Abstract
The biotin-tagged electrophiles 1-biotinamido-4-(4′-[maleimidoethylcyclohexane]-carboxamido)butane (BMCC) and N-iodoacetyl-N-biotinylhexylenediamine (IAB) have been used as model electrophile probes in complex proteomes to identify protein targets associated with chemical toxicity. Whereas IAB activates stress signaling and apoptosis in HEK293 cells, BMCC does not. Cysteine Michael adducts formed from BMCC and non-biotinylated analogs rapidly disappeared in the intact cells, whereas the adducts were stable in BMCC-treated subcellular fractions, even in the presence of the cellular reductants reduced glutathione, NADH and NADPH. In contrast, cysteine thioether adducts formed from IAB and its non-biotinylated analogs were stable in intact cells. Loss of the BMCC adduct in cells was reduced at 4 °C, which suggests the involvement of a metabolic process in adduct removal. Model studies with a glutathione-BMCC conjugate indicated rapid hydrolysis of the adducted imide ring, but neither the conjugate nor its hydrolysis product dissociated to release the electrophile in neutral aqueous buffer at significant rates. The results suggest that low BMCC toxicity reflects facile repair that results in transient adduction, which fails to trigger damage signaling pathways.
Reactive electrophilic metabolites of toxic chemicals and endogenous electrophilic products of cellular oxidative stress can covalently modify cellular macromolecules and cause cellular damage associated with drug and chemical toxicity (1-5). Proteins are major targets of reactive electrophiles due to their diverse nucleophilic side chains, including cysteine, histidine and lysine. Of these, cysteine sulfhydryl groups are particularly important because of their high reactivity toward electrophiles and oxidants. Modification of thiol groups on functional protein sensors is thought to be a primary mechanism of initiating signaling responses associated with cell toxicity and adaptive responses to electrophiles (6-8).
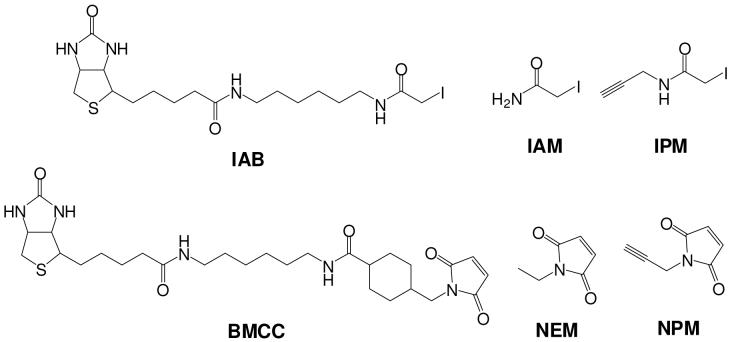
A major challenge in the study of protein damage and its consequences is the difficulty of selectively capturing and analyzing adducted proteins, which are often present as a small fraction of total protein (4). We and others have employed the biotin-tagged electrophiles BMCC1 and IAB (Scheme 1) as model electrophile probes to evaluate the characteristics of protein damage on a global scale (9-11), as well as to study the mechanisms by which site-specific damage induces specific effects (12-16). These probes display reaction chemistries relevant to several classes of electrophiles generated by metabolism of xenobiotics.
Scheme 1.
Structures of electrophiles.
Previous studies demonstrate that IAB and BMCC display surprising differences in both global protein adduction (9-11) and in the formation of site-specific adducts on individual proteins (12, 13, 17, 18). These two probes also produce different biological effects in cells. Most strikingly, IAB is toxic to HEK293 cells, whereas BMCC is not. IAB causes cell death accompanied by apoptotic markers including cytochrome c release, caspase-3 activation and poly(ADP-ribose)polymerase cleavage (11). IAB also induces endoplasmic reticulum stress (ER stress) (10) and induction of stress genes via the antioxidant response element and induction of the transcription factor NF-E2-related factor 2 (Nrf2) (12), whereas BMCC induces none of these effects.
We have previously speculated that these differences in toxicity are related to significant differences in protein adduction by IAB and BMCC observed in studies with subcellular fractions (10, 11). Only about 20% of the identified protein targets and target sites are adducted by both compounds (9-11). However, we also began studies of the binding of IAB and BMCC in intact HEK293 cells and discovered that relative levels of IAB and BMCC adducts appeared to differ depending on the time of sampling post-treatment. This suggested significant differences in the kinetics of adduct formation or the stability of the adducts.
Here we describe studies of the formation and stability of IAB and BMCC adducts in intact HEK293 cells. We report that IAB adducts accumulate and are stable in HEK293 cells, whereas BMCC adducts rapidly reach peak levels, but then decline. In contrast, BMCC adducts are stable in HEK293 cell lysates. Similar results were obtained with small alkynyl analogs of BMCC and IAB, which form protein adducts that can be biotinylated ex vivo using Click chemistry. Comparison between small alkynyl analogs and the larger biotin electrophile probes confirms the instability of the maleimide adducts and the corresponding lack of toxicity. We found that BMCC added to HEK293 cells undergoes rapid conjugation with GSH and that the maleimide ring in both BMCC and the GS-BMCC conjugate is rapidly hydrolyzed. However, neither the GS-BMCC conjugate nor its ring-opened hydrolysis product undergoes a facile retro-Michael reaction to release the corresponding electrophile and GSH. These observations collectively suggest that stability or persistence of protein covalent adducts may be critical determinants of toxicity.
EXPERIMENTAL PROCEDURES
General methods and materials
BMCC and IAB were obtained from Pierce (Rockford, IL). N-ethylmaleimic acid was from Alfa Aesar (Ward Hill, MA), and Alexa Fluor 680-conjugated streptavidin was from Molecular Probes (Eugene, OR). Mass spectrometry grade trypsin (Trypsin Gold) was from Promega (Madison, WI). 1H NMR (300 or 400 MHz) spectra were acquired with a Bruker DPX-300 or AV-400 instrument (Bruker Instruments, Billerica, MA) and tetramethylsilane or the solvent peak served as an internal standard for reporting chemical shifts, expressed on the δ scale.
Cell culture, lysate preparation and treatments with electrophiles
HEK293 cells were obtained frozen at low passage from Master Cell Bank cultures from GIBCO Life Technologies (Grand Island, NY). Cells were grown in DMEM supplemented with 10% fetal bovine serum. For lysate preparation, cells grown to 80-90% confluency in 150 mm plates were washed and harvested with cold PBS. The cells were pelleted by centrifugation at 1000g for 5 min and then buffer was aspirated. The ∼ 200 μL pellet was resuspended in 800 μL of lysis buffer (50 mM Tris base, pH 7.5, 150 mM NaCl, 1% Igepal, 1 mM EDTA) with protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN).
For studies of protein adduction in HEK293 cell lysates, lysate protein (1 mg mL-1) was reacted in lysis buffer at 37 °C with 10 μM IAB or BMCC for intervals ranging from 2 min to 24 h. For subsequent studies of the stability of BMCC adducts in lysates treated for 1 h with 10 μM BMCC, the treated lysate was further incubated with 1, 10, 100 μM, 1 and 10 mM of GSH, NADH or NADPH at pH 7.5 or 8.5 for 1 h at 37 °C.
For studies of adduction by electrophiles in intact HEK293 cells, cultures grown to approximately 90% confluency in 100 mm plates were washed with PBS and treated with 25 μM BMCC or IAB or 5 μM NPM or 5 μM IAM, or equal volumes of vehicle (DMSO at 0.3% of total volume) were delivered in 4 mL of DMEM with 5% fetal bovine serum. For toxicity studies, cells were washed once with PBS, treated with 1-100 μM IAM, IPM, NEM, or NPM delivered in DMEM with 5% fetal bovine serum for 30 min at 37 °C. Cytotoxicity was measured by LDH leakage determined as described in the LDH-based in vitro toxicology assay kit (Sigma) according to the manufacturer’s instructions.
Synthesis of NPM and IPM
NPM and IPM were prepared by chemical synthesis core of the Vanderbilt Institute of Chemical Biology. NPM: Maleimide (500mg, 5.16 mmol), propargyl alcohol (318mg, 5.7 mmol), triphenylphosphine (2.02g, 7.72mmol) and diethylazidodicarboxylate (1.35g, 7.72 mmol) were combined in 20 mL CH2Cl2 and stirred at 25°C overnight to afford NPM in 60% yield. 1H-NMR: ( 400 MHz, CDCl3) δ2.25 (t, 1H), 4.25 (t, 2H), 6.5 (d, 2H). IPM: Iodoacetic acid (1g, 5.4 mmol), propargylamide (326mg, 5.9 mmol), N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (1.03g, 5.4 mmol) and dimethylaminopyridine (66mg, 0.54 mmol) were dissolved together in 20 mL CH2Cl2 (20 ml) and stirred at 25°C overnight to produce IPM in 55% yield. 1H-NMR: ( 400 MHz, CDCl3) δ2.25 (t, 1H), 3.90 (t, 2H), 4.10 (s, 2H). The crude products of both the NPM and IPM syntheses were purified by chromatography on silica gel with 10% ethyl acetate in hexane.
Biotinylation of alkynyl protein adducts by Click chemistry
Following treatment of cells with alkynyl electrophiles, protein samples (2 mg mL-1 in PBS containing 1% Igepal CA 630, 200 μL) were treated with 4 μL biotin azide (10 mM in DMSO) and the sample was subjected to brief vortex mixing. TCEP (50 mM in H2O, 4 μL) and the Cu(I) ligand (tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (Scheme 2) (1.7 mM in DMSO:butanol 1:4, 12 μL) were added to each reaction. After vortexing, CuSO4 (50 mM in water, 4 μL) was added. Samples were then mixed and allowed to react for 2 h at room temperature. Proteins were precipitated by addition of trichloroacetic acid to final concentration of 20% (w/v) and the mixture was incubated on ice for 10 min and centrifuged at 14,000 rpm at 4 °C for 5 min. The supernatant was removed and 50 μL of ice-cold acetone were added to wash the pellet. The sample was incubated on ice for 5 min and centrifuged again as described above. The acetone-containing supernatant was removed and the pellet was air dried. The washed protein precipitate was dissolved in 100 μL pH 8.0 RIPA buffer (20 mM Tris, 500 mM NaCl, 1 mM EDTA, 1% Igepal CA-630, 0.1% SDS, 10% glycerol, 50 mM IAM, 1.0 mM PMSF, 10 μg mL-1 leupeptin, 10 μg mL-1 aprotinin, and 10 μg mL-1 pepstatin) and NuPAGE LDS sample buffer in a ratio of 1:3 (v/v) and incubated at 70 oC for 10 min.
Scheme 2.
Hydrolysis of GS-BMCC and GS-NEM conjugates.
SDS-PAGE and streptavidin blotting for analysis of biotinylated electrophile protein adducts
Protein samples (1-20 μg) were separated on a NuPAGE 10% SDS-PAGE gel (Invitrogen, Carlsbad, CA) using NuPAGE MOPS SDS running buffer. All gel lanes in each gel were loaded with same amount of protein in 4x LDS buffer. To visualize biotin-labeled proteins, the gels were transferred to PVDF membranes, blocked with TBS: blocking buffer (1:1, v/v) for near infrared fluorescence Western blotting (Rockland, Gilbertsville, PA) and then the biotin-labeled proteins were probed with Alexa Fluor 680-conjugated streptavidin (Molecular Probes, Junction City, OR). Immunoblot images were acquired and processed using the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
Synthesis of GS-NEM, GS-BMCC, Cys-Gly-BMCC and Cys-BMCC conjugates
To a solution of NEM (3.5 mg, 0.028 mmol) in d6-DMSO (200 μL) in an NMR tube was added GSH (8.6 mg, 0.028 mmol) in D2O (500 μL), to give a final concentration of NEM (40 mM). The reaction was monitored by 1H-NMR spectrometry. After 24 h at room temperature, the spectrum indicated 100% conversion to GS-NEM conjugate. GS-NEM: 1H NMR δ 1.04 (t, 3H J = 7.5 Hz), 2.02-2.10 (2H), 2.44 (t, 2H, J = 7.2 Hz), 2.54-3.30 (4H), 3.45 (q, 2H, J = 7.5 Hz), 3.69 (t, 2H, J = 6.0 Hz), 3.86 (s, 2H), 3.94-3.99 (1H), 4.54-4.64 (1H). The GS-BMCC conjugate was prepared in the similar way, except that the reaction was done in H2O containing 10% DMSO and the reactant concentrations were 1 mM and 10 mM for BMCC and GSH, respectively. Cys-BMCC and Cys-Gly-BMCC were prepared in the same manner as GS-BMCC. All BMCC conjugates were confirmed by positive electrospray LC-MS-MS, which indicated no other side reactions and the products were used as standard compounds without further purification.
LC-MS-MS analyses of BMCC metabolites
HEK 293 cells were seeded in 24 mm plates at different numbers (0, 0.4, 1, 2 and 4 x 105 cells) in DMEM with 10% FBS. The following morning, cells were washed twice with PBS followed by the addition of 100 μM BMCC in 100 μL DMEM with 5% FBS for 1 h at 37 °C. Cells then were scraped and the media were collected after centrifugation (1,000 rpms, 5 min, 4 °C). Media samples (5 μL) were subjected to chromatographic separation on a 250 x 2.0 mm YMCTM ODS-AQ 5 μm C18 column (Waters, Milford, MA) and eluted conjugates were analyzed with a Thermo LCQ DecaXP ion trap mass spectrometer (ThermoElectron, San Jose, CA). The mobile phase consisted of 5% acetonitrile, 95% H2O, and 0.1% formic acid (solvent A), and 95% acetonitrile, 5% H2O and 0.1% formic acid (solvent B). The flow rate was 400 μL/min and the gradient program was 100% solvent A from 0 to 5 min, 100% A to 40% A by 20 min, 40% A to 0% A by 22 min, then held at 0% A from 22 to 25 min, followed by a linear gradient to 100% A by 29 min, then held at 100% A from 29 to 34 min. MS-MS spectra of the MH+ ions of the conjugates were obtained to confirm the identities of the GS conjugates.
NMR studies of hydrolysis of BMCC and NEM
To a solution of NEM (0.875 mg, 0.007 mmol) or BMCC (0.373 mg, 0.0007 mmol) in d6-DMSO (70 μL) in an NMR tube was added 100 mM pD 8.0 deuterium phosphate buffer (630 μL), to give a final concentration of NEM (10 mM) or BMCC (1 mM). The reaction was monitored periodically by 1H-NMR. After 24 h, the spectra indicated 100% conversion to the acid forms. N-ethylmaleimic acid: 1H NMR δ 1.01 (t, 3H, J = 7.3 Hz), 3.12 (q, 2H, J = 7.3 Hz), 5.79 (d, 1H, J = 12.4 Hz), 6.18 (d, 1H, J = 12.4 Hz). BMCC-acid: 1H NMR δ 2.98 (d, 2H, J = 7.1 Hz), 5.78 (d, 1H, J = 12.5 Hz), 6.20 (d, 1H, J = 12.5 Hz).
Solution stability studies of the GS-BMCC conjugate
To a solution of GS-BMCC (1 mM in H2O containing 10% DMSO, 50 μL; prepared as described above) was added 100 mM, pH 8.0 phosphate buffer (950 μL), to give a final concentration of GS-BMCC (50 μM). The reaction was incubated at 37 °C and aliquots were analyzed by positive electrospray LC-MS-MS at different time points (0-96 h).
RESULTS
Formation and stability of IAB and BMCC adducts in intact HEK293 cells
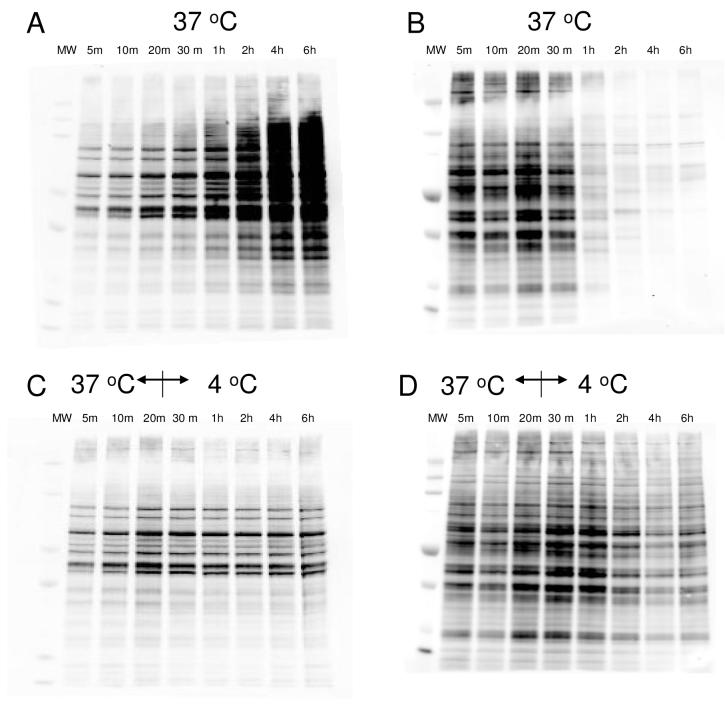
Treatment of HEK293 cells with 25 μM IAB or BMCC resulted in formation of protein adducts, which were detected by western blotting with Alexa-Fluor-conjugated streptavidin. Although both agents are thiol-reactive electrophiles, the kinetics of protein adduction differed markedly (Figure 1). Adducts gradually accumulated over 24 h in cells treated with IAB (Figure 1A), whereas BMCC adduct accumulation was maximal at 20-30 min and decreased quickly thereafter (Figure 1B). When cells were treated with BMCC at 37°C and then cooled to 4 °C beginning at 20 min, the decrease in adduct levels was significantly slowed (Figure 1D). Cooling to 4 °C beginning at 20 min did not affect the accumulation of IAB adducts (Figure 1C).
Figure 1.
Temperature-dependence of BMCC or IAB adduction in HEK293 cells in vivo. HEK293 cells were treated with 25 μM BMCC or IAB at 37 °C for 20 min, then cells were maintained at 37 °C (A: IAB; B: BMCC) or 4 °C (C: IAB; D: BMCC) up to 6 h.
Modification of lysate proteins by IAB and BMCC in HEK 293 cell lysates
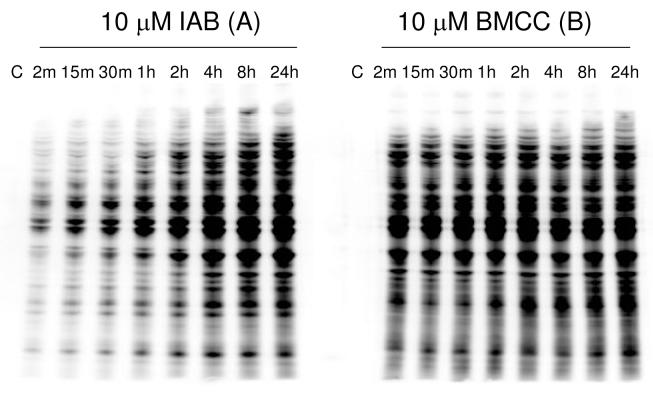
We then examined the kinetics of adduct formation in HEK293 cell lysates treated with IAB and BMCC. Incubation with 10 μM IAB and BMCC produced a time-dependent accumulation of adducts. IAB adducts accumulated in the lysate gradually, as in intact cells (Figure 2A). BMCC adducts accumulated more rapidly than did IAB adducts, but then remained stable, in contrast to the rapid loss of BMCC adducts in the intact cells. Modification by BMCC was maximal in 20-30 min, whereas maximal adduction by IAB took up to 8 h. The rapid initial formation of BMCC adducts was also observed in intact cells (see above) and is consistent with previous studies in microsomes (10). Consistent kinetics of adduct accumulation in both cells and lysates suggests that the differences between IAB and BMCC are not due to differences in uptake and distribution in cells, but may reflect instead differences in kinetic thiol reactivity between the SN2 acceptor iodoacetyl group in IAB and Michael addition acceptor maleimide group in BMCC (19).
Figure 2.
Time-dependent protein adduction in HEK293 cell lysate treated with 10 μM IAB (A) or BMCC (B). After exposure to the electrophiles for the times indicated, samples of lysate protein were analyzed by SDS-PAGE and western blotting with Alexa-Fluor-conjugated streptavidin.
Rapid loss of BMCC Michael adducts in cells could be due to chemical instability in the presence of cellular reductants. To test this hypothesis, we incubated HEK293 cell lysates with 10 μM BMCC for 1 h (see above) and then incubated the adducted lysates with different concentrations of GSH (Figure S1), NADH and NADPH (Figure S2) at either pH 7.5 or 8.5. Even at concentrations up to 10 mM, these reductants did not reverse protein-BMCC covalent binding at either pH 7.5 or 8.5. Thus, BMCC adduct loss in cells does not reflect simple chemical instability in the presence of cellular reductants.
Application of click chemistry to study cellular protein modification by NPM and IPM
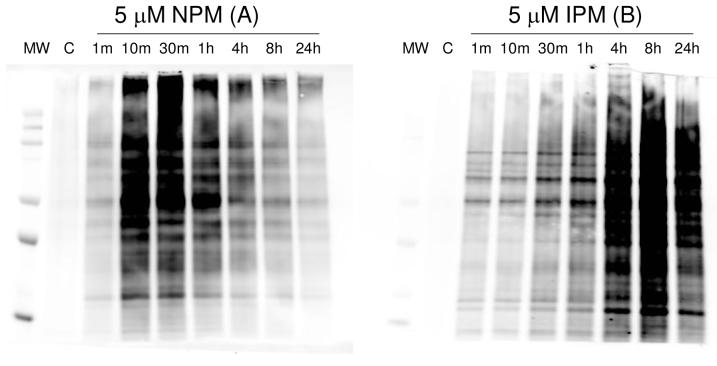
One possible explanation for the apparent loss of BMCC adducts in intact cells is a metabolism-based cleavage of the BMCC linker between the maleimide moiety and the biotin group or some other modification of the biotin tag that renders the BMCC adducts undetectable by streptavidin blotting. To address this possibility, we employed the small alkynyl BMCC and IAB analogs NPM and IPM (Figure 3). These compounds contain the same electrophilic groups as BMCC and IAB, respectively, but do not have linker and biotin moieties. However, during sample workup, adducts formed from these electrophiles were biotinylated using coppeRI)-catalyzed cycloaddition to an azidobiotin tag (“Click chemistry” (20, 21)) (Scheme S1) and the adducts were detected by streptavidin blotting as described above.
Figure 3.
Time-dependent covalent adduction of proteins by 5 μM NPM and IPM in HEK293 cells. After exposure to NPM and IPM for the times indicated, cells were lysed and the NPM/IPM-adducted proteins were biotinylated by copper (I)-catalyzed [3+2] cycloaddition with biotin-tagged azide following by SDS-PAGE analysis and Western blotting with streptavidin. Lanes labeled C represent lysate proteins biotinylated by copper (I)-catalyzed [3+2] cycloaddition with biotin-tagged azide without NPM/IPM treatment.
We used this approach to measure the time-dependent modification of HEK293 cell proteins by 5 μM NPM and IPM. As shown in Figure 3, maximal protein modification by NPM occurred at 30 min, but, as with BMCC, the adducts rapidly disappeared and only about 20% of the adducts remained after 24 h. Incubation with 5 μM IPM produced time-dependent accumulation of stable adducts, similar to what was observed with IAB. These results indicate that differences in adduction kinetics between IAB and BMCC were not due to differences in the linkers and strongly suggest that loss of BMCC adducts reflects rapid removal of the adducts rather than metabolic degradation of the biotin tag.
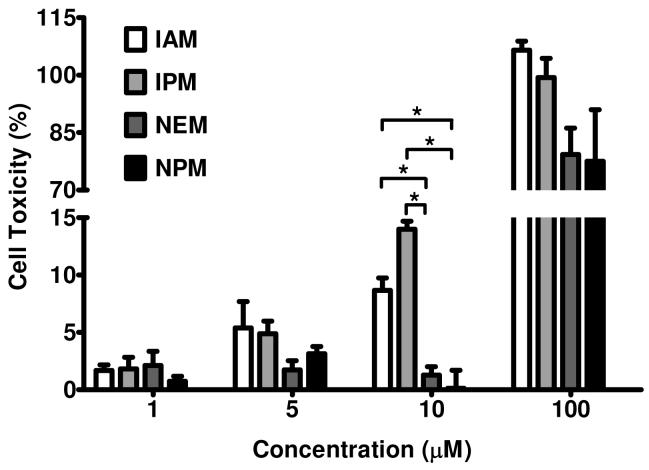
Cytotoxicity of electrophiles
Our results correlated lack of BMCC toxicity with rapid clearance of maleimide adducts and IAB toxicity with the formation of stable adducts. Because adducts of the small alkynyl analogs NPM and IPM exhibited the same differences in binding kinetics, we evaluated the toxicity of these alkynyl analogs and their non-alkynyl congeners NEM and IAM in HEK 293 cells. Cells were treated for 30 min with different concentrations of the maleimides NPM and NEM, and the iodoacetamides IAM and IPM. All four small molecules induced concentration-dependent toxicity, as measured by LDH leakage. However, IPM and IAM exhibited significantly greater toxicity than did NPM and NEM at exposure concentrations of 10 μM (Figure 4). The lack of NPM toxicity correlated with rapid reversal of NPM covalent binding at the 5 μM exposure concentration (Figure 3). These results further suggest that rapid clearance of maleimide adducts accounts for the lack of toxicity of both BMCC and NPM.
Figure 4.
Cytotoxicity of NEM, NPM, IAM and IPM in HEK293 cells. Cells were treated with indicated concentrations (1-100 μM) of electrophiles for 30 min. Cytotoxicity was measured as LDH leakage from the cells and is reported as a percent of total LDH in the cells. Results were determined from three independent experiments (n = 3). Statistical evaluation was performed applying one-way ANOVA (*p < 0.05).
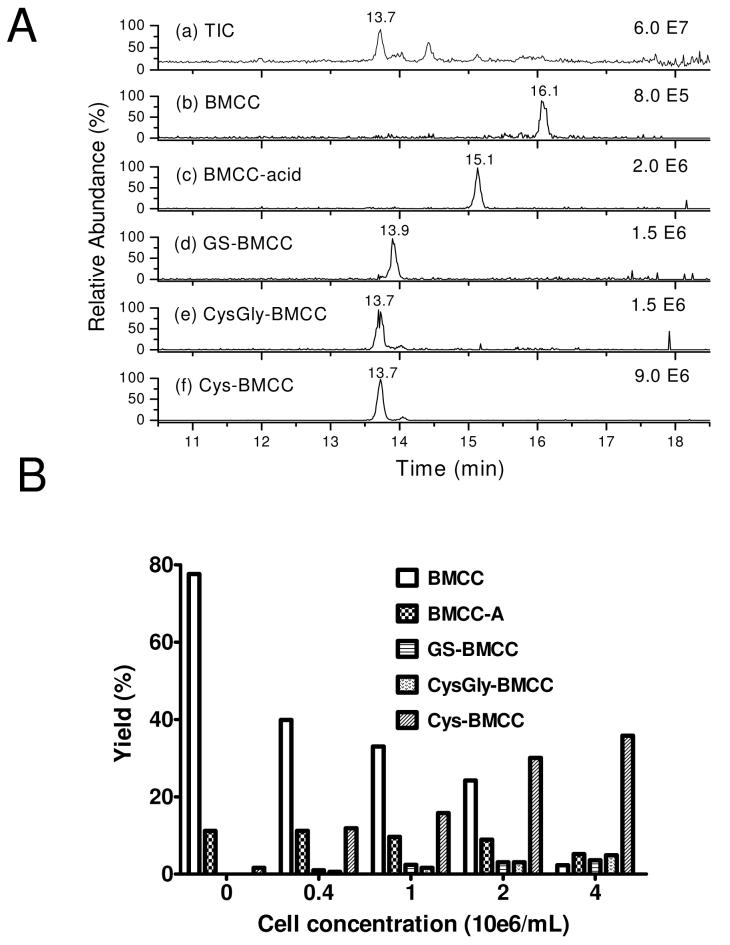
LC-MS analysis of BMCC metabolites formed in HEK293 cells
Because our covalent binding and toxicity data suggested that rapid maleimide adduct clearance was a key determinant of toxicity, we further investigated the fate of BMCC in HEK293 cells. We analyzed products of BMCC in HEK293 cell medium by LC-MS (Figure 6A). These analyses detected BMCC (16.1 min, MH+ m/z 534.3), BMCC-acid (15.1 min, MH+ m/z 552.3), GS-BMCC (13.9 min, MH+ m/z 841.3), Cys-Gly-BMCC (13.7 min, MH+ m/z 655.3) and Cys-BMCC (13.7 min, MH+ m/z 712.3) (Figure 5A). Yields of BMCC and metabolites were calculated from standard curves and from co-injection of standards for each compound, which enabled calibration of LC-MS signal intensities. The yields of all metabolites were calculated based on starting BMCC. Consumption of BMCC and formation of its metabolites were cell concentration-dependent when HEK293 cells were treated with 100 μM BMCC for 1 h (Figure 5B). The yields of all metabolites except BMCC-acid increased with cell concentration up to 4 x 106 cells mL-1. Cys-BMCC, which was formed in 36% yield at a cell concentration of 4 x 106 cells mL-1, was the major product, whereas Cys-Gly-BMCC (5%) and GS-BMCC (4%) were present in lower yields. Cys-BMCC may arise either from reaction of BMCC with extracellular cysteine or from catabolism of GS-BMCC (via Cys-Gly-BMCC).
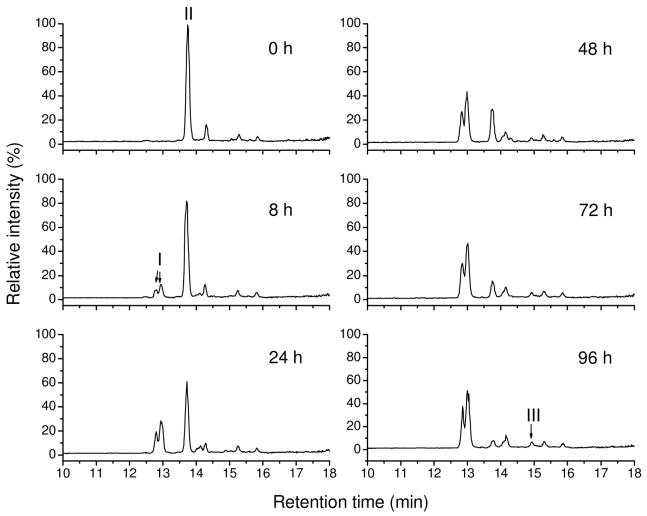
Figure 6.
Reversed phase LC-MS total ion chromatograms of 50 μM GS-BMCC in 100 mM, pH 8.0 phosphate buffer containing 0.5% DMSO at 37 °C for times ranging form 0 to 96 h. (I) GS-B MCC-A; (II) GS-BMCC; (III) BMCC-A.
Figure 5.
A. Reverse phase LC-MS analysis of the medium of HEK293 (4 x 105) cells treated for 1h with 100 μL 100 μM BMCC. (a) Total ion current (TIC) chromatogram; (b) Selected ion chromatogram (SIC) for MH+ (m/z 552.3) expected for BMCC-acid; (c) SIC for MH+ (m/z 552.3) expected for BMCC-acid; (d) SIC for MH+ (m/z 841.3) expected for GS-BMCC; (e) SIC for MH+ (m/z 712.3) expected for CysGly-BMCC; (f) SIC for MH+ (m/z 655.3) expected for Cys-BMCC. B. Effects of cell concentration on metabolism of 100 μM BMCC by HEK293 cells for 1 h. After exposure to BMCC, media were analyzed by LC-MS and the yield of BMCC-acid, GS-BMCC, Cys-Gly-BMCC and Cys-BMCC were calculated according to original BMCC.
We also found that only about 60% of total BMCC was accounted for as the starting material and the detected metabolites (2% BMCC, 5% BMCC-acid, 4% GS-BMCC, 5% Cys-Gly-BMCC and 36 % Cys-BMCC in the media of BMCC-treated 4 x 106 cell mL-1 for 1 h). This suggests that a maximum of 40% of the remaining BMCC could be covalently bound to cell proteins at 1 h.
Analysis of NEM and BMCC hydrolysis and GSH conjugation by NMR
Because maleimide hydrolysis and GSH conjugation were the two principal reactions that consumed BMCC in cells, we conducted NMR studies to further characterize the products. Hydrolysis of NEM was observed by 1H-NMR analysis of NEM in 100 mM pH 8.0 D2O-phosphate buffer containing 10% d6-DMSO. Analysis of time-dependent spectral changes in the downfield ppm region reveals ring-opened acid formation, as indicated by the chemical shift of N-ethyl CH2 protons at 3.4 ppm to 3.1 ppm (Figure S3). Similarly, hydrolysis of BMCC in 100 mM pH 8.0 D2O-phosphate buffer containing 10% d6-DMSO was accompanied by similar spectral changes. The chemical shift of the methylene protons next to the BMCC maleimide group decreased from 3.2 ppm to 2.9 ppm (Figure S4). No other significant changes in resonances were observed and hydrolysis to BMCC-acid was determined to be complete after 24 h.
The 1H-NMR spectrum of GS-NEM conjugate formed by the reaction of one equivalent each of GSH and NEM in D2O/d6-DMSO (5:2, v/v) was characterized by disappearance of the NEM olefinic protons in the downfield ppm region and a slight downfield shift of the cysteine α-proton in the glutathionyl moiety of the conjugate (Figure S5). Analogous spectral changes were observed during the formation of the GS-BMCC conjugate under similar conditions (Figure S6).
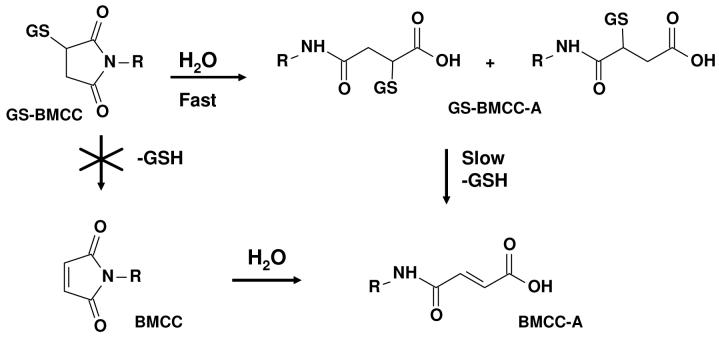
LC-MS analysis of stability of GS-BMCC in solution
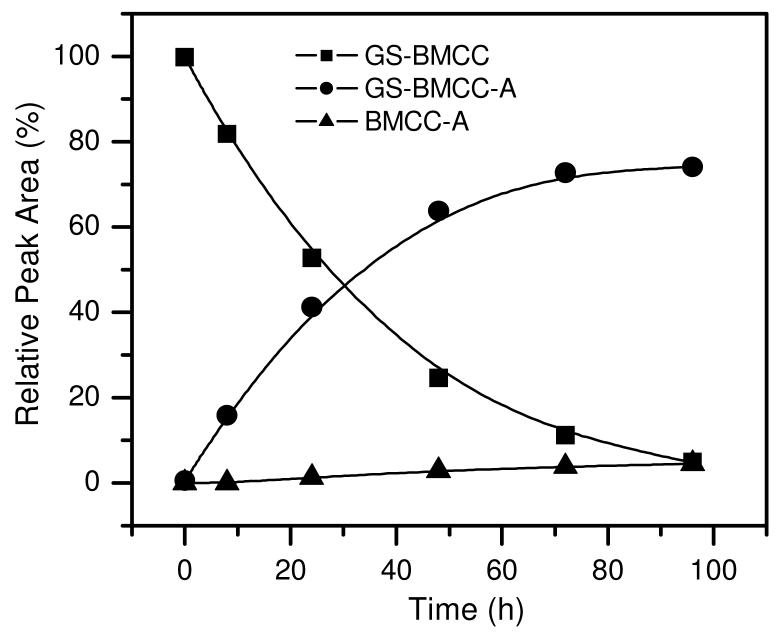
Because GS-BMCC is a potentially useful model for the behavior of BMCC-protein adducts, we studied the aqueous solution decomposition of GS-BMCC. These analyses were directed at assessing the relative rates of hydrolysis of GS-BMCC to the corresponding carboxylic acid adducts (GS-BMCC-A) and the elimination of BMCC from either GS-BMCC or GS-BMCC-acid (Scheme 2). The generation of BMCC and BMCC-acid via these elimination reactions would be analogous to a reaction in cells that results in clearance of BMCC-protein adducts. GS-BMCC was incubated in neutral aqueous buffer and samples were taken periodically for the analysis of GS-BMCC, GS-BMCC-acid, BMCC-acid and BMCC. Hydrolysis of GS-BMCC (MH+, m/z 841.3) resulted principally in the formation of GS-BMCC-acid, which eluted as two isomeric products at 12.8 and 13.0 min (MH+, m/z 859.3) (Figure 6). (MS-MS spectra of GS-BMCC and GS-BMCC-acid acquired in these analyses are presented in Figures S7 and S8.) We also detected another peak that appeared at 14.9 min with m/z 552.3, which corresponded to BMCC-acid. Based on the relative peak areas from the selected ion chromatograms for GS-BMCC, GS-BMCC-acid and BMCC-acid, it appears that approximately 95% of the GS-BMCC was hydrolyzed to GS-BMCC-acid over 96 h (Figure 7). BMCC was not detected as a product of GS-BMCC decomposition and BMCC-acid was formed only very slowly, which indicates that nonenzymatic elimination is much slower than the imide hydrolysis reaction.
Figure 7.
Time profile of the relative peak area from reversed phase LC selected ion chromatograms for GS-BMCC (■, MH+ (m/z 841.3)) and its decomposition products GS-BMCC-A (•, MH+ (m/z 859.3)) and BMCC-A (▲, MH+ (m/z 552.3)) in 50 μM GS-BMCC in 100 mM, pH 8.0 phosphate buffer containing 0.5% DMSO at 37 °C.
DISCUSSION
Protein thiol adducts formed by maleimide electrophiles are rapidly removed in intact cells and this rapid adduct clearance coincides with a lack of maleimide toxicity. In contrast, iodoacetamide electrophiles form stable adducts and are toxic. The instability of the maleimide adducts in cells contrasts markedly with the stability of these adducts in cell lysates, even in the presence of cellular reductants (Figures S1 and S2). Moreover, the inhibition of cellular maleimide adduct removal at low temperature (Figure 1) suggests that adduct clearance involves a metabolism-dependent process. These observations suggest that protein adduct stability is a critical determinant of the induction of toxic or adaptive responses by electrophiles.
We had previously reported that the protein targets adducted by the maleimide BMCC and the iodoacetamide IAB overlapped by only about 20% in subcellular fractions exposed to these probes (10, 11). Accordingly, differences in toxicity and stress responses between these compounds were attributed to differential covalent modification of critical targets. This interpretation is consistent with the longstanding view that toxic and adaptive responses may be driven by modification of only a small fraction of protein targets (4, 22). According to this view, the identities of protein targets govern the observed responses. However, to our knowledge, previous work has not considered the dynamic nature of protein covalent modification and the roles of adduct stability, repair and turnover in toxicity outcomes.
We attribute the rapid disappearance of maleimide adducts from cells in our studies to some type of repair process rather than to simple chemical instability or to rapid degradation of the adducted proteins. The stability of BMCC adducts in HEK293 cell lysates, even in the presence of reducing agents (Figures S1 and S2) argues that the maleimide adducts do not spontaneously decompose or dissociate during the time course when rapid adduct disappearance occurs in cells (1-4 hr). Although our studies with GS-BMCC document the hydrolysis of the thiol-conjugated maleimide (Figures 6 and 7), the nonenzymatic hydrolysis reaction occurs over a period of days, rather than minutes to hours. We also distinguish adduct repair (i.e., removal of the adduct from a protein) from turnover of the adducted proteins. Our studies of turnover of IAB-protein adducts in HEK293 cells indicates that this process occurs over a time period of 6-48 hrs, depending on the degree of adduction and factors that influence the activities of proteasomal and lysosomal degradation2. Although the factors that govern the degradation of damaged proteins are inadequately understood, it seems unlikely that sterically similar, bulky adducts such as those formed from IAB and BMCC would confer dramatic differences in protein turnover kinetics.
Our observations of the hydrolysis of NEM and BMCC and their thiol conjugates are consistent with previous work on maleimide-protein modification chemistry. Hydrolysis of maleimides to maleimic acids can compete with thiol modification (23). Hydrolysis of NEM was also reported to be catalyzed by antibody H11 (24) and the Raines group reported that both molybdate and chromate could catalyze the hydrolysis of the imido group in a maleimide conjugate (25). Although a maleimide hydrolase enzyme is not known to occur in human tissues, it should be noted that the reaction is similar to that catalyzed by dihydropyrimidinases in pyrimidine catabolism and that multiple forms of these enzymes catalyze amidohydrolase reactions (26).
Although we have no direct evidence for a specific repair mechanism for maleimide adducts, two of our observations provide some boundaries for consideration. First, our experiments with BMCC and the small alkynyl analog NPM yielded the same rapid adduction and disappearence kinetics for their maleimide protein adducts, which indicates that the loss of streptavidin reactivity did not simply reflect metabolism of the biotin tag in BMCC. Second, although the GS-BMCC conjugate hydrolyzed, this reaction was so slow that spontaneous BMCC-protein maleimide adduct hydrolysis appears to be unlikely to contribute to the adduct clearance process.
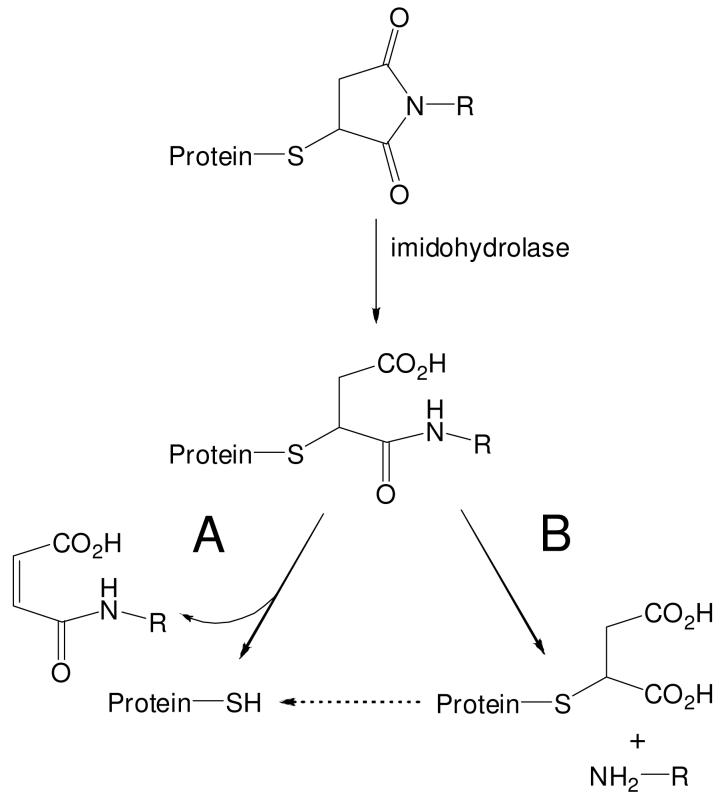
Based on these considerations, we can consider two possible mechanisms for the observed disappearance of BMCC and NPM adducts in HEK293 cells (Scheme 3). Protein thiol maleimide adducts may undergo enzymatic hydrolysis catalyzed by an aminohydrolase to form the ring-opened acid, which then undergoes either A) β-elimination to release the adduct moiety and regenerate the protein thiol or B) amidase-catalyzed cleavage of the alkylamide to release the corresponding N-alkylamine (including the alkyne from NPM or the biotin tag from BMCC) and leave a maleic acid-modified protein thiol. Pathway A would result in true adduct repair, whereas pathway B would leave a maleic acid-modified thiol that is not detected by streptavidin. Of course, two major uncertainties about these mechanisms should be noted. First, it is not clear that any amidohydrolase(s) would operate on maleimide adducts derived from bulky structures (e.g., BMCC). Second, an enzyme-catalyzed β-elimination of either the ring-opened acid in pathway A or the maleic acid adduct formed in pathway B is without clear precedent, although glutathione-S-transferase P1-1 was shown to catalyze a similar β-elimination from a glutathione derivative (27). Further studies certainly would be needed to establish the mechanism of maleimide adduct disappearance.
Scheme 3.
Possible repair mechanisms for maleimide adducts.
Whatever mechanism(s) that account for BMCC adduct disappearance may not necessarily apply to adducts formed from other electrophiles. Nevertheless, this work illustrates the potentially important relationship between protein adduct stability and the induction of toxic or adaptive responses. Although BMCC and IAB differ in target selectivity at both the protein and amino acid sequence level, the instability of BMCC adducts appears to confer protection against toxicity. If enzymatic repair reactions govern the stability of adducts from other electrophiles, then two implications should be considered. First, the assessment of covalent binding in intact cells or tissues, as opposed to subcellular fractions (e.g., microsomes) may provide information that is more relevant to interpreting the link between adducts and toxicity. The relative merits of different types of covalent binding analyses for prediction of drug toxicity have been discussed in recent reviews (5, 28). Second, factors that compromise repair of protein adducts (e.g., polymorphisms in genes coding for enzymes involved in adduct repair or drug interactions with repair enzymes) could sensitize certain individuals to toxicity.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge support of the Chemical Synthesis Core of the Vanderbilt Institute of Chemical Biology. This work was supported by NIH grants ES010056 and ES000267 and a postdoctoral fellowship to D.L. from Novartis. We thank Dr. Mark Kagan for helpful discussions.
Footnotes
- BCA
- bicinchoninic acid
- BMCC
- 1-biotinamido-4-(4′-[maleimidoethylcyclohexane]-carboxamido) butane
- BMCC-A
- BMCC acid
- DMEM
- Dulbecco’s modified Eagle’s medium
- Cys-BMCC
- S-cysteinyl-BMCC conjugate
- Cys-Gly-BMCC
- S-cysteinylglycine-BMCC conjugate
- DTT
- dithiothreitol
- GSH
- glutathione
- GS-BMCC
- S-glutathionyl-BMCC conjugate
- GS-NEM
- S-glutathionyl-NEM conjugate
- IAB
- N-iodoacetyl-N-biotinylhexylenediamine
- IAM
- iodoacetamide
- Igepal CA 630
- Octylphenylpolyethylene glycol
- IPM
- N-propynyliodoacetamide
- LC-MS-MS
- liquid chromatography-tandem mass spectrometry
- LDH
- lactate dehydrogenase
- MS-MS
- tandem mass spectrometry
- PBS
- phosphate-buffered saline
- NEM
- N-ethylmaleimide
- NPM
- N-propynylmaleimide
- TCEP
- tris(carboxyethyl)phosphine
Burnette, E.B. and Liebler, D.C., unpublished observations.
SUPPORTING INFORMATION AVAILABLE Scheme S1 and Figures S1 to S8. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Guengerich FP. Forging the links between metabolism and carcinogenesis. Mutat. Res. 2001;488:195–209. doi: 10.1016/s1383-5742(01)00059-x. [DOI] [PubMed] [Google Scholar]
- (2).Marnett LJ, Riggins JN, West JD. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J.Clin.Invest. 2003;111:583–593. doi: 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab. Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- (4).Liebler DC. Protein Damage by Reactive Electrophiles: Targets and Consequences. Chem. Res. Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Evans DC, Watt AP, Nicoll-Griffith DA, Baillie TA. Drug-protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem. Res. Toxicol. 2004;17:3–16. doi: 10.1021/tx034170b. [DOI] [PubMed] [Google Scholar]
- (6).Dinkova-Kostova AT, Holtzclaw WD, Kensler TW. The role of Keap1 in cellular protective responses. Chem. Res. Toxicol. 2005;18:1779–1791. doi: 10.1021/tx050217c. [DOI] [PubMed] [Google Scholar]
- (7).Nakamura T, Lipton SA. S-Nitrosylation and uncompetitive/fast off-rate (UFO) drug therapy in neurodegenerative disorders of protein misfolding. Cell Death Differ. 2007;14:1305–1314. doi: 10.1038/sj.cdd.4402138. [DOI] [PubMed] [Google Scholar]
- (8).Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic. Biol. Med. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Dennehy MK, Richards KAM, Wernke GW, Shyr Y, Liebler DC. Cytosolic and nuclear protein targets of thiol-reactive electrophiles. Chem. Res. Toxicol. 2006;19:20–29. doi: 10.1021/tx050312l. [DOI] [PubMed] [Google Scholar]
- (10).Shin NY, Liu Q, Stamer SL, Liebler DC. Protein targets of reactive electrophiles in human liver microsomes. Chem. Res. Toxicol. 2007;20:859–867. doi: 10.1021/tx700031r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wong HL, Liebler DC. Mitochondrial protein targets of thiol-reactive electrophiles. Chem. Res. Toxicol. 2008;21:796–804. doi: 10.1021/tx700433m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J. Biol. Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- (13).Codreanu SG, Adams DG, Dawson ES, Wadzinski BE, Liebler DC. Inhibition of protein phosphatase 2A activity by selective electrophile alkylation damage. Biochemistry. 2006;45:10020–10029. doi: 10.1021/bi060551n. [DOI] [PubMed] [Google Scholar]
- (14).Rachakonda G, Xiong Y, Sekhar KR, Stamer SL, Liebler DC, Freeman ML. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem. Res. Toxicol. 2008;21:705–710. doi: 10.1021/tx700302s. [DOI] [PubMed] [Google Scholar]
- (15).Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc.Natl.Acad.Sci.U.S.A. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Prior IA, Clague MJ. Detection of thiol modification following generation of reactive nitrogen species: analysis of synaptic vesicle proteins. Biochim. Biophys. Acta. 2000;1475:281–286. doi: 10.1016/s0304-4165(00)00078-7. [DOI] [PubMed] [Google Scholar]
- (17).Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, van Breemen RB. Sites of alkylation of human Keap1 by natural chemoprevention agents. J. Am. Soc. Mass Spectrom. 2007;18:2226–2232. doi: 10.1016/j.jasms.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Eggler AL, Luo Y, van Breemen RB, Mesecar AD. Identification of the highly reactive cysteine 151 in the chemopreventive agent-sensor Keap1 protein is method-dependent. Chem. Res. Toxicol. 2007;20:1878–1884. doi: 10.1021/tx700217c. [DOI] [PubMed] [Google Scholar]
- (19).Sturdik E, Drobnica L. Interaction of cytotoxic antibiotic dactylarin with glycolytic thiol enzymes in Ehrlich ascites carcinoma cells. J. Antibiot. (Tokyo) 1981;34:708–712. doi: 10.7164/antibiotics.34.708. [DOI] [PubMed] [Google Scholar]
- (20).Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- (21).Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem.Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- (22).Nelson SD, Pearson PG. Covalent and noncovalent interactions in acute lethal cell injury caused by chemicals. Annu. Rev. Pharmacol. Toxicol. 1990;30:169–195. doi: 10.1146/annurev.pa.30.040190.001125. [DOI] [PubMed] [Google Scholar]
- (23).Khan MN. Kinetics and mechanism of the alkaline hydrolysis of maleimide. J. Pharm. Sci. 1984;73:1767–1771. doi: 10.1002/jps.2600731227. [DOI] [PubMed] [Google Scholar]
- (24).Khalaf AI, Linaza S, Pitt AR, Stimson WH, Suckling CJ. On the specificity of reactions catalysed by the antibody H11. Tetrahedron. 2000;56:489–495. [Google Scholar]
- (25).Kalia J, Raines RT. Catalysis of imido group hydrolysis in a maleimide conjugate. Bioorg. Med. Chem. Lett. 2007;17:6286–6289. doi: 10.1016/j.bmcl.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hamajima N, Matsuda K, Sakata S, Tamaki N, Sasaki M, Nonaka M. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene. 1996;180:157–163. doi: 10.1016/s0378-1119(96)00445-3. [DOI] [PubMed] [Google Scholar]
- (27).Morgan AS, Sanderson PE, Borch RF, Tew KD, Niitsu Y, Takayama T, Von Hoff DD, Izbicka E, Mangold G, Paul C, Broberg U, Mannervik B, Henner WD, Kauvar LM. Tumor efficacy and bone marrow-sparing properties of TER286, a cytotoxin activated by glutathione S-transferase. Cancer Res. 1998;58:2568–2575. [PubMed] [Google Scholar]
- (28).Baillie TA. Future of toxicology-metabolic activation and drug design: challenges and opportunities in chemical toxicology. Chem. Res. Toxicol. 2006;19:889–893. doi: 10.1021/tx060062o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.