Abstract
Francisella tularensis, the etiological agent of tularemia, is one of the most infectious bacterial pathogens known. No vaccine is currently approved for public use. Previously, we identified epitopes recognized specifically by T cells obtained from individuals following infection with F. tularensis. Here, we report that a subunit vaccine constructed based upon these epitopes elicited protective immunity in “humanized” HLA class II (DRB1*0401) transgenic mice. Vaccinated mice challenged intratracheally with a lethal dose of F. tularensis (Live Vaccine Strain) exhibited a rapid increase in pro-inflammatory cytokine production and diminished number of organisms in the lungs, and a concurrent increased rate of survival. These results demonstrate the efficacy of an epitope-based tularemia vaccine and suggest that such an approach might be widely applicable to the development of vaccines specific for intracellular bacterial pathogens.
Keywords: Francisella tularensis, lungs, DNA vaccine
1. Introduction
Francisella tularensis, the etiological agent of tularemia, is a small, gram-negative, facultative intracellular coccobacillus [1]. Tularemia is a zoonotic disease; small mammals including mice, squirrels, rats and rabbits are natural reservoirs. Human infections usually occur through incidental environmental exposure, e.g., contact or ingestion of contaminated food, water or soil, bites by infected arthropods, inhalation of infective aerosols. Lymph nodes, lungs, spleen, liver and kidneys are major target organs [2]. F. tularensis is divided into two major subspecies: tularensis (type A), which is highly virulent and the most common form found in North America; and holarctica (type B), a less virulent form that causes most human illnesses in Europe and Asia [3]. Clinical presentations include glandular, ulceroglandular, intestinal and pneumonic forms dependent upon route of infection. The organism is difficult to identify microscopically; unfamiliarity with its presentation can delay proper diagnosis and treatment. In the absence of prompt antibiotic therapy, inhalation of as few as 10 organisms can cause severe pneumonia and 80% mortality [4,5]. As such, aerosolized F. tularensis represents a potentially dangerous biological weapon, classified by CDC as a category A select agent [6].
Cell-mediated immunity is the dominant factor in host resistance to tularemia; both CD4+ and CD8+ T cells are key elements in the efficient resolution of primary and secondary infections [1,7]. Mice inoculated intraperitoneally (i.p.) with immune sera exhibit FcγR-dependent protection from lethal intranasal (i.n.) challenge suggesting that B cells and antibody production might play an additional role [8,9]. Both interferon gamma (IFN-γ) and tumor necrosis alpha (TNF-α) are also critical factors in primary host defenses to systemic infections in mice [10]. It is speculated that IFN-γ and TNF-α synergize to promote nitric oxide production, and to regulate iron homeostasis and pH thus limiting F. tularensis survival within macrophages, the primary site of intracellular replication in vivo [11,12].
Until recently, immunization with an attenuated Live Vaccine Strain (LVS) of F. tularensis subsp. holarctica was used to protect laboratory personnel working with F. tularensis. Vaccination by scarification, however, provides human volunteers only incomplete protection against challenge with aerosolized F. tularensis type A [13]. Furthermore, several safety issues exist. For one, the molecular basis for the attenuation of F. tularensis LVS has never been established. Moreover, culture conditions contribute to variations in the pathogenicity of the attenuated strain rendering F. tularensis LVS an unreliable vaccine and potentially hazardous particularly to immunocompromised individuals [14,15]. Despite considerable need and years of research, no tularemia vaccine is currently approved for public use.
Efforts to develop a safe, effective vaccine against F. tularensis have focused on three different strategies: inactivated whole cell, attenuated and subunit vaccines. Heat- or chemically-inactivated whole-cell preparations have generally failed to elicit sufficient protective immunity in either humans or animal models though vaccination with paraformaldehyde-fixed F. tularensis LVS administered in conjunction with IL-12 or targeted to Fc receptors via anti-F. tularensis LPS mAb has shown promise in recent studies [16,17]. Live attenuated organisms are considered a logical alternative approach to vaccine development given the ability of F. tularensis LVS to reduce the incidence of laboratory-acquired infections [18]. Recent efforts have targeted virulence and metabolic genes to create weakened mutants capable of eliciting protective immunity, yet limited in their ability to survive, replicate, and cause disease [19,20]. Nonetheless, even the attenuated vaccines that meet these criteria represent considerable risk to immunocompromised individuals. Moreover, the failure of current live-attenuated F. tularensis vaccine developers to resolve issues regarding the basis of attenuation and mechanisms of action provides significant reason for concern about safety on the part of regulatory authorities, and creates a nearly insurmountable obstacle to licensure, marketing, and possible stockpile acquisition.
Subunit vaccines, consisting of a single antigen or set of antigens, constitute a third approach to immunizing against F. tularensis. To date, only the O-antigenic component of the LPS molecule derived from F. tularensis, was found to elicit protective immunity to systemic infections by Type B, but not Type A, strains of F. tularensis [21]. Undoubtedly, this is due in part to the failure of LPS to induce a T cell response required for protection. Additionally, the results suggest that a subunit vaccine must be composed of more than a single antigen in order to elicit protective immunity. Computer analysis of the microbial genome coupled with algorithms that predict T cell epitopes associated with a multiplicity of immunogenic proteins offers a means of generating an effective, low cost subunit vaccine capable of provoking a broad T cell response to a large number of antigens. Using this approach, we previously identified a number of epitopes that were subsequently recognized specifically by T cells obtained from individuals following infection with F. tularensis [22]. Here, we report that a subunit vaccine constructed based upon these clinically relevant epitopes elicited protective immunity in “humanized” HLA class II (DRB1*0401) transgenic mice.
2. Materials and Methods
2.1. Francisella tularensis live vaccine strain (LVS)
F. tularensis LVS was obtained from the Centers for Disease Control and Prevention (Division of Vector-Borne Infectious diseases, Fort Collins, CO). The bacteria were grown in Mueller Hinton broth (Difco, Detroit, MI) supplemented with 0.1% glucose, 0.025% ferric pyrophosphate (Sigma Chemical, St. Louis, MO), 2.0% Isovitalex (Becton Dickinson, Cockeysville, MD) and 2.5% FBS, harvested at mid-log phase of growth, centrifuged, resuspended in fresh broth mixed 1:1 with 2.6% gelatin, and stored frozen at −80°C until use. The numbers of bacteria in the lungs, spleens and livers of infected mice were estimated from the colonies that grew on supplemented Mueller-Hinton agar plates inoculated with an aliquot of organ homogenate and cultured for 4 days at 37°C in a humidified environment composed of 5% CO2 in air. In preliminary experiments, 1 LD50 was found equivalent to 1,280 CFUs F. tularensis LVS inoculated intratracheally as described below.
2.2. Animals
Specific pathogen-free HLA-DRB1 transgenic mice (DRB1*0401 on a B10.M background) were obtained from Dr. Dennis Zaller, Merck Research Laboratories, Rahway, NJ [23]; a colony was bred and maintained at Taconic Farms, Inc., Germantown, NY. HLA-DRB1 mice express recombinant MHC class II molecules in which the α1 and β1 domains of mouse I-E were replaced by the corresponding domains of human DRB1*0401 (DR4Dw4) molecules; as such, mouse MHC class II I-E are not expressed. Mice imported into the Rhode Island Hospital central animal facility were treated in accordance with NIH publications entitled "Principles for Use of Animals" and "Guide for the Care and Use of Laboratory Animals." The mice were housed in well-ventilated rooms maintained at 22°C and an alternating 12-hour light and dark cycle; food and water were provided ad libitum. All experiments were conducted using female animals between 6 and 8 weeks of age at the time of initiation. The mice were infected intratracheally in accordance with methods previously described [24]. Briefly, a 50µl aliquot of bacteria suspended in saline was delivered to the back of the oropharyngeal cavity of mice deeply anesthetized with isoflurane, temporarily occluding airflow; the bacteria were inhaled as the animals revived.
2.3. Epitope-based DNA vaccine construct
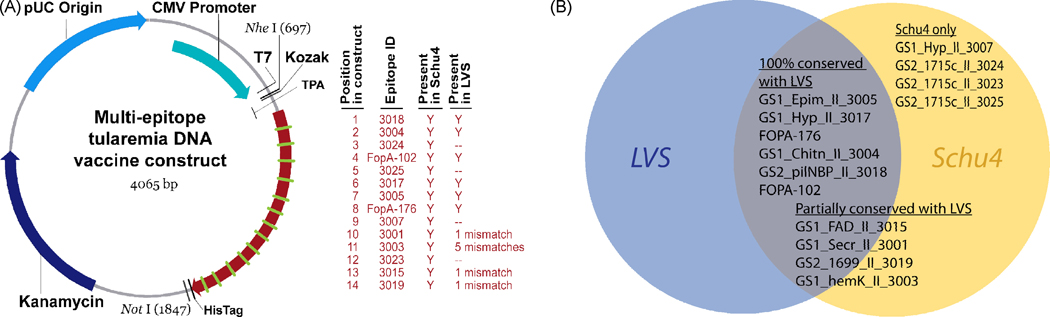
Prediction and validation of the HLA class II-restricted epitopes incorporated into the vaccine construct reported here were previously described in detail [22]. Briefly, computational tools were used to scan the fully annotated F. tularensis subsp. tularensis genome (Schu4 strain) [25] and to identify gene sequences most likely to encode secreted proteins (Genome Scan 1); 147 putative proteins were identified. An additional 53 proteins were gleaned from the literature (Genome Scan 2). The EpiMatrix System was then used to screen the combined set of 200 selected proteins for the presence of class II-restricted T cell epitope clusters. Twenty-seven promiscuous epitopes capable of binding a set of 8 archetype/supertype class II matrices including both HLA DRB1*0101 and HLA DRB1*0401 were synthesized as peptides. Twenty-four synthesized peptides elicited IFN-γ production in ELISpot assays using PBMCs derived from individuals previously infected with F. tularensis, thus validating their relevance. Of these, 14 peptides (shown in Table 1) were bound by HLA DRB1*0101 in an in vitro competitive binding assay. These fourteen epitopes were selected for inclusion in a candidate vaccine construct; 13 epitopes were immunogenic in peptide immunizations of DRB1*0401 transgenic mice (data not shown). One epitope (GS1_Secr_II_3001) not meeting any of the criteria was included as an internal negative control. The candidate epitopes were aligned in a string-of-beads conformation shown in Figure 1A.
Table 1.
Epitopes selected for vaccine construct
| Peptide ID | AA Sequence | Protein Location |
GenBank Accession |
Protein Description |
|---|---|---|---|---|
| FOPA 102 (AKA Fopa-2) |
IGYNINKYFAVQYNQLVGRVFAGLGE | 102–127 | GI-1332665 | immunogenic chaperone protein |
| FOPA 176 (AKA Fopa-4) |
SVDYRYIQTMAPSNISGA | 176–193 | GI-1332665 | immunogenic chaperone protein |
| GS1_Secr_II_3001 | SLGYELWRRYNGFPINLN | 273–290 | GI-56707198 | Secretion protein |
| GS1_hemK_II_3003 | PLAYILGYKYFWNQKLYVTKD | 51–71 | GI-56707339 | hemK protein homolog |
| GS1_Chitn_II_3004 | ADGKKYYLAIAVNGARSRIEAM | 292–313 | GI-56707834 | chitinase family 18 protein |
| GS1_Epim_II_3005 | KEKYYKINTQLTYDLAKQ | 46–63 | GI-56708503 | UDP-glucose 4-epimerase |
| GS1_Hyp_II_3007 | NIIMVKVFVYNLNNNINAIK | 24–43 | GI-56707430 | hypothetical membrane protein |
| GS1_FAD_II_3015 | GEQFAFRPNNKITIAMV | 314–330 | GI-56708305 | FAD binding family protein |
| GS1_Hyp_II_3017 | ENGIWKVNRPNPGPVTIA | 127–144 | GI-56708544 | hypothetical protein FTT1506 |
| GS2_pilNBP_II_3018 | AESLQIVISQRLLKRKGGGRVAAYEV | 258–283 | GI- 56707266 |
Type IV pili nucleotide- binding protein |
| GS2_1699_II_3019 | HKDFNFLLSPNQPILLDIQ | 247–265 | GI- 56708708 |
hypothetical protein FTT1699 |
| GS2_1715c_II_3023 | RGDVKSFIPLYLRISGKASSALF | 185–207 | GI- 56708724 |
hypothetical protein FTT1715c |
| GS2_1715c_II_3024 | FSDMEIRVRFGLYSKQSASKL | 419–439 | GI- 56708724 |
hypothetical protein FTT1715c |
| GS2_1715c_II_3025 | DRLVRGFYFLLRRMVKNRNTKVGSHN | 899–924 | GI- 56708724 |
hypothetical protein FTT1715c |
Column 1: peptide ID, composed of Genome Scan 1 (GS1; putative secreted protein) or GS2 (confirmed expressed protein), abbreviated description of the parent protein, MHC class predicted to bind the peptide, a four-digit ID number; Column 2: amino acid sequence; Column 3: GenBank assession of parent protein; and Column 4: protein description [22].
Figure 1. Multi-epitope DNA vaccine construct.
A synthetic gene encoding a concatamer of 14 validated tularemia T cell epitopes was inserted into the pVAX1 DNA vaccine vector (A). Expression of the concatamer was directed for secretion by the tissue plasminogen activator (TPA) N-terminal signal sequence. The synthetic gene was constructed of 6 sequences that were 100% conserved between F. tularensis LVS and the virulent Schu4 strain, 4 that were partially conserved, and 4 that were only found in F. tularensis Schu4 (B).
Of the sequences incorporated into the synthetic gene, 6 were 100% conserved between F. tularensis LVS and the virulent Schu4 strain, 4 were partially conserved, and 4 were only found in F. tularensis Schu4 (Figure 1B). A “breaker” sequence (GlyProGlyPro) was inserted between each epitope pair in order to minimize the introduction of MHC binding “neo-epitopes” at the junctions. A Kozak sequence (A/GCCACCATGG) was engineered upstream of the coding sequence to assure efficient translation initiation. The tissue plasminogen activator secretion sequence (QMSPALTCLVLGLALVFGEGSA) was inserted between the start codon and the start of the multi-epitope coding sequence to target the protein product to the extracellular space and the MHC class II processing pathway. Two stop codons were inserted following the multi-epitope coding sequence to assure translation termination. The gene was synthesized by GeneArt (Toronto, Canada) and subsequently cloned into the pVAX1 plasmid DNA vaccine vector (Invitrogen Corporation, Carlsbad, CA).
2.4. Vaccination
The HLA class II DNA construct was mixed with in vivo-JetPEI, a linear polyethylenimine derivative, to enhance transfection according to the manufacturer’s instructions (Polyplus-transfection Inc., New York, NY). To promote mucosal immunity, HLA-DRB1-transgenic mice were inoculated intratracheally (i.t.) three times at 2-week intervals (i.e, on days 0, 14 and 28) with 25 µg DNA/50 µl saline according to the method of Kim and co-workers described above [24]. On days 42 and 56, the mice were boosted i.t. with 25 µg/50 µl aliquots of the same class-II-restricted peptides encoded by the construct. The peptides, synthesized commercially, and phosphorothioate CpG ODN 1555 (Integrated DNA Technology; Coralville, IA) [26] were co-packaged in sterically stabilized cationic liposomes as previously reported by other investigators [27]. Briefly, small unilamellar liposomal vesicles were mixed with peptides and CpG ODN, freeze-dried, and resuspended in distilled water to internalize peptides and CpG ODN in liposomes. Non-vaccinated control mice were inoculated i.t. with unmodified pVAX1 vector and liposomes that contained CpG ODN only.
2.5. RNA extraction, purification, and quantitative real-time RT-PCR
Total cellular RNA in representative lung, spleen and liver samples was extracted and purified using TRIzol (Invitrogen Corporation, Carlsbad, CA). Real-time RT-PCR was conducted by a slight modification of the methods we described previously [28]. Briefly, purified RNA was reverse transcribed with QuantiTect reverse transcriptase (Qiagen Inc., Valencia, CA). Quantitative gene expression analysis was performed using the Eppendorf Mastercycle ep Realplex gradient S (Eppendorf, Westbury, NY) and SYBR green technology. iTaq SYBR Green Supermix with ROX (BioRad Laboratories, Hercules, CA) was mixed with an equal volume of RNase-free water containing 0.5 µM of both the forward and reverse primers, and cDNA corresponding to 100 ng of total RNA input. Subsequently, 40 cycles were run each consisting of 30 sec at 95°C followed by 1 min at 55°C and 30 sec at 72°C. At the end of the run, melting curves were constructed by heating the samples to 95°C with a ramp time of 20 min to verify that single PCR products were generated. Ribosomal RNA (18S) was used as the housekeeping standard. Copies of messenger and ribosomal RNAs were estimated from the number of PCR cycles required for the fluorescent signal to reach a fixed intensity, the threshold cycle (Ct). The mean number targeted gene mRNA copies/103 18S rRNA copies ± SE for samples derived from six mice treated comparably is reported. The PCR primers listed in Table 2 (designed from published sequences using Primer3 software, Whitehead Institute, Cambridge, MA) were purchased from Operon Biotechnologies, Inc. (Huntsville, AL).
Table 2.
Real-time RT-PCR Primers
| Gene | GenBank accession |
Orientation | Primer sequence |
Amplicon size |
|---|---|---|---|---|
| Interleukin-1β | NM_008361 | forward | TCACAGCAGCACATCAACAA | 112 |
| reverse | TGTCCTCATCCTGGAAGGT | |||
| Interleukin-2 | NM_008366 | forward | CCCACTTCAAGCTCCACTTC | 155 |
| reverse | ATCCTGGGGAGTTTCAGGTT | |||
| Interleukin-4 | NM_021283 | forward | CCTCACAGCAACGAAGAACA | 155 |
| reverse | ATCGAAAAGCCCGAAAGAGT | |||
| Interleukin-6 | NM_031168 | forward | AGTTGCCTTCTTGGGACTGA | 103 |
| reverse | CAGGTCTGTTGGGAGTGG | |||
| Interleukin-10 | NM_010548 | forward | CCAAGCCTTATCGGAAATGA | 162 |
| reverse | TTCACAGGGGAGAAATCG | |||
| Interleukin-12 (p40) | NM_008352 | forward | GGAAGCACGGCAGCAGAATA | 180 |
| reverse | AACTTGAGGGAGAAGTAGGAATG | |||
| IFN-γ | NM_008337 | forward | ACTGGCAAAAGGATGGTGAC | 211 |
| reverse | ACCTGTGGGTTGTTGACCTC | |||
| TNF-α | NM_013693 | forward | GTTCTGCAAAGGGAGAGTGG | 106 |
| reverse | GCACCTCAGGGAAGAGTCT | |||
| MIP-1α (Ccl3) | NM_011337 | forward | ACCATGACACTCTGCAACCA | 202 |
| reverse | CCCAGGTCTCTTTGGAGTCA | |||
| MCP-1 (Ccl2) | NM_011333 | forward | CCCACTCACCTGCTGCTACT | 164 |
| reverse | TCTGGACCCATTCCTTCTTG | |||
| KC (Cxcl1) | NM_008176 | forward | GCTGGGATTCACCTCAAGAA | 169 |
| reverse | TGGGGACACCTTTTAGCATC | |||
| 18S ribosomal | XR_000144 | forward | AATGGTGCTACCGGTCATTCC | 192 |
| RNA | reverse | ACCTCTCTTACCCGCTCTC |
2.6. Cytokine and chemokine analyses
The cytokines and chemokines present in representative tissue samples obtained from individual mice were quantified using the Bio-Plex cytokine array system. The samples (50–100 µg/ml) were homogenized in calcium- and magnesium-free HBSS containing 0.5% Triton X-100 and 2× complete protease inhibitor cocktail (Roche Diagnostics Corporation Indianapolis, IN). Homogenates were incubated for 30 minutes on ice then centrifuged at 13,500g for 5 minutes at 4°C to remove cell debris. The supernatants were stored at −20°C until assays are performed. Bio-Plex cytokine assay kits were purchased and used with the Bio-Plex 200 system in accordance with the manufacturer’s instructions (Bio-Rad Laboratories GmbH, Munich, Germany). The following panel of potentially relevant cytokines and chemokines were quantified: IL-1β, IL-2, IL-4, IL-6, IL-10, IL-12 (p40), TNF-α, IFN-γ, MIP-1α, MCP-1 and KC.
2.7. Statistical analysis
The data were analyzed using the SigmaStat statistics program (Jandel Scientific, San Rafael, CA). Individual means were compared using a non-paired Student's t test or a Mann-Whitney Rank Sum test. Data derived from 3 or more groups were compared by one-way analysis of variance (ANOVA) followed by a Tukey test to identify the groups that differed significantly (P <0.05).
3. Results
3.1. Epitope-based vaccination and LVS challenge
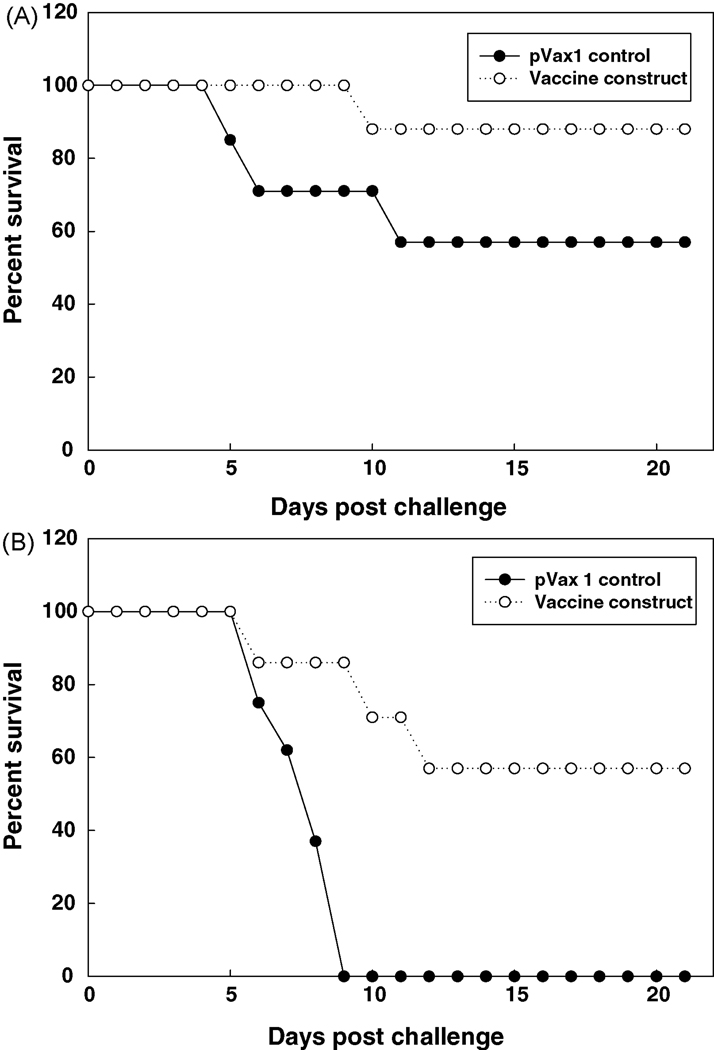
HLA class II (DRB1*0401) transgenic mice were immunized i.t. with the DNA vaccine construct shown in Figure 1, then boosted with the corresponding peptides formulated in liposomes with CpG ODN 1555; control mice received unmodified pVAX1 vector and liposomes that contained CpG ODN only. The mice were subsequently challenged i.t. with F. tularensis LVS on day 18 following the final peptide boost. The data derived from 2 separate experiments in which the mice were challenged with 2 LD50 (~2,560 CFUs; Figure 2A) or 5 LD50 (~6,400 CFUs; Figure 2B) are shown. Notably, while all the control mice challenged with 5 LD50 died, ~60% of the vaccinated animals survived over the course of the 3-week period following infection.
Figure 2. Epitope-based vaccine induces protective immunity to tularemia in HLA class II transgenic mice.
Groups of vaccinated and control mice were challenged i.t. with 2 LD50 (A) or 5 LD50 (B) F. tularensis LVS and survival was monitored over a 3-week period. Three experiments conducted under similar conditions yielded comparable results; the data derived from 2 of these experiments are shown.
Relative to non-immunized controls, vaccinated mice challenged i.t. with F. tularensis LVS exhibited a significant reduction in the bacterial burden of the lungs assessed on day 3 post-infection (Table 3). The numbers of organisms in the spleens and livers dissected from the same animals were extremely low compared to the lungs; no detectable differences between these organs obtained from vaccinated and control mice were determined.
Table 3.
The bacterial burden of the lungs is decreased in vaccinated micea
| Tissue | CFUs/mg tissue | |
|---|---|---|
| pVAX1 control | Vaccine construct | |
| Lungs | 27,352 ± 10,251 | 13,561 ± 1,428b |
| Liver | 53 ± 16 | 66 ± 55 |
| Spleen | 13 ± 5 | 17 ± 9 |
Groups (n=4) of control and vaccinated mice were challenged i.t. with 2 LD50 F. tularensis LVS. Values are the mean colony forming units ± SD/mg tissue dissected on day 3 postinfection.
Significantly less than pVAX1 control; P=0.029 (Mann-Whitney Rank Sum Test).
3.2. Vaccination promotes rapid, proinflammatory cytokine production in the lungs of mice challenged with F. tularensis LVS
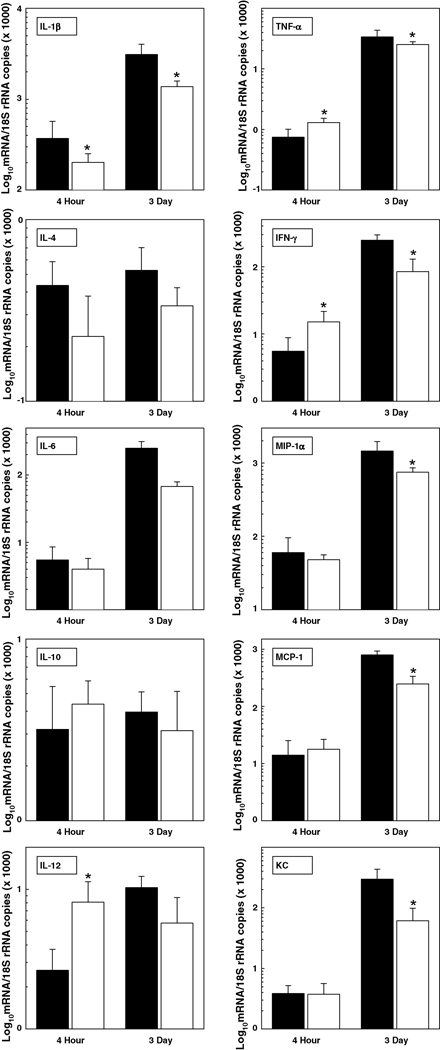
The expression of several proinflammatory cytokine mRNAs was up-regulated rapidly in the lungs of vaccinated mice challenged i.t. with 6 LD50 F. tularensis LVS on day 18 post-vaccination. Particularly, the expression of IL-12, TNF-α and IFN-γ messages was significantly higher in the lungs of vaccinated animals at 4 hours post-infection i.t. while average IL-4 mRNA expression was lower, albeit variation between samples prohibited finding statistical significance (Figure 3). By comparison, cytokine and chemokine mRNA expression was generally lower in the lungs of vaccinated mice than non-vaccinated control mice at 3 days post-infection correlating directly with the reduced number of organisms found at this time (shown in Table 3). Notably, cytokine/chemokine mRNA expression was generally higher in the lungs of both groups of mice on day 3 than at 4 hours post-infection.
Figure 3. Cytokine/chemokine mRNA expression in the lungs of vaccinated mice is rapidly up-regulated following i.t. challenge with F. tularensis LVS.
Groups of control (solid bar) and vaccinated (open bar) mice were infected i.t. with 6 LD50 F. tularensis LVS. The lungs were dissected at 4 hours and 3 days post-infection; cytokine and chemokine mRNA expression was quantified by real-time RT-PCR. Data are the means ± SD derived from 4 mice treated comparably. A second experiment yielded similar results. *Vaccinated group is significantly different from control: P<0.05.
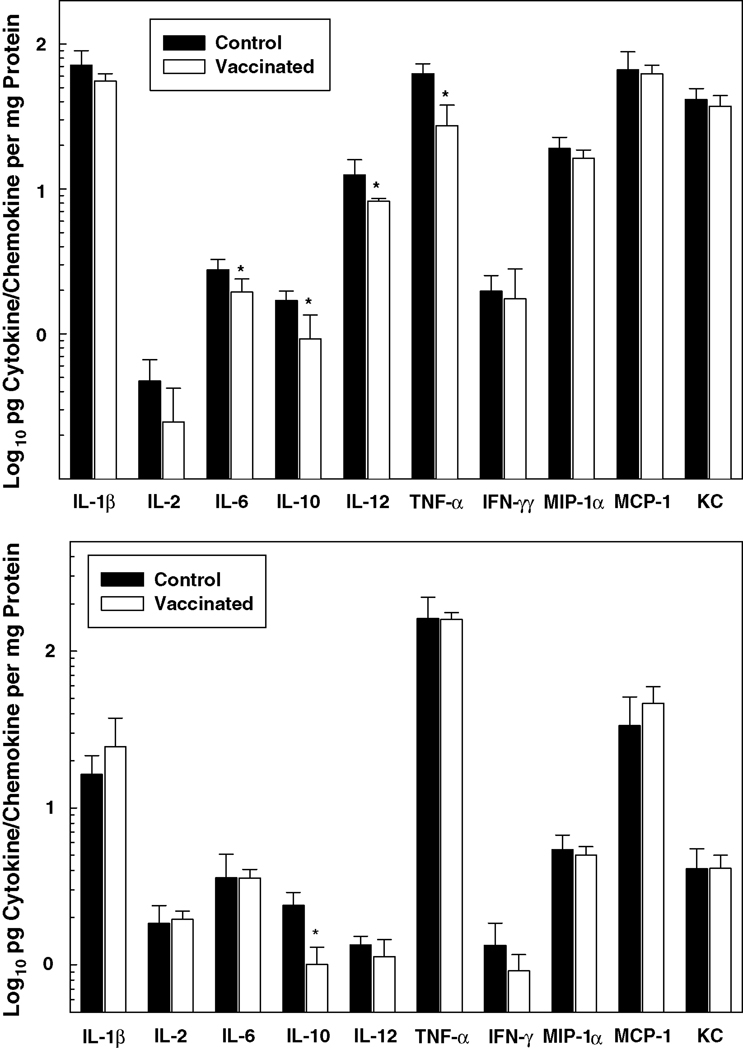
Pro-inflammatory cytokine and chemokine protein levels were also generally higher in the lungs of vaccinated, relative to control, mice at 4 hours post-infection i.t. with F. tularensis LVS (Figure 4). While variations among individual cytokines/chemokines occurred, protein levels overall were higher in the lungs at 3 days than at 4 hours post-infection. However, in contrast to 4 hours, no statistical difference was found in the levels of these proteins in the lungs of the vaccinated versus non-vaccinated groups at 3 days. Notably, a strict correlation between mRNA expression (illustrated in Figure 3) and protein production (Figure 4) was lacking, which probably relates to the relative turnover rate of these two entities and the stability of proteins.
Figure 4. Proinflammatory cytokine and chemokine protein production is up-regulated in the lungs of vaccinated mice shortly following i.t. inoculation of F. tularensis LVS.
Groups of control and vaccinated mice were challenged i.t. with 6 LD50 F. tularensis LVS. At 4 hours (top panel) and 3 days (bottom panel) post-infection, the lungs were dissected, and the cytokines and chemokines listed were quantified by Bio-Plex cytokine bead array analysis. Data are the means ± SD derived from 4 mice treated comparably in each group. A second experiment yielded similar results. *Vaccinated group is significantly greater than control: P<0.05. Notably, IL-4 concentrations (not shown) were below the level of detection.
3.3. Cytokine/chemokine levels in the spleens and livers of F. tularensis-infected mice
In contrast to the lungs in which there was no detectable difference on day 3 post-infection, cytokine and chemokine protein levels were generally lower in the spleens of vaccinated mice (Figure 5). In particular, the tissue concentrations of IL-6, IL-10, IL-12 and TNF-α were significantly decreased in the spleens of vaccinated, relative to non-vaccinated, animals. The cytokine/chemokine levels in the livers of control and vaccinated mice, on the other hand, were comparable; only the concentration of IL-10 was diminished in the latter group.
Figure 5.
Proinflammatory cytokine/chemokine protein production is down-regulated in the spleens of vaccinated mice challenged i.t. with F. tularensis LVS. Groups of control and vaccinated mice were challenged i.t. with 6 LD50 F. tularensis LVS. At 3 days post-infection, the spleens (top panel) and livers (bottom panel) were dissected from each group, and the cytokines and chemokines listed were quantified by Bio-Plex cytokine bead array analysis. Data are the means ± SD derived from 4 mice treated comparably. A second experiment yielded similar results. *Vaccinated group is significantly less than control: P<0.05. IL-4 concentrations were too low to be detected (not shown).
4. Discussion
Clinical studies conducted in the 1960s demonstrated the utility of F. tularensis LVS in immunizing humans against exposure to a virulent pathogenic strain [13,29]. Despite its development over 40 years ago, however, the basis for the attenuation of F. tularensis LVS, protective antigens, and immunological mechanisms that underlie its efficacy remain unresolved. These as well as other issues, e.g., variations in inherent pathogenicity, have instigated a search for an improved, better defined tularemia vaccine. Given the effectiveness achieved with F. tularensis LVS, strategies to develop a new generation vaccine have focused largely upon attenuated organisms that possess only limited ability to survive, replicate, and cause disease. Vaccination with a viable organism, however, continues to represent a potential risk for normal, as well as immunocompromised individuals. In a mouse model, for example, vaccination with a highly attenuated Type A strain of F. tularensis subspecies tularensis administered i.n. caused severe tissue injury in the lungs even in the absence of extensive bacterial replication [30]. Subunit vaccines offer an alternate approach with clear safety advantages.
The LPS-associated O-antigen is the only component of F. tularensis identified to date that, by itself, is capable of stimulating protective immunity against systemic infections by virulent Type B, or against respiratory challenge by either Type A or B strains [21,31]. Mice immunized subcutaneously with LVS endotoxin-BSA conjugate were partially protected against aerosol challenge with virulent type B, but were fully susceptible to aerosol challenge with virulent type A strain [21]. A number of highly immunogenic proteins have been identified (e.g., FopA and TUL4); none are able to elicit protective immunity [31,32].
The experiments reported herein document the efficacy of a F. tularensis subunit vaccine composed of HLA class II-restricted epitopes, predicted by computer analyses of the microbial genome and verified by the recognition of T cells obtained from individuals previously exposed to tularemia [22]. Mouse MHC class II I-E molecules were not expressed by HLA DRB1 transgenic mice immunized in the experiments described herein, negating their potential role in the results reported. This is notable inasmuch as promiscuous binding was a dominant criterion employed in selecting the T cell epitopes incorporated into the vaccine construct used [22]. Consequently, cross reactivity is expected and it is predicted that several epitopes would bind to mouse MHC class II molecules if the molecules were expressed. Vaccinated HLA DRB1 transgenic mice exhibited elevated resistance to a lethal dose of F. tularensis LVS inoculated i.t. This finding is remarkable in several respects. First, the vaccine lacked MHC class I-restricted epitopes required to elicit a CD8+ T cell response and maximum protection against F. tularensis infection [22]. Moreover, 4 of the 14 epitopes incorporated into the vaccine construct are expressed exclusively by the virulent, Schu4 strain of F. tularensis (i.e., not LVS illustrated in Figure 1B) and, thus, presumably failed to elicit T cell responses relevant to protective immunity in the challenge experiments reported here. It is pertinent to note that the vaccine was only effective when administered i.t.; intramuscular vaccination with the DNA vaccine construct alone failed to elicit protective immunity in mice against subsequent challenge (data not shown). This result correlates with findings obtained by other investigators who reported that vaccination by a mucosal route induced the greatest protection against pneumonic tularemia [33].
Relative to the controls, mice immunized with the epitope-based subunit vaccine described herein exhibited increases in IL-12, TNF-α and IFN-γ mRNA expression, and IL-1β, IL-6, IL-12, MIP-1α, MCP-1 and KC protein production in the lungs within a 4-hour period post-infection. IL-12, TNF-α and IFN-γ are critical factors in host resistance to F. tularensis infections in mice [34]. While such cytokines initially derive from NK cells, macrophages and/or dendritic cells during primary infection, they constitute an important component of the adaptive immune response in which memory T lymphocytes serve as a primary cell source during a secondary response to F. tularensis infection [33–35]. In this regard, immunized mice challenged intradermally with a lethal dose of F. tularensis LVS exhibited the rapid, elevated, local expression of IL-12, IFN-γ and TNF-α mRNAs correlating with a marked decrease in the replication of bacteria at the site of infection [36]. In more recent studies, both CD4+ and CD8+ T cells, and the production of IFN-γ were key factors in the vaccine-induced protection of mice against respiratory challenge with the virulent type A strain of F. tularensis [33,37]. These findings correlate with those reported here in which increases in cytokine/chemokine mRNA expression and/or protein production in vaccinated animals correlated with a diminished number of organisms enumerated in the lungs at 3 days post-infection, and an increased rate of survival.
The elevated expression of both IL-12 and IFN-γ mRNA and production of IL-12 in the lungs of vaccinated mice at 4 hours post-infection are particularly noteworthy. IL-12 is essential for the induction of IFN-γ production by T lymphocytes, and a critical factor in host defenses to respiratory F. tularensis LVS infections [38,39]. Mice deficient in either IL-12 or IFN-γ readily succumbed to infections that were sublethal for wild-type animals [39]. Moreover, exogenous IL-12 treatment prevented the death of mice infected with a lethal dose of F. tularensis LVS; protection was dependent upon IFN-γ. The elevated protein concentration of IL-12, but not IFN-γ, in the lungs of vaccinated mice at 4 hours post-infection supports the sequential synthesis of these two cytokines and the requisite production of IL-12 for the induction of IFN-γ synthesis in the lungs of mice infected i.n. with F. tularensis [39]. The rapid increase in IL-12 mRNA expression and protein production presumably by tissue macrophages in vaccinated, relative to naïve, mice challenged with F. tularensis has been reported [36]. The mechanisms that enable such an increase remain to be delineated.
TNF-α message expression was also elevated significantly in the lungs of vaccinated, relative to control, mice at 4 hours post-infection i.t. with F. tularensis LVS. Like IL-12 and IFN-γ, TNF-α is a key factor in host defenses to tularemia infections in mice [40]. Replication of the organism is increased markedly in organs of TNF-deficient mice or mice treated with neutralizing, anti-TNF monoclonal antibody [10,40]. As in the case of IFN-γ in the experiments reported here, TNF-α message expression seemingly preceded TNF-α production so that protein levels in the lungs of vaccinated and control mice were equivalent at 4 hours post-infection despite detecting elevated mRNA levels in the vaccinated group at this time.
While it is widely believed that alveolar macrophages constitute the principal target of F. tularensis infections of the lungs, they also represent a dominant cell population in host defenses [41]. In this regard, it has been suggested that the lack of alveolar macrophage activation and the production of cytokines contributes to the increased susceptibility of TLR2-deficient mice to pneumonic tularemia [42]. The elevated production of cytokines and chemokines normally associated with macrophages (i.e., IL-1β, IL-6, IL-12, MIP-1α, MCP-1 and KC) in the lungs of vaccinated mice within a 4-hour period following infection supports this purported role of alveolar macrophages in cytokine/chemokine production and host defenses to F. tularensis. As noted in the Results Section, the absence of a strict correlation between message expression (Figure 3) and protein production (Figure 4) conceivably derives from a difference in stability of the mRNA transcript and protein product [43]. As in the case of IL-12 discussed above, the factors that trigger a rapid increase in mRNA expression and/or the production of these cytokines/chemokines in the lungs of vaccinated mice challenged i.t. with F. tularensis are unclear.
Although cytokine and chemokine protein levels were frequently higher in the lungs of vaccinated animals at 4 hours post-infection, these same proteins were generally lower (albeit not significantly) in the lungs at 3 days (Figure 4). Similarly, these same cytokines and chemokines were lower in the spleens of vaccinated mice on day 3 post-infection (Figure 5). Other investigators report similar findings, that is, decreases in pro-inflammatory cytokine (TNF-α, IFN-γ, IL-6 and/or MCP-1) concentrations in tissues derived from vaccinated (relative to control) mice after an extended period following challenge i.n. with F. tularensis [16,20]. Indeed, the exaggerated production of pro-inflammatory cytokines such as MCP-1 and IL-6 in the tissues of mice infected i.n. with F. tularensis was found to correlate directly with critical illness, sepsis and organ failure [44].
In conclusion, the experiments reported here demonstrate the efficacy of a subunit vaccine constructed of multiple epitopes recognized by T cells derived from individuals previously exposed to tularemia and capable of eliciting protective immunity in “humanized” HLA class II (DRB1*0401) transgenic mice. Vaccinated mice challenged intratracheally with a lethal dose of F. tularensis LVS exhibited a rapid increase in pro-inflammatory cytokine production, a diminished number of organisms in the lungs, and an increased rate of survival. At later time points during the course of infection, proinflammatory cytokine/chemokine levels in the tissues of vaccinated animals are reduced for the most part correlating with the reduction in bacterial burden and critical illness. These results suggest that an epitope-based approach might be widely applicable to the development of vaccines against other intracellular bacterial pathogens. Experiments involving HLA-A2.1-/HLA-DRB1-double transgenic mice vaccinated with DNA constructs (and the corollary peptides) that encode both HLA class I- and class II-restricted F. tularensis-specific epitopes, and subsequent challenge i.t. with the virulent, Schu4 strain of F. tularensis are envisioned. Relative to the attenuated vaccines currently being developed, an epitope-based vaccine with demonstrated efficacy against aerosolized F. tularensis subtype tularensis would be safer, more readily approved by governing agencies, and easier to market.
Acknowledgments
The authors wish to acknowledge the efforts of Daniel S. Rivera, who helped conduct these experiments, and Miriam A. Goldberg, who played an instrumental role in the bioinformatics analysis underlying this research. This study was supported by National Institutes of Health Research Grants: R21AI055657 (PI: S.H. Gregory) and 1R43AI058326 (PI: A.S. De Groot). Anne S. De Groot and William Martin are senior officers and majority shareholders at EpiVax, Inc., a privately owned vaccine design company located in Providence, RI. These authors acknowledge that there is a potential conflict of interest related to their relationship with EpiVax and attest that the work contained in this report is free of any bias that might be associated with the commercial goals of the company.
References
- 1.Tarnvik A. Nature of protective immunity to Francisella tularensis. Rev.Infect.Dis. 1989;11(3):440–451. [PubMed] [Google Scholar]
- 2.Evans ME, Gregory DW, Schaffner W, McGee ZA. Tularemia: a 30-year experience with 88 cases. Medicine (Baltimore) 1985;64(4):251–269. [PubMed] [Google Scholar]
- 3.Johansson A, Ibrahim A, Goransson I, Eriksson U, Gurycova D, Clarridge JE, III, et al. Evaluation of PCR-based methods for discrimination of Francisella species and subspecies and development of a specific PCR that distinguishes the two major subspecies of Francisella tularensis. J.Clin.Microbiol. 2000;38(11):4180–4185. doi: 10.1128/jcm.38.11.4180-4185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill V, Cunha BA. Tularemia pneumonia. Semin.Respir.Infect. 1997;12(1):61–67. [PubMed] [Google Scholar]
- 5.Chin J, editor. American Public Health Association. Control of Communicable Diseases Manual. Washington, D.C: American Public Health Association; 2000. pp. 532–535. Tularemia. [Google Scholar]
- 6.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285(21):2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 7.Conlan JW, Sjostedt A, North RJ. CD4+ and CD8+ T-cell-dependent and -independent host defense mechanisms can operate to control and resolve primary and secondary Francisella tularensis LVS infection in mice. Infect.Immun. 1994;62:5603–5607. doi: 10.1128/iai.62.12.5603-5607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulop M, Mastroeni P, Green M, Titball RW. Role of antibody to lipopolysaccharide in protection against low- and high-virulence strains of Francisella tularensis. Vaccine. 2001;19(31):4465–4472. doi: 10.1016/s0264-410x(01)00189-x. [DOI] [PubMed] [Google Scholar]
- 9.Kirimanjeswara GS, Golden JM, Bakshi CS, Metzger DW. Prophylactic and therapeutic use of antibodies for protection against respiratory infection with Francisella tularensis. J.Immunol. 2007;179(1):532–539. doi: 10.4049/jimmunol.179.1.532. [DOI] [PubMed] [Google Scholar]
- 10.Sjostedt A, North RJ, Conlan JW. The requirement of tumour necrosis factor-α and interferon-γ for the expression of protective immunity to secondary murine tularaemia depends on the size of the challenge inoculum. Microbiology. 1996;142(Pt 6):1369–1374. doi: 10.1099/13500872-142-6-1369. [DOI] [PubMed] [Google Scholar]
- 11.Fortier AH, Leiby DA, Narayanan RB, Asafoadjei E, Crawford RM, Nacy CA, et al. Growth of Francisella tularensis LVS in macrophages: the acidic intracellular compartment provides essential iron required for growth. Infect.Immun. 1995;63(4):1478–1483. doi: 10.1128/iai.63.4.1478-1483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect.Immun. 2004;72(6):3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saslaw S, Eigelsbach HT, Prior JA, Wilson HE, Carhart S. Tularemia vaccine study, II: respiratory challenge. Arch.Int.Med. 1961;107:134–146. doi: 10.1001/archinte.1961.03620050068007. [DOI] [PubMed] [Google Scholar]
- 14.Cherwonogrodzky JW, Knodel MH, Spence MR. Increased encapsulation and virulence of Francisella tularensis live vaccine strain (LVS) by subculturing on synthetic medium. Vaccine. 1994;12(9):773–775. doi: 10.1016/0264-410x(94)90284-4. [DOI] [PubMed] [Google Scholar]
- 15.Oyston PC, Sjostedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat.Rev.Microbiol. 2004;2(12):967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 16.Baron SD, Singh R, Metzger DW. Inactivated Francisella tularensis live vaccine strain protects against respiratory tularemia by intranasal vaccination in an immunoglobulin A-dependent fashion. Infect.Immun. 2007;75(5):2152–2162. doi: 10.1128/IAI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rawool DB, Bitsaktsis C, Li Y, Gosselin DR, Lin Y, Kurkure NV, et al. Utilization of Fc receptors as a mucosal vaccine strategy against an intracellular bacterium, Francisella tularensis. J.Immunol. 2008;180(8):5548–5557. doi: 10.4049/jimmunol.180.8.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke DS. Immunization against tularemia: analysis of the effectiveness of live Francisella tularensis vaccine in prevention of laboratory-acquired tularemia. J.Infect.Dis. 1977;135(1):55–60. doi: 10.1093/infdis/135.1.55. [DOI] [PubMed] [Google Scholar]
- 19.Wayne CJ, Oyston PC. Vaccines against Francisella tularensis. Ann.N.Y.Acad.Sci. 2007;1105:325–350. doi: 10.1196/annals.1409.012. [DOI] [PubMed] [Google Scholar]
- 20.Bakshi CS, Malik M, Mahawar M, Kirimanjeswara GS, Hazlett KR, Palmer LE, et al. An improved vaccine for prevention of respiratory tularemia caused by Francisella tularensis SchuS4 strain. Vaccine. 2008;26(41):5276–5288. doi: 10.1016/j.vaccine.2008.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conlan JW, Shen H, Webb A, Perry MB. Mice vaccinated with the O-antigen of Francisella tularensis LVS lipopolysaccharide conjugated to bovine serum albumin develop varying degrees of protective immunity against systemic or aerosol challenge with virulent type A and type B strains of the pathogen. Vaccine. 2002;20(29–30):3465–3471. doi: 10.1016/s0264-410x(02)00345-6. [DOI] [PubMed] [Google Scholar]
- 22.McMurry JA, Gregory SH, Moise L, Rivera D, Buus S, De Groot AS. Diversity of Francisella tularensis Schu4 antigens recognized by T lymphocytes after natural infections in humans: Identification of candidate epitopes for inclusion in a rationally designed tularemia vaccine. Vaccine. 2007;25(16):3179–3191. doi: 10.1016/j.vaccine.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 23.Woods A, Chen HY, Trumbauer ME, Sirotina A, Cummings R, Zaller DM. Human major histocompatibility complex class II-restricted T cell responses in transgenic mice. J.Exp.Med. 1994;180(1):173–181. doi: 10.1084/jem.180.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Merry AC, Nemzek JA, Bolgos GL, Siddiqui J, Remick DG. Eotaxin represents the principal eosinophil chemoattractant in a novel murine asthma model induced by house dust containing cockroach allergens. J.Immunol. 2001;167(5):2808–2815. doi: 10.4049/jimmunol.167.5.2808. [DOI] [PubMed] [Google Scholar]
- 25.Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat.Genet. 2005 doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 26.Takeshita S, Takeshita F, Haddad DE, Ishii KJ, Klinman DM. CpG oligodeoxynucleotides induce murine macrophages to up-regulate chemokine mRNA expression. Cell Immunol. 2000;206(2):101–106. doi: 10.1006/cimm.2000.1735. [DOI] [PubMed] [Google Scholar]
- 27.Gursel I, Gursel M, Ishii KJ, Klinman DM. Sterically stabilized cationic liposomes improve the uptake and immunostimulatory activity of CpG oligonucleotides. J.Immunol. 2001;167(6):3324–3328. doi: 10.4049/jimmunol.167.6.3324. [DOI] [PubMed] [Google Scholar]
- 28.Gehring S, Dickson EM, San Martin ME, van Rooijen N, Papa EF, Harty MW, et al. Kupffer cells abrogate cholestatic liver injury in mice. Gastroenterology. 2006;130(3):810–822. doi: 10.1053/j.gastro.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Saslaw S, Eigelsbach HT, Wilson HE, Prior JA, Carhart S. Tularemia vaccine study, I: intracutaneous challenge. Arch.Int.Med. 1961;107:121–133. doi: 10.1001/archinte.1961.03620050055006. [DOI] [PubMed] [Google Scholar]
- 30.Pechous RD, McCarthy TR, Mohapatra NP, Soni S, Penoske RM, Salzman NH, et al. A Francisella tularensis Schu S4 purine auxotroph is highly attenuated in mice but offers limited protection against homologous intranasal challenge. PLoS.ONE. 2008;3(6):e2487. doi: 10.1371/journal.pone.0002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fulop M, Manchee R, Titball R. Role of two outer membrane antigens in the induction of protective immunity against Francisella tularensis strains of different virulence. FEMS Immunol.Med.Microbiol. 1996;13(3):245–247. doi: 10.1111/j.1574-695X.1996.tb00245.x. [DOI] [PubMed] [Google Scholar]
- 32.Fulop M, Manchee R, Titball R. Role of lipopolysaccharide and a major outer membrane protein from Francisella tularensis in the induction of immunity against tularemia. Vaccine. 1995;13(13):1220–1225. doi: 10.1016/0264-410x(95)00062-6. [DOI] [PubMed] [Google Scholar]
- 33.Conlan JW, Shen H, KuoLee R, Zhao X, Chen W. Aerosol-, but not intradermal-immunization with the live vaccine strain of Francisella tularensis protects mice against subsequent aerosol challenge with a highly virulent type A strain of the pathogen by an αβ T cell- and interferon gamma-dependent mechanism. Vaccine. 2005;23(19):2477–2485. doi: 10.1016/j.vaccine.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 34.Elkins KL, Cowley SC, Bosio CM. Innate and adaptive immunity to Francisella. Ann.N.Y.Acad.Sci. 2007;1105:284–324. doi: 10.1196/annals.1409.014. [DOI] [PubMed] [Google Scholar]
- 35.Cowley SC, Sedgwick JD, Elkins KL. Differential requirements by CD4+ and CD8+ T cells for soluble and membrane TNF in control of Francisella tularensis live vaccine strain intramacrophage growth. J.Immunol. 2007;179(11):7709–7719. doi: 10.4049/jimmunol.179.11.7709. [DOI] [PubMed] [Google Scholar]
- 36.Stenmark S, Sunnemark D, Bucht A, Sjostedt A. Rapid local expression of interleukin-12, tumor necrosis factor alpha, and gamma interferon after cutaneous Francisella tularensis infection in tularemia-immune mice. Infect.Immun. 1999;67(4):1789–1797. doi: 10.1128/iai.67.4.1789-1797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu TH, Hutt JA, Garrison KA, Berliba LS, Zhou Y, Lyons CR. Intranasal vaccination induces protective immunity against intranasal infection with virulent Francisella tularensis biovar A. Infect.Immun. 2005;73(5):2644–2654. doi: 10.1128/IAI.73.5.2644-2654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat.Rev.Immunol. 2003;3(2):133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 39.Duckett NS, Olmos S, Durrant DM, Metzger DW. Intranasal interleukin-12 treatment for protection against respiratory infection with the Francisella tularensis live vaccine strain. Infect.Immun. 2005;73(4):2306–2311. doi: 10.1128/IAI.73.4.2306-2311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowley SC, Goldberg MF, Ho JA, Elkins KL. The membrane form of tumor necrosis factor is sufficient to mediate partial innate immunity to Francisella tularensis live vaccine strain. J.Infect.Dis. 2008;198(2):284–292. doi: 10.1086/589620. [DOI] [PubMed] [Google Scholar]
- 41.Metzger DW, Bakshi CS, Kirimanjeswara G. Mucosal Immunopathogenesis of Francisella tularensis. Ann.N.Y.Acad.Sci. 2007;1105:266–283. doi: 10.1196/annals.1409.007. [DOI] [PubMed] [Google Scholar]
- 42.Malik M, Bakshi CS, Sahay B, Shah A, Lotz SA, Sellati TJ. Toll-like receptor 2 is required for control of pulmonary infection with Francisella tularensis. Infect.Immun. 2006;74(6):3657–3662. doi: 10.1128/IAI.02030-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4(9):117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiavolini D, Alroy J, King CA, Jorth P, Weir S, Madico G, et al. Identification of immunologic and pathologic parameters of death versus survival in respiratory tularemia. Infect.Immun. 2008;76(2):486–496. doi: 10.1128/IAI.00862-07. [DOI] [PMC free article] [PubMed] [Google Scholar]