Two constructs were synthesized (Fig. 2), each with a single 2-AP inserted at a specific position in the DIS24(GA) stem-loop: one construct with 2-AP substituted in the loop at position 12, DIS24(GA)-12ap, and a second with 2-AP inserted as a single bulged nucleotide in the stem at position 4, DIS24(GA)-4ap. The single 2-AP residues inserted into either the loop or the stem bulge in the DIS hairpin were found to be useful probes, respectively, of loop-loop helix formation in the ‘kissing’ interaction and stem strand exchange in the NCp7 mediated conversion. In both positions, 2-AP provided a high fluorescent signal that could be readily monitored in quenching experiments. It should be noted that for the detection of the strand exchange reaction, alternative approaches of placing 2-AP inside larger bulges (2 - 3 nts) or at the termini of the RNA did not allow for easy distinction between the unpaired and paired states associated with the strand exchange reaction since both states were found to be highly quenched. Two unlabeled DIS(UC) constructs, with hexanucleotide loop sequences complementary to the DIS24(GA) stem-loops, have also been designed: one construct with a stem sequence complementary to DIS24(GA)-4ap, that can specifically probe the DIS structural conversion of DIS24(UC), and a second construct with an exchanged stem sequence that is capable of forming only the ‘kissing’ dimer DIS23(HxUC). For the latter construct, the extended duplex form of the DIS dimer is made energetically disfavored due to multiple mismatches in each of the exchanged stem helices.
2.4 Incorporation of 2-AP labeled DIS stem-loops into larger HIV-1 genome sequences
To extend the application of fluorescent nucleotide base analog probes to studies of larger RNA sequences derived from the 5′ leader sequence of HIV-1, T4 RNA ligase can be employed to couple chemically synthesized, sequence fragments containing 2-AP bases, with unmodified RNA sequences to produce large RNA oligonucleotides that are site-specifically labeled with the fluorescent base analog. Using reaction conditions optimized for the joining of single stranded RNAs, T4 RNA ligase can be used to generate larger 5′ leader sequences of HIV-1 that contain single site 2-AP labeled DIS stem-loops and these chemical-biosynthesis hybrid methods have been described (30, 39, 40).
3. 2-AP Fluorescence detection methods for probing RNA structural transitions
3.1. Detection of the DIS ‘kissing’ dimer formation and determining equilibrium KDs
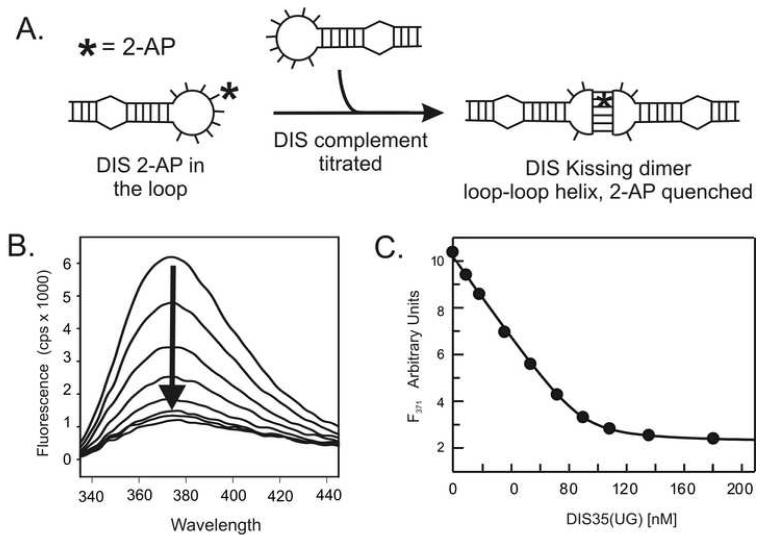
Steady-state fluorescence emission spectra of the AP-containing RNA oligonucleotide samples (100 nM) are measured on a SPEX Fluoromax-3 spectrofluorometer (Instruments SA, Edison, NJ) using a 3 mm square cuvette in 150 μL of standard buffer solution with 5 mM MgCl2. Temperature is regulated by a circulating water bath and is typically set between 10 °C and 40 °C. Emission spectra are recorded over the wavelength range of 330 to 450 nm with an excitation wavelength of 310 nm and a spectral band pass of 5 nm. In each set of experiments, the fluorescence spectrum of a 150 μL solution of buffer without RNA or protein is first recorded. This spectral blank, in which the strong Raman scatter is observed, is subtracted from all subsequent spectra acquired in an experimental dataset. Using the designed DIS constructs with 2-AP inserted within the complementary loop sequences, the equilibrium binding constant (KD) for the DIS loop-loop ‘kissing’ dimer formation is determined by following the decrease in fluorescence at 371 nm as a fixed concentration of the fluorescent DIS(GA) is titrated with increasing amounts of DIS(HxUC) complement (Fig. 3). In a typical titration experiment, a 10 μM stock solution of the titrated DIS(HxUC) RNA is used which allows addition of aliquots of < 2 μl for each point in the binding curve. After addition of the DIS(HxUC) RNA to the cuvette, the solution is mixed thoroughly using a transfer pipette before acquiring the fluorescence spectrum. In the course of a titration, the increase in the total volume is normally kept to less than 10% and the observed fluorescence intensities are corrected to account for this minor dilution effect before fitting.
Fig. 3.
(A) Incorporation of a 2-AP, indicated schematically by an asterisk in the loop sequence of the DIS, allows the ‘kissing’ interaction to be quantified as a function of quenching of the fluorescence of the 2-AP base as it forms a stacked, base pair in the ‘kissing’ dimer loop-loop helix. (B) An example of the observed fluorescence changes accompanying titration of 100 nM DIS24(GA)-12ap with increasing concentrations of DIS23(HxUC) in standard buffer with 5 mM MgCl2 at 25 °C. Emission spectra are plotted from 330 to 450 nm. (C) Representative fitted plot of the fluorescence decrease at 371 nm as a function of total DIS35(UC) concentration for a titration using 100 nM DIS35(AG)-19ap. The solid curve is fit (KD = 2.2 ± 0.5 nM) using a single site equilibrium binding equation (1).
In titrations with DIS stem-loop containing a 2-AP within the complementary loop sequence, the direction and magnitude of the fluorescence change is consistent with 2-AP•U base pairing and stacking that would accompany the formation of the loop-loop helix in the DIS ‘kissing’ dimer. The RNA-RNA binding interaction in these experiments can be fit using a single site equilibrium binding equation using the graphing program Grafit (Erithacus, Surrey, UK). For example, single site binding of DIS(HxUC) to 2-AP labeled DIS (GA) can be fit using equation 1 as follows,
| (1) |
where Fo and Ff are the initial and final fluorescence intensities, respectively, [DIS(GA)]tot is the total DIS(GA) concentration, and [DIS(UC)]tot is the total concentration of DIS(HxUC).
In the equilibrium binding experiments, the 2-AP labeled DIS(GA) concentration is normally varied from 25 to 200 nM, to obtain an average KD value. Using this approach, the stability of the DIS stem-loop ‘kissing’ dimer in the presence of different cations and buffering conditions can readily be measured and compared. For example, a concentration of 0.5 - 5 mM Mg+2 is found to stabilize the DIS ‘kissing’ dimer resulting in a kinetically trapped complex. In contrast, stable ‘kissing’ dimer formation was found to require monovalent cations, such as Na+, at concentrations of > 200 mM under standard buffer conditions.
3.2. Fluorescence detection methods for measuring the DIS stem-loop on- and off-rate kinetics
Using the same approach of incorporating a 2-AP probe into the complementary sequence of the DIS loop, the dissociation kinetics of the DIS loop-loop ‘kissing’ dimer can be directly measured in solution by following the increase in fluorescence of 2-AP labeled DIS(GA) over time as the DIS(GA)•DIS(HxUC) complex dissociates. This reaction is carried out using DIS ‘kissing’ dimer concentrations of 100 nM, in standard buffer solution with either NaCl (> 200 mM) or MgCl2 (0.5 - 5.0 mM) added to form stable ‘kissing’ dimers and made irreversible by trapping the free DIS(HxUC) with a 20-fold excess of unlabeled DIS24(GA). The time course of the dissociation of the DIS ‘kissing’ dimer is fit using the first order rate equation 2:
| (2) |
where F1 and k1 are the amplitude and observed off-rate, respectively, for the complex and C is a constant offset. Similarly, the kinetics of the DIS dimer association can be followed using pseudo-first order conditions, where DIS(HxUC) is present at concentrations of 20-fold greater than the 2-AP labeled DIS(GA). As with the dissociation kinetics experiment, a 2-AP labeled DIS(GA) concentration of 100 nM is typically used. The time course of the quenching of 2-AP fluorescence (Ft) as a result of loop-loop complex formation was fit using the first order rate equation 3,
| (3) |
where F1 and k1 are the amplitude and apparent on-rate, respectively, for the complex and C is a constant offset. The ratio of association to dissociation kinetic rate constants can be used to calculate the equilibrium KD and used to confirm the value derived from the fit of the equilibrium binding experiments.
3.3 Fluorescence detection methods for dissecting thermal unfolding transitions of the DIS ‘kissing’ dimer
Thermal denaturation of RNA structures are normally detected globally by monitoring the increase at 260 nm in the hyperchromicity of bases as an oligonucleotide unfolds (41). In complex RNA structures with more than one thermal melting transition (TM), the UV-detected thermal melting approach does not allow these transitions to be readily assigned. In principle, melting transitions can be assigned to bimolecular interactions by examining the concentration dependence of the transition, since melting transitions associated with bimolecular interactions are expected to show an increased TM at higher RNA concentrations. As an alternative to the UV-absorbance based detection of the thermal melting of the DIS RNA (38, 42-44) , changes in fluorescence of site specifically incorporated 2-AP probes, which are almost exclusively sensitive to local structural changes, can provide assignment of the melting transitions to the substructural elements within the RNA fold: e.g. 2-AP can be inserted within the sequence to detect the melting of individual helices in a multi-helical structure or the unfolding/unstacking of specific junction or bulge nucleotides which may precede the overall melting of the RNA structure (10).
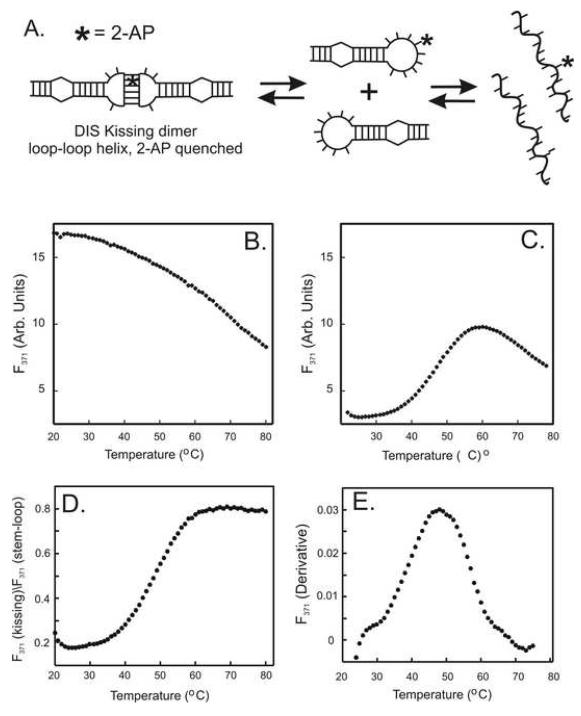
As an example (Fig. 4), the melting of the loop-loop helix in the DIS dimer can be selectively monitored through changes in fluorescence of a 2-AP probe inserted within the complementary loop. In this example, the change in 2-AP fluorescence intensity was measured as a function of temperature for the 2-AP labeled DIS(GA) stem-loop (2-AP inserted into the complementary loop sequence) (Fig. 4b) and for the same DIS stem-loop in complex with its DIS(UC) complement to form a ‘kissing’ dimer complex (Fig. 4c). In practice, thermal melts of the 2-AP labeled RNA are carried out in 1 cm path length cuvettes. All buffers and samples were degassed prior to thermal melting. Cuvettes were equilibrated at 15 °C for 15 minutes before initiation of a 1.0 °C/min temperature gradient to 90 °C. 2-AP fluorescence at 371 nm was monitored at 1.0 °C intervals using a FluoroMax2 fluorometer equipped with a programmable thermal water bath. Fluorescence spectra acquired at each point in the thermal denaturation curve are subtracted from the blank buffer spectrum. Using this series of experiments, the temperature dependence of 2-AP emission intensity in the ‘kissing’ dimer melting curve is normalized using the melting curve of the individual DIS stem-loop by taking the ratio of the intensity at each point in the melting curves (Fig. 4d). This approach allows the fluorescence changes resulting from the loop-loop thermal denaturation to be distinguished from thermal effects on fluorescence that result from the melting of the DIS stem-loop. The apparent melting temperature of the RNA loop-loop substructure transitions can then be estimated from a derivative plot of the normalized fluorescence intensities (Fig. 4e). In the example case shown for the 2-AP detected loop-loop thermal melting two transitions are observed in the derivative plot, one at ~ 28 °C and the second at 48 °C, with the observed lower temperature transitions suggesting a minor structural change in the loop-loop structure that precedes the dimer to monomer transition.
Fig. 4.
(A) Schematic for a simple two step thermal denaturation of the DIS ‘kissing’ dimer: dissociation of the loop-loop helix into individual RNA stem-loops followed by the unfolding of the stem-loops to single strands. An example of a 2-AP detected thermal denaturation of the RNA ‘kissing’ dimer, in which the 2-AP probe is placed within the loop to detect unfolding of the loop-loop helix. The blank corrected fluorescence of the DIS24(GA)-12ap stem-loop and the ‘kissing’ complex formed by DIS(24)-12ap and DIS23(HxUC) are shown in (B) and (C), respectively. Both fluorescence detection denaturation curves were acquired using standard buffer with 0.5 mM MgCl2 added. (D) A plot of the normalized fluorescence intensity values for the ‘kissing’ loop-loop denaturation obtained by taking the ratio of the fluorescence observed for the ‘kissing’ dimer denaturation and the individual DIS stem-loop at each temperature point. (E) Derivative plot of the normalized fluorescence as a function of temperature used to determine the melting temperature (Tm) for the loop-loop helix. Two thermal melting transitions can be extracted from the derivative plot, one at ~ 28 °C and a second at ~ 48 °C.
3.4 Fluorescence detection methods for probing local RNA structure and dynamics
In regions of RNA structure, like the highly conserved purine nucleotides on the 5′ and 3′ ends of the DIS loop, where structure is observed to exhibit temperature, metal and or pH dependence, insertion of 2-AP at these sites can provide a rapid probe to monitor local base stacking and dynamics. In the case of the DIS dimer, probing the dynamics and stacking of these purine nucleotides is of particular interest since the rate of NCp7 catalyzed maturation of the DIS ‘kissing’ dimer has been observed to directly correlate with a proton-coupled conformational dynamics localized around these bases (28). Monitoring the fluorescence of 2-AP bases that have been inserted into first or second position of the DIS loop in response to ‘kissing’ dimer formation and small molecule interactions (45) under various metal and buffer conditions, provides an approach to probing the structural role these bases play in stabilizing the ‘kissing’ dimer and facilitating NCp7 conversion. It should be noted that before using these constructs as probes of structure, these 2-AP labeled DIS stem-loops must be first tested to assess whether they maintain wild-type RNA binding properties and NCp7 interactions.
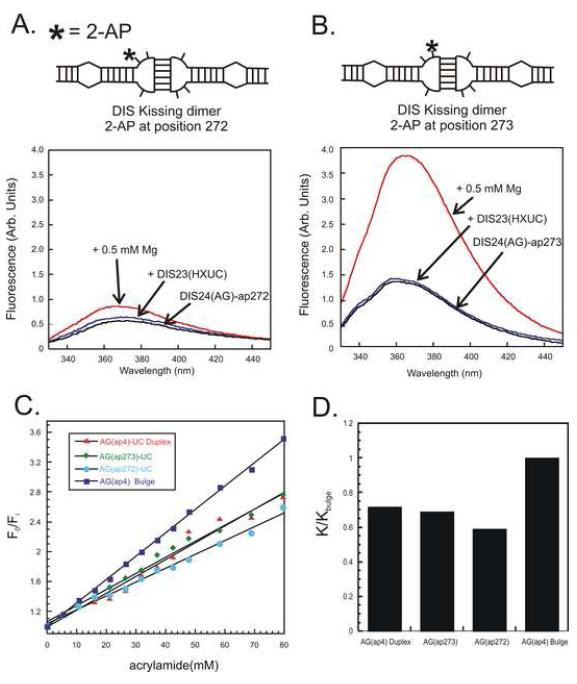
As an example, Fig. 5a and b shows the fluorescence response to ‘kissing’ dimer formation of 2-AP at the first (272) and second (273) position on the 5′ end of the DIS loop in the presence of 0.5 mM Mg+2. For each position, an increase is observed in the fluorescence emission spectrum which suggests that both bases become more unstacked in the complex when compared to the conformation found in the stem-loop. The increase in fluorescence intensity, however, is much more profound for the 2-AP inserted at the second position of the loop, showing the exquisite sensitivity of 2-AP to the substituted nucleotide position in the structure. In this respect, it is important to note that the NMR (46-48) and X-ray structures (49) have reported different stacked conformations for these bases. In the solution state structures, the bulge nucleotides are reported to be stacked into the helix, while in the crystal the same bases are found to bulge out from the helix. Considering the previous reported conformations for these bases, the changes in the fluorescence emission observed for 2-AP in these positions of the DIS loop suggest that the bases are in a dynamic exchange.
Fig. 5.
Observed changes in the fluorescence spectral intensity of the DIS stem-loop with a 2-AP label at position 1 (A) or position 2 (B) in the loop in response to addition of 1:1 stoichiometric amount of the DIS complement and then the addition of 0.5 mM MgCl2. Emission spectra are plotted from 330 to 450 nm. (C) An example of an acrylamide quenching titration assay aimed at determining the accessibility of the unpaired purines in the loop of the DIS ‘kissing’ RNA complex. The relative fluorescence with respect to increasing acrylamide concentration for a DIS ‘kissing’ RNA dimers with 2-AP at position 1 (blue circles) and position 2 (green diamonds) of the loop are plotted with similar titrations shown for two control structures, one in which the 2-AP is in a bulged position (blue squares) and a second (red triangles) where 2-AP is base paired within a helix. (D) Slopes of the Stern–Volmer plots for each 2-AP labeled RNA relative to that of the (AG)-4ap bulged position. The relative sensitivity to the quench (K/Kbulge) of the 2-AP position within the DIS dimer is defined as the slope for the indicated 2-AP position (K) divided by the slope measured for the (AG)-4ap bulged position (Kbulge).
In this respect, to further assess the environments of the 2-AP bases placed within the bulge of DIS, acrylamide quenching analysis (10, 50) can be performed to assess the solvent accessibility of the 2-AP residues in these structures. As an example of acrylamide quenching data, the measured ratio F0/Fi as a function of the concentration of acrylamide of two constructs with 2-AP inserted at either first of second position in the DIS loop together and two control sequences (one in which 2-AP is in a known bulged position and one where it is well stacked in a helix) is shown in Fig. 5c. In these plots, F0 and Fi represent the fluorescence emission intensities in the presence and absence of quenching agent. For the titration, 1 M acrylamide stocks were made in standard buffers. The titration stock was added and manually mixed with a transfer pipette before the fluorescence emission at 371 nm was read. The RNA concentration was 100 nM. The plotted ratios of F0/ Fi versus acrylamide concentration can be fit using the standard Stern-Volmer equation (4):
| (4) |
where F0 is the steady state fluorescence in the absence of acrylamide, Fi is the fluorescence of the sample in the presence of acrylamide at a concentration [Q] and the slope of the line (K) is the Stern-Volmer constant describing the average sensitivity of the fluorophore to acrylamide. The measured slopes (K) of these Stern-Volmer plots are proportional to the accessibility of the quenching agent to the 2-AP base, as well as the lifetime of the 2-AP excited states. By comparison with the control samples, the relative accessibility of 2-AP inserted at both first and second positions of the DIS loop to quencher can be estimated (Fig. 5d). While these examples of acrylamide quenching could be fit with the simple Stern-Volmer equation, in certain cases where non-linearity is observed in the Stern-Volmer plot (51), the data can be fit assuming a two state modification to the fitting equation, where only fraction of the 2-AP population is considered to be accessible to solvent.
3.5 Fluorescence methods for measuring NCp7 chaperoned DIS dimer maturation
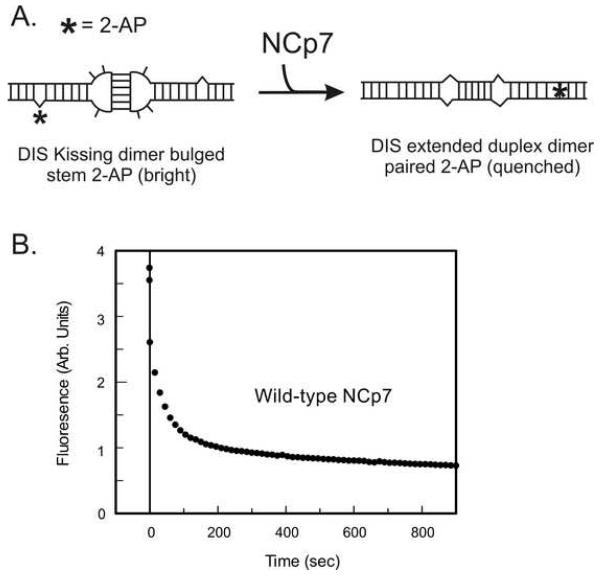
The 2-AP fluorescence detection method for measuring NCp7 chaperone activity was designed to measure the time dependent fluorescence quenching of the 2-AP reported in the DIS24(GA)-4ap stem-loop after addition of NCp7 to the DIS ‘kissing’ complexes preformed between the DIS24(GA)-4ap and complementary DIS24(UC) stem-loops. As described above, the DIS24(UC) stem-loop contains a single stem bulge uridine that can form a 2-AP•U base pair in the mature duplex dimer with the 2-AP base from DIS24(GA)-4ap. Thus, the environment of the reporter 2-AP is expected to change from a bulged, highly fluorescent state in the ‘kissing’ dimer to a stacked, quenched state in the mature dimer, assuming that the stem strands are exchanged during the reaction. For typical experiments, the NCp7 catalyzed structural conversion of the DIS dimer is measured at 25 °C using a SPEX Fluoromax-3 spectrofluorometer by following the decrease in fluorescence emission at 371 nm as the ‘kissing’ dimer linkage is converted to the extended duplex conformation. In these experiments, the ‘kissing’ dimer complex is first formed in standard buffer solution at pH 6.5 in the presence of 5 mM MgCl2 and equilibrated for approximately 15 minutes. NCp7 protein is then added to the cuvette and mixed manually with a transfer pipette, then rapidly inserted into the spectrofluorometer for the 2-AP fluorescence measurements. The rates of NCp7 conversion are typically slow enough (seconds to minutes), to allow manual mixing and insertion of the cuvette into the fluorometer. In practice, a rapid mixing kinetics device could be used to detect faster time scales and reduce the dead time of the manual experiments, which is typically a few seconds. The concentration of the ‘kissing’ complex in each case is normally between 100 and 200 nM and NCp7 protein is added in approximate stoichiometric ratios of slightly greater than 2 NCp7 per ‘kissing’ RNA stem-loop dimer. For larger RNA constructs, the required concentration for NCp7 activity is determined empirically, but usually is added in a ratio of ~ 1 NCp7 per seven nucleotides of RNA sequence.
As shown as an example in Fig. 6, a time-dependent decrease in 2-AP fluorescence emission is observed in this experiment which can be fit to determine the kinetic rates of chaperone activity of NCp7 protein. As with the 2-AP quenching assay for detecting the kissing interaction, the direction and magnitude of the observed fluorescence change indicates 2-AP base pairing and stacking that would accompany exchange of the stem strands during the conversion process. To confirm that the change in fluorescence observed in the experiment is due to DIS structural conversion and exchange of stem strands, the fluorescence response of NCp7 addition to a ‘kissing’ complex formed between DIS24(GA)-4ap and DIS23(HxUC), which contains a mismatched stem sequence incapable of forming proper Watson-Crick base pairs in the mature duplex dimer, is measured. In this case, only a static change in the fluorescence of the 2-AP probe would be expected if there was a slight change in fluorescence due to protein binding. A time-dependent change would however, not result. In this respect, changes in the addition of Mg+2 directly to the individual DIS24(GA)-4ap stem-loop must also be considered. As with NCp7, only slight static increase in the 2-AP fluorescence emission would be expected if the 2-AP probe was minimally influenced by binding of Mg+2 to the RNA.
Fig. 6.
(A) Schematic of the structural conversion of the DIS24(GA)-ap4•DIS23(UC) from a ‘kissing’ to extended duplex conformation. The location of the 2-AP probe on the DIS stem-loop is indicated by an asterisk. (B) An example of the 2-AP detected maturation of the [DIS24(GA)-ap4]•[DIS24(UC)] ‘kissing’ complex monitored as a function of quenching of 2-AP fluorescence emission after addition of NCp7 at pH 6.5 for wild-type NCp7 protein. Structural isomerization rates (kconv, karr) can be fit from these data using a double exponential rate equation and comparison of the rates can be used to dissect the relative importance of different amino acids as well as structural elements in NCp7’s chaperone activity.
In most cases, the time course of the conversion of the DIS ‘kissing’ dimer is best fit using the double exponential equation,
| (5) |
where kconv is the observed isomerization rate for conversion of the DIS ‘kissing’ to extended mature duplex dimer and a second slower, lower amplitude rate, karr, is attributed to the rate of rearrangement of the 2-AP probe subsequent to its stacking in the duplex conformation.
4. Limitations and alternatives to 2-AP Fluorescence detection Approaches
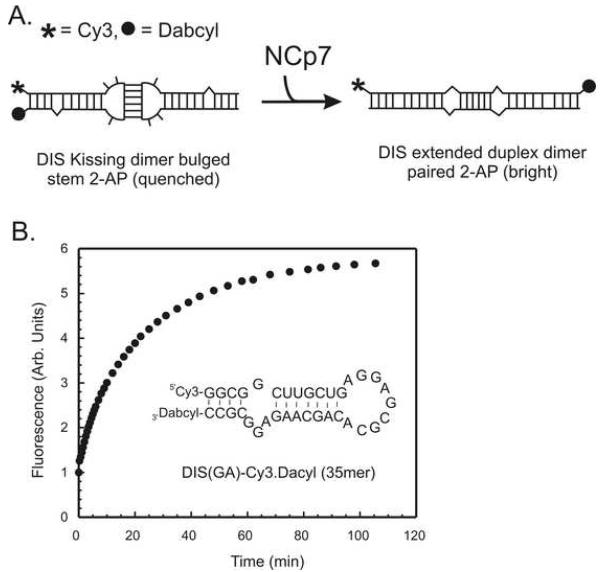
While 2-AP has many advantages as a fluorescent probe of nucleic acid interactions, which include being almost exclusively sensitive to only local environmental changes, facile insertion within the oligonucleotide sequences and limited perturbations to the native structure, this fluorescent base analog does present a few limitations in its application. One limitation is the relative sensitivity of the fluorescence of the 2-AP base when compared to more conventional fluorescence dyes, which have also been used to detect structural changes in nucleic acids and have enabled detection down to the level of single molecules (52). Due to the limits in sensitivity, the 2-AP fluorescence detection approaches described here are not readily adaptable to fluorescent plate reader formats. Additionally, while the red shifted absorption spectrum of 2-AP allows differential excitation in the presence of nucleic acids and proteins (without interference for example from intrinsic protein tryptophan fluorescence), use of 2-AP assays to screen for small molecule interactions can sometimes be complicated by the overlapping background absorption and fluorescence of the small molecules, particular for molecules that contain aromatic moieties. In this respect, fluorescence quenching experiments are considered less robust for detection of small molecule binders/inhibitors when compared to those detection schemes that are based on a fluorescence burst response. In all cases, however, the possibility for artifact quenching of the fluorescence reporter has to be considered. As an alternative to the 2-AP fluorescence detection of RNA structural rearrangements, fluorescence resonance energy transfer (FRET) or molecular beacon (53) style RNA stem–loops provide potentially more useful approaches for the detection of the structural transition associated with conversion from kissing to duplex dimer (44) as well as the interaction of small molecule antagonists. These fluorescence detection schemes are more easily adapted to high-throughput screening formats. An example of the use of a Cy3/Dabcyl molecular beacon version of the DIS(AG) stem-loop to detect the NCp7 chaperoned conversion reaction is shown in Fig. 7. In this experiment, the fluorescence of Cy3 is found to increase in a time dependent fashion after addition of NCp7 as would be expected as the stem strands exchange and the Cy3 and Dabcyl are separated to opposite ends of the extended duplex dimer. Unlike 2-AP, however, these dyes appear more prone to direct quenching by NCp7, which can complicate the interpretation and analysis of the data.
Fig. 7.
(A) Schematic of the structural conversion of the DIS35(AG)-Cy3.Dabcyl• DIS35(UC) from a ‘kissing’ to extended duplex conformation. The Cy3 dye is covalently attached to the 5′ end of the strand and the Dabcyl quencher to the 3′ end via phosphoramidite linkages. (B) An example of the molecular beacon detected maturation of the DIS35(AG)-Cy3.Dabcyl•DIS35(UC) ‘kissing’ complex monitored as a function of a burst of fluorescence emission after addition of NCp7 at pH 6.5 for wild-type NCp7 protein. The sequence of the DIS35(AG)-Cy3.Dabcyl is shown inside the plot. As with the 2-AP detected methods, the rates of NCp7 chaperone activity can be fit from these data using either single, or double exponential rate equation.
5. Summary
The application of 2-AP fluorescence methods to monitor local structure and dynamics in nucleic acids provides significant advantages in parsing the analysis of structural transitions and local dynamics that may be associated with RNA folding. In particular, the sensitivity of 2-AP primarily to its local environment reduces or eliminates the complication of globally merged signals measured in other spectroscopic methods, like UV/Vis spectroscopy. The 2-AP fluorescence methods described here for the detection of structural changes are generally applicable to studies of other analogous RNA structural interactions and folding. In addition, as other fluorescent base analogs become commercially available in phosphoramidite form, like pyrrolo-cytosine (54), similar fluorescence methods for RNA structure and dynamics analyses can be designed that take advantages of the unique photophysical properties of these bases.
6. Acknowledgements
This work is supported by NIH Grant GM59107 awarded to J.P.M. We acknowledge the contributions of R. Mihailescu, M. Rist and C. Zhao, who were involved in the development and application of many of these methods. We thank F. Song for some of the oligonucleotide synthesis. Fluorescence instrumentation was supported in part by the National Institute of Standards and Technology.
Footnotes
1Certain commercial equipment, instruments, and materials are identified in this paper in order to specify the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the material or equipment identified is necessarily the best available for the purpose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Millar DP. Curr. Opin. Struct. Biol. 1996;6:322–26. doi: 10.1016/s0959-440x(96)80050-9. [DOI] [PubMed] [Google Scholar]
- 2.Rist MJ, Marino JP. Curr. Organ. Chem. 2002;6:775–93. [Google Scholar]
- 3.Ward DC, Reich E, Stryer L. J. Biol. Chem. 1969;244:1228. [PubMed] [Google Scholar]
- 4.Rachofsky EL, Osman R, Ross JBA. Biochemistry. 2001;40:946–56. doi: 10.1021/bi001664o. [DOI] [PubMed] [Google Scholar]
- 5.Hardman SJO, Thompson KC. Biochemistry. 2006;45:9145–55. doi: 10.1021/bi060479t. [DOI] [PubMed] [Google Scholar]
- 6.Menger M, Eckstein F, Porschke D. Biochemistry. 2000;39:4500–07. doi: 10.1021/bi992297n. [DOI] [PubMed] [Google Scholar]
- 7.Bharill S, Sarkar P, Ballin JD, Gryczynski I, Wilson GM, Gryczynski Z. Anal. Biochemistry. 2008;377:141–49. doi: 10.1016/j.ab.2008.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clerte C, Hall KB. Biochemistry. 2004;43:13404–15. doi: 10.1021/bi049117g. [DOI] [PubMed] [Google Scholar]
- 9.Hall KB, Williams J. RNA. 2004;10:34–47. doi: 10.1261/rna.5133404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballin JD, Bharill S, Fialcowitz-White EJ, Gryczynski I, Gryczynski Z, Wilson GM. Biochemistry. 2007;46:13948–60. doi: 10.1021/bi7011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blouin S, Lafontaine DA. RNA. 2007;13:1256–67. doi: 10.1261/rna.560307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris DA, Rueda D, Walter NG. Biochemistry. 2002;41:12051–61. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 13.Menger M, Tuschl T, Eckstein F, Porschke D. Biochemistry. 1996;35:14710–16. doi: 10.1021/bi960440w. [DOI] [PubMed] [Google Scholar]
- 14.Walter NG, Harris DA, Pereira MJB, Rueda D. Biopolymers. 2001;61:224–42. doi: 10.1002/bip.10144. [DOI] [PubMed] [Google Scholar]
- 15.Stephens OM, Yi-Brunozzi HY, Beal PA. Biochemistry. 2000;39:12243–51. doi: 10.1021/bi0011577. [DOI] [PubMed] [Google Scholar]
- 16.Austin RJ, Xia TB, Ren JS, Takahashi TT, Roberts RW. Biochemistry. 2003;42:14957–67. doi: 10.1021/bi0351312. [DOI] [PubMed] [Google Scholar]
- 17.DeJong ES, Chang CE, Gilson MK, Marino JP. Biochemistry. 2003;42:8035–46. doi: 10.1021/bi034252z. [DOI] [PubMed] [Google Scholar]
- 18.Kaul M, Barbieri CM, Pilch DS. J. Am. Chem. Soc. 2004;126:3447–53. doi: 10.1021/ja030568i. [DOI] [PubMed] [Google Scholar]
- 19.Bradrick TD, Marino JP. RNA. 2004;10:1459–68. doi: 10.1261/rna.7620304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao PW, Chow CS. Bioorg. & Med. Chem. 2007;15:3825–31. doi: 10.1016/j.bmc.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaul M, Barbieri CM, Pilch DS. J. Mol. Biol. 2005;346:119–34. doi: 10.1016/j.jmb.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 22.Lang K, Rieder R, Micura R. Nucl. Acids Res. 2007;35:5370–78. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avilov SV, Piemont E, Shvadchak V, de Rocquigny H, Mely Y. Nucl. Acids Res. 2008;36:885–96. doi: 10.1093/nar/gkm1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacourciere KA, Stivers JT, Marino JP. Biochemistry. 2000;39:5630–41. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 25.Yan ZH, Baranger AM. Bioorg. & Med. Chem. Lett. 2004;14:5889–93. doi: 10.1016/j.bmcl.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 26.Barbieri C, Kaul M, Pilch D. Tetrahedron. 2007;63:3567–6574. doi: 10.1016/j.tet.2006.08.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirk SR, Luedtke NW, Tor Y. Bioorg. & Med. Chem. 2001;9:2295–301. doi: 10.1016/s0968-0896(01)00123-7. [DOI] [PubMed] [Google Scholar]
- 28.Mihailescu MR, Marino JP. Proc. Natl. Acad. Sci. USA. 2004;101:1189–94. doi: 10.1073/pnas.0307966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rist MJ, Marino JP. Biochemistry. 2002;41:14762–70. doi: 10.1021/bi0267240. [DOI] [PubMed] [Google Scholar]
- 30.Zhao C, Marino JP. Tetrahedron. 2007;63:3575–84. doi: 10.1016/j.tet.2006.08.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Souza V, Summers MF. Nat. Rev. Microbiol. 2005;3:643–55. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 32.Paillart JC, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Biochimie. 1996;78:639–53. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 33.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Nat. Rev. Microbiol. 2004;2:461–72. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 34.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Nucl. Acids Res. 1987;15:8783–98. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milligan JF, Uhlenbeck OC. Methods in Enzymology. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 36.Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. J. Mol. Biol. 1998;279:633–49. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 37.St Louis DC, Gotte D, Sanders-Buell E, Ritchey DW, Salminen MO, Carr JK, McCutchan FE. J. Virol. 1998;72:3991–98. doi: 10.1128/jvi.72.5.3991-3998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi KI, Baba S, Chattopadhyay P, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. RNA. 2000;6:96–102. doi: 10.1017/s1355838200991635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rieder R, Lang K, Graber D, Micura R. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 40.Lang K, Micura R. Nat. Protoc. 2008;3:1457–66. doi: 10.1038/nprot.2008.135. [DOI] [PubMed] [Google Scholar]
- 41.Puglisi JD, Tinoco I. Meth. Enzymol. 1989;180:304–25. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 42.Theilleux-Delalande V, Girard F, Huynh-Dinh T, Lancelot G, Paoletti J. Eur. J. Biochem. 2000;267:2711–19. doi: 10.1046/j.1432-1327.2000.01292.x. [DOI] [PubMed] [Google Scholar]
- 43.Weixlbaumer A, Werner A, Flamm C, Westhof E, Schroeder R. Nucl. Acids Res. 2004;32:5126–33. doi: 10.1093/nar/gkh841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernacchi S, Ennifar E, Toth K, Walter P, Langowski J, Dumas P. J. Biol. Chem. 2005;280:40112–21. doi: 10.1074/jbc.M503230200. [DOI] [PubMed] [Google Scholar]
- 45.Tam VK, Kwong D, Tor Y. J. Am. Chem. Soc. 2007;129:3257–66. doi: 10.1021/ja0675797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Nat. Struct. Bio. 1998;5:432–36. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 47.Baba S, T. K, Noguchi S, Takaku H, Koyanagi Y, Yamamoto N, Kawai G. J Biochem. 2005;138:583–92. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 48.Kieken F, Paquet F, Brulé F, Paoletti J, Lancelot G. Nucl. Acids Res. 2006;34:343–52. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Nat. Struct. Biol. 2001;8:1064–68. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 50.Stivers JT. Nucl. Acids Res. 1998;26:3837–44. doi: 10.1093/nar/26.16.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ballin JD, Prevas JP, Bharill S, Gryczynski I, Gryczynski Z, Wilson GM. Biochemistry. 2008;47:7043–52. doi: 10.1021/bi800487c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss S. Science. 1999;283:1676–83. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Zhou X, Ye D. Biochem. Biophys. Res. Commun. 2008;373:457–61. doi: 10.1016/j.bbrc.2008.05.038. [DOI] [PubMed] [Google Scholar]
- 54.Tinsley RA, Walter NG. RNA. 2006;12:522–29. doi: 10.1261/rna.2165806. [DOI] [PMC free article] [PubMed] [Google Scholar]