Abstract
Context
Continuing advances in genotyping technologies and the inclusion of DNA collection in observational studies have resulted in an increasing number of genetic association studies.
Objective
To evaluate the overall progress and contribution of candidate gene association studies to current understanding of the genetic susceptibility to cancer.
Data Sources
We systematically examined the results of meta- and pooled analyses for genetic polymorphisms and cancer risk published through March 2008.
Study Selection
We identified 161 meta- and pooled analyses, encompassing 18 cancer sites and 99 genes. Analyses had to meet the following criteria: 1) at least 500 cases, 2) cancer risk as outcome, 3) not focused on HLA genetic markers, and 4) published in English.
Data Extraction
Information on cancer site, gene name, variant, point estimate and 95% confidence interval, allelic frequency, number of studies and cases, tests of study heterogeneity and publication bias were extracted by one investigator and reviewed by other investigators.
Results
These 161 analyses evaluated 344 gene-variant/cancer associations and included on average 7.3 studies and 3,551 cases (range: 508–19,729 cases) per investigated association. The summary OR for 98 (28%) statistically significant associations (p-value <0.05) were further evaluated by estimating the false-positive report probability (FPRP) at a given prior probability and statistical power. At a prior probability level of 0.001 and statistical power to detect an OR of 1.5, thirteen gene-variant/cancer associations remained noteworthy (FPRP<0.2). Assuming a very low prior probability of 0.000001, similar to a probability assumed for a randomly selected SNP in a genome-wide association study, and statistical power to detect an OR of 1.5, four associations were considered noteworthy as denoted by a FPRP value < 0.2: 1) GSTM1 null and bladder cancer (OR:1.5, 95% CI: 1.3–1.6, p-value=1.9×10−14), 2) NAT2 slow acetylator and bladder cancer (OR: 1.46, 95% CI:1.26–1.68, p-value=2.5×10−7), 3) MTHFR C677T and gastric cancer (OR: 1.52, 95% CI: 1.31–1.77, p-value=4.9×10−8), and 4) GSTM1 null and acute leukemia (OR: 1.20, 95% CI: 1.14–1.25, p-value=8.6×10−15). When the OR used to determine statistical power was lowered to 1.2, two of the four noteworthy associations remained so: GSTM1 null with bladder cancer and acute leukemia.
Conclusions
Phase II enzymes, which are key enzymes involved in the detoxification and excretion of carcinogens (and particularly deletion of GSTM1), were among the most consistent and highly significant associations.
Introduction
During the last few decades, extensive effort has been invested in identifying sources of genetic susceptibility to cancer. Both the International Human Genome Sequencing Project and the International HapMap Project have generated a very large amount of data on the location, quantity, type, and frequency of genetic variants in the human genome.1–4 Facilitated by continuing technological advances that allow faster and cheaper genotyping results, a large and increasing number of observational studies investigating the association between variants in candidate genes and cancer risk have emerged.5
This growing number of studies prompted us to assess the overall contribution of these studies to our current understanding of the genetic susceptibility to cancer. One of the main criticisms of genetic epidemiology has been a lack of replication. There are several examples of studies exploring a previously published statistically significant finding for a genetic variant and failing to reproduce those findings, suggesting a large number of “false positive” reports.6, 7 The size of these genetic association studies is also an important methodologic concern, which has prompted the utilization of meta- and pooled analyses to combine both statistically significant and non-significant results from individual studies and weighting these results by their precision (a function of sample size).8–10
To evaluate the overall progress of candidate gene association studies in identifying genetic variants associated with cancer risk, we systematically examined the results of all published meta- and pooled analyses on genetic polymorphisms and risk of cancer and report observed point estimates, 95% confidence intervals and p-values. Just as three parameters are needed to fully evaluate medical diagnostic tests (specificity, sensitivity, and predictive value of a positive test), three analogous parameters are needed to evaluate fully statistical tests of an association (e.g., between a genetic variant and cancer).11 The p-value, the probability of obtaining a more extreme estimate than the one observed when the null hypothesis of no association (OR=1.0) is true, is analogous to 1 minus specificity (the likelihood of a test classifying a person as having the condition when they truly do not have the condition). Study power, the likelihood of detecting an association when one exists, is analogous to sensitivity (the likelihood of a test classifying someone as having the condition when they truly have it.) However it is well established in medical diagnostics that specificity and sensitivity can be high, but the predictive value of a positive test can still be low. This is because, if the condition is rare, positive diagnostic tests will mostly be false positives. This is less appreciated but also important in evaluating statistical tests of hypothesized associations: when the prior probability is small that an exposure-disease hypothesis is true, then a statistically significant finding has a high chance of being a false positive. The false-positive report probability (FPRP) is defined as “the probability of no association given a statistically significant finding”12 and is analogous to 1 minus the predictive value of a positive test. Thus, it is the FPRP rather than the p-value that answers the question of how probable the hypothesis, as tested, actually is.
In this paper, we evaluate the results of candidate gene-cancer association studies by presenting the p-value, power, and FPRP for all statistically significant associations as reported in meta- or pooled analyses. The FPRP is calculated from the statistical power of the test, the observed p-value, and a given prior probability for the association.12 Because the prior probabilities are not easily determined, we calculated the FPRP for two levels of prior probabilities that are appropriate for a range of hypotheses, from low probabilities, appropriate for polymorphisms with known functional consequences in important candidate genes to very low probabilities, appropriate for randomly selected variants as used in a genome-wide association studies.
This review presents information on knowledge generated thus far by candidate gene association studies conducted to identify cancer susceptibility genes, and can also be used to direct future studies towards areas that remain unclear. Furthermore, results from this analysis provide information on the allelic frequency and expected effect size (strictly speaking, strength of association), which can be helpful for planning (genome-wide) association studies.
Methods
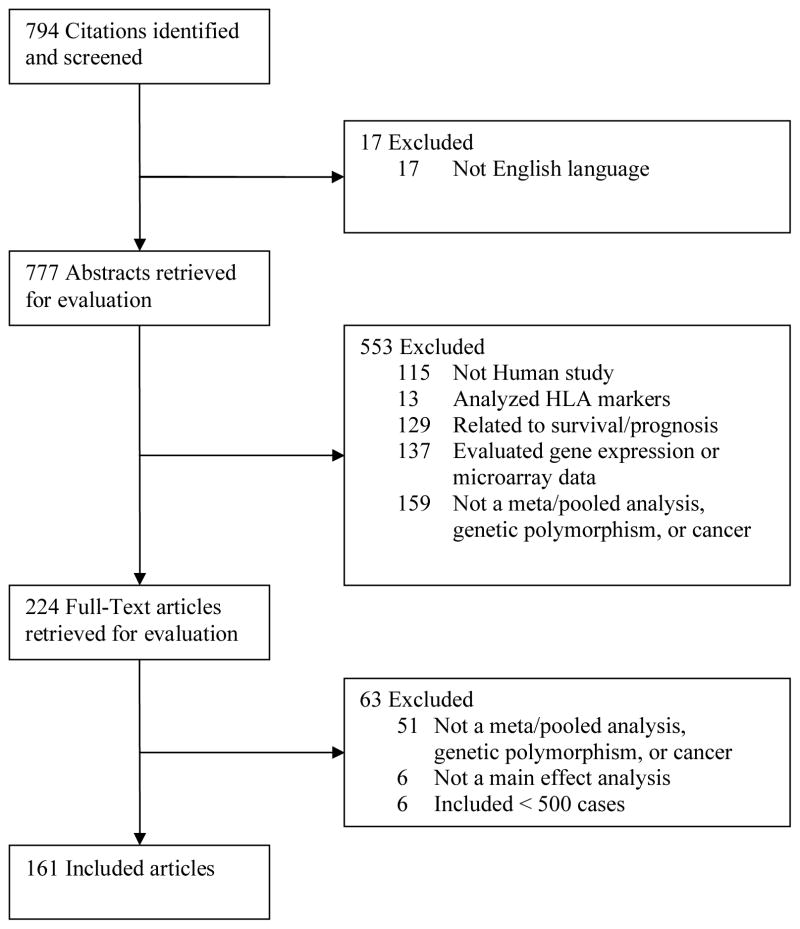
We identified all published meta- and pooled analyses that had evaluated the association between genetic polymorphisms and cancer risk in observational studies (i.e. case-control and nested case-control studies) through March 15, 2008. Meta- and pooled analyses are defined as tools that integrate results from individual studies that, alone, may not have sufficient power to detect a statistically significant association.8–10 In brief, the data (i.e. crude and adjusted odds ratios) used for a meta-analysis are extracted from published results, whereas original datasets acquired from a number of independent studies are used for a pooled analysis. We performed a literature search of the PubMed database using the following search terms for our literature searches: the keyword combinations of “cancer + meta + gene,” “cancer + pooled + gene,” “cancer + consortium + gene,” and the keyword combinations of “gene + cancer” and “genetic + cancer” restricted to publication type “meta-analysis.” We considered 794 articles identified through our search methods, screened in detail 224 articles, for a final 161 articles included (Figure 1). Studies included in our review had to meet all of the following criteria: 1) included at least 500 cases combined from all summarized studies, 2) evaluated cancer risk as the outcome (analyses of survival, neoplastic markers or precursors, such as polyps, were excluded), 3) excluded HLA genetic markers, and 4) published in English. Furthermore, as this review focuses on common variants, meta-and pooled analysis of low-frequency, high-penetrance genes, such as APC and BRCA1/2 were excluded. In addition, although statistically significant associations were reported for HRAS1 polymorphisms and risks of breast and lung cancer, these associations have been questioned because of flawed genotyping methods. Thus, these are not reported with other statistically significant associations in Table 2. To avoid duplication of results from more than one meta- or pooled analysis addressing the same association, we selected the most recent one, which typically had the largest number of cases (sometimes smaller, due to stricter inclusion criteria). Data extracted from each meta- or pooled analysis included cancer site, gene name, genetic variant, point estimate (i.e. relative risk [RR] or odds ratio [OR]) and 95% confidence interval (CI), allelic frequency (if provided), number of studies, number of cases, test of study heterogeneity (e.g. Q test), and test of publication bias (including Begg’s test, Egger’s test and funnel plots). Random-effect estimates from meta-analyses were presented, unless only fixed-effect estimates were available.
Figure 1.
Selection of Studies
Table 2.
Statistically Significant Gene-variant/Cancer Associations and False Positive Report Probabilities (FPRP)
| Cancer Site |
Gene | Variant | Comparison | MAF or Freq at Risk1 |
OR | 95%CI | p-value | Evid for Pub Bias2 |
Evid for Het2 |
Studies | Cases | Power3 | Power3 | FPRP values at Prior Probability | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | OR:1.5 |
OR:1.2 |
OR:1.5 |
OR:1.2 |
Ref | |||||||||||||
| 0.001 | 0.000001 | 0.001 | 0.000001 | |||||||||||||||
| Bladder | ||||||||||||||||||
| GSTM1 | null | vs. present | 0.51W, 0.53S | 1.5 | 1.3–1.6 | 1.9×10−14 | No | No | 28 | 5072 | 1.0 | 0.017 | <0.001 | <0.001 | <0.001 | <0.001 | 23 | |
| GSTP1 | Ile105Val | GG+GA vs. AA | 0.14W | 1.23 | 1.04–1.57 | 0.0488 | Yes | No | 7 | 1903 | 0.944 | 0.421 | 0.990 | 1.000 | 0.996 | 1.000 | 82 | |
| NAT2 | acetylator | Slow vs. rapid/intermed | 0.56W | 1.46 | 1.26–1.68 | 2.5×10−7 | No | No | 36 | 5747 | 0.647 | 0.003 | <0.001 | 0.163 | 0.039 | 0.976 | 19 | |
| NQO1 | Pro187Ser | CT+TT vs. CC | 0.13–.0.20W | 1.204 | 1.00–1.43 | 0.0457 | No | No | 6 | 1066 | 0.994 | 0.500 | 0.977 | 1.000 | 0.988 | 1.000 | 83 | |
| XPC | Ala499Val | TT vs. CT+CC | 0.25W, 0.31S | 1.324 | 1.06–1.63 | 0.0114 | No | No | 4 | 2765 | 0.883 | 0.188 | 0.918 | 1.000 | 0.981 | 1.000 | 84 | |
| XPD | Asp312Asn | GA+AA vs. GG | 0.32–0.71 | 1.244 | 1.07–1.45 | 0.0055 | No | No | 3 | 789 | 0.991 | 0.341 | 0.876 | 1.000 | 0.954 | 1.000 | 85 | |
| XRCC3 | Thr241Met | TT vs. CC | 0.37 | 1.17 | 1.00–1.36 | 0.0409 | No | No | 7 | 3112 | 0.999 | 0.629 | 0.976 | 1.000 | 0.985 | 1.000 | 86 | |
| Breast6 | ||||||||||||||||||
| ATP1B2 | −8852T>C | TC vs. TT | 0.23–0.33 | 0.88 | 0.77–0.99 | 0.0462 | - | Yes | 2 | 2692 | 1.0 | 0.818 | 0.971 | 1.000 | 0.976 | 1.000 | 87 | |
| CASP8 | Asp302His | GC vs. GG | 0.13 | 0.89 | 0.85–0.94 | 5.7×10−6 | - | No | 14 | 16423 | 1.0 | 0.991 | 0.028 | 0.967 | 0.029 | 0.967 | 18 | |
| CHEK2 | *1100delC | heterozygotes vs. non-carrier | 0.002–0.02W | 2.4 | 1.8–3.2 | 2.5×10−9 | No | No | 12 | 18329 | 0.001 | 1×10−6 | 0.004 | 0.782 | 0.678 | 1.000 | 21 | |
| COMT | Met108/158V al | GG vs. AA | 0.45–0.47 | 1.144 | 1.03–1.26 | 0.0108 | - | No | 11 | 6398 | 1.0 | 0.842 | 0.911 | 1.000 | 0.924 | 1.000 | 88 | |
| CYP17 | rs4919687 | AA vs. GG | 0.26 | 1.17 | 1.03–1.34 | 0.0193 | - | No | 5 | 5166 | 1.0 | 0.643 | 0.959 | 1.000 | 0.973 | 1.000 | 89 | |
| CYP17 | rs4919682 | TT vs. CC | 0.25 | 1.16 | 1.01–1.33 | 0.0345 | - | No | 5 | 5146 | 1.0 | 0.686 | 0.971 | 1.000 | 0.980 | 1.000 | 89 | |
| CYP19 | TTTA10 | carrier vs. non-carrier | 0.01–0.02W | 1.59 | 1.01–2.48 | 0.0430 | - | - | 5 | 3934 | 0.399 | 0.107 | 0.990 | 1.000 | 0.997 | 1.000 | 90 | |
| CYP1A1 | A2455G | GG vs. AA | 0.25S | 0.72 | 0.53–0.99 | 0.0393 | No | No | 3 | 2938 | 0.682 | 0.184 | 0.984 | 1.000 | 0.996 | 1.000 | 91 | |
| CYP1B1 | Leu432Val | GG +GC vs. CC | 0.43W | 1.5 | 1.1–2.1 | 0.0140 | No | No | 6 | 2176 | 0.500 | 0.097 | 0.973 | 1.000 | 0.995 | 1.000 | 92 | |
| GATA3 | rs570613 | CT vs. TT | 0.40–0.45w | 0.85 | 0.75–0.95 | 0.007 | - | No | 2 | 2590 | 1.0 | 0.636 | 0.807 | 1.000 | 0.868 | 1.000 | 93 | |
| IGFBP3 | −202 C>A | AA vs. CC | 0.45W | 0.92 | 0.86–0.99 | 0.0202 | - | No | 10 | 13101 | 1.0 | 0.996 | 0.963 | 1.000 | 0.963 | 1.000 | 18 | |
| NBS1 | 657del5 | carrier vs. non-carrier | 0.02W | 3.13 | 1.40–7.00 | 0.0055 | - | - | 2 | 786 | 0.037 | 0.010 | 0.993 | 1.000 | 0.998 | 1.000 | 94 | |
| POR | Gly5Gly | GG vs. AA | 0.21–0.25A | 1.58 | 1.04–2.41 | 0.0329 | - | No | 4 | 1038 | 0.405 | 0.101 | 0.988 | 1.000 | 0.997 | 1.000 | 95 | |
| PR | PROGINS | T2/T2 vs. T1/T1 | 0.14 | 0.32 | 0.16–0.65 | 0.0014 | - | - | 4 | 1106 | 0.021 | 0.004 | 0.987 | 1.000 | 0.998 | 1.000 | 90 | |
| PTGS2 | Ex10+837 | CC vs. TT | 0.35W | 0.80 | 0.66–0.97 | 0.0231 | - | No | 3 | 2194 | 0.968 | 0.339 | 0.960 | 1.000 | 0.986 | 1.000 | 96 | |
| TGFB1 | Leu10Pro | TT vs. CC | 0.38W | 1.16 | 1.08–1.25 | 6.9×10−5 | - | No | 11 | 12946 | 1.0 | 0.813 | 0.090 | 0.990 | 0.108 | 0.992 | 18 | |
| TGFBR1 | *6A | *9A/*6A + *6A/*6A vs. *9A/*9A | 0.12–0.15 | 1.38 | 1.14–1.67 | 0.0009 | No | - | 7 | 1420 | 0.804 | 0.075 | 0.537 | 0.999 | 0.925 | 1.000 | 97 | |
| WDR79 | Arg68Gly | GG vs. CC | 0.21–0.23W | 1.60 | 1.04–2.47 | 0.0332 | - | No | 2 | 2692 | 0.385 | 0.097 | 0.989 | 1.000 | 0.997 | 1.000 | 87 | |
| WDR79 | Phe150Phe | CT vs. CC | 0.15–0.16W | 1.15 | 1.00–1.32 | 0.0485 | - | No | 2 | 2655 | 1.0 | 0.727 | 0.979 | 1.000 | 0.985 | 1.000 | 87 | |
| XRCC1 | Arg399Gln | AA vs. GG | 0.30S | 1.6 | 1.1–2.3 | 0.0125 | No | No | 4 | 1567 | 0.364 | 0.060 | 0.968 | 1.000 | 0.995 | 1.000 | 98 | |
| Colorectal | ||||||||||||||||||
| CCND1 | G870A | GA vs. GG | 0.12–0.64 | 1.18 | 1.06–1.32 | 0.0031 | No | No | 12 | 4618 | 1.0 | 0.616 | 0.792 | 1.000 | 0.861 | 1.000 | 99 | |
| GSTT1 | null | null vs. present | 0.21W, 0.44S | 1.37 | 1.17–1.60 | 8.1×10−5 | - | - | 11 | 1490 | 0.874 | 0.047 | 0.074 | 0.988 | 0.598 | 0.999 | 16 | |
| MTHFR | 677 C/T | TT vs. CC | 0.32W0.40S | 0.83 | 0.75–0.93 | 0.0007 | No | No | 25 | 12261 | 1.0 | 0.472 | 0.570 | 0.999 | 0.737 | 1.000 | 100 | |
| MTHFR | A1298C | CC vs. CA+AA | 0.29W, 0.22S | 0.81 | 0.69–0.96 | 0.0124 | No | No | 14 | 4764 | 0.988 | 0.372 | 0.938 | 1.000 | 0.976 | 1.000 | 101 | |
| NAT2 | acetylator5 | rapid vs. slow | 0.32–0.77W, 0.46–0.95S | 1.084 | 1.00–1.16 | 0.0421 | - | No | 18 | 6741 | 1.0 | 0.998 | 0.972 | 1.000 | 0.972 | 1.000 | 47 | |
| NQO1 | Pro187Ser | CT+TT vs. CC | 0.11–0.23W | 1.184 | 1.02–1.35 | 0.0206 | No | No | 5 | 1637 | 1.0 | 0.597 | 0.941 | 1.000 | 0.964 | 1.000 | 83 | |
| XPC | Lys939Gln | CA vs. AA | 0.65S,0.61W | 1.324 | 1.11–1.56 | 0.0014 | No | No | 2 | 1060 | 0.933 | 0.132 | 0.546 | 0.999 | 0.895 | 1.000 | 102 | |
| Esophageal | ||||||||||||||||||
| ALDH2 | *2*2 | *2*2 vs. *1*1 | 0.10–0.33S | 0.36 | 0.16–0.80 | 0.0128 | No | No | 5 | 705 | 0.065 | 0.020 | 0.995 | 1.000 | 0.998 | 1.000 | 103 | |
| CYP1A1 | Ile462Val | GG vs. AA | 0.21–0.25S | 2.52 | 1.62–3.91 | 3.9×10−5 | No | No | 9 | 754 | 0.010 | 0.0 | 0.783 | 1.000 | 0.988 | 1.000 | 104 | |
| XPD | Lys751Gln | AC+CC vs. AA | 0.07–0.37 | 1.394 | 1.15–1.68 | 0.0007 | No | No | 4 | 1053 | 0.785 | 0.064 | 0.456 | 0.999 | 0.911 | 1.000 | 85 | |
| Gastric | ||||||||||||||||||
| CDH1 | -160C>A | CA+AA vs. CC | 0.14–0.16S | 0.81 | 0.67–0.99 | 0.0343 | No | No | 7 | 1174 | 0.971 | 0.391 | 0.976 | 1.000 | 0.990 | 1.000 | 105 | |
| GSTT1 | null | null vs. present | 0.13–0.26W | 1.27 | 1.03–1.56 | 0.0240 | No | No | 8 | 835 | 0.944 | 0.294 | 0.960 | 1.000 | 0.987 | 1.000 | 106 | |
| IL1RN | VNTR | *2 carrier vs. LL | 0.22–0.29W | 1.30 | 1.09–1.54 | 0.0029 | No | No | 16 | 2293 | 0.951 | 0.177 | 0.716 | 1.000 | 0.931 | 1.000 | 107 | |
| MTHFR | C677T | TT vs. CT+CC | 0.35–0.57M | 1.52 | 1.31–1.77 | 4.9×10−8 | No | No | 16 | 2727 | 0.432 | 0.001 | <0.001 | 0.140 | 0.057 | 0.984 | 20 | |
| P53 | Arg72Pro | GG vs. CC | 0.41–0.49S | 0.84 | 0.72–0.99 | 0.0319 | No | - | 8 | 1295 | 0.997 | 0.538 | 0.974 | 1.000 | 0.986 | 1.000 | 108 | |
| TNF-A | -308G>A | AA vs. GG | 0.04–0.16M | 1.49 | 1.11–1.99 | 0.0074 | No | No | 19 | 3660 | 0.518 | 0.071 | 0.930 | 1.000 | 0.990 | 1.000 | 109 | |
| Glioma | ||||||||||||||||||
| ATR | rs11920625 | AG vs. GG | 0.08W | 1.40 | 1.11–1.77 | 0.0047 | - | No | 5 | 1010 | 0.718 | 0.099 | 0.873 | 1.000 | 0.980 | 1.000 | 110 | |
| CHAF1A | rs243356 | CT vs. CC | 0.21W | 1.33 | 1.10–1.60 | 0.0028 | - | No | 5 | 1010 | 0.899 | 0.138 | 0.735 | 1.000 | 0.948 | 1.000 | 110 | |
| CHAF1A | rs243341 | CT vs. TT | 0.26W | 1.25 | 1.04–1.50 | 0.0169 | - | No | 5 | 1010 | 0.975 | 0.330 | 0.944 | 1.000 | 0.980 | 1.000 | 110 | |
| CHAF1A | rs105038 | CT vs. CC | 0.26W | 1.25 | 1.04–1.50 | 0.0169 | - | No | 5 | 1010 | 0.975 | 0.330 | 0.944 | 1.000 | 0.980 | 1.000 | 110 | |
| CHAF1A | rs2992 | GG vs. AA | 0.26W | 1.47 | 1.05–2.04 | 0.023 | - | No | 5 | 1010 | 0.548 | 0.112 | 0.975 | 1.000 | 0.995 | 1.000 | 110 | |
| DCLRE1B | rs3761936 | CC vs. TT | 0.19W | 0.36 | 0.20–0.65 | 0.0007 | - | No | 5 | 1010 | 0.020 | 0.003 | 0.972 | 1.000 | 0.996 | 1.000 | 110 | |
| DCLRE1B | rs12022378 | TT vs. CC | 0.19W | 0.36 | 0.20–0.65 | 0.0007 | - | No | 5 | 1010 | 0.020 | 0.003 | 0.972 | 1.000 | 0.996 | 1.000 | 110 | |
| ERCC1 | rs3212986 | GT vs. GG | 0.27W | 0.76 | 0.63–0.92 | 0.0045 | - | No | 5 | 1010 | 0.911 | 0.172 | 0.842 | 1.000 | 0.966 | 1.000 | 110 | |
| ERCC1 | rs3212955 | AG vs. AA | 0.26W | 0.79 | 0.66–0.96 | 0.0137 | - | No | 5 | 1010 | 0.956 | 0.296 | 0.949 | 1.000 | 0.984 | 1.000 | 110 | |
| IL4 | rs2243248 | TG vs. TT | 0.05W | 1.44 | 1.05–1.97 | 0.0231 | - | No | 2 | 654 | 0.601 | 0.127 | 0.974 | 1.000 | 0.994 | 1.000 | 111 | |
| IL6 | rs1800795 | GG vs. CC | 0.45W | 0.70 | 0.51–0.96 | 0.0271 | - | No | 2 | 654 | 0.619 | 0.140 | 0.977 | 1.000 | 0.995 | 1.000 | 111 | |
| NEIL3 | rs12645561 | CT vs. CC | 0.13W | 1.29 | 1.05–1.59 | 0.0161 | - | No | 5 | 1010 | 0.921 | 0.249 | 0.949 | 1.000 | 0.986 | 1.000 | 110 | |
| MSH5 | rs707938 | CC vs. TT | 0.36W | 0.67 | 0.50–0.89 | 0.0065 | - | No | 5 | 1010 | 0.514 | 0.066 | 0.917 | 1.000 | 0.989 | 1.000 | 110 | |
| POLD1 | rs1673041 | AA vs. CC | 0.26W | 0.53 | 0.35–0.79 | 0.0022 | - | No | 5 | 1010 | 0.130 | 0.013 | 0.933 | 1.000 | 0.993 | 1.000 | 110 | |
| RPA3 | rs4140805 | GG vs. TT | 0.40W | 1.43 | 1.11–1.85 | 0.0061 | - | No | 5 | 1010 | 0.642 | 0.091 | 0.910 | 1.000 | 0.986 | 1.000 | 110 | |
| RPA3 | rs2160138 | CC vs. TT | 0.44W | 1.46 | 1.14–1.88 | 0.003 | - | No | 5 | 1010 | 0.583 | 0.064 | 0.852 | 1.000 | 0.981 | 1.000 | 110 | |
| RPA3 | rs6947203 | TT vs. CC | 0.33W | 1.47 | 1.11–1.94 | 0.0068 | - | No | 5 | 1010 | 0.557 | 0.076 | 0.921 | 1.000 | 0.988 | 1.000 | 110 | |
| TP53 | rs8079544 | CT vs. CC | 0.06W | 1.34 | 1.04–1.72 | 0.0226 | - | No | 5 | 1010 | 0.812 | 0.193 | 0.964 | 1.000 | 0.991 | 1.000 | 110 | |
| Head/neck | ||||||||||||||||||
| GSTM1 | null | null vs. present | 0.47–0.54W | 1.16 | 1.01–1.33 | 0.0345 | Yes | No | 11 | 3754 | 1.0 | 0.686 | 0.971 | 1.000 | 0.980 | 1.000 | 112 | |
| GSTT1 | null | null vs. present | 0.11–0.53S, 0.14–0.52W | 1.08 | 1.02–1.14 | 0.0067 | No | No | 23 | 3974 | 1.0 | 1.0 | 0.840 | 1.000 | 0.840 | 1.000 | 113 | |
| MGMT | Leu84Phe | CT+ TT vs. CC | 0.16M | 0.74 | 0.55–1.00 | 0.0483 | - | No | 3 | 514 | 0.752 | 0.220 | 0.985 | 1.000 | 0.996 | 1.000 | 114 | |
| MGMT | Ile143Val | AG + GG vs. AA | 0.13M | 0.73 | 0.53–1.00 | 0.0520 | - | No | 3 | 536 | 0.714 | 0.205 | 0.986 | 1.000 | 0.996 | 1.000 | 114 | |
| XPC | PAT+/− | + vs − | 0.67S,0.59W | 1.294 | 1.04–1.59 | 0.0187 | No | No | 2 | 720 | 0.921 | 0.249 | 0.949 | 1.000 | 0.986 | 1.000 | 102 | |
| Leukemia (acute) | ||||||||||||||||||
| GSTM1 | null | null vs. present | 0.28–0.57W | 1.20 | 1.14–1.25 | 8.6×10−15 | No | No | 19 | 3532 | 1.0 | 0.841 | <0.001 | <0.001 | <0.001 | <0.001 | 22 | |
| GSTP1 | Ile105Val | GA + GG vs. AA | 0.25–0.35W | 1.09 | 1.01–1.16 | 0.0147 | No | No | 8 | 1571 | 1.0 | 0.999 | 0.869 | 1.000 | 0.869 | 1.000 | 22 | |
| GSTT1 | null | null vs. present | 0.08–0.32W | 1.19 | 1.14–1.29 | 3.5×10−8 | No | No | 17 | 3484 | 1.0 | 0.581 | 0.023 | 0.960 | 0.039 | 0.976 | 22 | |
| MTHFR | C677T | TT vs. CT+CC | 0.27–0.43W | 0.84 | 0.71–0.99 | 0.0398 | No | No | 13 | 2191 | 0.997 | 0.538 | 0.974 | 1.000 | 0.986 | 1.000 | 115 | |
| Lung6 | ||||||||||||||||||
| CYP1A1 | MspI (T3801C) | MspI/MspI vs. not present | 0.22W | 2.36 | 1.16–4.81 | 0.0180 | No | No | 17 | 1759 | 0.106 | 0.031 | 0.994 | 1.000 | 0.998 | 1.000 | 116 | |
| CYP1A1 | exon7 poor | AG+ GG vs. AA | 0.26S | 1.61 | 1.24–2.08 | 0.0003 | No | Yes | 11 | 1176 | 0.294 | 0.012 | 0.477 | 0.999 | 0.956 | 1.000 | 117 | |
| CYP2D6 | poor vs. extensive | 0.08W | 0.69 | 0.52–0.90 | 0.0080 | - | No | 17 | 7504 | 0.600 | 0.082 | 0.912 | 1.000 | 0.987 | 1.000 | 118 | ||
| GSTT1 | metabolizer5 null | null vs. present | 0.54S | 1.28 | 1.10–1.49 | 0.0014 | No | No | 8 | 1364 | 0.980 | 0.203 | 0.596 | 0.999 | 0.877 | 1.000 | 119 | |
| MDM2 | SNP309 | GG vs. TT | 0.37W, 0.48S | 1.274 | 1.12–1.44 | 0.0002 | - | Yes | 7 | 4276 | 0.995 | 0.188 | 0.162 | 0.995 | 0.505 | 0.999 | 13 | |
| mEH | His113Tyr | CC vs. TT | 0.36W, 0.43S | 0.70 | 0.51–0.96 | 0.0271 | No | Yes | 8 | 986 | 0.619 | 0.140 | 0.977 | 1.000 | 0.995 | 1.000 | 120 | |
| MPO | G463A | AA vs. GG | 0.23M | 0.71 | 0.57–0.88 | 0.0020 | No | No | 10 | 3688 | 0.717 | 0.072 | 0.711 | 1.000 | 0.961 | 1.000 | 121 | |
| XPA | G23A | GA vs. AA | 0.37M | 0.73 | 0.61–0.89 | 0.0011 | No | No | 7 | 1913 | 0.815 | 0.095 | 0.694 | 1.000 | 0.951 | 1.000 | 14 | |
| XPC | Lys939Gln | CC vs. CA+AA | 0.38W,0.36S, 0.28A | 1.304 | 1.11–1.53 | 0.0014 | No | No | 6 | 2580 | 0.957 | 0.168 | 0.625 | 0.999 | 0.905 | 1.000 | 84 | |
| XPD | Lys751Gln | CC vs. AA | 0.30M | 1.30 | 1.13–1.49 | 0.0002 | No | No | 15 | 5004 | 0.980 | 0.125 | 0.143 | 0.994 | 0.566 | 0.999 | 14 | |
| XRCC1 | Arg399Gln | AA vs. GG | 0.27–0.46S | 1.34 | 1.16–1.54 | 5.2×10−5 | No | No | 6 | 1702 | 0.944 | 0.060 | 0.038 | 0.975 | 0.383 | 0.998 | 17 | |
| Non-melanoma skin | ||||||||||||||||||
| XRCC3 | Thr241Met | TT vs. CC +CT | 0.30–0.41W | 0.76 | 0.62–0.93 | 0.0080 | No | No | 4 | 1599 | 0.898 | 0.186 | 0.896 | 1.000 | 0.976 | 1.000 | 122 | |
| Meningioma | ||||||||||||||||||
| BRIP1 | rs4968451 | AC vs. AA | 0.15W | 1.61 | 1.26–2.06 | 0.0001 | - | No | 5 | 631 | 0.287 | 0.010 | 0.347 | 0.998 | 0.940 | 1.000 | 123 | |
| Non-Hodgkin Lymphoma | ||||||||||||||||||
| IL10 | −3575T>A | TA+AA vs. TT | 0.36W | 1.11 | 1.01–1.23 | 0.0379 | - | No | 8 | 3030 | 1.0 | 0.932 | 0.979 | 1.000 | 0.980 | 1.000 | 124 | |
| IL1RN | 9589A>T | AT +TT vs. AA | 0.27W | 1.08 | 1.00–1.17 | 0.0547 | - | No | 8 | 3020 | 1.0 | 0.995 | 0.983 | 1.000 | 0.984 | 1.000 | 124 | |
| MTHFR | 677C>T | TT vs. CC | 0.29–0.45W | 1.17 | 1.02–1.34 | 0.0241 | No | No | 11 | 4121 | 1.0 | 0.643 | 0.959 | 1.000 | 0.973 | 1.000 | 125 | |
| TNF | −308G>A | GA+AA vs. GG | 0.14W | 1.19 | 1.05–1.33 | 0.0039 | - | No | 8 | 2718 | 1.0 | 0.559 | 0.685 | 1.000 | 0.795 | 1.000 | 124 | |
| Ovarian | ||||||||||||||||||
| CDK6 | IVS2–4184C>T | CT vs. CC | 0.21W | 1.09 | 1.00–1.19 | 0.0521 | - | No | 11 | 3597 | 1.0 | 0.984 | 0.982 | 1.000 | 0.982 | 1.000 | 126 | |
| CDKN1B | Val109Gly | GG vs. TT | 0.25W | 0.79 | 0.65–0.95 | 0.0149 | - | Yes | 11 | 3618 | 0.964 | 0.285 | 0.927 | 1.000 | 0.977 | 1.000 | 126 | |
| CDKN2A/2 B | *4780C>T | CT vs. CC | 0.27W | 0.89 | 0.81–0.97 | 0.0113 | - | Yes | 11 | 3601 | 1.0 | 0.933 | 0.888 | 1.000 | 0.895 | 1.000 | 126 | |
| Prostate | ||||||||||||||||||
| AR | CAG21 | >CAG21vs. ≤CAG21 | 0.50M | 1.19 | 1.07–1.31 | 0.0008 | No | No | 22 | 4274 | 1.0 | 0.568 | 0.279 | 0.997 | 0.405 | 0.999 | 127 | |
| AR | GGN16 | ≤GGN16 vs. >GGN16 | 0.50W | 1.31 | 1.06–1.61 | 0.0113 | No | No | 8 | 1918 | 0.901 | 0.202 | 0.919 | 1.000 | 0.981 | 1.000 | 127 | |
| CDH1 | −160C>A | AA+CA vs. CC | 0.14–0.61M | 1.31 | 1.08–1.60 | 0.0071 | No | Yes | 8 | 2633 | 0.908 | 0.195 | 0.899 | 1.000 | 0.977 | 1.000 | 105 | |
| CYP17 | rs2486758 | TC vs. TT | 0.21M | 1.07 | 1.00–1.14 | 0.043 | - | No | 7 | 7914 | 1.0 | 1.0 | 0.973 | 1.000 | 0.973 | 1.000 | 89 | |
| CYP17 | rs6892 | AG vs. AA | 0.18M | 1.08 | 1.00–1.15 | 0.0309 | - | No | 7 | 8013 | 1.0 | 0.999 | 0.942 | 1.000 | 0.942 | 1.000 | 89 | |
| RNASEL | Asp541Glu | GT +GG vs. TT | 0.42–0.57W | 1.27 | 1.13–1.44 | 0.0001 | - | No | 6 | 3038 | 0.995 | 0.188 | 0.162 | 0.995 | 0.505 | 0.999 | 15 | |
| Upper digestive tract | ||||||||||||||||||
| XRCC1 | Arg399Gln | AG+GG vs. AA | 0.28–0.47M | 0.85 | 0.75–0.98 | 0.0172 | No | No | 7 | 1672 | 1.0 | 0.607 | 0.962 | 1.000 | 0.976 | 1.000 | 128 | |
| Urothelial | ||||||||||||||||||
| CDH1 | −160C>A | AA vs. CC | 0.23–0.43W, 0.14–0.61S | 2.57 | 1.55–4.24 | 0.0002 | No | No | 3 | 558 | 0.018 | 0.001 | 0.926 | 1.000 | 0.994 | 1.000 | 105 | |
Minor allelic frequency for WWhites, SAsians, MMixedor AAfricans or frequency at risk for variants that are not SNPs.
Evidence for publication bias and heterogeneity represent whether quality assessment of studies was performed and the corresponding results from those tests as reported by each published meta- or pooled analysis. “−” Dash indicates unclear whether test was performed.
Statistical power to detect an OR of 1.5 (0.67=1/1.5) or an OR of 1.2 (0.83=1/1.2)
Fixed effect estimate
Phenotype and genotype methods of detection.
Although HRAS1 alleles were significantly associated with breast cancer this finding is the consequence of an error in genotyping - see text.
Shading indicates noteworthiness of association at 0.2 level
We calculated summary estimates to describe published reports identified through our search. Differences in the number of studies and cases were evaluated by t-test. Associations were considered statistically significant if the reported p-value was <0.05 or if the 95 % CI excluded 1.0. P-values were determined by first calculating a Z-score based on the reported OR and 95% CI: Z-score= ln(OR)/[(ln(upper CI) − ln(lower CI))/(2*1.96)], and then comparing it to a normal distribution.
For each statistically significant association reported, we estimated the FPRP using methods described by Wacholder et al.12 The FPRP value is determined by the p-value, the given prior probability for the association, and the statistical power of the test. Assigning a prior probability should be determined before obtaining results from a study and should be independent of any data used in the analysis. Prior probabilities are subjective and are influenced by both previous epidemiologic findings and experimental evidence about known functions of a genetic variant. Therefore, we chose to calculate FPRP values for two levels of prior probabilities: at a low prior that would be similar to what would be expected for a candidate gene (0.001) and at a very low prior that would be similar to what would be expected for a random SNP (0.000001), thus allowing the reader to evaluate the association using their own judgment about the supporting evidence for a given loci. Wacholder et al.12 suggests estimating statistical power based on the ability to detect an OR of 1.5 (or its reciprocal 0.67=1/1.5 for ORs less than 1.0), with an alpha level equal to the observed p-value.12 But given the recent attention to much smaller ORs this estimate may be too conservative, thus we have chosen to present results for both an OR of 1.5 and 1.2 (or its reciprocal 0.83=1/1.2). To evaluate whether an association is “noteworthy”, we used a FPRP cut-off value of 0.2, as suggested by the authors12 for summary analyses. Hence, FPRP values less than 0.2 indicate an association that remained robust for a given prior probability and will be referred to as noteworthy in the present paper. Statistical power and FPRP were computed by the Excel spreadsheet provided by Wacholder et al.12
Results
We identified 161 published meta- and pooled analyses, encompassing 18 cancer sites and 99 different genes. These 161 meta- and pooled analyses addressed 344 gene-variant/cancer associations with an average of 7.3 studies and 3,551 cases per investigated association (range: 508–19,729 cases). As expected, most analyses were conducted for common cancers, such as breast (n=119), prostate (n=42), and lung (n=34) cancer; there are very few evaluations of genetic associations in rare cancers, such as cervical and esophageal (Table 1). Across all cancer sites, variants in genes involved in DNA repair (e.g. XRCC1 and XPD; n=81) and genes encoding metabolizing enzymes (e.g. cytochrome P450 (CYP) variants, n=58; or glutathione S-transferases (GSTs), n=31) were most often evaluated. Meta- and pooled analyses that found a statistically significant association evaluated a higher number of studies but included a lower number of cases than those that found a non-significant association (p=0.02 and p=0.05, respectively; Table 1). A complete table that lists all data extracted from each of the 344 associations identified in our search is included in the Appendix (Table A1).
Table 1.
Significance of Gene-variant/Cancer Associations Demonstrated by Meta- and Pooled Analyses by Cancer Site
| Cancer Site | Total | Significant Associations | Non-significant Associations | ||||||
|---|---|---|---|---|---|---|---|---|---|
| # of associations |
Average cases per association |
Average studies per association |
# of associations |
Average cases per association |
Average studies per association |
# of associations |
Average cases per association |
Average studies per association |
|
| Bladder | 13 | 2574 | 9.8 | 7 | 2922 | 13 | 6 | 2168 | 6.2 |
| Breast | 119 | 5246 | 6.0 | 21 | 5111 | 6 | 98 | 5275 | 6.0 |
| Cervical | 1 | 14999 | 70.0 | 0 | - | - | 1 | - | 70.0 |
| Colorectal | 24 | 2589 | 7.0 | 7 | 4653 | 12.4 | 17 | 1738 | 4.8 |
| Esophageal | 5 | 839 | 6.6 | 3 | 837 | 6.0 | 2 | 843 | 7.5 |
| Gastric | 18 | 1559 | 9.6 | 6 | 1997 | 12.3 | 12 | 1339 | 8.2 |
| Glioma | 31 | 913 | 3.9 | 18 | 970 | 4.7 | 13 | 832 | 2.8 |
| Head & Neck | 12 | 1582 | 7.3 | 5 | 1900 | 8.4 | 7 | 1356 | 6.6 |
| Hepatocellular | 2 | 2469 | 13.5 | 0 | - | - | 2 | 2469 | 13.5 |
| Leukemia | 8 | 1930 | 9.8 | 4 | 2695 | 14.3 | 4 | 1165 | 5.3 |
| Lung | 34 | 3073 | 9.8 | 12 | 2738 | 9.9 | 22 | 3256 | 9.8 |
| Meningioma | 1 | 631 | 5.0 | 1 | 631 | 5.0 | 0 | - | - |
| Non-Hodgkin Lymphoma | 13 | 3051 | 8.2 | 4 | 3222 | 8.8 | 9 | 2975 | 8.0 |
| Ovarian | 8 | 3737 | 11 | 3 | 3605 | 11.0 | 5 | 3815 | 11.0 |
| Prostate | 42 | 4557 | 7.1 | 6 | 4632 | 9.5 | 36 | 4544 | 6.7 |
| Skin | 8 | 1599 | 4.6 | 1 | 1599 | 4.0 | 7 | 1599 | 4.7 |
| Upper digestive tract | 4 | 1364 | 5.0 | 1 | 1672 | 7.0 | 3 | 1261 | 4.3 |
| Urothelial | 1 | 558 | 3.0 | 1 | 558 | 3.0 | 0 | - | - |
| 344 | 3551 | 7.3 | 100 | 3014 | 8.4 | 244 | 3772 | 6.8 | |
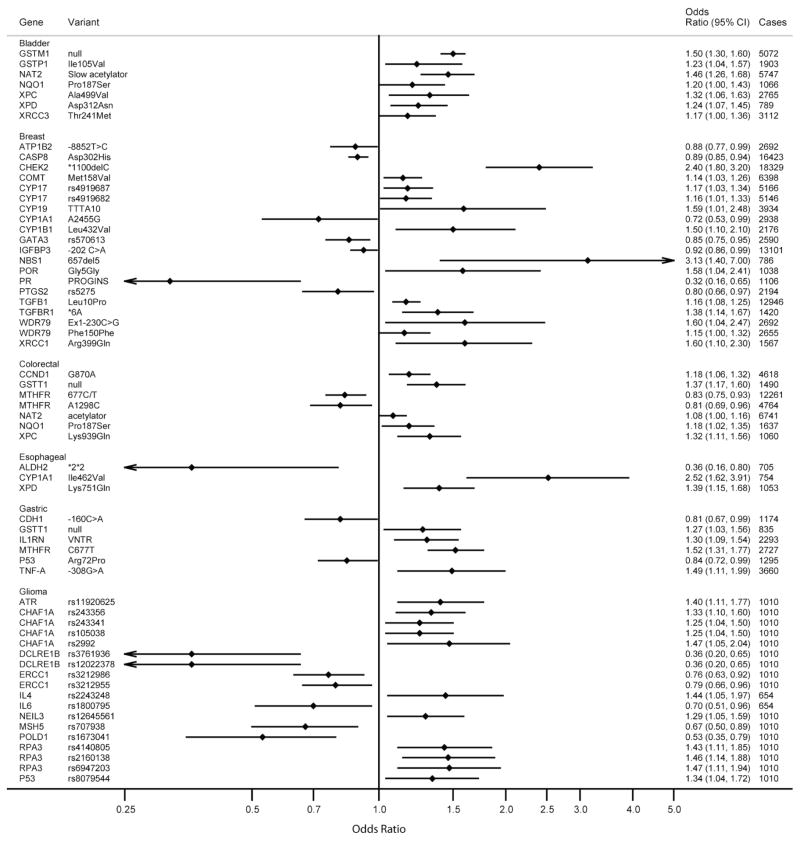
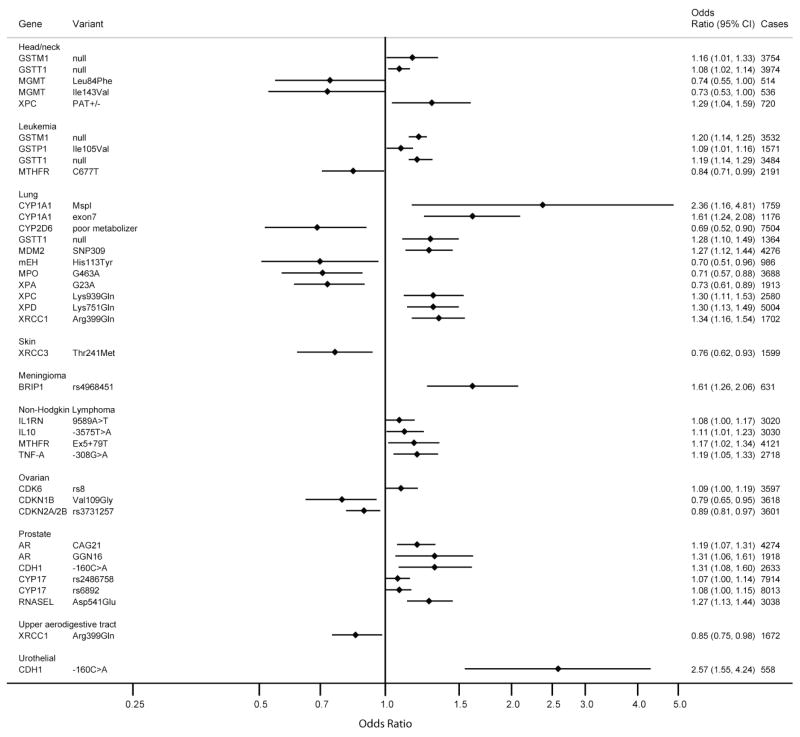
Among the 344 gene-variant/cancer associations evaluated, the summary OR for 98 (28%) associations (excluding those involving HRAS1) were statistically significant (p-values between 0.05 to 8.6×10−15; Figure 2a, 2b and Table 2). Thirty of these 98 associations were inverse for the variant, with a mean OR of 0.73 (median: 0.75; range: 0.32–0.92). The other 68 analyses reported ORs above 1.0, with a mean of 1.47 (median: 1.34; range 1.07–3.13). Statistically significant associations were found among 16 cancer sites, predominantly among studies investigating breast, glioma and lung cancer.
Figure 2.
Figure 2a and Figure 2b. Summary ORs and 95% CIs for Cancer Risk by Genetic Variants – Limited to Meta- and Pooled Analyses With Significant Summary Risk Estimates
In order to evaluate the robustness of these findings, we calculated FPRP values at two levels of prior probabilities (Table 2). Among the 98 associations, 85 gene-variant/cancer associations had FPRP values higher than 0.2 across the pre-specified prior probabilities (0.001 and 0.000001); these results are not considered noteworthy. For example, although the summary OR from the pooled analysis for XRCC1 Arg399Gln indicated a statistically significant positive association with risk of breast cancer (OR, 1.6; 95% CI, 1.1–2.3), FPRP values were higher than 0.2, at any of the two prior probabilities; hence, the finding is not considered noteworthy.
At a prior probability level of 0.001 and statistical power to detect an OR of 1.5, 13 gene-variant/cancer associations remained noteworthy (FPRP ≤0.2) for: 1) MDM2 SNP309 and lung cancer (OR, 1.27; p-value=0.0002)13; 2) XPD Lys751Gln and lung cancer (OR, 1.30; p-value=0.0002)14; 3) RNASEL Asp541Glu and prostate cancer (OR, 1.27; p-value=0.0001)15; 4) GSTT1 null and colorectal cancer (OR, 1.37; p-value=8.1×10−5)16; 5) XRCC1 Arg399Gln and lung cancer (OR, 1.34; p-value=5.2×10−5)17; 6) TGFB1 Leu10Pro and breast cancer (OR, 1.16; p-value=6.9×10−5)18; 7) CASP8 Asp302His and breast cancer (OR, 0.89; p-value=5.7×10−6)18; 8) NAT2 slow acetylator and bladder cancer (OR, 1.46; p-value=2.5×10−7)19; 9) MTHFR C677T and gastric cancer (OR, 1.52; p-value=4.9×10−8)20; 10) CHEK2 *1100delC and breast cancer (OR, 2.4; p-value=2.5×10−9)21; 11) GSTT1 null and acute leukemia (OR, 1.19; p-value=3.5×10−8)22; 12) GSTM1 null and bladder cancer (OR, 1.5; p-value=1.9×10−14)23; and 13) GSTM1 null and acute leukemia (OR, 1.20; p-value=8.6×10−15).22 At a very low prior probability of 0.000001, four of these thirteen gene-variant/cancer associations remained noteworthy: MTHFR C677T, NAT2 slow acetylator, and GSTM1 null (Table 2). This number further reduced to two (GSTM1 null with bladder cancer and GSTM1 null with leukemia) when we calculated statistical power based on a lower OR of 1.2. Consistent with the FPRP, associations noteworthy at a very low prior probability were highly statistically significant (p-values between 10−7 to 10−15).
Discussion
Overall, close to one-third of all gene-variant/cancer associations from published meta- and pooled analyses were reported to be statistically significant. Thirteen of these associations were noteworthy at a prior probability of 0.001 and statistical power to detect an OR of 1.5, of which four remained noteworthy at even a lower prior probability similar to one appropriate for a randomly selected SNP in a genome-wide association study (1/1,000,000=0.000001) with p-values between 10−7 to 10−15. These associations are thus less likely to be false positives and have a high likelihood of being true associations with cancer risk. Specifically, we observed that, among the noteworthy associations, genes encoding for phase II metabolizing enzymes made up the majority of noteworthy associations.
Continuing advances in genotyping technologies have led to the feasibility of testing a large number of genetic variants; with this has come the potential for the publication of a large number of false positives due to the widely used strategy of declaring significance based on a p-value <0.05. A key feature of the Bayesian approach using the FPRP is that it is based, not only on the observed p-value, but also on both the power and prior probability of the hypothesis, allowing the user to incorporate prior knowledge, including functional information, of the specifically tested variants. Although the FPRP calculation allows an evaluation at different scenarios of prior probability, statistical power, and noteworthiness criterion, the choice for these parameters should be determined a priori using empirical evidence from past studies. Accordingly, it may be reasonable to claim that SNPs of relevant candidate genes with known or predicted function (based on experimental studies or in silico tests) are more likely to be associated with cancer risk and hence justify higher prior probabilities. However, choice of a single prior probability will be subject to debate; hence, here, we provide readers with the opportunity to use their own judgment about the body of evidence for a given candidate gene or variant. In this paper, we chose a more agnostic approach to evaluating associations by applying two levels of prior probability (0.001 and 0.000001) and statistical power (OR of 1.5, recommended by Wacholder et al. and similar to the average reported OR in our review; as well as OR of 1.2, close to the median reported OR in our review) to all statistically significant associations. As suggested by Thomas and Clayton 24, the prior probability for studies evaluating candidate genes will usually exceed 1000:1 (or 0.001). Thus, at a prior probability of 0.001, thirteen associations were noteworthy and may plausibly be true associations. The likelihood of being a true association, however, is even greater for the four associations that remain noteworthy at a very low prior probability (0.000001).
GSTM1 and GSTT1 belong to a family of phase II enzymes, the glutathione S-transferases, that are involved in the metabolism and biotransformation of toxic xenobiotics and endobiotics.25 Deletion of GSTT1 was associated with an increased risk of colorectal cancer16 and acute leukemia22 and the GSTM1 deletion was statistically significantly associated with risk of bladder cancer23 and acute leukemia22; and the latter two were found to be among the most noteworthy findings across all meta- and pooled analyses. Individual studies conducted subsequent to the meta analyses continue to support findings for GSTT126–31 and GSTM132–37, except for one study that reported a statistically significant inverse association between GSTT1 null and colorectal cancer38 and a few small studies on GSTT1 and leukemia providing inconsistent results.35, 37, 39, 40 The prevalence of GSTT1 null ranges from 20% in Caucasians to 60% among Asians,41 and approximately 50% of humans (ranging from 22% in Africa to 62% in Europe) are GSTM1 null.42 GSTT1 and GSTM1 are involved in the elimination of carcinogens in the body, such as products of oxidative stress and polycyclic aromatic hydrocarbons from tobacco smoke.43 Deletion of the GSTT1 and GSTM1 gene results in the variant called GSTT1/GSTM1 null and a complete loss of enzymatic activity.44 An individual with the null variants is thus expected to have an impaired ability to detoxify carcinogens and an increased risk of cancer, potentially affecting multiple cancer sites. This and the fact that GSTT1 and GSTM1 result in noteworthy associations with risk of various cancers lends support to the theory that these two variants, in particular GSTM1 are functional and truly impact cancer risk.
Another finding that was among the most noteworthy was the association between NAT2 slow acetylator phenotype and bladder cancer.19 This meta-analysis was published recently, thus no additional studies were identified subsequent to the meta-analysis. NAT2 is one of two N-acetyl transferase isoforms expressed in humans, which are involved in the detoxification of heterocyclic or aromatic amines and their metabolites.45 NAT2 is highly polymorphic and several non-synonymous polymorphisms result in poor expression, an unstable protein, or decreased catalytic activity, all of which result in the slow acetylator phenotype.46 The prevalence of NAT2 slow acetylators in European whites is about 56% and approximately 11% among Asians.23 The change in the rate of acetylation is expected to alter the effect of carcinogens on cancer risk, but the effect of this change may differ by cancer site. The NAT2 slow-acetylator phenotype is associated with an increased risk of bladder cancer (due to decreased detoxification of carcinogens from tobacco smoke), but has been associated with decreased risk of colorectal cancer (due to reduced activation of carcinogens).45–47 Taken together, the strong evidence supporting a functional effect of the NAT2 slow acetylator and the highly statistically significant association with bladder cancer supports the hypothesis that this variant is likely to modify cancer risk.
The recently published association between MTHFR C677T and gastric cancer was also among the most noteworthy associations.20 MTHFR, 5,10-methyletetetrahydrofolate reductase, plays a key role in the one-carbon metabolism pathway. Specifically, MTHFR converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate which then allows for the metabolism of homocysteine and the provision of methyl groups. Enzyme activity among individuals homozygous for MTHFR C677T is much reduced, approximately 30% of expected enzyme activity, compared with those who are homozygous for the common variant. 48, 49 Consequently, the reduced ability of MTHFR has been associated with alteration in methylation patterns and potentially aberrant DNA synthesis, repair, and chromosomal instability.50 Due to its role in a key pathway, the MTHFR C677T variant may have a true impact on cancer risk.
Among associations noteworthy at prior probabilities of 0.001 were three genes associated with DNA repair (CHEK2, XPD, and XRCC1). Pathways involving these genes are responsible for repairing DNA damage and errors that may occur during DNA replication. There have been no studies published subsequent to the meta-analysis on CHEK2 *1100delC and breast cancer.21 Studies conducted subsequent to the meta-analysis on XPD Lys751Gln and lung cancer51, 52 have drawn the same conclusions as our review. The statistically significant finding for XRCC1 was present among Asians only, and one of the three subsequent studies conducted among Asians53–55 found a statistically significant association between XRCC1 Arg399Gln and lung cancer. Overall, it is biologically plausible that genes associated with DNA repair have an impact on the risk of cancer and our review lends support towards the likelihood of these associations.
RNASEL Asp541Glu, MDM2 SNP309, TGFB1 Leu10Pro and CASP8 Asp302His are additional variants identified through our review as being noteworthy; they belong to key pathways plausibly influencing cancer susceptibility. RNASEL plays an important role in the inflammatory response pathway and was first identified as a candidate gene for prostate cancer risk due to its location within the hereditary prostate cancer 1 (HPC1) region.56, 57 As the meta-analysis has been published recently, only three subsequently published studies were identified but with conflicting results for prostate cancer.58–60 MDM2 encodes for the human homolog of mouse double minute 2, a nuclear phospholipoprotein that binds and inhibits p53, a tumor suppressor.61 A further study published after the meta-analysis lend support when analysis was restricted to never smokers.62 TGFB1, which encodes transforming growth factor beta 1, has been implicated as both a tumor suppressor and a tumor promoter.63, 64 An additional study published subsequent did not find an association.65 CASP8 encodes for Caspase 8 which plays a central role in the initiation and activation of a cascade of caspases leading to apoptosis.66 The decreased risk with CASP8 Asp302His for breast cancer observed in the pooled analysis is further supported by findings from a recent association study.67
Very recently, results from the first genome-wide association studies of cancer have become available, in which hundreds of thousands of variants were genotyped across the entire genome. These studies detected several highly statistically significant variants in the human chromosome 8q24 region that were associated with prostate, colorectal, and breast cancer susceptibility; however, there are no known characterized genes within this region.68–75 Variants located within SMAD774, a gene involved with cell signaling, and DAB2IP76, a putative tumor suppressor gene, have also been associated with colorectal and prostate cancer, respectively. Three follow-up genome wide-scans in prostate cancer have confirmed the previously identified loci and identified several additional loci that may be associated with prostate cancer risk.77–79 The loci which were identified in at least two of the studies were as follows: 8q24, HNF1B (17q12), MSMB (10q11), NUDT10/11 (Xp11.22), and 17q24. Six highly statistically significant variants associated with breast cancer susceptibility have also been identified through genome-wide studies, of which three are located within genes associated with control of cell growth or cell signaling (TNRC9, MAP3K1 and LSP1).75, 80, 81 Two variants were located in the 8q24 and 2q35 regions, and the sixth within FGFR2, a tumor suppressor gene overexpressed in breast cancer. The substantial evidence supporting these variants, including sizeable power and replication in large samples, indicates that these associations are likely to be true and yet none of the statistically significant variants had been previously identified because most did not reside in “interesting” candidate regions. Genome-wide association studies of cancer have also demonstrated that the effect size of statistically significant genetic variants is overall quite modest (point estimates between 1.1–1.5 for an additive mode of inheritance), which is consistent with the weak associations found in most meta- and pooled analyses.
We attempted to review all published meta- and pooled analyses covering the topic of genetic variants and cancer risk through several iterations of search criterion; however, it is possible that we have missed some studies. Many of the noteworthy variants identified were deletions (which may not be well captured by genome-wide association studies) and non-synonymous SNPs, but this may be due to the fact that these types of mutations tend to be the most commonly studied. Our focus was strictly on results from candidate-gene association studies and did not take into account results from linkage studies to identify high-penetrance genes. A further potential limitation of this review is that associations were confined to those summarized in a meta- or pooled analysis. We are aware of individual studies with potentially much larger sample sizes and hence more power to find a statistically significant association than some meta- and pooled analyses; some of these studies have been conducted subsequent to the meta- or pooled analyses and some prior. To address this issue in part, we reviewed studies conducted subsequent to the latest meta- or pooled analysis for associations considered noteworthy at a low prior probability to determine whether evidence continued to support the previously observed associations. Another limitation of our review is that our results are susceptible to reduced quality and breadth of the meta- or pooled analysis as a result of publication bias. However, most analyses included here tested for publication bias and heterogeneity, as noted in the accompanying tables. As the power to assess gene-gene and gene-environment interactions is even lower than that to assess main effects and most meta- and pooled analyses focused on main effects, we only reported on main effects of genetic variants. Therefore, we may have missed important subgroup effects, as it is possible that certain genetic variants may only be relevant when “the system is under stress,” e.g. smoking, concurrent illness, or malnutrition. Most analyses evaluated single candidate polymorphisms; however, because genotyping has become increasingly affordable in recent years, this now allows investigators to test for genetic variants across entire candidate genes and pathways and most recently across the entire genome. Although results from single SNPs are easy to compare, this approach is certainly less comprehensive and does not rule out that other SNPs in the same gene may be related to cancer risk. As the number of articles on genetic variants published in the past decade has increased considerably and continues to grow, we accept that this review will not long remain current but does provide a snapshot of progress in the field.
In summary, we observed 98 statistically significant gene-variant/cancer associations, of which thirteen were considered noteworthyat a prior probability of 0.001. At at very low prior probability (0.000001), four remained noteworthy of which all were highly statistically significant (p-values between 10−7 to 10−15). A majority of the most noteworthy associations identified are not SNPs but deletions, four involve GST variants. Results from meta-and pooled analyses were helpful in synthesizing published results and may guide future genetic studies toward areas that require further clarification and away from those that do not.
Acknowledgments
Funding/Support: This research was supported in part by NIH R25 CA94880.
Footnotes
Author contributions: Drs. Dong and Peters had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Peters
Acquisition of data: Dong, Peters
Analysis and interpretation of data: Dong, Potter, White, Ulrich, Cardon, Peters
Drafting of the manuscript: Dong, Peters
Critical revision of the manuscript for important intellectual content: Dong, Potter, White, Ulrich, Cardon, Peters
Statistical analysis: Dong
Study supervision: Peters, Potter, White
Financial Disclosures: Dr. Cardon has served as a consultant to Illumnia. No other financial disclosures were reported
Role of the Sponsor: The sponsor had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript.
Additional Contributions: We thank Nat Rothman, MD, MPH, MHS (Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD) for his helpful comments on the FPRP. None of these individuals received compensation for their contributions.
Contributor Information
Linda M Dong, Email: donglm@mail.nih.gov.
John D Potter, Email: jpotter@fhcrc.org.
Emily White, Email: ewhite@fhcrc.org.
Cornelia M Ulrich, Email: nulrich@fhcrc.org.
Lon R Cardon, Email: lcardon@fhcrc.org.
Ulrike Peters, Email: upeters@fhcrc.org.
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001 Feb 15;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001 Feb 16;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Finishing the euchromatic sequence of the human genome. Nature. 2004 Oct 21;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 4.The International HapMap Project. Nature. 2003 Dec 18;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 5.Lin BK, Clyne M, Walsh M, et al. Tracking the epidemiology of human genes in the literature`the HuGE Published Literature database. Am J Epidemiol. 2006 Jul 1;164(1):1–4. doi: 10.1093/aje/kwj175. [DOI] [PubMed] [Google Scholar]
- 6.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001 Nov;29(3):306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 7.Morgan TM, Krumholz HM, Lifton RP, Spertus JA. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007 Apr 11;297(14):1551–1561. doi: 10.1001/jama.297.14.1551. [DOI] [PubMed] [Google Scholar]
- 8.Dickersin K, Berlin JA. Meta-analysis: state-of-the-science. Epidemiol Rev. 1992;14:154–176. doi: 10.1093/oxfordjournals.epirev.a036084. [DOI] [PubMed] [Google Scholar]
- 9.Mosteller F, Colditz GA. Understanding research synthesis (meta-analysis) Annu Rev Public Health. 1996;17:1–23. doi: 10.1146/annurev.pu.17.050196.000245. [DOI] [PubMed] [Google Scholar]
- 10.Morris RD. Meta-analysis in cancer epidemiology. Environ Health Perspect. 1994 Nov;102( Suppl 8):61–66. doi: 10.1289/ehp.94102s861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poole C. Low P-values or narrow confidence intervals: which are more durable? Epidemiology. 2001 May;12(3):291–294. doi: 10.1097/00001648-200105000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004 Mar 17;96(6):434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkening S, Bermejo JL, Hemminki K. MDM2 SNP309 and cancer risk: a combined analysis. Carcinogenesis. 2007 Nov 2007;28(11):2262–2267. doi: 10.1093/carcin/bgm191. [DOI] [PubMed] [Google Scholar]
- 14.Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: a meta-analysis. Int J Med Sci. 2007;4(2):59–71. doi: 10.7150/ijms.4.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Tai BC. RNASEL gene polymorphisms and the risk of prostate cancer: a meta-analysis. Clin Cancer Res. 2006 Oct 1;12(19):5713–5719. doi: 10.1158/1078-0432.CCR-05-2799. [DOI] [PubMed] [Google Scholar]
- 16.de Jong MM, Nolte IM, te Meerman GJ, et al. Low-penetrance genes and their involvement in colorectal cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2002 Nov;11(11):1332–1352. [PubMed] [Google Scholar]
- 17.Kiyohara C, Takayama K, Nakanishi Y. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis. Lung Cancer. 2006 Dec;54(3):267–283. doi: 10.1016/j.lungcan.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Cox A, Dunning AM, Garcia-Closas M, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007 Mar;39(3):352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 19.Sanderson S, Salanti G, Higgins J. Joint Effects of the N-Acetyltransferase 1 and 2 (NAT1 and NAT2) Genes and Smoking on Bladder Carcinogenesis: A Literature-based Systematic HuGE Review and Evidence Synthesis. Am J Epidemiol. 2007 Aug 4; doi: 10.1093/aje/kwm167. [DOI] [PubMed] [Google Scholar]
- 20.Boccia S, Hung R, Ricciardi G, et al. Meta- and Pooled Analyses of the Methylenetetrahydrofolate Reductase C677T and A1298C Polymorphisms and Gastric Cancer Risk: A Huge-GSEC Review. Am J Epidemiol. 2007 Dec 27; doi: 10.1093/aje/kwm344. [DOI] [PubMed] [Google Scholar]
- 21.Weischer M, Bojesen SE, Ellervik C, Tybjaerg-Hansen A, Nordestgaard BG. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol. 2008 Feb 1;26(4):542–548. doi: 10.1200/JCO.2007.12.5922. [DOI] [PubMed] [Google Scholar]
- 22.Ye Z, Song H. Glutathione s-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and meta-analysis. Eur J Cancer. 2005 May;41(7):980–989. doi: 10.1016/j.ejca.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Closas M, Malats N, Silverman D, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005 Aug 20–26;366(9486):649–659. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas DC, Clayton DG. Betting odds and genetic associations. J Natl Cancer Inst. 2004 Mar 17;96(6):421–423. doi: 10.1093/jnci/djh094. [DOI] [PubMed] [Google Scholar]
- 25.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30(6):445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 26.Yeh CC, Sung FC, Tang R, Chang-Chieh CR, Hsieh LL. Association between polymorphisms of biotransformation and DNA-repair genes and risk of colorectal cancer in Taiwan. J Biomed Sci. 2007 Mar;14(2):183–193. doi: 10.1007/s11373-006-9139-x. [DOI] [PubMed] [Google Scholar]
- 27.Martinez C, Martin F, Fernandez JM, et al. Glutathione S-transferases mu 1, theta 1, pi 1, alpha 1 and mu 3 genetic polymorphisms and the risk of colorectal and gastric cancers in humans. Pharmacogenomics. 2006 Jul;7(5):711–718. doi: 10.2217/14622416.7.5.711. [DOI] [PubMed] [Google Scholar]
- 28.Little J, Sharp L, Masson LF, et al. Colorectal cancer and genetic polymorphisms of CYP1A1, GSTM1 and GSTT1: a case-control study in the Grampian region of Scotland. Int J Cancer. 2006 Nov 1;119(9):2155–2164. doi: 10.1002/ijc.22093. [DOI] [PubMed] [Google Scholar]
- 29.Rajagopal R, Deakin M, Fawole AS, et al. Glutathione S-transferase T1 polymorphisms are associated with outcome in colorectal cancer. Carcinogenesis. 2005 Dec;26(12):2157–2163. doi: 10.1093/carcin/bgi195. [DOI] [PubMed] [Google Scholar]
- 30.Ates NA, Tamer L, Ates C, et al. Glutathione S-transferase M1, T1, P1 genotypes and risk for development of colorectal cancer. Biochem Genet. 2005 Apr;43(3–4):149–163. doi: 10.1007/s10528-005-1508-z. [DOI] [PubMed] [Google Scholar]
- 31.Bolufer P, Collado M, Barragan E, et al. The potential effect of gender in combination with common genetic polymorphisms of drug-metabolizing enzymes on the risk of developing acute leukemia. Haematologica. 2007 Mar;92(3):308–314. doi: 10.3324/haematol.10752. [DOI] [PubMed] [Google Scholar]
- 32.Vineis P, Veglia F, Garte S, et al. Genetic susceptibility according to three metabolic pathways in cancers of the lung and bladder and in myeloid leukemias in nonsmokers. Ann Oncol. 2007 May 11; doi: 10.1093/annonc/mdm109. [DOI] [PubMed] [Google Scholar]
- 33.Cengiz M, Ozaydin A, Ozkilic AC, Dedekarginoglu G. The investigation of GSTT1, GSTM1 and SOD polymorphism in bladder cancer patients. Int Urol Nephrol. 2007 Mar 6; doi: 10.1007/s11255-007-9179-9. [DOI] [PubMed] [Google Scholar]
- 34.Bolufer P, Barragan E, Collado M, Cervera J, Lopez JA, Sanz MA. Influence of genetic polymorphisms on the risk of developing leukemia and on disease progression. Leuk Res. 2006 Dec;30(12):1471–1491. doi: 10.1016/j.leukres.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Aydin-Sayitoglu M, Hatirnaz O, Erensoy N, Ozbek U. Role of CYP2D6, CYP1A1, CYP2E1, GSTT1, and GSTM1 genes in the susceptibility to acute leukemias. Am J Hematol. 2006 Mar;81(3):162–170. doi: 10.1002/ajh.20434. [DOI] [PubMed] [Google Scholar]
- 36.Shao J, Gu M, Zhang Z, Xu Z, Hu Q, Qian L. Genetic variants of the cytochrome P450 and glutathione S-transferase associated with risk of bladder cancer in a south-eastern Chinese population. Int J Urol. 2008 Mar;15(3):216–221. doi: 10.1111/j.1442-2042.2007.01915.x. [DOI] [PubMed] [Google Scholar]
- 37.Majumdar S, Mondal BC, Ghosh M, et al. Association of cytochrome P450, glutathione S-transferase and N-acetyl transferase 2 gene polymorphisms with incidence of acute myeloid leukemia. Eur J Cancer Prev. 2008 Apr;17(2):125–132. doi: 10.1097/CEJ.0b013e3282b6fd68. [DOI] [PubMed] [Google Scholar]
- 38.Huang K, Sandler RS, Millikan RC, Schroeder JC, North KE, Hu J. GSTM1 and GSTT1 polymorphisms, cigarette smoking, and risk of colon cancer: a population-based case-control study in North Carolina (United States) Cancer Causes Control. 2006 May;17(4):385–394. doi: 10.1007/s10552-005-0424-1. [DOI] [PubMed] [Google Scholar]
- 39.Clavel J, Bellec S, Rebouissou S, et al. Childhood leukaemia, polymorphisms of metabolism enzyme genes, and interactions with maternal tobacco, coffee and alcohol consumption during pregnancy. Eur J Cancer Prev. 2005 Dec;14(6):531–540. doi: 10.1097/00008469-200512000-00007. [DOI] [PubMed] [Google Scholar]
- 40.Pakakasama S, Mukda E, Sasanakul W, et al. Polymorphisms of drug-metabolizing enzymes and risk of childhood acute lymphoblastic leukemia. Am J Hematol. 2005 Jul;79(3):202–205. doi: 10.1002/ajh.20404. [DOI] [PubMed] [Google Scholar]
- 41.Nelson HH, Wiencke JK, Christiani DC, et al. Ethnic differences in the prevalence of the homozygous deleted genotype of glutathione S-transferase theta. Carcinogenesis. 1995 May;16(5):1243–1245. doi: 10.1093/carcin/16.5.1243. [DOI] [PubMed] [Google Scholar]
- 42.Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1997 Sep;6(9):733–743. [PubMed] [Google Scholar]
- 43.Hayes JD, Strange RC. Potential contribution of the glutathione S-transferase supergene family to resistance to oxidative stress. Free Radic Res. 1995 Mar;22(3):193–207. doi: 10.3109/10715769509147539. [DOI] [PubMed] [Google Scholar]
- 44.Parl FF. Glutathione S-transferase genotypes and cancer risk. Cancer Lett. 2005 Apr 28;221(2):123–129. doi: 10.1016/j.canlet.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 45.Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002 Sep 30;506–507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 46.Hein DW, Doll MA, Fretland AJ, et al. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev. 2000 Jan;9(1):29–42. [PubMed] [Google Scholar]
- 47.Chen K, Jiang QT, He HQ. Relationship between metabolic enzyme polymorphism and colorectal cancer. World J Gastroenterol. 2005 Jan 21;11(3):331–335. doi: 10.3748/wjg.v11.i3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frosst P, Blom HJ, Milos R, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995 May;10(1):111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 49.Rozen R. Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR) Thromb Haemost. 1997 Jul;78(1):523–526. [PubMed] [Google Scholar]
- 50.Choi SW, Mason JB. Folate status: effects on pathways of colorectal carcinogenesis. J Nutr. 2002 Aug;132(8 Suppl):2413S–2418S. doi: 10.1093/jn/132.8.2413S. [DOI] [PubMed] [Google Scholar]
- 51.Lopez-Cima MF, Gonzalez-Arriaga P, Garcia-Castro L, et al. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of Northern Spain. BMC Cancer. 2007;7:162. doi: 10.1186/1471-2407-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Ruyck K, Szaumkessel M, De Rudder I, et al. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007 Jul 28;631(2):101–110. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Pachouri SS, Sobti RC, Kaur P, Singh J. Contrasting impact of DNA repair gene XRCC1 polymorphisms Arg399Gln and Arg194Trp on the risk of lung cancer in the north-Indian population. DNA Cell Biol. 2007 Mar;26(3):186–191. doi: 10.1089/dna.2006.9999. [DOI] [PubMed] [Google Scholar]
- 54.Yin J, Vogel U, Ma Y, Qi R, Sun Z, Wang H. The DNA repair gene XRCC1 and genetic susceptibility of lung cancer in a northeastern Chinese population. Lung Cancer. 2007 May;56(2):153–160. doi: 10.1016/j.lungcan.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 55.Sreeja L, Syamala VS, Syamala V, et al. Prognostic importance of DNA repair gene polymorphisms of XRCC1 Arg399Gln and XPD Lys751Gln in lung cancer patients from India. J Cancer Res Clin Oncol. 2007 Oct 19; doi: 10.1007/s00432-007-0328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun J, Turner A, Xu J, Gronberg H, Isaacs W. Genetic variability in inflammation pathways and prostate cancer risk. Urol Oncol. 2007 May–Jun;25(3):250–259. doi: 10.1016/j.urolonc.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Smith JR, Freije D, Carpten JD, et al. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996 Nov 22;274(5291):1371–1374. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- 58.Shook SJ, Beuten J, Torkko KC, et al. Association of RNASEL variants with prostate cancer risk in Hispanic Caucasians and African Americans. Clin Cancer Res. 2007 Oct 1;13(19):5959–5964. doi: 10.1158/1078-0432.CCR-07-0702. [DOI] [PubMed] [Google Scholar]
- 59.Cybulski C, Wokolorczyk D, Jakubowska A, et al. DNA variation in MSR1, RNASEL and E-cadherin genes and prostate cancer in Poland. Urol Int. 2007;79(1):44–49. doi: 10.1159/000102913. [DOI] [PubMed] [Google Scholar]
- 60.Shea PR, Ishwad CS, Bunker CH, Patrick AL, Kuller LH, Ferrell RE. RNASEL and RNASEL-inhibitor variation and prostate cancer risk in Afro-Caribbeans. Prostate. 2008 Mar 1;68(4):354–359. doi: 10.1002/pros.20687. [DOI] [PubMed] [Google Scholar]
- 61.Momand J, Wu HH, Dasgupta G. MDM2--master regulator of the p53 tumor suppressor protein. Gene. 2000 Jan 25;242(1–2):15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 62.Liu G, Wheatley-Price P, Zhou W, et al. Genetic polymorphisms of MDM2, cumulative cigarette smoking and nonsmall cell lung cancer risk. Int J Cancer. 2008 Feb 15;122(4):915–918. doi: 10.1002/ijc.23178. [DOI] [PubMed] [Google Scholar]
- 63.Glick AB. TGFbeta1, back to the future: revisiting its role as a transforming growth factor. Cancer Biol Ther. 2004 Mar;3(3):276–283. doi: 10.4161/cbt.3.3.849. [DOI] [PubMed] [Google Scholar]
- 64.Rosfjord EC, Dickson RB. Growth factors, apoptosis, and survival of mammary epithelial cells. J Mammary Gland Biol Neoplasia. 1999 Apr;4(2):229–237. doi: 10.1023/a:1018789527533. [DOI] [PubMed] [Google Scholar]
- 65.Cox DG, Penney K, Guo Q, Hankinson SE, Hunter DJ. TGFB1 and TGFBR1 polymorphisms and breast cancer risk in the Nurses’ Health Study. BMC Cancer. 2007;7:175. doi: 10.1186/1471-2407-7-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grenet J, Teitz T, Wei T, Valentine V, Kidd VJ. Structure and chromosome localization of the human CASP8 gene. Gene. 1999 Jan 21;226(2):225–232. doi: 10.1016/s0378-1119(98)00565-4. [DOI] [PubMed] [Google Scholar]
- 67.Sigurdson AJ, Bhatti P, Doody MM, et al. Polymorphisms in apoptosis- and proliferation-related genes, ionizing radiation exposure, and risk of breast cancer among U.S. Radiologic Technologists. Cancer Epidemiol Biomarkers Prev. 2007 Oct;16(10):2000–2007. doi: 10.1158/1055-9965.EPI-07-0282. [DOI] [PubMed] [Google Scholar]
- 68.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007 May;39(5):631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 69.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007 May;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 70.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007 May;39(5):638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007 Jul 8; doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 72.Haiman CA, Le Marchand L, Yamamato J, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007 Aug;39(8):954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007 Aug;39(8):989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 74.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007 Nov;39(11):1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 75.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007 Jun 28;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duggan D, Zheng SL, Knowlton M, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007 Dec 19;99(24):1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 77.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008 Mar;40(3):316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 78.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008 Mar;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 79.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008 Mar;40(3):281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007 Jul;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007 Jul;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 82.Kellen E, Hemelt M, Broberg K, et al. Pooled analysis and meta-analysis of the glutathione S-transferase P1 Ile 105Val polymorphism and bladder cancer: a HuGE-GSEC review. Am J Epidemiol. 2007 Jun 1;165(11):1221–1230. doi: 10.1093/aje/kwm003. [DOI] [PubMed] [Google Scholar]
- 83.Chao C, Zhang ZF, Berthiller J, Boffetta P, Hashibe M. NAD(P)H:quinone oxidoreductase 1 (NQO1) Pro187Ser polymorphism and the risk of lung, bladder, and colorectal cancers: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006 May;15(5):979–987. doi: 10.1158/1055-9965.EPI-05-0899. [DOI] [PubMed] [Google Scholar]
- 84.Francisco G, Menezes PR, Eluf-Neto J, Chammas R. XPC polymorphisms play a role in tissue-specific carcinogenesis: a meta-analysis. Eur J Hum Genet. 2008 2008 Feb 20;0(0):1–11. doi: 10.1038/ejhg.2008.6. [DOI] [PubMed] [Google Scholar]
- 85.Wang F, Chang D, Hu FL, et al. DNA Repair Gene XPD Polymorphisms and Cancer Risk: A Meta-analysis Based on 56 Case-Control Studies. Cancer Epidemiol Biomarkers Prev. 2008 Mar;17(3):507–517. doi: 10.1158/1055-9965.EPI-07-2507. [DOI] [PubMed] [Google Scholar]
- 86.Figueroa JD, Malats N, Rothman N, et al. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007 Aug;28(8):1788–1793. doi: 10.1093/carcin/bgm132. [DOI] [PubMed] [Google Scholar]
- 87.Garcia-Closas M, Kristensen V, Langerod A, et al. Common genetic variation in TP53 and its flanking genes, WDR79 and ATP1B2, and susceptibility to breast cancer. Int J Cancer. 2007 Aug 7; doi: 10.1002/ijc.22985. [DOI] [PubMed] [Google Scholar]
- 88.Onay UV, Aaltonen K, Briollais L, et al. Combined effect of CCND1 and COMT polymorphisms and increased breast cancer risk. BMC Cancer. 2008;8:6. doi: 10.1186/1471-2407-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Setiawan VW, Schumacher FR, Haiman CA, et al. CYP17 genetic variation and risk of breast and prostate cancer from the National Cancer Institute Breast and Prostate Cancer Cohort Consortium (BPC3) Cancer Epidemiol Biomarkers Prev. 2007 Nov;16(11):2237–2246. doi: 10.1158/1055-9965.EPI-07-0589. [DOI] [PubMed] [Google Scholar]
- 90.de Jong MM, Nolte IM, te Meerman GJ, et al. Genes other than BRCA1 and BRCA2 involved in breast cancer susceptibility. J Med Genet. 2002 Apr;39(4):225–242. doi: 10.1136/jmg.39.4.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen C, Huang Y, Li Y, Mao Y, Xie Y. Cytochrome P450 1A1 (CYP1A1) T3801C and A2455G polymorphisms in breast cancer risk: a meta-analysis. J Hum Genet. 2007;52(5):423–435. doi: 10.1007/s10038-007-0131-8. [DOI] [PubMed] [Google Scholar]
- 92.Paracchini V, Raimondi S, Gram IT, et al. Meta- and pooled analyses of the cytochrome P-450 1B1 Val432Leu polymorphism and breast cancer: a HuGE-GSEC review. Am J Epidemiol. 2007 Jan 15;165(2):115–125. doi: 10.1093/aje/kwj365. [DOI] [PubMed] [Google Scholar]
- 93.Garcia-Closas M, Troester MA, Qi Y, et al. Common genetic variation in GATA-binding protein 3 and differential susceptibility to breast cancer by estrogen receptor alpha tumor status. Cancer Epidemiol Biomarkers Prev. 2007 Nov 2007;16(11):2269–2275. doi: 10.1158/1055-9965.EPI-07-0449. [DOI] [PubMed] [Google Scholar]
- 94.Steffen J, Nowakowska D, Niwinska A, et al. Germline mutations 657del5 of the NBS1 gene contribute significantly to the incidence of breast cancer in Central Poland. Int J Cancer. 2006 Jul 15;119(2):472–475. doi: 10.1002/ijc.21853. [DOI] [PubMed] [Google Scholar]
- 95.Haiman CA, Setiawan VW, Xia LY, et al. A variant in the cytochrome p450 oxidoreductase gene is associated with breast cancer risk in African Americans. Cancer Res. 2007 Apr 15;67(8):3565–3568. doi: 10.1158/0008-5472.CAN-06-4801. [DOI] [PubMed] [Google Scholar]
- 96.Cox DG, Buring J, Hankinson SE, Hunter DJ. A polymorphism in the 3′ untranslated region of the gene encoding prostaglandin endoperoxide synthase 2 is not associated with an increase in breast cancer risk: a nested case-control study. Breast Cancer Res. 2007;9(1):R3. doi: 10.1186/bcr1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pasche B, Kaklamani V, Hou N, et al. TGFBR1*6A and cancer: a meta-analysis of 12 case-control studies. J Clin Oncol. 2004 Feb 15;22(4):756–758. doi: 10.1200/JCO.2004.99.271. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Y, Newcomb PA, Egan KM, et al. Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006 Feb;15(2):353–358. doi: 10.1158/1055-9965.EPI-05-0653. [DOI] [PubMed] [Google Scholar]
- 99.Tan XL, Nieters A, Kropp S, Hoffmeister M, Brenner H, Chang-Claude J. The association of cyclin D1 G870A and E-cadherin C-160A polymorphisms with the risk of colorectal cancer in a case control study and meta-analysis. Int J Cancer. 2008 Jan 14;0(0):0. doi: 10.1002/ijc.23363. [DOI] [PubMed] [Google Scholar]
- 100.Hubner RA, Houlston RS. MTHFR C677T and colorectal cancer risk: A meta-analysis of 25 populations. Int J Cancer. 2007 Mar 1;120(5):1027–1035. doi: 10.1002/ijc.22440. [DOI] [PubMed] [Google Scholar]
- 101.Huang Y, Han S, Li Y, Mao Y, Xie Y. Different roles of MTHFR C677T and A1298C polymorphisms in colorectal adenoma and colorectal cancer: a meta-analysis. J Hum Genet. 2007;52(1):73–85. doi: 10.1007/s10038-006-0082-5. [DOI] [PubMed] [Google Scholar]
- 102.Zhang D, Chen C, Fu X, et al. A meta-analysis of DNA repair gene XPC polymorphisms and cancer risk. J Hum Genet. 2008 Jan 2008;53(1):18–33. doi: 10.1007/s10038-007-0215-5. [DOI] [PubMed] [Google Scholar]
- 103.Lewis SJ, Smith GD. Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 2005 Aug;14(8):1967–1971. doi: 10.1158/1055-9965.EPI-05-0196. [DOI] [PubMed] [Google Scholar]
- 104.Yang CX, Matsuo K, Wang ZM, Tajima K. Phase I/II enzyme gene polymorphisms and esophageal cancer risk: a meta-analysis of the literature. World J Gastroenterol. 2005 May 7;11(17):2531–2538. doi: 10.3748/wjg.v11.i17.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang GY, Lu CQ, Zhang RM, Hu XH, Luo ZW. The E-cadherin gene polymorphism 160C->A and cancer risk: A HuGE review and meta-analysis of 26 case-control studies. Am J Epidemiol. 2008 Jan 1;167(1):7–14. doi: 10.1093/aje/kwm264. [DOI] [PubMed] [Google Scholar]
- 106.Boccia S, La Torre G, Gianfagna F, Mannocci A, Ricciardi G. Glutathione S-transferase T1 status and gastric cancer risk: a meta-analysis of the literature. Mutagenesis. 2006 Mar;21(2):115–123. doi: 10.1093/mutage/gel005. [DOI] [PubMed] [Google Scholar]
- 107.Wang P, Xia HH, Zhang JY, Dai LP, Xu XQ, Wang KJ. Association of interleukin-1 gene polymorphisms with gastric cancer: a meta-analysis. Int J Cancer. 2007 Feb 1;120(3):552–562. doi: 10.1002/ijc.22353. [DOI] [PubMed] [Google Scholar]
- 108.Zhou Y, Li N, Zhuang W, et al. P53 codon 72 polymorphism and gastric cancer: a meta-analysis of the literature. Int J Cancer. 2007 Oct 1;121(7):1481–1486. doi: 10.1002/ijc.22833. [DOI] [PubMed] [Google Scholar]
- 109.Gorouhi F, Islami F, Bahrami H, Kamangar F. Tumour-necrosis factor-A polymorphisms and gastric cancer risk: a meta-analysis. Br J Cancer. 2008 Mar 4; doi: 10.1038/sj.bjc.6604277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bethke L, Webb E, Murray A, et al. Comprehensive analysis of the role of DNA repair gene polymorphisms on risk of glioma. Hum Mol Genet. 2008 Mar 15;17(6):800–805. doi: 10.1093/hmg/ddm351. [DOI] [PubMed] [Google Scholar]
- 111.Brenner AV, Butler MA, Wang SS, et al. Single-nucleotide polymorphisms in selected cytokine genes and risk of adult glioma. Carcinogenesis. 2007 Dec;28(12):2543–2547. doi: 10.1093/carcin/bgm210. [DOI] [PubMed] [Google Scholar]
- 112.Tripathy CB, Roy N. Meta-analysis of glutathione S-transferase M1 genotype and risk toward head and neck cancer. Head Neck. 2006 Mar;28(3):217–224. doi: 10.1002/hed.20295. [DOI] [PubMed] [Google Scholar]
- 113.Ye Z, Song H, Guo Y. Glutathione S-transferase M1, T1 status and the risk of head and neck cancer: a meta-analysis. J Med Genet. 2004 May;41(5):360–365. doi: 10.1136/jmg.2003.016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang WY, Olshan AF, Schwartz SM, et al. Selected genetic polymorphisms in MGMT, XRCC1, XPD, and XRCC3 and risk of head and neck cancer: a pooled analysis. Cancer Epidemiol Biomarkers Prev. 2005 Jul;14(7):1747–1753. doi: 10.1158/1055-9965.EPI-05-0162. [DOI] [PubMed] [Google Scholar]
- 115.Pereira TV, Rudnicki M, Pereira AC, Pombo-de-Oliveira MS, Franco RF. 5,10-Methylenetetrahydrofolate reductase polymorphisms and acute lymphoblastic leukemia risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006 Oct;15(10):1956–1963. doi: 10.1158/1055-9965.EPI-06-0334. [DOI] [PubMed] [Google Scholar]
- 116.Vineis P, Veglia F, Benhamou S, et al. CYP1A1 T3801 C polymorphism and lung cancer: a pooled analysis of 2451 cases and 3358 controls. Int J Cancer. 2003 May 1;104(5):650–657. doi: 10.1002/ijc.10995. [DOI] [PubMed] [Google Scholar]
- 117.Shi X, Zhou S, Wang Z, Zhou Z, Wang Z. CYP1A1 and GSTM1 polymorphisms and lung cancer risk in Chinese populations: A meta-analysis. Lung Cancer. 2008 Feb 2008;59(2):155–163. doi: 10.1016/j.lungcan.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 118.Rostami-Hodjegan A, Lennard MS, Woods HF, Tucker GT. Meta-analysis of studies of the CYP2D6 polymorphism in relation to lung cancer and Parkinson’s disease. Pharmacogenetics. 1998 Jun;8(3):227–238. doi: 10.1097/00008571-199806000-00005. [DOI] [PubMed] [Google Scholar]
- 119.Raimondi S, Paracchini V, Autrup H, et al. Meta- and pooled analysis of GSTT1 and lung cancer: a HuGE-GSEC review. Am J Epidemiol. 2006 Dec 1;164(11):1027–1042. doi: 10.1093/aje/kwj321. [DOI] [PubMed] [Google Scholar]
- 120.Lee WJ, Brennan P, Boffetta P, et al. Microsomal epoxide hydrolase polymorphisms and lung cancer risk: a quantitative review. Biomarkers. 2002 May–Jun;7(3):230–241. doi: 10.1080/13547500210121882. [DOI] [PubMed] [Google Scholar]
- 121.Taioli E, Benhamou S, Bouchardy C, et al. Myeloperoxidase G-463A polymorphism and lung cancer: a HuGE genetic susceptibility to environmental carcinogens pooled analysis. Genet Med. 2007 Feb;9(2):67–73. doi: 10.1097/gim.0b013e31803068b1. [DOI] [PubMed] [Google Scholar]
- 122.Han S, Zhang HT, Wang Z, et al. DNA repair gene XRCC3 polymorphisms and cancer risk: a meta-analysis of 48 case-control studies. Eur J Hum Genet. 2006 Jun 21; doi: 10.1038/sj.ejhg.5201681. [DOI] [PubMed] [Google Scholar]
- 123.Bethke L, Murray A, Webb E, et al. Comprehensive analysis of DNA repair gene variants and risk of meningioma. J Natl Cancer Inst. 2008 Feb 20;100(4):270–276. doi: 10.1093/jnci/djn004. [DOI] [PubMed] [Google Scholar]
- 124.Rothman N, Skibola CF, Wang SS, et al. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol. 2006 Jan;7(1):27–38. doi: 10.1016/S1470-2045(05)70434-4. [DOI] [PubMed] [Google Scholar]
- 125.Lee KM, Lan Q, Kricker A, et al. One-carbon metabolism gene polymorphisms and risk of non-Hodgkin lymphoma in Australia. Hum Genet. 2007 Dec;122(5):525–533. doi: 10.1007/s00439-007-0431-2. [DOI] [PubMed] [Google Scholar]
- 126.Gayther SA, Song H, Ramus SJ, et al. Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007 Apr 1;67(7):3027–3035. doi: 10.1158/0008-5472.CAN-06-3261. [DOI] [PubMed] [Google Scholar]
- 127.Zeegers MP, Kiemeney LA, Nieder AM, Ostrer H. How strong is the association between CAG and GGN repeat length polymorphisms in the androgen receptor gene and prostate cancer risk? Cancer Epidemiol Biomarkers Prev. 2004 Nov;13(11 Pt 1):1765–1771. [PubMed] [Google Scholar]
- 128.Hung RJ, Hall J, Brennan P, Boffetta P. Genetic Polymorphisms in the Base Excision Repair Pathway and Cancer Risk: A HuGE Review. Am J Epidemiol. 2005 Nov 15;162(10):925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]