Abstract
17-α-Ethynylestradiole (17EE) is a mechanism-based inactivator of CYP 2B1 and CYP 2B6 in the reconstituted system. The loss in enzymatic activity was due to the binding of a reactive intermediate of 17EE to the apoprotein. CYP 2B1 and CYP 2B6 were inactivated by 17EE and digested with trypsin. The peptides obtained following digestion with trypsin of 17EE-inactivated CYP 2B1 and CYP 2B6 were separated by liquid chromatography and analyzed by ESI-MS. Adducted peptides exhibiting an increase in mass consistent with the addition of the mass of the reactive intermediate of 17EE were identified for each enzyme. Analysis of these modified peptides by ESI-MS/MS and precursor ion scanning facilitated the identification of the S360 in both enzymes as the site that had been adducted by a reactive intermediate of 17EE. A CYP 2B1 mutant where S360 was replaced by alanine was constructed, expressed and purified. Activity and inactivation studies indicated that mutation of the S360 residue to alanine did not prevent inactivation of the mutant enzyme by 17EE. These observations suggest that S360 is not critical for the catalytic function of these P450s. Spectral binding studies of the 17EE-inactivated CYP 2B1 and CYP 2B6 indicated that modification of the enzymes by the reactive intermediate of 17EE resulted in an enzyme that was no longer capable of binding substrates. These results suggest that the inactivation by 17EE may be due to modification of an amino acid residue in the substrate access channel near the point of entry into the active site.
Introduction
The membrane-bound nature of mammalian CYPs has presented a particularly difficult challenge to the elucidation of their three dimensional structures. Recently, exciting progress has been made in crystallizing some members of the CYP family in both substrate-free and substrate-bound forms (Williams at al., 2000, Scott et al., 2003, Scott et al., 2004, Zhao et al., 2006). These studies have illustrated that CYP enzymes undergo dramatic conformational changes upon binding different substrates (Zhao et al., 2006, Zhao and Halpert, 2006). The ability of the CYP active site to mold itself to not only accommodate but also to metabolize different substrates to give, in some cases, several products has generated renewed emphasis on the identification of amino acid residues that are important in the process of substrate binding, orientation, catalysis, and product release.
17-α-Ethynylestradiol (17EE) was developed in 1938 and continues to be the major synthetic steroid in many oral contraceptives (Innhofen and Holweg, 1938, Bolt, 1979). Incorporation of an acetylenic moiety into the estradiol molecule resulted in an increase in the oral availability of the drug. However, it is well known that acetylenic functional groups may serve as sites of metabolism for CYP enzymes leading to their inactivation (Correia and Ortiz de Montellano, 2005). 17EE was shown to be a mechanism-based inactivator for CYPs 3A4, 2B1, and 2B6 (Guengerich 1988, Kent et al, 2002, Lin et al., 2002). Whereas the primary loss in activity of CYP 3A4 is due to heme modification, CYP 2B1 and CYP 2B6 are inactivated due to the formation of a covalent adduct to the apo-protein. The toxicological relevance of the inactivation of CYP enzymes has not been established in vivo. Yet, it is known that apo-protein modification of cellular macromolecules by reactive intermediates can lead to different disease states ranging from autoimmune diseases to neurotoxicity (LoPachin and DeCaprio, 2005).
A combination of approaches has been utilized to locate a) the site of adduct formation on the protein and b) the mass of the reactive intermediate of 17EE that was responsible for the loss in enzymatic activity during mechanism-based inactivation of the 2B P450s (Kent et al., 2006). Initial studies with four members of the CYP 2B family revealed that only CYP 2B1 and CYP 2B6 were inactivated by 17EE when incubated with NADPH in the reconstituted system. Isolation, N-terminal sequencing, and mass spectrometric analysis of peptides derived from CNBr digestion of CYP 2B1 and CYP 2B6 that had been inactivated using radio-labeled 17EE indicated that a peptide located between P347 and M376 (P347-M365 in 2B6) was the likely region that was modified. Because of the high sequence conservation between CYP 2B1 and CYP 2B2 for that peptide it was possible to narrow the region of interest to the relatively small sequence spanning S360 to V367. Trapping of the reactive 17EE intermediates with GSH from reaction mixtures containing either CYP 2B1 or CYP 2B6 resulted in the identification of glutathione conjugates with m/z values of 620 which would be indicative of glutathione trapping of a reactive 17EE intermediate with a mass of 312 Da. This value was also in very close agreement with the increase in mass of the CYP 2B1 apo-protein that was seen using whole protein mass spectrometric analysis.
This report now describes the identification of the amino acid residue (S360) in CYPs 2B1 and 2B6 that was modified by the reactive intermediate of 17EE using tandem mass spectral techniques. Mutation of the S360 residue to alanine and substrate binding studies indicate that the S360 residue is not required for or involved in catalytic activity. Rather, the inactivation is due to the location of S360 in the substrate access channel and when this residue is modified by a large bulky molecule such as the reactive 17EE intermediate further entry of substrates in to the CYP 2B1/2B6 active sites is blocked.
Experimental Procedures
Chemicals
Glutathione, catalase, NADPH, and L-α-dilauroyl-phosphatidylcholine were purchased from Sigma-Aldrich (St. Louis, MO). All other chemicals and solvents were of the highest purity available from commercial sources.
Purification of Enzymes
His-tagged CYP 2B1 was generated by modifying the CYP 2B1 expression plasmid. The CYP 2B1 containing pCW plasmid was digested with XbaI and KpnI and the sequence CATCACCACCAT was inserted to code for 4 His residues prior to the stop codon at the C-terminal end of CYP 2B1. The expression plasmid for His-tagged P450 2B6 was a generous gift from Dr. James R. Halpert. The enzymes were expressed in Escherichia coli Topp 3 cells (CYP 2B6) or MV1304 (CYP 2B1) and purified using Ni++-Agarose affinity chromatography as previously described (Domanski et al., 2001, Scott et al., 2001). Reductase was expressed and purified as described by Hanna et al. (Hanna et al., 1998).
Site-directed Mutagenesis and Purification of the S360A Mutant of 2B1
Construction of the S360A site-specific mutant of CYP 2B1 was performed using Stratagene’s Quik-Change site-directed mutagenesis-kit (Stratagene, La Jolla, CA). The upstream and downstream primers that were used for the PCR were 5′-CACGAGATTCAGAGGTTTGCAGATCTTGTCCCTATTGGAG-3′ and 5′-CTCCAATAGGGACAAGATCTGCAAACCTCTGAATCTCGTG-3′. The products of the PCR reaction were confirmed for the correct mutation at The University of Michigan DNA-sequencing facility (Ann Arbor, MI). The mutant plasmid was then transformed into MV1304 cells for expression. The expression and purification was carried out essentially according to published protocol (Hanna et al., 1998, 2000).
Enzyme Assays
CYP 2B6 (1 μM) was reconstituted with reductase (2 μM) on ice for 45 min. The CYP 2B6 primary reaction mixture contained 2B6 (1 μM), reductase (2 μM), catalase (110 units), 100 μM 17EE and 100 mM potassium phosphate buffer (pH 7.5) in a total volume of 1 mL. CYP 2B1 (1 μM) was reconstituted with reductase (2 μM) and DLPC (100 μg/ml). The CYP 2B1 reaction mixture received catalase (110 units), and 50 μM 17EE in a final volume of 1 ml of 100 mM phosphate buffer (pH 7.5). Each of the reconstitution mixtures was divided into two samples and incubated at 37 °C for 5 min. One sample received 1 mM NADPH and the control sample received an equal volume of water. The mixtures were incubated for 30 min at 37 °C. The CYP 2B1 S360A mutant was reconstituted with reductase and lipid as described for the wild type enzyme above. Samples received 1, 5, 10, 25 or 50 μM 17EE and were assayed for 7EFC activity at 2, 4, 8, and 16 min after adding NADPH to initiate the reactions as previously described (Kent et al., 2005). The loss in enzymatic activity was compared to a control sample that was incubated with solvent (DMSO) instead of 17EE.
The metabolism of testosterone was determined as previously described (Waxman et al., 1983; Wood et al., 1983). CYP 2B1 as well the CYP S360A mutant were reconstituted with reductase as described above. Each sample contained 25 pmol of reconstituted CYP enzymes and was incubated with 1 mL of testosterone assay buffer (0.2 mM testosterone, 2 mM NADPH in 50 mM HEPES buffer) at 37 °C for 20 min. The reactions were terminated by adding 1 mL of ethyl acetate. The metabolites were extracted twice with 1 mL of ethyl acetate and the organic phases were pooled and dried under N2. The products were dissolved in 65% methanol and resolved using isocratic HPLC analysis with 65% methanol as the solvent at a flow rate of 0.85 mL/min on a Varian Microsorb-MV C18 reversed phase column (5 μm, 4.6 mm ×150 mm, Walnut Creek, CA). Metabolites were detected using UV absorption at 254 nm. The area under the peaks for the major metabolites for testosterone which are 16α-hydroxytestosterone, 16β-hydroxytestosterone and androstenedione were integrated to compare the quantities that were formed for each enzyme.
Digestions with Trypsin
Control and 17EE-inactivated CYP 2B1 or CYP 2B6 (1 nmol in 1 ml each) were separated from the components of the reconstitution mixture by first adding a 10% cholate solution to a final concentration of 0.5% and incubating for 15 min at room temperature. Each sample then received 100 μL of a 50% suspension of Ni++-agarose beads that had been washed once with 0.5 M KPi, pH 7.4, followed by two washes with 100 mM KPi, pH 7.4, containing 0.5% cholate and 20% glycerol (wash buffer). The washed beads were suspended in a 2-fold volume of this wash buffer. The CYP samples were incubated with the Ni++-agarose beads for 2 hr at RT on a rocking platform to allow for binding of the His-tagged CYPs to the affinity matrix. The beads were pelleted by spinning the samples in a microfuge for 2 min at maximum speed and washed three times with 1 mL of wash buffer. The wash buffer was removed and the CYPs were eluted from the beads in a minimal volume of elution buffer (usually 50 - 100 μL of 100 mM KPi, pH 7.4, 20% glycerol, 0.73 M NaCl, and 400 mM imidazole). The eluted CYPs were dialyzed for 4 hr against 1 L of 100 mM KPi, pH 7.5, containing 20 % glycerol, 0.1 mM DTT followed by dialysis against 100 mM ammonium bicarbonate pH 7.5, and 0.1 mM DTT. Each sample received 1 μL of trypsin (Promega, 1 μg/10 μL suspension buffer) and was incubated over night at 37 °C with agitation. The digest was then acidified with 1/5 of the sample volume of a solution of 20% CH3CN and 5% formic acid. Approximately 1/5 of the sample was injected and analyzed by ESI-LC-MS/MS.
ESI-LC-MS/MS Analysis
Samples were analyzed on a C18 reverse phase column (Luna, 3 micron, 100 × 4.6 mm, Phenomenex, Torrance, CA) using a gradient of 20 - 30% B (A: 0.5 % formic acid in water, B: 0.5 % formic acid in acetonitrile) in 5 min followed by a linear increase to 40% B by 15 min and to 90% B by 30 min at a flow of 0.2 mL/min. The column effluent was directed into a LCQ mass analyzer. The ESI conditions were: Sheath gas 90 arbitrary units; auxiliary gas 30 arbitrary units; capillary temperature 170 °C; spray voltage 30 V. Data were acquired in positive mode using the Excalibur software package (Thermo-Finnegan, Schaumburg, IL) with one full scan followed by two data-dependent scans of the second most intense and the third most intense ion.
Alternatively, samples were analyzed using a LTQ mass analyzer (Thermo-Finnegan, Shaumburg, IL) equipped with a photodiode array detector under the same chromatography conditions as described above for the LCQ system. The analyzer conditions were optimized with a deuterated synthetic CYP 2B1 peptide (D5-FSDLVPIGVPHR) and the settings were: Sheath gas, 40 arbitrary units; auxiliary gas, 20 arbitrary units; capillary temperature, 350 °C; spray voltage, 4kV, capillary voltage, 40 V; tube lens offset, 30 arbitrary units.
Neutral loss scanning for ions with a mass of 310 -312 for confirmation purposes was carried out on a TSQ mass analyzer (Thermo-Finnegan, Shaumburg, IL) with a normalized collision energy of 35. The column and the liquid chromatography conditions were as described above for the LCQ.
Binding Spectra
CYP 2B1 or 2B6 (1 nmol/1 mL sample) were reconstituted with reductase as described above for the activity assays. Samples were incubated with 17EE in the absence or presence of 1 mM NADPH at 30 °C for 20 - 30 min. An aliquot was removed and assayed for residual activity. The remaining samples were dialyzed against 1 L of dialysis buffer containing 50 mM KPi, pH 7.4, 20 % glycerol, 0.5 mM EDTA for 4-5 h at 4 °C followed by overnight dialysis against another 1 L of dialysis buffer without EDTA. The samples were removed and the volume was adjusted so that the concentration of CYP was 1 μM. The absolute spectrum at 418 nm was used to ensure that the control and the inactivated sample contained the same amount of CYP. The sample and the reference cuvette received 0.5 nmol of CYP each and a baseline scan was taken between 500 and 350 nm. Aliquots (1 μL each) of either benzphetamine or n-octylamine in water were added to the sample cuvette. The change in absorbance between 418 and 395 nm (for benzphetamine, Type I) or between 430 and 410 nm (for n-octylamine, Type II) was recorded for each control or 17EE-inactivated CYP. The maximum change in absorbance observed with the control sample was assigned 100% and the relative amount for the inactivated sample was calculated.
Results
ESI-LC-MS/MS Analysis of 17EE-Inactivated P450 2B1 Following Digestion with Trypsin
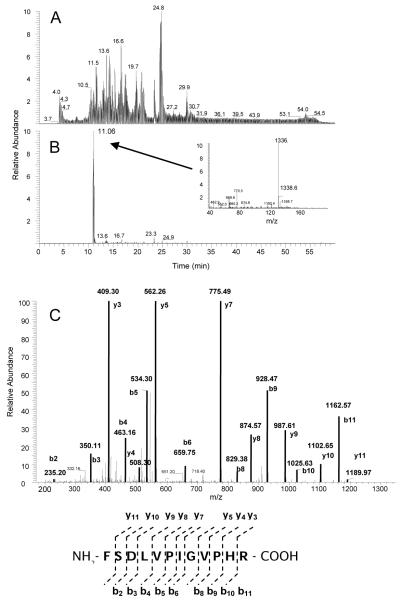
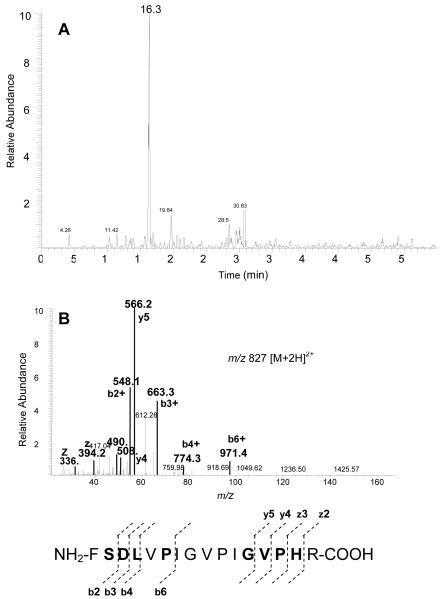
Figure 1A shows the total ion chromatogram (TIC) for the tryptic digest of CYP 2B1 that was inactivated by 17EE. Previously, a peptide spanning residues P347 - M376 isolated following digestion with CNBr was identified as containing the 17EE adduct (Kent et al., 2006). Digestions with trypsin of control, non-inactivated CYP 2B1 would have generated an un-adducted peptide from this region of the protein with a mass of 1337 Da spanning residues F359 - R370. A peptide with this mass (Fig. 1B inset) and sequence (Fig. 1C) was found to elute at 11.06 min (Fig. 1B). Previous studies identified a glutathione conjugate of 17EE that indicated that the mass of the reactive intermediate was 312 Da (Kent et al., 2006). A search of the digest for a peptide with a mass of 1648 Da revealed a doubly charged peptide that eluted at 16.32 min (Fig. 2A). A peptide with this mass was not observed in control digests of CYP 2B1 that had been incubated with 17EE in the absence of NADPH (data not shown). MS/MS analysis of this peptide showed that the increase in mass occurred at the b2 ion or at S360 in the sequence of 2B1 (Fig. 2B). As would be expected for a modification located on this S360, no increases in the masses of the y-ions from the c-terminus were observed.
Figure 1.
ESI-LC-MS analysis of 17EE-inactivated CYP 2B1. CYP 2B1 was reconstituted with reductase, inactivated with 17EE, purified and digested with trypsin as described in Experimental Procedures. (A) TIC of the peptides obtained from the trypsin digest of CYP 2B1 that was inactivated by 17EE, (B) Extracted ion chromatogram (XIC) of the non-adducted peptide with a m/z of 1336.7 (the inset is the mass spectrum of the peptide peak eluting at 11.06 min), and (C) MS/MS analysis and sequence of the m/z 1336.7 peptide. Residues in bold were positively identified by MS/MS.
Figure 2.
ESI-LC-MS/MS analysis of the modified CYP 2B1 peptide spanning residues F359-R370. (A) XIC of the predicted peptide with a m/z of 1648 Da (1336 + 312) and (B) MS/MS analysis of the doubly charged form of this peptide showed that the increase in mass occurred at the b2 ion or at S360 in the 2B1 sequence. Residues in bold were positively identified by MS/MS.
ESI-LC-MS/MS Analysis of 17EE-Inactivated P450 2B6 Following Digestion with Trypsin
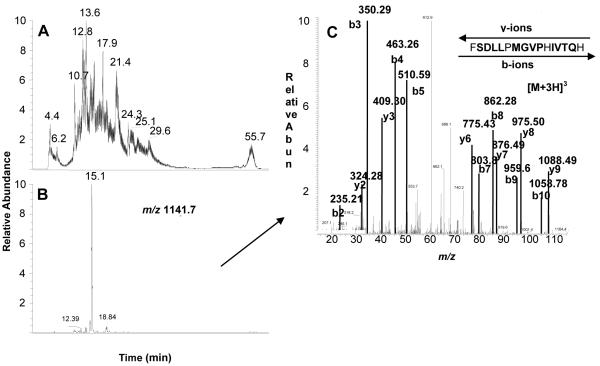
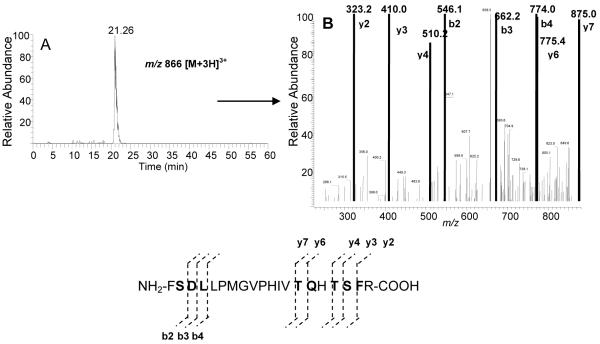
The TIC of the peptide digest obtained from digesting 17EE-inactivated CYP 2B6 with trypsin is shown in Fig 3A. A modification derived from a reactive 17EE intermediate had previously been located to a peptide spanning residues P347 - M365 isolated from a CNBr digestion of CYP 2B6 inactivated by 17EE. Digestion with trypsin in this region of the control, non-inactivated CYP 2B6 protein would be expected to yield an un-modified peptide with a mass of 2284 Da. The mass of this peptide is too large for it to be observed in the singly charged state but it was found as the doubly charged peptide with a m/z value of 1141.7 [M+2H]2+ and it eluted at 15.12 min (Fig. 3B). The sequence result obtained from MS/MS analysis of this peptide is shown in Fig. 3C and indicates that the peptide corresponds to the CYP 2B6 peptide comprising residues F359 - R378. A modified peptide with a mass increase of 312 Da was seen to elute at 21.26 min (Fig. 4A). The m/z of this peptide was 866 for [M+3H]3+ or 1297 for [M+2H]2+ which would correspond to the mass of the F359 - R378 peptide plus the mass of the reactive 17EE intermediate. A peptide with this mass was only seen in digests from the 17EE-inactivated samples and not in control samples (data not shown). The triply charged peptide gave the clearest MS/MS results (Fig. 4B) and was therefore used to obtain the sequence information shown in Fig. 4C. As was seen with CYP 2B1 above, a mass increase corresponding to the mass of the reactive 17EE intermediate occurred starting with the b2 ion. Additional residues are located in this sequence that could have been modified including T373, T375, or S376 since they also contain an OH-side chain that could be involved in forming an ester linkage. However, the y-ion series through this region indicated that none of these residues had undergone modification.
Figure 3.
ESI-LC-MS analysis of 17EE-inactivated CYP 2B6. CYP 2B6 was reconstituted with reductase, inactivated with 17EE, purified and digested with trypsin as described in Experimental Procedures. (A) TIC of the peptides obtained from the trypsin digest of CYP 2B6 inactivated by 17EE, (B) Extracted ion chromatogram of the doubly charged, non-adducted peptide with a m/z of 1141.7, and (C) MS/MS analysis and sequence of the m/z 1141.7 peptide. Residues in bold were positively identified by MS/MS.
Figure 4.
ESI-LC-MS/MS analysis of the CYP 2B6 peptide spanning residues F359-R378 following inactivation by 17EE. (A) XIC of a triply charged peptide with a predicted m/z of 2596 (2284 + 312) (B) MS/MS analysis of the triply charged form of this peptide showed that the increase in mass occurred at the b2 ion or at S360 in the 2B6 sequence. Residues in bold were positively identified by MS/MS.
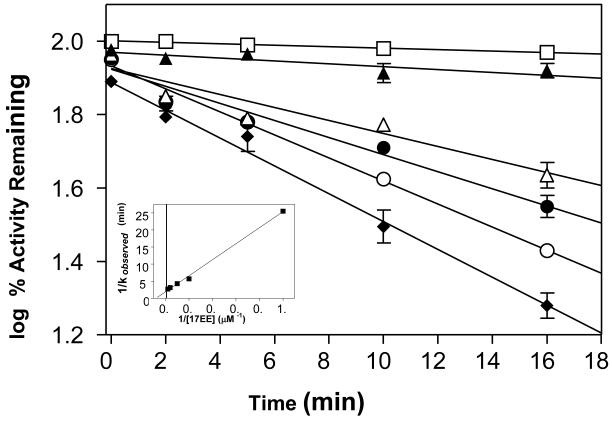
Effect of 17EE on the Activity of CYP 2B1 S360A
The S360 residue in CYP 2B1 that became modified by the reactive intermediate of 17EE was replaced with alanine. The resulting mutant was expressed, purified and analyzed for its ability to metabolize benzphetamine and to become inactivated by 17EE. Metabolism of benzphetamine by the S360A mutant resulted in the formation of 36 pmol of formaldehyde/min/pmol CYP. This was 40% lower, although still reasonably similar to the wild type enzyme which formed 60 pmol of formaldehyde/min/pmol CYP. The testosterone metabolite profile of the mutant was nearly identical to that of the wild type enzyme (data not shown). Because catalytic activity was retained, this suggested that S360 did not play a critical role in catalytic activity. When the CYP 2B1 S360A mutant was incubated with 17EE in the presence of NADPH, activity loss, as measured using either 7EFC or testosterone as the substrate, was still observed. Table 1 shows that after incubating the CYPs with 17EE and NADPH for 20 min there was a significant reduction in the formation of the 16α- and 16β-hydroxytestosterone, and androstenedione products by the wild type and S360A mutant. The extent to which the formation of 16α-hydroxytestosterone and androstenedione decreased was similar for both enzymes. However, the 17EE-inactivated S360A mutant was able to form considerably more (65% vs 8%) of the 16β product as compared to the wild type enzyme. The effect of incubating the enzyme with 17EE in the presence of NADPH on the 7-EFC O-deethylation activity of the S360 mutant of CYP 2B1 is shown in Figure 5. A time-, concentration-, and NADPH-dependent loss in the enzymatic activity was observed. The kinetic constants that were calculated for the mutant enzyme were: KI = 14 uM, kinact = 0.05 min-1, and t1/2 = 14 min. Although the KI for 17EE inactivation of the S360A mutant was similar to that observed with the wild type 2B1, the rate of inactivation was approximately 4-fold slower than what had been observed previously with the wild-type enzyme (KI = 11 μM, kinact = 0.2 min-1, t1/2 = 4 min).
Table 1. Effect of 17 EE on T estosterone Metabolism bythe Wild Type and S360A Mutant of CYP 2B1a.
| CYP | % of metabolite formed relative to Control | ||
|---|---|---|---|
| 16α-Hydroxytestosterone | 16β-Hydroxytestosterone | Androstenedione | |
| WT | 24 | 8 | 41 |
| S360A | 35 | 65 | 50 |
CYPs 2B1 or 2B1 S360A were reconstituted and inactivated with 17EE for 20 min as described in Experimental Procedures. Aliquots were removed and analyzed for metabolite formation using HPLC as described in Experimental Procedures. The area under each metabolite was integrated and used to calculate the percentage of metabolite formed compared to controls that were incubated with 17EE in the absence of NADPH.
Figure 5.
Time- and concentration-dependent inactivation of CYP 2B1 S360A. CYP 2B1 S360A was reconstituted with reductase and incubated with different concentrations of 17EE in the absence or presence of NADPH as described in Experimental Procedures. Aliquots were removed at 0, 2, 4, 8, and 16 min and assayed for residual 7-EFC activity. (□), 0 μM; (■) 1 μM; (△) 2 μM; (▲) 4 μM; (○) 8 μM and (●) 16 μM 17EE. The inset shows the double reciprocal plot of the inverse of the rates of inactivation vs the inverse of the substrate concentration which was used to estimate the kinetic constants describing the inactivation reaction.
Binding Spectra
Spectral binding studies of the control or 17EE-inactivated CYPs 2B1 and 2B6 enzymes were performed using two different size compounds that give different spectral changes, n-octylamine (Type II) and benzphetamine (Type I). Table 2 shows that in each case, the extent of loss in the ability of the 17EE-inactivated CYP to form a binding spectrum was similar to the level of inactivation. CYP 2B1 that had 34% of its activity remaining was able to form approximately 45% and 39% of the maximum n-octylamine and benzphetamine binding spectra that were possible with the non-inactivated enzyme. With CYP 2B6 a 79% loss in activity resulted in a loss of approximately 81% and 66% in the maximum spectral binding n-octylamine and benzphetamine, respectively.
Table 2. Effect of Inactivation by 17EE on the Ability of CYPs 2B1 and 2B6 to Metabolize and Bind Substratesa.
| % Remaining compared to Non-Inactivated Controls | |||
|---|---|---|---|
| CYP | Activity | n-Octylamine 431 nm- 410 nm |
Benzphetamine 385 nm- 418 nm |
| 2B1 | 34 | 45 | 39 |
| 2B6 | 21 | 19 | 34 |
CYPs 2B1 or 2B6 were reconstituted with reductase and incubated with 17EE in the presence or absence of NADPH as described in Experimental Procedures. Aliquots were removed and assayed for residual 7EFC activity and for spectral binding changes upon addition of the substrates indicated.
Discussion
In general, the isolation and identification of CYP peptides that have been adducted by reactive intermediates has been challenging. Multiple reasons account for these difficulties. With most mechanism-based inactivators the inactivation does not result in 100% of the molecules becoming modified. As the CYP enzymes undergo inactivation, the heme moiety can also become modified by the reactive intermediate or the damaged enzyme may generate other radicals that result in heme destruction and cross-linking as well as further modification of the protein. The inactivated CYPs generally have a greater tendency to aggregate and therefore, at each step in the isolation and analytical procedures the amount of the modified protein may decrease significantly. The adducted peptide will generally be more hydrophobic, also increasing the potential for a disproportional loss of the peptide of interest at each step. In addition, the chemical bond formed by the interaction of the reactive intermediate with an amino acid side chain may be labile to the purification or digestion conditions used. This may result in loss of adduct thereby resulting in the recovery of only the native peptide. The presence of the adduct at the site of adduction may result in incomplete digestion of the CYP. Finally, the adduct may prevent complete ionization of the peptide of interest or result in a fragmentation pattern that is difficult to interpret. Although all of the points mentioned above apply to a greater or lesser extent to 17EE-adducted CYPs, the relative selectivity and completeness of apo-protein adduct formation in a short period of time with the 2B family of enzymes made 17EE-adducted peptides more amenable to identification and sequencing. The use of His-tagged enzymes greatly facilitated the rapid isolation of only the CYPs from the reaction mixtures using affinity beads. This separation and purification technique allowed for direct enzymatic digestion of the desired protein without the potential losses of adduct or modified CYPs that can be encountered using reverse-phase chromatography and subsequent drying steps or the effects of SDS when gel electrophoresis is employed as a means of separation.
Previous observations were invaluable in the search for the adducted residues (Kent at al., 2002, Kent et al., 2006). N-terminal sequencing of [3H]-peptides obtained after cyanogen bromide cleavage of CYPs 2B1 and 2B6 that had been inactivated with [3H]17EE and purified by reverse-phase chromatography yielded the first 5-10 amino acid residue sequences corresponding to peptides P347 - M376 (for CYP 2B1) and P347 - M365 (for CYP 2B6). Comparison of the CYP 2B1 and 2B6 peptides narrowed the site of adduction to the 17 residues located between P347 and M365. The mass of the CNBr-derived CYP 2B1 peptide plus an adduct could be confirmed by MALDI-MS. Base treatment of the adducted CNBr-derived peptide resulted in the partial removal of the adduct suggesting that the adduct was linked to an amino acid residue via an ester linkage. This observation suggested that the 17EE derived adduct was either on Y348, T349, or S360. Sequence comparisons between the 2B enzymes (2B1 and 2B6) that were inactivated by 17EE and those that were not inactivated (2B2 and 2B4) indicated that S360 may be the most likely site of modification. Identification of the metabolites of 17EE that were produced by CYPs 2B1 and 2B6 and identification of 17EE-GS conjugates (m/z of 620, GSH + 312 Da) indicated that the size of the adduct (i.e. the increase in mass of the peptide of interest) would be 312 Da. In combination, the above information allowed for mass specific searches, sequence specific searches, and neutral loss confirmation by LC-MS/MS of tryptic digests that made it possible to identify S360 as the amino acid residue in CYPs 2B1 and 2B6 that was modified by a reactive 17EE intermediate.
The metabolism of substrates by CYP enzymes involves at least nine separate steps that require a two electron reduction of molecular oxygen that usually results in oxidation of the substrate with the concurrent formation of water (Guengerich FP, 2001). Covalent binding of a reactive intermediate to a CYP has been found to correlate with one or more defects in the different steps of the catalytic cycle (Halpert et al., 1985; Parkinson et al., 1986; Roberts et al., 1995, Kent et al., 2004). The functional significance of the S360 residue was investigated by first generating a S360A mutant enzyme and then by evaluating this mutant for its ability to a) metabolize substrates and b) to become inactivated by 17EE. Comparative metabolism studies of three common 2B substrates, 7EFC, benzphetamine and testosterone, showed virtually no difference in the amount or the type of product(s) that were generated by the mutant when compared to the wild type enzyme. The studies on the inactivation of the S360A mutant by 17EE indicated that not only was the mutant able to metabolize CYP 2B substrates but, it could also be inactivated by 17EE. Interestingly, the testosterone metabolite profile of the 17EE-inactivated mutant suggested that although all the metabolites produced by the wild type enzyme were also generated by the mutant, the mutant yielded a larger amount of the 16β-hydroxy product. This suggests that the location of the 17EE adduct in the mutant enzyme is at a site in the enzyme where it may affect the orientation of the testosterone molecule while bound in the active site. The kinetic data for the inactivation of the CYP 2B1 S360A mutant by 17EE was similar to that for the wild type enzyme in the reconstituted system. These data suggest that replacing the Ser residue at position 360 with Ala did not greatly affect the overall activity of the enzyme or its ability to become inactivated. Rather, it was the modification of the serine OH group by the bulky 17EE reactive intermediate that was responsible for the loss in activity. Examination of a model of CYP 2B6 that was constructed based on the CYP 2B4 crystal structure indicated that S360 was located greater than 5 Å from the active site iron with its OH side chain pointing away from the active site. This observation raises the possibility that modification at this location by a bulky group such as 17EE may prevent substrates from reaching the active site, thereby preventing metabolism and essentially inactivating the enzyme. This hypothesis was confirmed experimentally by performing substrate binding studies on 17EE-inactivated CYP 2B1 or 2B6 using two typical 2B substrates (large - benzphetamine, and small - n-octylamine). The results clearly demonstrated that only the fraction of the enzyme that had not been inactivated was able to bind either of the two substrates. The inability of substrates to reach the heme iron of the 17EE-modified CYPs was confirmed with phenyldiazine. When the 17EE-inactivated CYPs were incubated with phenyldiazine and analyzed for the phenyldiazine heme conjugates by LC-MS, the data indicated again that only the residual fractions of the enzymes that were still active were able to form the expected complexes of phenyldiazine with the heme (data not shown). These results suggest that the inactivation of P450s 2B1 and 2B6 by 17EE is due to the presence of the covalent adduct of 17EE on the serine 360 residue which is in the susbstrate access channel and therefore it blocks the entry of substrates into the P450 active sites, thereby preventing substrate metabolism by the modified P450.
Acknowledgements
The authors express their appreciation to Dr. Kathryn Noon of the Department of Pharmacology’s mass spectrometry facility for her assistance with the use of the LTQ mass analyzer and her advice and analytical help in the neutral loss experiments.
Footnotes
- 17EE
- 17-α-ethynylestradiol
- reductase
- NADPH-cytochrome P450 reductase
- GSH
- glutathione
- 7-EFC
- 7-ethoxy-4-(trifluoromethyl)coumarin
- ESI-LC-MS
- electrospray ionization liquid chromatography mass spectrometry
- MS/MS
- tandem mass spectrometry
- TIC
- total ion chromatogram
- XIC
- extracted ion chromatogram
References
- Bolt HM. Metabolism of estrogens - natural and synthetic. Pharmacol Ther. 1979;4:155–181. doi: 10.1016/0163-7258(79)90018-4. [DOI] [PubMed] [Google Scholar]
- Domanski TL, Finta C, Halpert JR, Zaphiropoulos PG. cDNA cloning and initial characterization of CYP3A43, a novel human cytochrome P450. Mol Pharmacol. 2001;59:386–392. doi: 10.1124/mol.59.2.386. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Oxidation of 17α-ethynylestradiol by human liver cytochrome P450. Mol Pharmacol. 1988;33:500–508. [PubMed] [Google Scholar]
- Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:612–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- Halpert JR, Miller NE, Gorsky LD. On the mechanism of the inactivation of the major phenobarbital-inducible isozyme of rat liver cytochrome P-450 by chloramphenicol. J Biol Chem. 1985;260:8397–8403. [PubMed] [Google Scholar]
- Hanna IH, Teiber JF, Kokones KL, Hollenberg PF. Role of alanine in position 363 of cytochrome P450 2B2 in influencing NADPH- and hydrogenperoxide-supported activities. Arch Biochem Biophys. 1998;350:324–332. doi: 10.1006/abbi.1997.0534. [DOI] [PubMed] [Google Scholar]
- Hanna IH, Reed JR, Guengerich FP, Hollenberg PF. Expression of human cytochrome P450 2B6 in Escherichia coli: characterization of catalytic activity and expression in human liver. Arch Biochem Biophys. 2000;376:206–216. doi: 10.1006/abbi.2000.1708. [DOI] [PubMed] [Google Scholar]
- Innhoffen HH, Holweg W. Neue per os-wirksame weibliche Keimdrüsen-hormon-Derivate. 17-Äthinylöstradiol und Pregnene-in-on-3-ol-17. Naturwissenschaften. 1938;26:96. [Google Scholar]
- Kent UM, Mills DE, Rajarayanan RV, Alworth WL, Hollenberg PF. Effect of 17-α-ethynylestradiol on activities of cytochrome P450 2B (P450 2B) enzymes: characterization of inactivation of P450s 2B1 and 2B6 and identification of metabolites. J Pharmacol Exp Therap. 2002;300:549–558. doi: 10.1124/jpet.300.2.549. [DOI] [PubMed] [Google Scholar]
- Kent UM, Pascual L, Roof RA, Ballou DP, Hollenberg PF. Mechanistic studies with N-benzyl-1-aminobenzotriazole-inactivated CYP 2B1: differential effects on metabolism of 7-ethoxy-4-(trifluoromethyl)coumarin, testosterone, and benzphetamine. Arch Biochm Biophys. 2004;423:277–287. doi: 10.1016/j.abb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Kent UM, Lin H-L, Mills DE, Regal KA, Hollenberg PF. Identification of 17-α-ethynylestradiol-modified active site peptides and glutathione conjugates formed during metabolism and inactivation of P450s 2B1 and 2B6. Chem Res Toxicol. 2006;19:279–287. doi: 10.1021/tx050256o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoPachin RM, DeCaprio AP. Protein adduct formation as a molecular mechanism in neurotoxicity. Toxicol Sci. 2005;86:214–225. doi: 10.1093/toxsci/kfi197. [DOI] [PubMed] [Google Scholar]
- Correia MA, Ortiz de Montellano PR. Inhibition of Cytochrome P450 Enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. Third Edition Kluiwer Academic/Plenum Publishers; New York: 2005. pp. 247–322. [Google Scholar]
- Parkinson A, Thomas PE, Ryan DE, Gorsky LD, Shively JE, Sayer JM, Jerina DM, LeAn W. Mechanism of inactivation of rat liver microsomal cytochrome P-450c by 2-bromo-4′-nitroacetophenone. J Biol Chem. 1986;261:11487–11495. [PubMed] [Google Scholar]
- Roberts ES, Ballou DP, Hopkins NE, Alworth WL, Hollenberg PF. Mechanistic studies of 9-ethynylphenanthrene-inactivated cytochrome P450 2B1. Arch Biochem Biophys. 1995;323:303–312. doi: 10.1006/abbi.1995.9960. [DOI] [PubMed] [Google Scholar]
- Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc Natl Acad Sci (USA) 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structrue of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Ko A, Walsh C. Regioselectivity and stereoslectivity of androgen hydroxylations catalyzed by cytochrome P450 isozymes purified from phenobarbital-induced rat liver. J Biol Chem. 1983;258:11937–11947. [PubMed] [Google Scholar]
- Wood AW, Ryan DE, Thomas PE, Levin W. Regio- and stereoselective metabolism of two C19 steroids by five highly puried and reconstituted rat hepatic cytochrome P450 isozymes. J Biol Chem. 1983;256:8839–8847. [PubMed] [Google Scholar]
- Wester MR, Johnson EF, Marques-Soares C, Dansette PM, Mansuy D, Stout CD. Structure of a substrate complexed of mammalian cytochrome P450 2C5 at 2.3 A resolution: evidence for multiple substrate binding modes. Biochemistry. 2003;42:6370–6379. doi: 10.1021/bi0273922. [DOI] [PubMed] [Google Scholar]
- Wester MR, Johnson EF, Marques-Soares C, Dijols S, Dansette PM, Mansuy D, Stout CD. Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1 A resolution: evidence for an induced fit model of substrate binding. Biochemistry. 2003;42:9335–9345. doi: 10.1021/bi034556l. [DOI] [PubMed] [Google Scholar]
- Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J Biol Chem. 2006;281:5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Halpert JR. Structure-function analysis of cytochromes P450 2B. Biochem Biophys Acta. 2006;1770:402–412. doi: 10.1016/j.bbagen.2006.07.006. [DOI] [PubMed] [Google Scholar]