Abstract
The spectrum of human immunodeficiency virus type 1 (HIV-1) protease and reverse transcriptase (RT) mutations selected by antiretroviral (ARV) drugs requires ongoing reassessment as ARV treatment patterns evolve and increasing numbers of protease and RT sequences of different viral subtypes are published. Accordingly, we compared the prevalences of protease and RT mutations in HIV-1 group M sequences from individuals with and without a history of previous treatment with protease inhibitors (PIs) or RT inhibitors (RTIs). Mutations in protease sequences from 26,888 individuals and in RT sequences from 25,695 individuals were classified according to whether they were nonpolymorphic in untreated individuals and whether their prevalence increased fivefold with ARV therapy. This analysis showed that 88 PI-selected and 122 RTI-selected nonpolymorphic mutations had a prevalence that was fivefold higher in individuals receiving ARVs than in ARV-naïve individuals. This was an increase of 47% and 77%, respectively, compared with the 60 PI- and 69 RTI-selected mutations identified in a similar analysis that we published in 2005 using subtype B sequences obtained from one-fourth as many individuals. In conclusion, many nonpolymorphic mutations in protease and RT are under ARV selection pressure. The spectrum of treatment-selected mutations is changing as data for more individuals are collected, treatment exposures change, and the number of available sequences from non-subtype B viruses increases.
Identifying the mutations responsible for human immunodeficiency virus type 1 (HIV-1) drug resistance has implications for drug resistance surveillance, HIV-1 genotypic resistance testing, and the biophysical mechanisms by which HIV-1 escapes from selective drug pressure. Many mutations in HIV-1 protease and reverse transcriptase (RT) are considered drug resistance mutations by virtue of emerging during antiretroviral (ARV) selection pressure in vitro or in vivo, reducing drug susceptibility in vitro, or reducing the virological response to therapy. As more sequenced HIV-1 isolates from ARV-exposed individuals are reported, more ARVs are licensed, and a greater proportion of published sequences of HIV-1 protease and RT belong to non-B subtypes, it is expected that new treatment-selected mutations will be identified.
We previously identified nonpolymorphic treatment-selected mutations in an analysis of subtype B protease and RT sequences from ∼6,000 individuals in the HIV Drug Resistance Database (HIVDB) (26). Here, we describe the results of a similar analysis that includes non-B group M sequences and about four times as many individuals than in the 2005 study.
MATERIALS AND METHODS
Patients, viruses, and mutations.
HIV-1 RT and protease sequences were compiled from published studies in the HIVDB (http://hivdb.stanford.edu) (27) and from previously unpublished sequences from HIV-1-infected individuals in Northern and Southern California as part of an Institutional Review Board-approved protocol. For the new virus sequences, treatment histories were obtained from patient charts and pharmacy records. We included sequences from individuals from whom the complete ARV drug class history was available.
Protease positions 1 to 99 and RT positions 1 to 350 were analyzed. Mutations were defined as amino acid differences from the HIV-1 group M consensus B sequence. In sequences from patients with multiple virus isolates, mutations occurring in more than one isolate were counted only once. When multiple clones were available from the same virus isolates, only the consensus of the clones was used.
To reduce the impact of sequencing errors, a sequence quality score was assigned to all sequences. This score equaled the total number of stop codons, highly ambiguous nucleotides (B, D, H, V, and N), and highly unusual mutations (defined as mutations occurring at a frequency of below 1 in 2,000 in pooled treated and untreated group M sequences). Protease sequences with a sequence quality score of 4 or higher and RT sequences with a sequence quality score of 6 or higher were excluded from the data set. Sequences containing an APOBEC3G-induced G-to-A hypermutation were also excluded (11).
Each mutation was also characterized by its presence on five published mutation lists, from the Agence Nationale de Recherche sur le SIDA (ANRS) (1), HIVDB (24), IAS-USA (20), Los Alamos National Laboratory (8), and Rega Institute (32).
Nonpolymorphic mutations.
We defined nonpolymorphic mutations using criteria similar to that outlined in two recent publications as being present at a frequency of ≤0.5% in ARV-naïve individuals infected with all subtypes for which >1,000 sequences were available and at levels of >0.5% in no more than one subtype for which fewer than 1,000 sequences were available (3, 29). In contrast to the definition used in these two recent publications, we did not exclude nonpolymorphic mutations occurring at positions that also contained polymorphic mutations.
Two steps were taken to reduce the influence of transmitted drug resistance on our current analysis: isolates from persons with primary HIV-1 infection in U.S. and European studies published after the year 2000 were excluded, and isolates from untreated persons that had two or more established nonpolymorphic drug-related mutations were excluded.
Treatment-selected mutations.
To identify protease inhibitor (PI)-selected mutations, we compared the prevalence of protease mutations in PI-treated individuals to the prevalence in PI-naïve individuals. To identify RT inhibitor (RTI)-selected mutations, we compared the prevalences of RT mutations in RTI-treated and RTI-naïve individuals. For each drug class, treatment-selected mutations were defined as being nonpolymorphic mutations that occurred more than five times more frequently in treated than in untreated HIV-1 isolates and that were significantly associated with treatment by Fisher's exact test using Holm's method to control the family-wise error rate for multiple-hypothesis testing at a P value of <0.01 (15). The P values were ranked in descending order. Starting from the smallest P value (rank r = n, where n is the number of hypotheses), we compared each p of rank r with a significance cutoff of 0.01/r as long as pr was ≤0.01/r. All P values of pr…pn were considered to be statistically significant.
Distinguishing nucleoside RTI (NRTI)- from nonnucleoside RTI (NNRTI)-selected mutations.
Whereas the association between an RT mutation and RTI therapy could be made unequivocally, the association with NRTI or NNRTI treatment was more difficult because nearly all individuals who received NNRTIs also received NRTIs, with the exception of 1,055 women receiving a single dose of nevirapine (NVP) to prevent mother-to-child HIV-1 transmission.
Two indexes were therefore created to assess the impact of NRTI and NNRTI selection pressure: (i) the NRTI selection index was defined as the prevalence of a mutation in NRTI-experienced but NNRTI-naïve individuals divided by the mutation's prevalence in RTI-naïve individuals, and (ii) the NNRTI selection index was defined as the prevalence of a mutation in NNRTI-experienced individuals (nearly all of whom were also NRTI experienced) divided by the mutation's prevalence in NRTI-experienced but NNRTI-naïve individuals.
When 0, 1, or 2 mutations were detected in NRTI-experienced (but NNRTI-naïve) individuals, the NRTI selection index was considered to be 1.0 because of the possibility that such mutations may have resulted from a virus transmitted from a treated person rather than having been selected by NRTI therapy. When 0 mutations were detected in RTI-naïve or NRTI-experienced (but NNRTI-naïve) individuals, 1 was added to avoid division by 0 in downstream calculations.
RTI-selected mutations were grouped into five categories based on their NRTI and NNRTI selection indexes: (i) definite NRTI-selected mutations, defined as mutations with statistically significant NRTI selection indexes (P < 0.05 following Holm's correction) or for which there was phenotypic evidence of decreased NRTI susceptibility; (ii) definite NNRTI-selected mutations, defined as mutations for which the NNRTI selection index but not the NRTI selection index was statistically significant (P < 0.05 following Holm's correction) or for which there was phenotypic evidence of decreased NNRTI susceptibility; (iii) probable NRTI-selected mutations, defined as mutations at established NRTI positions (positions 41, 62, 65, 67, 69, 70, 74, 75, 77, 115, 116, 151, 184, 208, 210, 215, and 219) that did not have statistically significant NRTI selection indexes after controlling for multiple comparisons; (iv) probable NNRTI-selected mutations, defined as mutations at established NNRTI positions (positions 98, 100, 101, 103, 106, 108, 138, 179, 181, 188, 190, 221, 225, 227, 230, 236, 238, and 318) that did not have statistically significant NNRTI selection indexes after controlling for multiple comparisons; and (v) undifferentiated RTI-selected mutations, for which neither the NRTI nor NNRTI selection indexes were statistically significant.
HIV-1 RT structural analyses.
For each of the new RTI-selected residues, we calculated the shortest interatomic distance between the residue and the incoming deoxynucleoside triphosphate, the NNRTI binding pocket, the established NRTI resistance residues, and the established NNRTI resistance residues. Distances between RT residues and the incoming deoxynucleoside triphosphate and established NRTI resistance residues were calculated using the coordinates from the X-ray crystallographic structure (Protein Data Bank [PDB] accession number 1RTD) described previously by Huang et al. (16). Distances between RT residues and the NNRTI NVP and the established NNRTI resistance positions were calculated using the coordinates from the X-ray crystallographic structure (PDB accession number 3HVT) described previously by Kohlstaedt et al. (23).
RESULTS
HIV-1 sequences by individual, ARV class, and subtype.
There were 32,530 HIV-1 group M protease sequences from 26,888 individuals, including 18,787 sequences (58%) from 17,581 PI-naïve individuals and 13,743 sequences (42%) from 9,307 PI-treated individuals. Eighty-six percent of individuals had a single sequence, 9% had two sequences, and 5% had three or more sequences. Individuals with non-B viruses accounted for 47% of PI-naïve and 12% of PI-treated sequences.
There were 32,151 HIV-1 group M RT sequences from 25,695 individuals, including 12,930 sequences (40%) from 12,730 RTI-naïve individuals and 19,221 sequences (60%) from 12,965 RTI-treated individuals. The RTI-treated individuals included 4,598 individuals who had received NRTIs but not NNRTIs, 7,312 who had received NRTIs and NNRTIs, and 1,055 who had received a single dose of NVP. Eighty-four percent of individuals had a single sequence, 10% had two sequences, and 6% had three or more sequences. Individuals with non-subtype B RT sequences accounted for 50% of RTI-naïve and 21% of RTI-treated sequences. Every RT sequence encompassed most amino acids between positions 1 and 250, 50% of the sequences also encompassed positions 250 to 300, and 28% also encompassed positions 300 to 350.
Twenty-three percent of sequences were obtained prior to 1999, 38% were obtained between 1999 and 2002, and 39% were obtained between 2003 and 2008. Plasma HIV-1 RNA and peripheral blood mononuclear cell DNA were the sources of genetic material for 93% and 7% of sequences, respectively. Approximately 88% of sequences were from previously published studies in the HIVDB; 12% of sequences were previously unpublished sequences from individuals in California.
PI-selected mutations.
Table 1 shows 88 nonpolymorphic PI-selected mutations at 40 protease positions that were more than five times more prevalent in PI-treated than in PI-naïve individuals. This number represents an increase of 47% compared with the 60 mutations at 34 positions identified in our 2005 study. Compared with the 2005 analysis of 8,426 sequences from 5,867 individuals, seven mutations (V11I, K20I/V, L33F, E35G, and T74A/S) were not considered to be PI-selected mutations because they were polymorphic in one or more non-B subtypes. The current analysis, however, identified 35 additional nonpolymorphic PI-selected mutations.
TABLE 1.
Prevalences of PI-selected nonpolymorphic mutations in PI-naïve and PI-treated individualsa
| Consensus B reference amino acid | Position | Amino acid change | Mutation prevalence (%) |
Presence of mutation on expert list fromb: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| PI naïve (n = 17,581) | PI treated (%) (n = 9,307) | H | A | R | I | L | |||
| L | 10 | F | 0.24 | 12 | √ | √ | √ | √ | √ |
| R | 0.02 | 0.65 | √ | √ | √ | √ | |||
| Y | 0 | 0.42 | √ | √ | |||||
| V | 11 | L | 0.05 | 1.1 | √ | ||||
| I | 13 | M | 0.01 | 0.69 | |||||
| K | 20 | T | 0.06 | 5.4 | √ | √ | √ | √ | √ |
| A | 22 | V | 0.11 | 1.3 | √ | ||||
| L | 23 | I | 0.05 | 1.8 | √ | √ | √ | ||
| L | 24 | I | 0.01 | 8.1 | √ | √ | √ | √ | √ |
| F | 0.08 | 0.92 | √ | √ | |||||
| M | 0 | 0.24 | |||||||
| D | 30 | N | 0.01 | 8.8 | √ | √ | √ | √ | √ |
| V | 32 | I | 0.01 | 7.5 | √ | √ | √ | √ | √ |
| L | 33 | M | 0 | 0.26 | √ | ||||
| E | 34 | Q | 0.09 | 4.4 | √ | √ | |||
| D | 0.05 | 0.49 | |||||||
| N | 0 | 0.14 | |||||||
| L | 38 | W | 0.01 | 0.36 | √ | ||||
| K | 43 | T | 0.12 | 7.6 | √ | √ | √ | √ | |
| I | 0.02 | 0.33 | |||||||
| K | 45 | I | 0.01 | 0.18 | √ | √ | |||
| V | 0 | 0.15 | |||||||
| M | 46 | I | 0.30 | 30 | √ | √ | √ | √ | √ |
| L | 0.27 | 13 | √ | √ | √ | √ | √ | ||
| V | 0.06 | 0.68 | √ | ||||||
| I | 47 | V | 0.05 | 7.2 | √ | √ | √ | √ | √ |
| A | 0 | 0.45 | √ | √ | √ | √ | √ | ||
| G | 48 | V | 0 | 6.3 | √ | √ | √ | √ | √ |
| M | 0 | 0.82 | √ | √ | √ | ||||
| A | 0 | 0.52 | √ | √ | |||||
| L | 0 | 0.16 | |||||||
| Q | 0 | 0.13 | |||||||
| I | 50 | V | 0.02 | 2.8 | √ | √ | √ | √ | √ |
| L | 0.01 | 0.64 | √ | √ | √ | √ | √ | ||
| G | 51 | A | 0.01 | 0.38 | |||||
| F | 53 | L | 0.04 | 8.4 | √ | √ | √ | √ | √ |
| Y | 0.03 | 0.74 | √ | √ | √ | √ | |||
| I | 54 | V | 0 | 31 | √ | √ | √ | √ | √ |
| L | 0.01 | 4.7 | √ | √ | √ | √ | √ | ||
| M | 0 | 3.9 | √ | √ | √ | √ | √ | ||
| A | 0 | 1.8 | √ | √ | √ | √ | √ | ||
| T | 0.02 | 1.5 | √ | √ | √ | √ | √ | ||
| S | 0 | 1.2 | √ | √ | √ | √ | √ | ||
| K | 55 | R | 0.25 | 9.6 | √ | ||||
| N | 0.03 | 0.3 | |||||||
| Q | 58 | E | 0.24 | 8.8 | √ | √ | √ | √ | √ |
| I | 66 | F | 0.03 | 2.3 | √ | ||||
| V | 0.08 | 1.8 | |||||||
| L | 0.01 | 0.64 | |||||||
| C | 67 | F | 0.03 | 1.8 | |||||
| L | 0.01 | 0.24 | |||||||
| A | 71 | I | 0.07 | 4.6 | √ | √ | √ | √ | √ |
| L | 0 | 0.64 | √ | √ | √ | √ | |||
| G | 73 | S | 0.03 | 13 | √ | √ | √ | √ | √ |
| T | 0 | 4.1 | √ | √ | √ | √ | |||
| C | 0.01 | 2.0 | √ | √ | √ | √ | |||
| A | 0 | 1.1 | √ | √ | √ | √ | |||
| V | 0.01 | 0.24 | √ | ||||||
| T | 74 | P | 0.06 | 2.6 | √ | √ | √ | √ | |
| K | 0.04 | 0.4 | |||||||
| E | 0 | 0.19 | √ | ||||||
| L | 76 | V | 0.01 | 4.4 | √ | √ | √ | √ | √ |
| P | 79 | A | 0.12 | 1.4 | |||||
| V | 82 | A | 0.03 | 29 | √ | √ | √ | √ | √ |
| T | 0.01 | 4.3 | √ | √ | √ | √ | √ | ||
| F | 0 | 2.4 | √ | √ | √ | √ | √ | ||
| S | 0.01 | 1.8 | √ | √ | √ | √ | √ | ||
| C | 0 | 0.93 | √ | √ | √ | ||||
| L | 0.02 | 0.35 | √ | √ | √ | √ | |||
| M | 0 | 0.28 | √ | √ | √ | √ | |||
| N | 83 | D | 0.02 | 1.0 | √ | √ | √ | ||
| S | 0.02 | 0.32 | |||||||
| I | 84 | V | 0.01 | 21 | √ | √ | √ | √ | √ |
| A | 0 | 0.27 | √ | √ | √ | √ | |||
| C | 0 | 0.24 | √ | √ | |||||
| I | 85 | V | 0.12 | 6.9 | √ | √ | √ | √ | |
| N | 88 | D | 0.05 | 6.8 | √ | √ | √ | √ | √ |
| S | 0.05 | 2.1 | √ | √ | √ | √ | √ | ||
| T | 0.01 | 0.20 | √ | ||||||
| G | 0 | 0.19 | √ | √ | |||||
| L | 89 | V | 0.10 | 5.9 | √ | √ | √ | √ | |
| T | 0.01 | 0.36 | √ | √ | √ | ||||
| L | 90 | M | 0.23 | 43 | √ | √ | √ | √ | √ |
| T | 91 | S | 0.01 | 2.5 | √ | ||||
| Q | 92 | R | 0.13 | 1.3 | |||||
| C | 95 | F | 0.02 | 2.6 | √ | √ | |||
| L | 0.01 | 0.20 | |||||||
| V | 0.01 | 0.20 | |||||||
Nonpolymorphic PI-selected mutations identified in the current analysis that were not identified in our 2005 analysis (26) are shown in boldface type.
A, ANRS; H, HIVDB; I, IAS-USA; L, Los Alamos National Laboratory; R, Rega. A checkmark indicates that the mutation is present on the expert list indicated.
Among the 35 new nonpolymorphic PI-selected mutations, 12 were at nonpolymorphic PI resistance positions, including L24F/M, G48A/L/Q, G73V, V82C/M/L, N83D/S, and N88G; 10 were at established polymorphic PI resistance positions, including L10Y, V11L, I13M, L33M, K43I, A71L, T74E/K, L89T, and T91S; and 13 were at positions not generally considered to be associated with PI resistance, including A22V, E34D/N, L38W, K45I/V, G51A, K55N, I66V/L, C67L, and C95V/L.
Of the 88 PI-selected mutations, 35%, 13%, 7%, 9%, 12%, and 24% were present on five, four, three, two, one, and none of the five expert mutation lists, respectively. Of the 35 new mutations, 15 were on one or more expert lists, including four mutations that were previously reported to reduce susceptibility or to be selected by one or more PIs: V82L (2, 10), V82M (5), N83D (2), and T91S (6, 10, 21). In other studies, A22V (30), L24M (2), and E34D (2) were significantly associated with PI exposure.
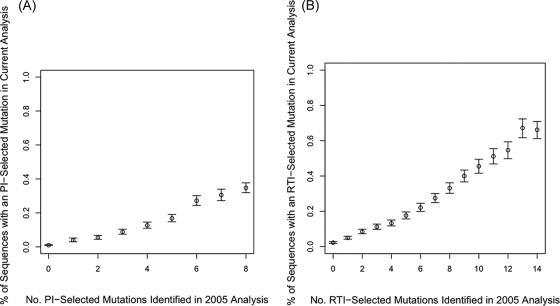
The proportion of sequences with a new PI-selected mutation was highly correlated with the number of previously published nonpolymorphic PI-selected mutations (Fig. 1A). Some of the new PI-selected mutations rarely occurred in viruses containing less than three previously published PI-selected mutations (e.g., G48A/L/Q, A71L, V82C, and T91S); other new PI-selected mutations occurred frequently in viruses containing less than three previously published PI-selected mutations (e.g., V82L and L89T). Although only 12% of the PI-treated viruses belonged to a non-B subtype, for four mutations, V82M, N83D, N88G, and L89T, non-subtype B isolates contributed more than 33% of the total number of new PI-selected mutations.
FIG. 1.
Correlation between the number of previously identified treatment-selected mutations (26) and the likelihood of having a new treatment-selected mutation. (A) PI-selected mutations. (B) RTI-selected mutations. Vertical bars are 95% confidence intervals.
RTI-selected mutations.
There were 122 nonpolymorphic RTI-selected mutations at 63 positions that occurred more than five times more frequently in RTI-treated than RTI-naive individuals. This represented an increase of 77% compared with the 69 RTI-selected mutations identified in a similar analysis that we reported in 2005. Compared with the 2005 analysis, seven RT mutations were not considered to be nonpolymorphic RTI-selected mutations because they were polymorphic in one or more subtypes: K43E, E44D, A62V, T69S/N, V75I, and V108I. Therefore, the current analysis identified 60 additional nonpolymorphic RTI-associated mutations.
NRTI-selected mutations.
Table 2 shows 67 nonpolymorphic NRTI-selected mutations at 30 positions. Compared with the 2005 analysis, we identified 31 new nonpolymorphic NRTI-selected mutations, including 19 mutations at 7 established NRTI resistance positions (D67T/S/H/del, T69E/G, K70E/G/S/T/N/Q, V75A/S, H208F, T215C, and K219W/H/D) and 12 mutations at 10 new positions (I31L, E40F, K64H/N/Y, T165L, I167V, E203V, R211D, K223E, A304G, and N348I).
TABLE 2.
Prevalences of NRTI-selected nonpolymorphic mutations in RTI-naïve, NRTI-treated (NNRTI-naïve), and NNRTI-treated individualsa
| Consensus B reference amino acid | Position | Amino acid change | Mutation prevalence (%) |
Selection indexc |
NNRTI index/ NRTI index ratio | Presence of mutation on expert list fromd: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RTI naïve (n = 12,730) | NRTI (n = 4,598) | NNRTI (n = 8,367)b | NRTI | NNRTI | H | A | R | I | L | ||||
| I | 31 | L | 0.1 | 0.8 | 1.5 | 7.1 | 1.8 | 0.3 | |||||
| E | 40 | F | 0 | 0.4 | 1.8 | 59 | 4.3 | 0.1 | |||||
| M | 41 | L | 0.3 | 36 | 47 | 120 | 1.3 | 0.0 | √ | √ | √ | √ | √ |
| K | 43 | Q | 0.3 | 3.4 | 5.9 | 11 | 1.7 | 0.2 | √ | ||||
| N | 0.2 | 1.8 | 3.3 | 8.6 | 1.8 | 0.2 | |||||||
| E | 44 | A | 0 | 1.1 | 3.5 | 163 | 3.0 | 0.0 | √ | √ | √ | √ | |
| K | 64 | N | 0.04 | 1.1 | 0.7 | 23 | 0.6 | 0.0 | |||||
| H | 0 | 0.8 | 1.0 | 119 | 1.1 | 0.0 | |||||||
| Y | 0 | 0.5 | 0.2 | 67 | 0.4 | 0.0 | |||||||
| K | 65 | R | 0.04 | 2.5 | 4.2 | 50 | 1.7 | 0.0 | √ | √ | √ | √ | √ |
| D | 67 | N | 0.01 | 30 | 41 | 2105 | 1.4 | 0.0 | √ | √ | √ | √ | √ |
| G | 0.10 | 1.3 | 3.8 | 13 | 2.8 | 0.2 | √ | √ | √ | √ | |||
| E | 0.01 | 0.6 | 0.9 | 31 | 1.4 | 0.0 | √ | √ | √ | ||||
| S | 0 | 0.2 | 0.7 | 27 | 3.5 | 0.1 | √ | √ | |||||
| T | 0 | 0.07 | 0.1 | 13 | 1.8 | 0.1 | √ | ||||||
| H | 0 | 0.04 | 0.4 | 9.0 | 5.9 | 0.7 | √ | ||||||
| Del | 0 | 0 | 0.2 | 3.0 | 9.5 | 3.2 | √ | √ | √ | ||||
| T | 69 | D | 0.05 | 5.8 | 12 | 102 | 2.0 | 0.0 | √ | √ | √ | √ | |
| Ins | 0 | 0.5 | 2.1 | 66 | 4.5 | 0.1 | √ | √ | √ | √ | √ | ||
| G | 0.01 | 0.09 | 0.7 | 7.5 | 6.9 | 0.9 | √ | √ | √ | ||||
| E | 0 | 0.06 | 0.4 | 12.0 | 4.7 | 0.4 | √ | ||||||
| K | 70 | R | 0.13 | 26 | 24 | 193 | 0.9 | 0.0 | √ | √ | √ | √ | √ |
| E | 0.01 | 0.4 | 0.6 | 27 | 1.5 | 0.1 | √ | √ | √ | √ | √ | ||
| G | 0 | 0.3 | 0.7 | 39 | 2.7 | 0.1 | √ | √ | |||||
| S | 0 | 0.1 | 0.2 | 21 | 1.5 | 0.1 | √ | √ | |||||
| T | 0.04 | 0.1 | 0.5 | 2.5 | 5.0 | 2.0 | √ | ||||||
| N | 0.04 | 0.2 | 0.4 | 4.5 | 2.1 | 0.5 | √ | ||||||
| Q | 0.01 | 0.1 | 0.3 | 7.5 | 3.1 | 0.4 | |||||||
| L | 74 | V | 0.01 | 3.9 | 17 | 185.8 | 4.3 | 0.0 | √ | √ | √ | √ | √ |
| I | 0.02 | 1.0 | 8.3 | 36.8 | 7.9 | 0.2 | √ | √ | √ | √ | |||
| V | 75 | T | 0 | 1.3 | 2.5 | 180.6 | 2.0 | 0.0 | √ | √ | √ | √ | |
| M | 0.04 | 1.2 | 5.3 | 29.1 | 4.3 | 0.1 | √ | √ | √ | √ | |||
| A | 0.02 | 0.61 | 1.3 | 22.2 | 2.1 | 0.1 | √ | √ | √ | ||||
| S | 0 | 0.20 | 0.50 | 30.6 | 2.3 | 0.1 | √ | √ | √ | ||||
| F | 77 | L | 0.04 | 1.8 | 2.7 | 42.3 | 1.5 | 0.0 | √ | √ | √ | ||
| Y | 115 | F | 0 | 1.1 | 2.9 | 158.2 | 2.6 | 0.0 | √ | √ | √ | √ | √ |
| F | 116 | Y | 0.01 | 1.9 | 2.9 | 136.4 | 1.5 | 0.0 | √ | √ | √ | ||
| Q | 151 | M | 0 | 2.4 | 3.9 | 345.5 | 1.6 | 0.0 | √ | √ | √ | √ | √ |
| T | 165 | L | 0.03 | 0.29 | 0.54 | 8.6 | 1.8 | 0.2 | |||||
| I | 167 | V | 0.04 | 0.42 | 1.1 | 8.9 | 2.5 | 0.3 | |||||
| M | 184 | V | 0.20 | 50 | 47 | 240.2 | 0.9 | 0.0 | √ | √ | √ | √ | √ |
| I | 0.05 | 1.1 | 2.7 | 19.8 | 2.4 | 0.1 | √ | √ | √ | √ | √ | ||
| E | 203 | K | 0.17 | 3.4 | 10 | 20.0 | 2.9 | 0.1 | √ | ||||
| V | 0.01 | 0.22 | 0.75 | 16.7 | 3.2 | 0.2 | |||||||
| H | 208 | Y | 0.21 | 5.3 | 17 | 24.4 | 3.1 | 0.1 | √ | ||||
| F | 0 | 0.24 | 0.73 | 36.8 | 2.8 | 0.1 | |||||||
| L | 210 | W | 0.02 | 21 | 34 | 717.0 | 1.7 | 0.0 | √ | √ | √ | √ | √ |
| R | 211 | D | 0.02 | 0.35 | 0.55 | 13.3 | 1.5 | 0.1 | |||||
| T | 215 | Y | 0.04 | 33 | 39 | 757.3 | 1.2 | 0.0 | √ | √ | √ | √ | √ |
| F | 0.01 | 9.0 | 13 | 418.4 | 1.5 | 0.0 | √ | √ | √ | √ | √ | ||
| I | 0.04 | 1.6 | 2.1 | 31.1 | 1.4 | 0.0 | √ | √ | √ | ||||
| V | 0.01 | 0.81 | 0.96 | 57.9 | 1.2 | 0.0 | √ | √ | √ | ||||
| C | 0.12 | 0.50 | 1.7 | 4.2 | 3.3 | 0.8 | √ | √ | √ | √ | |||
| D | 218 | E | 0.07 | 4.9 | 11 | 61.8 | 2.3 | 0.0 | √ | ||||
| K | 219 | Q | 0.10 | 16 | 15 | 144.3 | 1.0 | 0.0 | √ | √ | √ | √ | √ |
| E | 0.01 | 3.0 | 8.8 | 140.1 | 2.9 | 0.0 | √ | √ | √ | √ | √ | ||
| N | 0.09 | 1.0 | 7.2 | 11.4 | 6.7 | 0.6 | √ | √ | √ | √ | |||
| R | 0.09 | 1.3 | 5.6 | 13.4 | 4.2 | 0.3 | √ | √ | √ | √ | |||
| W | 0 | 0.11 | 0.46 | 18.1 | 3.6 | 0.2 | √ | √ | |||||
| D | 0 | 0.04 | 0.77 | 9.1 | 12.0 | 1.3 | |||||||
| H | 0 | 0.04 | 0.59 | 9.1 | 9.3 | 1.0 | √ | ||||||
| K | 223 | Q | 0 | 1.3 | 4.7 | 172 | 3.7 | 0.0 | |||||
| E | 0.04 | 0.57 | 4.0 | 13 | 6.8 | 0.5 | |||||||
| L | 228 | H | 0.24 | 5.4 | 17 | 22 | 3.1 | 0.1 | |||||
| R | 0.05 | 2.3 | 7.5 | 37 | 3.2 | 0.1 | |||||||
| A | 304 | G | 0.05 | 3.0 | 1.4 | 43 | 0.5 | 0.0 | |||||
| N | 348 | I | 0.18 | 5.5 | 9.7 | 26 | 1.7 | 0.1 | √ | ||||
Nonpolymorphic NNRTI-selected mutations identified in the current analysis that were not identified in our 2005 analysis (26) are shown in boldface type. The number of individuals for whom a sequence was available varies by position, with fewer individuals having sequences encompassing positions 1 to 40 and positions 240 to 350.
A total of 7,312 of 8,367 individuals also received NRTIs.
The NRTI selection index is the ratio of the prevalence of the mutation in NRTI-treated but NNRTI-naïve patients to the prevalence of the mutation in RTI-naïve patients. The NNRTI selection index is the ratio of the prevalence of the mutation in NNRTI-treated patients to the prevalence of the mutation in NRTI-treated patients.
A, ANRS; H, HIVDB; I, IAS-USA; L, Los Alamos National Laboratory; R, Rega. A checkmark indicates that the mutation is present on the expert list indicated.
Of the 67 NRTI-selected mutations, 24%, 14%, 13%, 6%, 16%, and 27% were on five, four, three, two, one, and none of the five expert mutation lists, respectively. Of the 30 new mutations, 11 were on one or more expert lists, including 6 mutations which were previously reported to reduce susceptibility or to be selected by one or more NRTIs: E40F (18), D67del (19), T69G (19), K70E/G (4, 9), and N348I (12, 36). In an independent study of 457 RTI-naïve and 1,247 RTI-experienced individuals, I31L, K64H/N/Y, T165L, E203V, R211D, and K223E were observed in five or more RTI-treated but in no RTI-naïve individuals (7). In addition, I31L (25), K64N (25), T165L (25), and K223E (28) were previously reported to possibly be associated with RTI therapy in other studies.
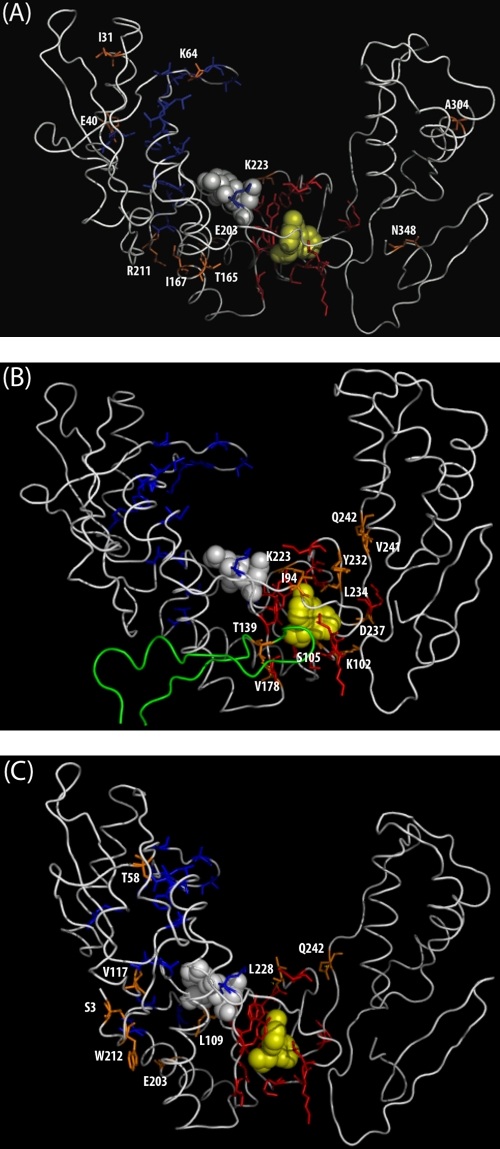
Figure 2A shows the locations of the 10 new NRTI positions within the three-dimensional structure of the polymerase coding region of p66 HIV-1 RT. I31, E40, and K64 are in the finger subdomain. T165, I167, E203, R211, and K223 are in the palm subdomain. A304 is in the thumb subdomain, and N348 is in the connection subdomain. Other than T165 (7 Å from Y181), K223 (3 Å from V108 and P225), and N348 (4 Å from L318), none of the new NRTI positions are within 10 Å of any established NNRTI resistance residue.
FIG. 2.
Three-dimensional structure (PDB accession number 3HVT) of HIV-1 RT bound to nevirapine showing residues 1 to 350 of the p66 monomer. The active-site residues D110, D185, and D186 are shown in space-fill mode in white. Nevirapine is shown in space-fill mode in yellow. Established NRTI resistance residues are shown in sticks mode in blue. Established NNRTI resistance residues are shown in sticks mode in red. Newly identified RTI-selected mutations are shown in sticks mode in orange. (A) Newly identified NRTI-selected mutations. (B) Newly identified NNRTI-selected mutations. Residues 120 to 148 in the p51 monomer are shown in green. (C) Newly identified undifferentiated mutations.
NNRTI-associated mutations.
Table 3 shows 46 nonpolymorphic NNRTI-selected mutations at 28 positions. Compared with the 2005 analysis, we identified 20 new nonpolymorphic NNRTI-associated mutations, including 9 mutations at known NNRTI-associated positions (K101H, K103H/T, V179F, G190C, H221C, F227Y, K238N, and Y318F) and 11 mutations at new positions (I94L, K102N, S105T, T139R, I178F, K223T, Y232H, L234I, D237E, V241M, and Q242L).
TABLE 3.
Prevalences of NNRTI-selected nonpolymorphic mutations in RTI-naïve, NRTI-treated (NNRTI-naïve), and NNRTI-treated individualsa
| Consensus B reference amino acid | Position | Amino acid change | Mutation prevalence (%) |
Selection indexc |
NNRTI index/NRTI index ratio | Presence of mutation on expert list fromd: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RTI naïve (n = 12,730) | NRTI (n = 4,598) | NNRTI (n = 8,367)b | NRTI | NNRTI | H | A | R | I | L | ||||
| I | 94 | L | 0.02 | 0.07 | 0.9 | 2.8 | 14 | 5 | |||||
| A | 98 | G | 0.20 | 2.1 | 8.5 | 10 | 4.1 | 0.4 | √ | √ | √ | √ | √ |
| L | 100 | I | 0.01 | 0.02 | 6.6 | 1.0 | 303 | 303 | √ | √ | √ | √ | √ |
| K | 101 | E | 0.1 | 0.4 | 8.5 | 3.8 | 19 | 5 | √ | √ | √ | √ | √ |
| P | 0 | 0 | 2.0 | 1.0 | 93 | 93 | √ | √ | √ | √ | √ | ||
| H | 0 | 0.09 | 1.9 | 4.0 | 21 | 5 | √ | ||||||
| N | 0.02 | 0.07 | 0.7 | 2.8 | 11 | 4 | √ | √ | |||||
| K | 102 | N | 0.02 | 0.02 | 0.5 | 1.5 | 12 | 8 | |||||
| K | 103 | N | 0.4 | 0.4 | 43 | 1.0 | 121 | 121 | √ | √ | √ | √ | √ |
| S | 0.01 | 0.02 | 2.2 | 1.0 | 49 | 49 | √ | √ | √ | √ | |||
| H | 0 | 0 | 0.2 | 1.0 | 8.7 | 9 | √ | √ | √ | √ | |||
| T | 0.02 | 0.1 | 0.3 | 4.1 | 3.2 | 0.8 | √ | √ | √ | √ | |||
| S | 105 | T | 0 | 0.04 | 0.6 | 1.0 | 15 | 15 | |||||
| V | 106 | A | 0.01 | 0.02 | 2.7 | 1.0 | 124 | 124 | √ | √ | √ | √ | √ |
| M | 0 | 0 | 2.2 | 1.0 | 101 | 101 | √ | √ | √ | √ | √ | ||
| E | 138 | Q | 0.01 | 0.1 | 1.0 | 6.2 | 7.6 | 1 | |||||
| T | 139 | R | 0.06 | 0.1 | 0.9 | 1.7 | 9.0 | 5 | |||||
| I | 178 | F | 0 | 0.04 | 0.5 | 1.0 | 10 | 10 | |||||
| V | 179 | F | 0 | 0 | 0.5 | 1.0 | 22 | 22 | √ | √ | √ | √ | √ |
| Y | 181 | C | 0.08 | 0.3 | 27 | 3.6 | 90 | 25 | √ | √ | √ | √ | √ |
| I | 0.01 | 0 | 1.2 | 1.0 | 56 | 56 | √ | √ | √ | √ | √ | ||
| V | 0 | 0 | 0.7 | 1.0 | 30 | 30 | √ | √ | √ | √ | √ | ||
| Y | 188 | L | 0.07 | 0.09 | 6.5 | 1.4 | 59 | 42 | √ | √ | √ | √ | √ |
| C | 0.02 | 0.02 | 1.3 | 1.0 | 30 | 30 | √ | √ | √ | √ | √ | ||
| H | 0.01 | 0.02 | 1.1 | 1.0 | 25 | 25 | √ | √ | √ | √ | √ | ||
| G | 190 | A | 0.04 | 0.09 | 20 | 2.2 | 180 | 81 | √ | √ | √ | √ | √ |
| S | 0 | 0 | 3.5 | 1.0 | 350 | 350 | √ | √ | √ | √ | √ | ||
| E | 0.01 | 0 | 0.6 | 1.0 | 33 | 33 | √ | √ | √ | √ | |||
| Q | 0 | 0 | 0.3 | 1.0 | 16 | 16 | √ | √ | √ | √ | |||
| C | 0 | 0 | 0.2 | 1.0 | 13 | 13 | √ | √ | √ | √ | |||
| H | 221 | Y | 0.2 | 0.7 | 8.6 | 4.4 | 12 | 3 | √ | ||||
| C | 0 | 0 | 0.3 | 1.0 | 12 | 12 | |||||||
| K | 223 | T | 0.02 | 0.07 | 1.1 | 2.8 | 16 | 6 | |||||
| P | 225 | H | 0.01 | 0.02 | 3.7 | 1.0 | 80 | 80 | √ | √ | √ | √ | √ |
| F | 227 | L | 0.05 | 0.2 | 3.3 | 3.5 | 20 | 6 | √ | √ | √ | ||
| Y | 0 | 0 | 0.3 | 1.0 | 14 | 14 | √ | ||||||
| M | 230 | L | 0.02 | 0.1 | 1.3 | 5.4 | 9.9 | 2 | √ | √ | √ | ||
| Y | 232 | H | 0.02 | 0 | 0.3 | 1.0 | 20 | 20 | |||||
| L | 234 | I | 0.01 | 0 | 0.3 | 1.0 | 21 | 21 | √ | √ | |||
| P | 236 | L | 0.03 | 0.1 | 0.3 | 3.3 | 3.1 | 0.9 | √ | √ | √ | ||
| D | 237 | E | 0.1 | 0.3 | 1.6 | 2.9 | 4.9 | 2 | |||||
| K | 238 | T | 0.05 | 0.2 | 3.4 | 3.2 | 18 | 6 | √ | √ | √ | ||
| N | 0.03 | 0.08 | 0.7 | 2.5 | 6.4 | 3 | √ | ||||||
| V | 241 | M | 0 | 0 | 0.3 | 1.0 | 20 | 20 | |||||
| Q | 242 | L | 0.02 | 0.02 | 0.5 | 1.0 | 12 | 12 | |||||
| Y | 318 | F | 0.1 | 0.1 | 1.8 | 1.4 | 9.4 | 7 | √ | √ | √ | ||
Nonpolymorphic NNRTI-selected mutations identified in the current analysis that were not identified in our 2005 analysis (26) are shown in boldface type.
A total of 7,312 of 8,367 individuals also received NRTIs.
The NRTI selection index is the ratio of the prevalence of the mutation in NRTI-treated but NNRTI-naïve patients to the prevalence of the mutation in RTI-naïve patients. The NNRTI selection index is the ratio of the prevalence of the mutation in NNRTI-treated patients to the prevalence of the mutation in NRTI-treated patients.
A, ANRS; H, HIVDB; I, IAS-USA; L, Los Alamos National Laboratory; R, Rega. A checkmark indicates that the mutation is present on the expert list indicated.
Of the 46 NNRTI-selected mutations, 37%, 11%, 13%, 4%, 9%, and 26% were on five, four, three, two, one, and none of the five expert mutation lists, respectively. Of the 20 new mutations, 7 were on one or more of the expert mutation lists, including 6 mutations which were previously reported to reduce susceptibility or to be selected by one or more NNRTIs: K101H (25, 34), K103H (13), V179F (33), G190C (17), K238N (31), and Y318F (14). In addition, L234I was reported to emerge during etravirine selection pressure in vitro (33), I94L and K102N were observed in five or more RTI-treated compared with no untreated individuals, and T139R was observed in 14 RTI-treated compared with 2 untreated individuals in a study reported previously by Ceccherini-Silberstein et al. (7).
Figure 2B shows that the new NNRTI resistance positions are either close to the NNRTI binding pocket (NVP) or close to one of the established NNRTI resistance positions: I94 (8 Å from NVP and 5 Å from Y181), K102 (4 Å from NVP and 1.3 Å from K101 and K103), S105 (6 Å from NVP and 1.3 Å from V106), T139 in the p51 monomer (8 Å from NVP and 5 Å from Y181), I78 (7 Å from NVP and 1.3 Å from V179), K223 (9 Å from NVP and 3 Å from V108 and P225), Y232 (6 Å from NVP and 3 Å from F227 and M230), L234 (3 Å from NVP and 4 Å from K238), D237 (6 Å from NVP and 1.3 Å from P236 and K238), V241 (12 Å from NVP and 7 Å from M230), and Q242 (13 Å from NVP and 7 Å from F227, M230, and K238) are adjacent to the NNRTI resistance residues that comprise the NNRTI binding pocket.
Mutations possibly associated with NRTI and NNRTI therapy.
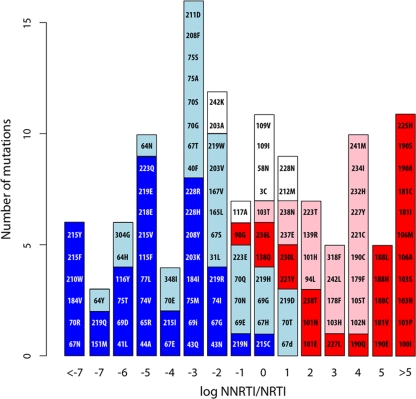
There were nine undifferentiated RTI-selected mutations: S3C, T58N, L109I/V, V117A, E203A, W212M, L228N, and Q242K. None of these nine mutations were present on any of the expert lists. However, S3C, L109I, and E203A were previously reported for five or more RTI-treated but no untreated individuals in the study reported previously by Ceccherini-Silberstein et al. (7). Figure 3 illustrates the ratios of the NNRTI selection index divided by the NRTI selection index for each of the 122 RTI-selected mutations. Mutations are colored according to whether they meet the criteria for being a newly identified or previously reported NRTI- or NNRTI-selected mutation or an undifferentiated RTI-selected mutation.
FIG. 3.
Ratios of the NRTI-selected index divided by the NNRTI-selected index for all 122 RTI-selected mutations. Blue indicates NRTI-selected mutations identified in the 2005 analysis (26). Light blue indicates newly identified NRTI-selected mutations. Red indicates NNRTI-selected mutations identified in the previous analysis. Pink indicates newly identified NNRTI-selected mutations. White indicates undifferentiated mutations. The x axis shows the log (base 2) of the ratio of the NRTI selection index divided by the NNRTI selection index. The y axis indicates the number of mutations having a specific NRTI-NNRTI selection ratio. 69i indicates insertions at codon 69, and 67d indicates deletions at codon 67.
Figure 2C shows that for the mutations S3C, T58N, V117A, K203A, and W212M, the shortest interatomic distance to an established NRTI resistance residue was much smaller than the distance to an established NNRTI resistance residue. In contrast, for the mutations L228 and Q242, the shortest interatomic distance to the established NNRTI resistance mutation was much smaller than the shortest distances to each of the established NRTI resistance residues. Residue L109 is adjacent to the active-site aspartate D110 and to NNRTI resistance position V108 and is 7 Å from M184. None of the undifferentiated mutations occurred at positions in the RT p51 monomer close to the NNRTI binding pocket.
The median number of studies reporting each of the nine undifferentiated mutations was 7 (range, 5 to 14), and the median nucleotide distance among sequences with each of the mutations was 8.4% (range, 7.3% to 11.1%). The fact that these mutations were reported in several studies and were present in highly divergent isolates excludes the possibility that their association with treatment was a result of a founder effect.
DISCUSSION
We analyzed >30,000 RT and >30,000 protease sequences from >25,000 HIV-1-infected individuals and identified 122 nonpolymorphic RT mutations that occurred more than five times more frequently in RTI-treated than in RTI-naïve individuals and 88 nonpolymorphic protease mutations that occurred more than five times more frequently in PI-treated than in PI-naïve individuals. The 122 RTI- and the 88 PI-selected nonpolymorphic mutations represented increases of 77% and 47%, respectively, compared with the 69 RTI- and 60 PI-selected mutations identified in a similar analysis that we previously reported in 2005 (26).
In the 2005 study, treatment-selected mutations were defined as occurring more than two times more frequently in ARV-experienced than in ARV-naïve individuals. However, in the 2005 study, all but three of the 129 treatment-selected mutations occurred more than five times more commonly among treated individuals. The control for multiple-hypothesis testing used in this study and in the 2005 study made it unlikely that nonpolymorphic mutations with a less-than-fivefold-increased prevalence in treated patients would be found to be significantly associated with ARV therapy. Therefore, in the present study, we defined treatment-selected mutations as those occurring more than five times more commonly among treatment-experienced than among treatment-naïve individuals.
In the 2005 study, we analyzed only subtype B sequences and defined nonpolymorphic mutations as those occurring in ≤0.5% of untreated individuals infected with subtype B viruses. In the current study, we analyzed all group M subtypes and defined nonpolymorphic mutations as those occurring in >0.5% of treatment-naïve individuals in no more than two subtypes or in a single subtype with >1,000 published naïve sequences. The definition for nonpolymorphism in the current study was similar to that used in a recently reported study that developed a list of 93 nonpolymorphic drug resistance mutations deemed suitable for identifying transmitted drug resistance (surveillance drug resistance mutations [SDRMs]) (3).
There are two differences, however, between the 210 treatment-selected mutations in this study compared with the 93 SDRMs. The SDRM list included only those nonpolymorphic mutations that were present on three or more expert mutation lists, whereas this study included all nonpolymorphic treatment-selected mutations regardless of the number of expert lists in which a mutation appeared. In addition, the SDRM list excluded nonpolymorphic mutations that occurred at positions that commonly contained polymorphisms. For example, the nonpolymorphic protease-selected mutations L10F/R/Y and A71I/L were excluded from the SDRM list because L10I/V and A71V/T are common polymorphisms. All but three SDRMs (the T215 revertant mutations T215S/D/E) were among the 210 treatment-selected mutations in this study.
This study contained non-subtype B RT sequences from about 6,500 RTI-naïve and 4,500 RTI-experienced individuals and from about 8,000 PI-naïve and 1,500 PI-experienced individuals. The non-subtype B data influenced the list of nonpolymorphic treatment-selected mutations in two ways. First, several mutations, including the PI-resistance-associated mutation L33F and the RTI-resistance-associated mutations E44D, V75I, and V108I in RT, were not classified as treatment-selected mutations because they had a prevalence of >0.5% in one or more subtypes. Second, non-subtype B sequences contributed to the statistical significance of several treatment-selected mutations. As the number of non-subtype B sequences from untreated individuals increases, it is likely that non-subtype B viruses will further influence the spectrum of treatment-selected mutations.
The vast majority of new treatment-selected mutations occurred in the presence of previously described treatment-selected mutations, suggesting that the increase in the number of new treatment-selected mutations since 2005 is a consequence of the increased number of sequences in the current data set, particularly from heavily treated individuals. Although two new PIs (tipranavir and darunavir) and one new RTI (the NNRTI etravirine) were licensed between the 2005 study and the current study, fewer than 300 individuals in this study received one of these new ARVs.
For several of the uncommon newly identified RTI-selected mutations, it was not possible to determine whether the mutation was selected by NRTIs, NNRTIs, or both classes of RT inhibitors. Moreover, certain mutations at positions 203, 223, and 228 have been strongly associated with NRTI therapy (E203KV, K223QE, and L228HR), whereas others at the same position have been strongly associated with NNRTI therapy (K223T) or have been undifferentiated (E203A and L228N). Likewise, Q242L was strongly associated with NNRTI therapy, whereas Q242K was undifferentiated. Of note, the distances of these residues from known NRTI and NNRTI resistance positions did not conclusively support an association with either drug class. The concept that one mutation might contribute to both NRTI resistance and NNRTI resistance is not new. The NRTI resistance mutations L74V/I and V75I were reported to occasionally emerge during NNRTI selection pressure (22, 35), and the RT mutation N348I was reported to be associated with decreased susceptibilities to both zidovudine and nevirapine (12, 36).
Several of the newly identified treatment-selected mutations have already become established drug resistance mutations, whereas others have been noted in one or more independent data sets. The phenotypic and/or clinical significance of the remaining novel mutations has not been studied, which explains why these mutations are often not present on existing expert mutation lists developed for genotypic resistance interpretation or for educational purposes. Several of these mutations, however, may be of clinical significance and should be considered for inclusion on current genotypic resistance reports because they occur at positions at which other mutations markedly reduce susceptibility to one or more drugs. Examples of these types of mutations include G48A/L/Q and V82C/M in protease and K70S/T/N/Q, V75A/S, K101H, K103H, G190C, F227Y, L234I, and K238N in RT. The effect of most of these mutations on drug susceptibility is not known, but they are likely to be associated with a decreased susceptibility to one or more ARVs in the ARV class responsible for their selection.
Acknowledgments
R.S., S.Y.R., T.L., and R.W.S. were supported by NIAID grant AI068581. S.H. acknowledges support from NIH grant R01GM086884-2.
Footnotes
Published ahead of print on 31 August 2009.
REFERENCES
- 1.Agence Nationale de Recherche sur le SIDA. 2007. ANRS genotypic resistance guidelines (version 13). ANRS, Paris, France. http://www.hivfrenchresistance.org/2007.
- 2.Baxter, J. D., J. M. Schapiro, C. A. Boucher, V. M. Kohlbrenner, D. B. Hall, J. R. Scherer, and D. L. Mayers. 2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80:10794-10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, D. E., R. J. Camacho, D. Otelea, D. R. Kuritzkes, H. Fleury, M. Kiuchi, W. Heneine, R. Kantor, M. R. Jordan, J. M. Schapiro, A. M. Vandamme, P. Sandstrom, C. A. Boucher, D. van de Vijver, S. Y. Rhee, T. F. Liu, D. Pillay, and R. W. Shafer. 2009. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS ONE 4:e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradshaw, D., S. Malik, C. Booth, M. Van Houtte, T. Pattery, A. Waters, J. Ainsworth, and A. M. Geretti. 2007. Novel drug resistance pattern associated with the mutations K70G and M184V in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 51:4489-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camacho, R., A. Godinho, P. Gomes, A. Abecasis, A.-M. Vandamme, C. Palma, A. P. Carvalho, J. Cabanas, and J. Goncalves. 2005. Different substitutions under drug pressure at protease codon 82 in HIV-1 subtype G compared to subtype B infected individuals including a novel I82M resistance mutations. Antivir. Ther. 10:S151. [Google Scholar]
- 6.Carrillo, A., K. D. Stewart, H. L. Sham, D. W. Norbeck, W. E. Kohlbrenner, J. M. Leonard, D. J. Kempf, and A. Molla. 1998. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 72:7532-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceccherini-Silberstein, F., F. Gago, M. Santoro, C. Gori, V. Svicher, F. Rodríguez-Barrios, R. d'Arrigo, M. Ciccozzi, A. Bertoli, A. d'Arminio Monforte, J. Balzarini, A. Antinori, and C.-F. Perno. 2005. High sequence conservation of human immunodeficiency virus type 1 reverse transcriptase under drug pressure despite the continuous appearance of mutations. J. Virol. 79:10718-10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, S., C. Calef, and J. Mellors. 2007. Mutations in retroviral genes associated with drug resistance. HIV Sequence Compendium, Los Alamos National Laboratory, Los Alamos, NM.
- 9.Delaugerre, C., L. Roudiere, G. Peytavin, C. Rouzioux, J. P. Viard, and M. L. Chaix. 2005. Selection of a rare resistance profile in an HIV-1-infected patient exhibiting a failure to an antiretroviral regimen including tenofovir DF. J. Clin. Virol. 32:241-244. [DOI] [PubMed] [Google Scholar]
- 10.Doyon, L., S. Tremblay, L. Bourgon, E. Wardrop, and M. G. Cordingley. 2005. Selection and characterization of HIV-1 showing reduced susceptibility to the non-peptidic protease inhibitor tipranavir. Antivir. Res. 68:27-35. [DOI] [PubMed] [Google Scholar]
- 11.Gifford, R. J., S. Y. Rhee, N. Eriksson, T. F. Liu, M. Kiuchi, A. K. Das, and R. W. Shafer. 2008. Sequence editing by apolipoprotein B RNA-editing catalytic component-B and epidemiological surveillance of transmitted HIV-1 drug resistance. AIDS 22:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hachiya, A., E. N. Kodama, S. G. Sarafianos, M. M. Schuckmann, Y. Sakagami, M. Matsuoka, M. Takiguchi, H. Gatanaga, and S. Oka. 2008. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 82:3261-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrigan, P. R., T. Mo, B. Wynhoven, J. Hirsch, Z. Brumme, P. McKenna, T. Pattery, J. Vingerhoets, and L. T. Bacheler. 2005. Rare mutations at codon 103 of HIV-1 reverse transcriptase can confer resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 19:549-554. [DOI] [PubMed] [Google Scholar]
- 14.Harrigan, P. R., M. Salim, D. K. Stammers, B. Wynhoven, Z. L. Brumme, P. McKenna, B. Larder, and S. D. Kemp. 2002. A mutation in the 3′ region of the human immunodeficiency virus type 1 reverse transcriptase (Y318F) associated with nonnucleoside reverse transcriptase inhibitor resistance. J. Virol. 76:6836-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm, S. 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6:65-70. [Google Scholar]
- 16.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 17.Huang, W., A. Gamarnik, K. Limoli, C. J. Petropoulos, and J. M. Whitcomb. 2003. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J. Virol. 77:1512-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huigen, M., P. Van Ham, L. de Graaf, C. Boucher, and M. Nijhuis. 2006. A novel and rare amino acid substitution E40F in HIV-1 reverse transcriptase increases zidovudine resistance and decreases replication capacity, abstr. 603. 13th Conf. Retrovir. Opportun. Infect., Denver, CO, 5 to 8 February 2006.
- 19.Imamichi, T., M. A. Murphy, H. Imamichi, and H. C. Lane. 2001. Amino acid deletion at codon 67 and Thr-to-Gly change at codon 69 of human immunodeficiency virus type 1 reverse transcriptase confer novel drug resistance profiles. J. Virol. 75:3988-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2008. Update of the drug resistance mutations in HIV-1. Top. HIV Med. 16:138-145. [PubMed] [Google Scholar]
- 21.Kagan, R. M., P. K. Cheung, T. K. Huard, and M. A. Lewinski. 2006. Increasing prevalence of HIV-1 protease inhibitor-associated mutations correlates with long-term non-suppressive protease inhibitor treatment. Antivir. Res. 71:42-52. [DOI] [PubMed] [Google Scholar]
- 22.Kleim, J. P., M. Rosner, I. Winkler, A. Paessens, R. Kirsch, Y. Hsiou, E. Arnold, and G. Riess. 1996. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-74→Val or Ile and Val-75→Leu or Ile) HIV-1 mutants. Proc. Natl. Acad. Sci. USA 93:34-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 24.Liu, T. F., and R. W. Shafer. 2006. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin. Infect. Dis. 42:1608-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkin, N. T., S. Gupta, C. Chappey, and C. J. Petropoulos. 2006. The K101P and K103R/V179D mutations in human immunodeficiency virus type 1 reverse transcriptase confer resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 50:351-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee, S. Y., W. J. Fessel, A. R. Zolopa, L. Hurley, T. Liu, J. Taylor, D. P. Nguyen, S. Slome, D. Klein, M. Horberg, J. Flamm, S. Follansbee, J. M. Schapiro, and R. W. Shafer. 2005. HIV-1 protease and reverse-transcriptase mutations: correlations with antiretroviral therapy in subtype B isolates and implications for drug-resistance surveillance. J. Infect. Dis. 192:456-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhee, S. Y., M. J. Gonzales, R. Kantor, B. J. Betts, J. Ravela, and R. W. Shafer. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saracino, A., L. Monno, L. Scudeller, D. C. Cibelli, A. Tartaglia, G. Punzi, C. Torti, S. Lo Caputo, F. Mazzotta, G. Scotto, G. Carosi, and G. Angarano. 2006. Impact of unreported HIV-1 reverse transcriptase mutations on phenotypic resistance to nucleoside and non-nucleoside inhibitors. J. Med. Virol. 78:9-17. [DOI] [PubMed] [Google Scholar]
- 29.Shafer, R. W., S. Y. Rhee, D. Pillay, V. Miller, P. Sandstrom, J. M. Schapiro, D. R. Kuritzkes, and D. Bennett. 2007. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS 21:215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svicher, V., F. Ceccherini-Silberstein, F. Erba, M. Santoro, C. Gori, M. C. Bellocchi, S. Giannella, M. P. Trotta, A. Monforte, A. Antinori, and C. F. Perno. 2005. Novel human immunodeficiency virus type 1 protease mutations potentially involved in resistance to protease inhibitors. Antimicrob. Agents Chemother. 49:2015-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tambuyzer, L., H. Azijn, L. T. Rimsky, J. Vingerhoets, P. Lecocq, G. Kraus, G. Picchio, and M. P. de Bethune. 2009. Compilation and prevalence of mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors. Antivir. Ther. 14:103-109. [PubMed] [Google Scholar]
- 32.Van Laethem, K., A. De Luca, A. Antinori, A. Cingolani, C. F. Perna, and A. M. Vandamme. 2002. A genotypic drug resistance interpretation algorithm that significantly predicts therapy response in HIV-1-infected patients. Antivir. Ther. 7:123-129. [PubMed] [Google Scholar]
- 33.Vingerhoets, J., H. Azijn, E. Fransen, I. De Baere, L. Smeulders, D. Jochmans, K. Andries, R. Pauwels, and M. P. de Bethune. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vingerhoets, J., M. Peeters, H. Azjin, L. Tambuyzer, A. Hoogstoel, S. Nijs, M. de Bethune, and G. Picchio. 2008. An update of the list of NNRTI mutations associated with decreased virological response to etravirine: multivariate analysis on the pooled DUET-1 and DUET-2 clinical trial data. Antivir. Ther. 13(Suppl. 3):A26. [Google Scholar]
- 35.Wirden, M., S. Lambert-Niclot, A. G. Marcelin, L. Schneider, H. Ait- Mohand, C. Brunet, F. Angleraud, S. Amard, C. Katlama, and V. Calvez. 2009. Antiretroviral combinations implicated in emergence of the L74I and L74V resistance mutations in HIV-1-infected patients. AIDS 23:95-99. [DOI] [PubMed] [Google Scholar]
- 36.Yap, S. H., C. W. Sheen, J. Fahey, M. Zanin, D. Tyssen, V. D. Lima, B. Wynhoven, M. Kuiper, N. Sluis-Cremer, P. R. Harrigan, and G. Tachedjian. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]