Abstract
The Pseudomonas aeruginosa PAO1 gene pvdQ encodes an acyl-homoserine lactone (AHL) acylase capable of degrading N-(3-oxododecanoyl)-l-homoserine lactone by cleaving the AHL amide. PvdQ has been proven to function as a quorum quencher in vitro in a number of phenotypic assays. To address the question of whether PvdQ also shows quorum-quenching properties in vivo, an infection model based on the nematode Caenorhabditis elegans was explored. In a fast-acting paralysis assay, strain PAO1(pMEpvdQ), which overproduces PvdQ, was shown to be less virulent than the wild-type strain. More than 75% of the nematodes exposed to PAO1(pMEpvdQ) survived and continued to grow when using this strain as a food source. Interestingly, in a slow-killing assay monitoring the survival of the nematodes throughout a 4-day course, strain PAO1-ΔpvdQ was shown to be more virulent than the wild-type strain, confirming the role of PvdQ as a virulence-reducing agent. It was observed that larval stage 1 (L1) to L3-stage larvae benefit much more from protection by PvdQ than L4 worms. Finally, purified PvdQ protein was added to C. elegans worms infected with wild-type PAO1, and this resulted in reduced pathogenicity and increased the life span of the nematodes. From our observations we can conclude that PvdQ might be a strong candidate for antibacterial therapy against Pseudomonas infections.
Pseudomonas aeruginosa is an opportunistic gram-negative pathogen of vertebrates and a primary pathogen of insects (17). It mainly infects individuals who are immunocompromised, such as human immunodeficiency virus-infected patients, as well as those who have cystic fibrosis. In addition, those having disruptions in normal barriers caused by severe burns or indwelling medical devices are at risk. Hospital-acquired P. aeruginosa pneumonias and septicemias are frequently lethal (2, 3). To facilitate the establishment of infection, P. aeruginosa produces an impressive array of both cell-associated and extracellular virulence factors, such as proteases and phospholipases, and also small molecules, including rhamnolipid, phenazines, and cyanide (17). Expression of many of the extracellular factors is cell density controlled, does not occur until the late logarithmic phase of growth, and is mediated through specific quorum-sensing signal molecules (23). Two of these molecules, N-butanoyl-l-homoserine lactone (C4-HSL) and N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL), have been studied in great detail. In our laboratory, we previously demonstrated that PA2385(pvdQ) from P. aeruginosa PAO1 encodes an acyl-homoserine lactone (AHL) acylase. Analysis of the gene product showed that the posttranslational processing of the acylase as well as the hydrolysis reaction type are similar to those of the beta-lactam acylases, strongly suggesting that the PvdQ protein is a member of the N-terminal nucleophile hydrolase superfamily. The main AHL signaling molecule of P. aeruginosa PAO1, 3-oxo-C12-HSL, is degraded by PvdQ (16). Addition of the purified protein to PAO1 cultures completely inhibited accumulation of 3-oxo-C12-HSL and production of the signal molecule 2-heptyl-3-hydroxy-4(1H)-quinolone and reduced production of the virulence factors elastase and pyocyanin. Similar results were obtained when pvdQ was overexpressed in P. aeruginosa (16). These results demonstrate that this protein has in situ quorum-quenching activity. This AHL acylase may enable P. aeruginosa PAO1 to modulate its own quorum-sensing-dependent pathogenic potential and, moreover, offers possibilities for novel antipseudomonal therapies.
To test our hypothesis that PvdQ can exert its beneficial functions also in vivo, we chose to study its effect on the infection of the nematode Caenorhabditis elegans. This model has been used before in multiple pathogenicity studies of Cryptococcus neoformans (13) and gram-positive (6) and gram-negative (9, 10, 11) bacteria. Infection with P. aeruginosa strain PA14 was found to result in fast (hours) or slow (days) killing, depending on the growth medium used (19, 20). When Darby and colleagues (2) used the system to study P. aeruginosa PAO1, a lethal paralysis of the worms was observed, indicating another mechanism by which P. aeruginosa can kill C. elegans. It was shown that quorum-sensing-dependent hydrogen cyanide production on rich medium by P. aeruginosa PAO1 is the causative agent for the fast paralysis (5). Under the same conditions, an attenuation of paralysis by an AHL acylase from Ralstonia sp. strain XJ12B upon expression in P. aeruginosa PAO1 was observed (12). Those authors performed the assays with a mixed population of worms only and did not test for slow killing. In this study we show that P. aeruginosa PAO1 can also elicit a slow-killing response when grown in low-nutrient medium. Moreover, we report that not only overexpression of the gene from its host but also external addition of the purified PvdQ renders P. aeruginosa PAO1 less pathogenic to C. elegans and increases the life span of the infected animals in both slow- and fast-killing assays. As the correlation between the bacterial virulence factors required for pathogenesis in mammals and in C. elegans has been shown to be very high (20), we propose that the external addition of purified AHL acylases may be developed into a novel quorum-quenching therapy.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The plasmids and bacterial strains used in this study are listed in Table 1. Bacterial cells were routinely grown at 37°C in either Luria-Bertani (LB) medium or brain-heart infusion (BHI) medium, as indicated for the assays, and on LB or BHI agar plates. Expression of PvdQ was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (6, 15, 16). P. aeruginosa competent cells were prepared as described previously (1). Plasmids were maintained by addition of tetracycline (60 mg/liter).
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Description | Reference |
|---|---|---|
| E. coli strains | ||
| DH10B | pMCT-pvdQ | 16 |
| S17-1 | galU galK rpsL(Strr) endA1 nupG thi pro hsdR hsdM+recA (RP4-2Tc::Mu Km::Tn7) λpir | 8 |
| P. aeruginosa strains | ||
| PAO1 | Wild type, Marseille strain | |
| PAO1ΔpvdQ | ΔpvdQ chromosomal deletion mutant derived from PAO1 | This study |
| PAO1(pMEpvdQ) | pvdQ overexpressing strain derived from PAO1 | 16 |
| Plasmids | ||
| pME6032 | lacIQ-Ptac expression vector; pVS1-p15A shuttle vector TetR | 7 |
| pME6032-pvdQ | pvdQ in pME6032 | 16 |
| pSB1075 | lasR lasI′ (P. aeruginosa PAO1)::luxCDABE (P. luminescens [ATCC 29999]) fusion in pUC18 Apr, with acyl-HSL biosensor producing bioluminescence | 24 |
DNA manipulations.
DNA manipulations were performed using standard techniques (15). PCR fragments were purified by using the QIAquick PCR purification kit (Qiagen, Valencia, CA). DNA fragments were extracted and purified from agarose gels by using a QIAquick gel extraction kit according to the manufacturer's instructions. Genomic DNA from Pseudomonas strains was isolated using a genomic DNA isolation kit (GenElute bacterial genomic DNA kit; Sigma-Aldrich, St. Louis, MO). Plasmid isolation was performed using the Nucleospin plasmid isolation kit from Macherey-Nagel (Dueren, Germany). DNA sequencing was carried out by Macrogen (Seoul, Korea).
Caenorhabditis elegans.
The C. elegans strain used in this study was the wild-type Bristol N2 strain. Standard conditions were used for C. elegans propagation at 23°C. Worms were synchronized by hypochlorite bleaching, hatched overnight, and subsequently cultured on nematode growth medium (NGM) (18) plates with Escherichia coli OP50. Stocks were maintained by transferring 1-cm2 pieces of NGM agar with nematodes onto fresh NGM plates seeded with E. coli OP50.
Construction of PAO1(pME6032pvdQ).
The construction of the PAO1(pvdQ) strain has been previously described (16). Briefly, chromosomal DNA was isolated from an overnight culture of PAO1. The open reading frame encoding the acylase gene (nucleotides 2638805 to 2636517 [obtained from www.pseudomonas.com]) was amplified from chromosomal DNA by PCR and cloned into the plasmid pMcTNde under the control of the tac promoter, resulting in plasmid pMc-pvdQ. For this, the 3 nucleotides preceding the putative start codon were altered to create an NdeI restriction site. Subsequently, the open reading frame was amplified from plasmid pMc-pvdQ using primers that added an EcoRI restriction site in front of the putative start codon and a BglII restriction site after the putative stop codon. The resulting PCR product was cloned into the P. aeruginosa-E. coli shuttle vector pME6032 (7) by using these restriction sites, putting the gene under the control of a tac promoter, which is repressed in the absence of IPTG by lacIq.
Construction of the ΔpvdQ deletion mutant.
The ΔpvdQ deletion mutant was constructed as follows. The flanking regions of the target gene were amplified from PAO1 chromosomal DNA using primer pair A (ForA, 5′-GACAAGCTTGGTGTCGCAGAGCGAGTT-3′, containing a HindIII restriction site [underlined]; RevA, 5′-CATGAGACACGCGTCCCCATCGATGTCGTTTC-3′) and primer pair B (ForB, 5′-GGGACGCGTGTCTCATGATAAGCAATGCCTATC-3′; RevB, 5′-CAGGAATTCGGCCATCGGTAGCA-3′, containing an EcoRI restriction site [underlined]). Subsequently, these two fragments were joined together in a second-round PCR using ForA and RevB primers, digested with HindIII and EcoRI, and cloned into similarly digested pEX18-Gm (8). The resulting plasmid, pEX18-ΔPA2385, was transformed into E. coli S17-1 λpir and introduced into PAO1 cells by conjugation (8) to generate a mutant containing an in-frame deletion of the PA2385 gene. Gentamicin-resistant, sucrose-sensitive P. aeruginosa colonies were selected on Vogel-Bonner minimal medium plates containing 5% sucrose (21). This deletion was confirmed by PCR of the PA2385 gene and Southern blot analysis of digested genomic DNA by using the DIG High Prime DNA labeling and detection starter kit I (Roche) according to the manufacturer's instructions (data not shown).
Purification of PvdQ.
A two-step purification protocol was adapted from the one published by Sio et al. (16). Briefly, E. coli DH10B harboring the plasmid pMCT-pvdQ was grown for 48 h at 30°C in 2xTY medium (15) supplemented with 50 μg/ml chloramphenicol and 0.1% glycerol. Cells were harvested by centrifugation, resuspended in ca. 3 ml of Tris-EDTA buffer (50 mM Tris-HCl, 2 mM Na-EDTA, pH 8.8)/g of wet cells, lysed by sonication, and centrifuged at high speed to remove cell debris. Anion exchange chromatography (Q-Sepharose) was used as a first purification step. The flowthrough, containing PvdQ, was collected, adjusted to a 0.7 M final concentration of ammonium sulfate, and purified by hydrophobic interaction chromatography using a 5-ml HiTrap phenyl Sepharose column. PvdQ eluted at 0% ammonium sulfate with a purity of ≥95%.
C. elegans slow-killing assays.
Slow-killing kinetics of C. elegans by PAO1 and its derivatives were determined by using the following procedure adapted from Tan and Ausubel (19). Pseudomonas strains were grown overnight at 37°C in LB broth supplemented with appropriate antibiotics and then diluted 100-fold into fresh broth. NGM plates (59-mm diameter) were then spread with 80 μl of the respective culture. After the plates were incubated at 37°C for 24 h and allowed to equilibrate to room temperature for 30 min, 40 to 50 larval stage 4 (L4) nematodes from stock plates were transferred onto the P. aeruginosa lawn. The plates were then incubated at 24°C and scored for living and dead worms every 3 to 4 h for 7 days. For statistical purposes, a minimum of four replicates per trial was performed. E. coli OP50 was used as a negative control to evaluate background levels of worm death. A worm was considered to be dead when it failed to respond to plate tapping or gentle touch with a platinum wire. Worms that died as a result of getting stuck to the wall of the plate were excluded from the analysis. Results are presented as the percentage of living nematodes on the killing plates compared to their survival on the E. coli OP50 control strain.
C. elegans standard paralysis assay.
P. aeruginosa PAO1 and its derivatives were grown overnight in BHI broth and then diluted 100-fold into fresh broth. BHI agar plates (59-mm diameter) were spread with 80 μl each of the respective dilution and then incubated for 24 h at 37°C to form lawns of bacteria (2, 12). Nematodes were then transferred directly from stock plates (50 worms per plate) to the PAO1 bacterial lawns with a wire pick and incubated at 24°C. In general the animals were examined every hour for 5 h and scored as paralyzed if they did not move and did not respond detectably to mechanical stimulation. The percentage of dead worms after 4 h of incubation was calculated.
Studying capabilities of the purified protein to interfere with the virulence of P. aeruginosa in the C. elegans model.
Synchronized larvae were grown in 96-well plates as previously described (14). When the worms reached the L4 developmental stage they were screened against our PAO1 and its pvdQ knockout mutant in the following method. The strains were grown overnight in LB and then diluted 100-fold in fresh medium. They were then allowed to grow to an optical density at 600 nm of 0.4, at which stage 5 μg of purified protein was added to both PAO1 wild type and PAO1-ΔpvdQ. Cultures were incubated thereafter 6 h at 37°C. Fifteen L4 developmental stage worms (previously synchronized as described above and allowed to hatch overnight in M9 buffer) were transferred to a well of a 96-well plate and mixed with 80 μl overnight bacterial culture and 50 μl M9 buffer containing 10 mg/ml cholesterol. Each strain was tested four times in each assay, and three independent experiments were performed. The worms were then incubated at 20°C while shaking at 150 rpm to prevent suffocation. Each well was then scored at 20-min intervals under a dissecting microscope.
Detection and quantification of 3-oxo-C12-HSL levels present in the different strains at the time of killing.
A liquid assay using the conditions described above was performed. After the completion of the assay, cell-free supernatant fractions of PAO1, PAO1-pvdQ, PAO1(pvdQ), and PAO1 wild type, to which purified PvdQ protein was added, were obtained in the following way. The culture mixture from the wells was collected and centrifuged at 11,000 × g for 10 min to remove the worms and cell debris from cultures, thus yielding cell extracts that were then filtered using a 0.2-μm-pore-size filter (Whatman, Dassel, Germany). The optical density at 600 nm at this harvest point was measured. A bioassay was then performed at 37°C by adding 100-μl samples of extracellular medium fractions to the wells of a white 96-well plate together with 100 μl of a 1/100 dilution of an overnight culture of the biosensor strain, E. coli JM109(pSB1075) (24), which produces light in response to long AHLs (15). All samples were tested in triplicate in each assay. The response of the biosensor after an incubation period of 4 h was analyzed with a ChemiGeniusXE imaging system (Syngene UK) and quantified using a LumiCount microplate luminometer (Packard Bioscience).
RESULTS
Killing of C. elegans by a diffusible toxin.
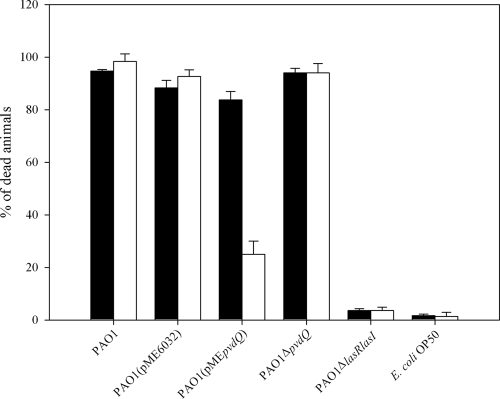
Under certain conditions PAO1 rapidly paralyzes and kills the nematode C. elegans. This killing is mediated by hydrogen cyanide that is under the control of both the LasR and RhlR quorum-sensing regulators (2, 12). We performed the standard paralysis assay under similar conditions to the ones previously described by Darby et al. (2), with a slight modification. We chose to expose only L4-stage and young adult worms to the P. aeruginosa strains, as these were the developmental stages used before by Tan et al. for the C. elegans killing assays. However, under these conditions there were only slight differences observed between the various PAO1 strains (Fig. 1). PAO1 and the control strain PAO1(pME6032), as well as the pvdQ-overexpressing strain and the ΔpvdQ mutant, resulted in paralysis of the animals after 4 h of exposure. The worms experienced a dramatic reduction in their pharyngeal pumping rates, ceased processes such as defecation and egg laying, and failed to respond to mechanical stimulus within minutes after exposure. Transferring the paralyzed worms to new plates seeded with E. coli OP50 failed to recover them from their paralysis, thus confirming their death. Only the quorum-sensing-negative PAO1 ΔlasR ΔlasI mutant failed to induce paralysis under these conditions. The latter was used as a negative control to confirm our hypothesis that the signaling molecule 3-oxo-C12-HSL produced by PAO1 is a key factor for the virulence of the bacterium (Fig. 1). When following exactly the protocol previously described (12) and exposing a large number of a mixed population of the worms to the different bacterial lawns, the results were dramatically different (Fig. 1). We observed a much lower rate of paralysis when PvdQ was overexpressed compared to the wild type and the ΔpvdQ mutant. Overexpression led to survival of more than 75% of the worms fed on the bacterial lawn for 4 h and allowed them to complete their life cycle, showing unaffected locomotion, pharyngeal pumping, and egg-laying processes. We kept monitoring the worms that survived and followed growth rates of their progeny. A healthy brood consisting of hundreds of progeny was produced by day 4 from an initial founding population of 50 worms, and the entire lawn of bacteria was consumed. The second generation seemed to be growing slightly slower than the naïve worms initially exposed to the strain. It is very important to stress here that from the lethal paralysis only the young larvae (L1 to L3) were able to escape (Fig. 1). The above observations suggest that PvdQ influences the toxicity level of P. aeruginosa PAO1 against at least L1- to L3-stage nematodes. All the L4 and young adults, regardless of the strain that they were allowed to feed upon, started showing sluggish locomotion, within minutes, and twitching movements. There was only one exception: no matter which developmental stage we selected for, the quorum-sensing-negative PAO1 ΔlasR ΔlasI mutant did not result in nematode paralysis in any of the assays. The observations made under standard paralytic conditions confirm our hypothesis that PvdQ is capable of functioning as a quorum quencher in vivo. This assay did not reveal differences in the killing of ΔpvdQ and the parental PAO1. It should be noted here that all of the strains grew at the same rate as the wild-type strain in both media, indicating that the attenuated pathogenicity phenotypes observed were not simply a result of growth defects of the mutants.
FIG. 1.
Effects of pvdQ overexpression and deletion in P. aeruginosa virulence in the C. elegans infection model under standard paralytic conditions. The virulence of the PAO1 wild type and ΔpvdQ and pvdQ-overexpressing mutants was tested on C. elegans by screening only L4 and young adults (black bars) and a mixed population of all developmental stages (white bars). In both cases the PAO1 wild type, the control strain PAO1(pME6032), and the ΔpvdQ mutant resulted in rapid paralysis of the worms with almost a 100% mortality of the population screened. More than 95% of the worms fed with E. coli OP50 survived the 4-h exposure. The results were significantly different for PAO1(pMEpvdQ). When only L4 larvae were used, only 20% of the worms fed on this strain survived. However, in the mixed population more than 75% of the nematodes survived and continued to use this strain as a food source. This mainly included the L1 and L2 developmental stages, with a small number of L3 larvae also surviving.
Higher-sensitivity assay to determine the killing kinetics of the different mutants.
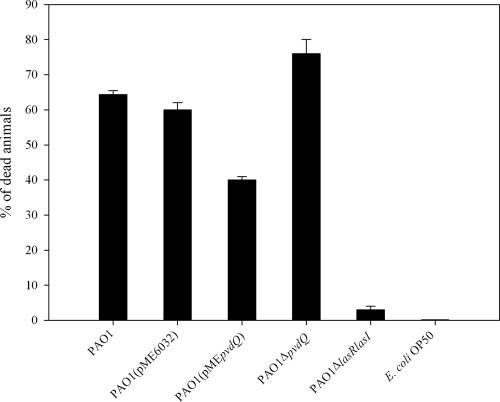
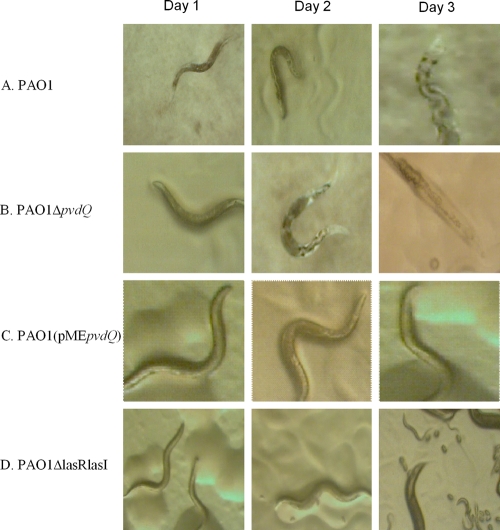
To be able to observe any differences between the killing kinetics of PAO1 and its ΔpvdQ mutant we followed an alternative approach, using a slow-killing assay previously described by Tan et al. (19, 20) in which the time required for 50% of the worms to die (LT50) for PAO1 was shown to be around 68 h. We chose to screen our mutants with a slow-killing assay, as this method offers the advantage of a higher sensitivity and thus the opportunity to identify strains that are only slightly impaired in their ability to kill C. elegans. During the first 24 h there were no significant differences observed among any of the mutants screened. All nematodes exposed to the different lawns showed normal behavior. However, between 48 and 72 h postexposure, the harmful effects of the more-pathogenic strains were easily detected on the animals. We calculated the LT50 of the PAO1 wild type to be 60 h (data not shown). More specifically, 65% of the worms fed on the wild type were already dead at 72 h postexposure (Fig. 2), and the remainder showed a very sluggish locomotion and distended intestine (Fig. 3A). In addition, in most cases, the animals became laden with eggs and embryos hatched internally, suggesting an egg-laying defect. The amount of eggs laid by day 3 was also very limited when compared to the negative control, E. coli OP50. The same observations were made for PAO1(pME6032). PAO1 ΔpvdQ reached its LT50 significantly faster, in 48 h. The surviving animals had a severely sick appearance and did not move unless mechanically stimulated. By 72 h of exposure, 75% mortality was found (Fig. 2), and the dead animals had almost completely dissolved in the agar (Fig. 3B). In addition, the very few progeny that were seen in the population of worms fed on the ΔpvdQ mutant eventually died and the bacterial lawn remained intact. Different results were obtained when overproducing PvdQ. Up until the first 48 h the worms did not show any symptoms of disease (Fig. 3C). On the contrary, they were moving around the plate continuously, feeding as expected and maintaining a normal pharyngeal pumping rate. The egg-laying process was not affected, and hundreds of eggs were present on the agar. The larvae emerging went through their normal 15-day life cycle without experiencing any complications. After 72 h postexposure, only 40% of the worms were dead (Fig. 2) and the survivors appeared to continue feeding upon cells of PAO1(pMEpvdQ) as a food source. Although overproducing the protein does not render PAO1 nonpathogenic, it seems to delay its killing course. The progeny of these animals was monitored up until the appearance of the third-generation eggs. The animals grew normally through each developmental stage. PAO1 ΔlasR ΔlasI, which produces no 3-oxo-C12, showed mortality rates similar to that of the negative control, E. coli OP50 (less than 3%), and the animals survived throughout the duration of the assay without any complications or any altered behavior (Fig. 2). Plates containing PAO1 ΔlasR ΔlasI bacterial lawns were overcrowded after 60 h (Fig. 3D).
FIG. 2.
Killing kinetics of PAO1, PAO1(pME6032), PAO1 Δpvd, PAO1(pMEpvdQ), and PAO1 ΔlasR ΔlasI. A higher-sensitivity assay was used to study the killing rates of the different mutants. Here we present the percentage of dead animals exposed for a period of 4 days to the respective mutant. After 72 h of exposure to the wild-type strain, 65% of the animals died from accumulation of toxic bacteria in their intestines. The control strain PAO1(pME6032) resulted in 61% of the animals dying, in contrast to PAO1(pMEpvdQ), which killed only about 40% at the same time point. Interestingly, more than 65% of the worms survived the exposure to this strain and continued to use it as a food source, showing a healthy growth rate with no alterations in their behavior. The ΔpvdQ mutant proved to be more pathogenic than the wild type, resulting in the death of about 76% of the population. The quorum-sensing-negative mutant PAO1 ΔlasR ΔlasI and the E. coli OP50 used as a background control did not exert any toxicity to the animals.
FIG. 3.
Morphological appearance of the worms exposed to the different strains under slow-killing conditions. The morphological appearance of the worms at each of the 3 days postexposure to PAO1 wild type (A), PAO1-ΔpvdQ (B), PAO1(pMEpvdQ) (C), and PAO1-ΔlasRlasI (D). It is very clear from these pictures that in the case of the worms shown in both panels A and B there is progressive distension of the gut of the animals due to the colonization of toxic bacteria. In the case shown in panel B at day 3, the animal was already dead and started dissolving in the agar. When PvdQ is overproduced more than 65% of the animals exposed to it remain and look healthy even at the end of the third day (C) and continue to go through their life cycle at a normal rate. Lastly, all of the worms exposed to the quorum-sensing-negative strain survived and started laying eggs by day 2. After 72 h (D) the plate was already getting crowded with more eggs and many larvae (L1).
Use of purified protein to attenuate the virulence of P. aeruginosa PAO1.
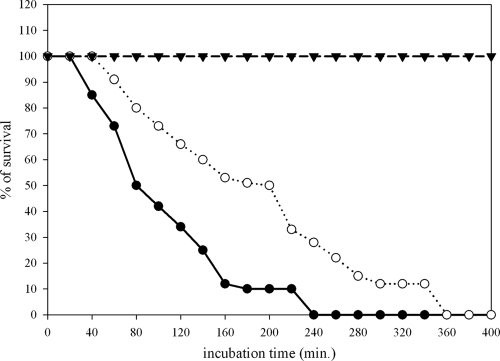
To test for the capabilities of purified PvdQ to interfere with quorum sensing in our model organism, we had to develop an assay that allowed the protein to be added exogenously to our P. aeruginosa strains. To allow homogenous administration of the protein, we replaced the time-consuming multiple steps in an agar-based assay with a liquid pathogenicity assay in which the steps required are reduced to a minimum. By taking advantage of the liquid culture with the simple media normally used for propagation of C. elegans feeding on E. coli OP50, we were able to monitor our strains side by side by visual inspection. Ten to 15 L4 larvae were transferred to each well of a flat-bottom 96-well microtiter plate. The worms were suspended in 50 μl of M9 buffer containing cholesterol (10 μg/ml). To this we added 100 μl per well of our overnight bacterial cultures and incubated the mixtures at room temperature while shaking at 150 rpm to prevent suffocation. It was interesting to see that even though the worms placed in the nonpathogenic E. coli OP50 strain did survive for almost 7 days and could be further propagated by addition of more E. coli OP50 to the wells, the worms that were exposed to PAO1 were already dead after 3 h (Fig. 4). To confirm that the worms were indeed dead, we transferred them to plates seeded with E. coli OP50. No movement or feeding was detected. PAO1 seems to be very pathogenic under these conditions, with worms showing similar life spans as seen in the standard paralytic assay, which has a killing course of 4 h (Fig. 1). Once we established the toxicity of PAO1 under these conditions, we then used this strain as a control culture to compare its virulence with and without addition of purified PvdQ. Exogenously added PvdQ did delay the lethal effects of PAO1 on C. elegans. After 3 h of incubation, in contrast to 100% mortality detected for PAO1, 45% survival was measured for the worms feeding on the P. aeruginosa PAO1 with purified protein added to it. The LT50 value for the wild type was calculated to be 80 min, whereas adding the protein resulted in an increased LT50 of 200 min (an additional 2 h) (Fig. 4).
FIG. 4.
Addition of the purified protein to the culture reduces the pathogenicity of P. aeruginosa PAO1 in the C. elegans infection model. Purified PvdQ was added to a PAO1 wild-type culture and incubated at 37°C for 6 to 8 h before being used as a food source in the liquid assay to detect any reduction or inhibition of pathogenicity. This graph demonstrates the number of worms alive in the wells at 20-min intervals when exposed to E. coli OP50 (inverted triangles), PAO1 wild type (open circles), and PAO1 with purified protein added to it (filled circles). When PvdQ is added to the wild type there is a slower progression of the infection, resulting in an increased LT50 of about 1.3 h for the animals before they eventually die.
Quantification of 3-oxo-C12-HSL levels.
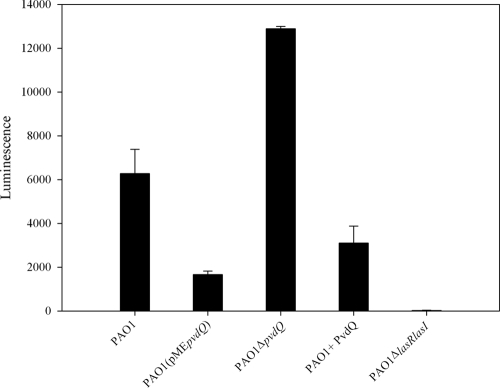
As an additional confirmation that the effects seen in these experiments are due to the differences in the concentrations of the signaling molecule 3-oxo-C12-HSL, we quantified the levels of this molecule released in the assay medium by PAO1 with or without addition of purified PvdQ and by the PvdQ-overexpressing strain as well as by the ΔpvdQ mutant. After the completion of the assay performed in liquid, worms were removed by centrifugation (11,000 × g for 10 min) and the supernatant was tested against the biosensor strain E. coli JM109(pSB1075) (24), which in the presence of 3-oxo-C12-HSL produces light. The response of the biosensor after incubation of 4 h with the supernatant samples was analyzed with a ChemiGenusXE imaging system (Syngene) and quantified using a LumiCount microplate luminometer (Packard Bioscience). Bioluminescence quantification revealed that the ΔpvdQ mutant accumulated more 3-oxo-C12-HSL than the parental strain. Addition of the protein to the wild type resulted in a significantly lower accumulation of 3-oxo-C12-HSL (Fig. 5). The levels of 3-oxo-C12-HSL detected in PAO1(pMEpvdQ) were also significantly lower than in either the wild type or in the knockout mutant.
FIG. 5.
Quantification of 3-oxo-C12-HSL. Cell extracts taken at the end of the liquid assay were analyzed using the biosensor strain pSB1075. Light produced in response to 3-oxo-C12-HSL was quantified. All values are the means from at least three independent experiments, with standard errors shown. Higher levels of 3-oxo-C12-HSL were detected in PAO1 ΔpvdQ compared to the PAO1 wild type. Both overexpression of the protein (IPTG induced) and external addition of the purified protein resulted in reduced 3-oxo-C12-HSL levels, correlating to the better survival of the worms under these conditions.
DISCUSSION
It has only been 10 years since the nematode C. elegans was first suggested as and proven to be a predictive model for studying pathogenesis in mammals (20). Since then C. elegans has been extensively described in the literature as a model to study host-pathogen interactions (6, 9, 10, 11, 13). There are many reasons that make this nematode an excellent study model for bacterial infections. It is easily propagated on petri dishes, feeding on the slow-growing E. coli OP50 strain. The nematode has a well-characterized genome, a small size, and a transparent body which allows easy visualization of the internal organs. Many gram-positive and gram-negative bacteria have been tested for their pathogenicity against this model by simply replacing the normal food source given to the worm (E. coli OP50) with the pathogen of interest. For the gram-negative bacterium P. aeruginosa, it was also established that it is pathogenic for C. elegans. Depending on the experimental conditions, P. aerginosa PA14 kills the nematodes over a period of days (slow killing) or hours (fast killing) (19, 20). Using a different P. aeruginosa strain (P. aeruginosa PAO1) Darby and coworkers (2) reported another mechanism by which P. aeruginosa can result in the death of C. elegans. The lethal effect is associated with a rapid neuromuscular paralysis and is caused by the action of primarily hydrogen cyanide, whose production requires the functionality of both the Las and the Rhl quorum-sensing system (2). Our present study shows that P. aeruginosa PAO1 results in paralysis of C. elegans on BHI agar and leads to slow killing when using NGM plates. Both assays have been used to test quorum-quenching abilities of the PvdQ AHL acylase. In our first setup of the fast-killing assay, following the protocol of Tan et al. (19, 20), synchronized C. elegans L4-stage worms were used. When these worms are placed on a lawn of the P. aeruginosa PAO1 wild-type strain, the pumping of the pharyngeal muscles dramatically slows down and completely ceases after some minutes. After about 4 to 5 h of exposure the worms become completely paralyzed. The L4-stage worms reacted similarly when fed with PAO1(pME6032) or the ΔpvdQ mutant. However, a few animals feeding on the PAO1(pMEpvdQ) strain were rescued from the lethal paralysis, suggesting a quorum-quenching effect resulting from the overproduction of PvdQ. This is in line with earlier observations that, upon overproduction of the AHL acylase in liquid cultures, the concentration of the signaling molecule 3-oxo-C12-HSL is strongly reduced (16).
Following the protocol first described by Darby et al. (2) for screening a large mixed population of the worms for the pathogenicity of PAO1(pMEpvdQ), the results were dramatically different (Fig. 1). Over 75% of the worms, all in early developmental stages, survived the 4 h of exposure to the lawn of PAO1(pMEpvdQ) and were feeding upon the cells of this strain as a food source. We conclude from our findings that the animals that are most likely to be rescued by an overproduction of an N-acyl-homoserine lactone acylase are the L1 to L3 larvae. This is in line with the increased survival rate (80%) observed with an N-acyl homoserine lactone acylase, AiiD, derived from Ralstonia sp. when expressed in P. aeruginosa PAO1 (12). A mixed population was also used in this case, although there was no reference as to the developmental stage of the surviving worms (12). Since the fast-killing assay was not sensitive enough for detecting significant differences between PAO1 and the PAO1 ΔpvdQ, the slow-killing assay was used to determine differences between the kinetics of the various PAO1 strains. The more time required for the hermaphrodite worms to be killed, the more eggs laid and the more progeny produced, resulting in an increased population of young larvae that are more resistant to infection by P. aeruginosa. Indeed, the slow-killing assay was found to show an increased sensitivity and allowed differentiation among strains that are only slightly impaired in their ability to kill C. elegans. About 35% of the nematode population fed on the wild type survived, as opposed to the 60% survival detected for the nematode population that fed on the PAO1(pMEpvdQ) strain. The pathogenic effects of PAO1 ΔpvdQ were more pronounced, with only 15% of the nematode population surviving. In addition, the onset of the killing could be seen 24 h earlier. The slow-killing assay as well as the fast-killing assay on the mixed population of nematodes prove that PvdQ overproduction reduces virulence of P. aeruginosa PAO1. Deletion of the gene results in a mutant with an increased virulence in the slow-killing assay.
We believe that degradation of 3-oxo-C12-HSL is the major factor that results in the reduced virulence of P. aeruginosa PAO1. This was demonstrated by quantifying the levels of 3-oxo-C12-HSL present in all the strains, and indeed, the PAO1 wild type accumulated more 3-oxo-C12-HSL than PAO1(pMEpvdQ) but less than PAO1 ΔpvdQ. This was also confirmed by the results obtained with the quorum-sensing-negative strain (PAO1 ΔlasR ΔlasI), which is incapable of producing any 3-oxo-C12-HSL (Fig. 5) and also lacks the receptor to which this molecule binds. PAO1 ΔlasR ΔlasI failed to cause any infection-like symptoms in the worms in all the assays performed for this project.
To further test for the capabilities of purified PvdQ protein to interfere with quorum sensing in our model organism, we designed an assay that allowed the protein to be added exogenously to our P. aeruginosa strains. To avoid potential destabilization of the protein during the time-consuming incubation steps required for the agar assays, we developed a liquid culture in which the steps required to perform the assay were reduced to a minimum. This assay showed a high correlation with the fast-killing assay and under these conditions the LT50 value for PAO1 was calculated to be 80 min, whereas external addition of the protein to the parental strain resulted in an increased LT50 of 200 min.
External addition of the purified protein to PAO1 also resulted in lower 3-oxo-C12-HSL levels. We can therefore conclude that our enzyme can act as a quorum quencher in vivo in an infection model such as the C. elegans model. Quorum-quenching effects result from both overexpressing the gene and from externally adding the purified protein.
Even though either overexpressing the gene in its host or externally adding the purified protein to P. aeruginosa PAO1 results in attenuated toxicity and a delay in the onset of infection, the infection itself cannot be completely prevented. We believe that this is due to remaining amounts of 3-oxo-C12-HSL, as illustrated in Fig. 5. Since PvdQ is a periplasmic protein, it cannot access and degrade all the 3-oxo-C12-HSL molecules present; the latter will eventually accumulate to the threshold levels required to turn on quorum sensing. However, this will happen with a delay in time. We strongly believe that the PvdQ protein shows a high potential to be administered as an antibiotic, since external addition of the protein resulted in an increased life span of the nematodes by reducing the virulence of the bacterium. Delaying the onset of the infection is not as effective as preventing the onset altogether, but it gives a chance to the infected organism to clear the pathogen before the latter starts to become too toxic to its host. It will be very interesting to test whether these effects also take place in a mammalian organism, such as mice, and to detect whether the protein can be delivered as an antibiotic to help cure Pseudomonas infections or can be used in a combination therapy in parallel with other medicines. The pulmonary delivery (4, 22) of stabilized acylase together with colistin is a challenging experiment that may be taken up in the near future.
Acknowledgments
We gratefully acknowledge Miguel Cámara and Paul Williams (University of Nottingham) for providing the biosensor strain pSB1075. We thank S. Heeb for the gift of pEX18-Gm and Steve Diggle and Sue Dodson for E. coli S17-1 λpir (University of Nottingham). Also we thank Ellen A. A. Nollen, Karen L. Thijssen, and Tjakko J. van Ham (University of Groningen) for kindly providing us with the C. elegans wild-type Bristol N2 strain. We also acknowledge Steve Atkinson (University of Nottingham) and Steve Garvis (CNRS Marseille) for the helpful discussions and technical advice and Robbert H. Cool (University of Groningen) for critical reading of the manuscript.
This research was partly funded by EU grant ANTIBIOTARGET MEST-CT-2005-020278 to P.N.J., G.K., and E.P.
Footnotes
Published ahead of print on 31 August 2009.
REFERENCES
- 1.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 2.Darby, C., C. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans, E. A., T. Kawli, and M. W. Tan. 2008. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 4:e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frijlink, H. W., and A. H. de Boer. 2004. Dry powder inhalers for pulmonary drug delivery. Expert Opin. Drug Deliv. 1:67-86. [DOI] [PubMed] [Google Scholar]
- 5.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. 1998. A broad-host-range Flp-FTR recombination system for the site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 9.Jansen, W. T., M. Bolm, R. Balling, G. S. Chhatwal, and R. Schnabel. 2002. Hydrogen peroxide-mediated killing of Caenorhabditis elegans by Streptococcus pyogenes. Infect. Immun. 70:5202-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurz, C. L., and J. J. Ewbank. 2000. Caenorhabditis elegans for the study of host-pathogen interactions. Trends Microbiol. 8:142-144. [DOI] [PubMed] [Google Scholar]
- 11.Labrousse, A., S. Chauvet, C. Couillault, C. L. Kurz, and J. J. Ewbank. 2000. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 10:1543-1545. [DOI] [PubMed] [Google Scholar]
- 12.Lin, Y. H., J. L. Xu, J. Hu, L. H. Wang, S. L. Ong, J. R. Leadbetter, and L. H. Zhang. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47:849-860. [DOI] [PubMed] [Google Scholar]
- 13.Mylonakis, E., F. M. Ausubel, J. R. Perfect, J. Heitman, and S. B. Calderwood. 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 99:15675-15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nollen, E. A., S. M. Garcia, H. G. van, S. Kim, A. Chavez, R. I. Morimoto, and R. H. Plasterk. 2004. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA 101:6403-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2008. Molecular cloning: a laboratory manual, 4th ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Sio, C. F., L. G. Otten, R. H. Cool, S. P. Diggle, P. G. Braun, R. Bos, M. Daykin, M. Camara, P. Williams, and W. J. Quax. 2006. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect. Immun. 74:1673-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, R. S., and B. H. Iglewski. 2003. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J. Clin. Investig. 112:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stiernagel, T. 2006. Maintenance of C. elegans. In Wormbook. The C. elegans Research Community. www.wormbook.org.
- 19.Tan, M. W., and F. M. Ausubel. 2000. Caenorhabditis elegans: a model genetic host to study Pseudomonas aeruginosa pathogenesis. Curr. Opin. Microbiol. 3:29-34. [DOI] [PubMed] [Google Scholar]
- 20.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 22.Westerman, E. M., H. G. Heijerman, and H. W. Frijlink. 2007. Dry powder inhalation versus wet nebulisation delivery of antibiotics in cystic fibrosis patients. Expert Opin. Drug Deliv. 4:91-94. [DOI] [PubMed] [Google Scholar]
- 23.Williams, P., K. Winzer, W. C. Chan, and M. Camara. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B 362:1119-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Construction and analysis of LuxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]