Abstract
Biofilms gain resistance to various antimicrobial agents, and the presence of antibiotic resistance genes is thought to contribute to a biofilm-mediated antibiotic resistance. Here we showed the interplay between the tetracycline resistance efflux pump TetA(C) and the ampicillin resistance gene (blaTEM-1) in biofilms of Escherichia coli harboring pBR322 in the presence of the mixture of ampicillin and tetracycline. E. coli in the biofilms could obtain the high-level resistance to ampicillin, tetracycline, penicillin, erythromycin, and chloramphenicol during biofilm development and maturation as a result of the interplay between the marker genes on the plasmids, the increase of plasmid copy number, and consequently the induction of the efflux systems on the bacterial chromosome, especially the EmrY/K and EvgA/S pumps. In addition, we characterized the overexpression of the TetA(C) pump that contributed to osmotic stress response and was involved in the induction of capsular colanic acid production, promoting formation of mature biofilms. However, this investigated phenomenon was highly dependent on the addition of the subinhibitory concentrations of antibiotic mixture, and the biofilm resistance behavior was limited to aminoglycoside antibiotics. Thus, marker genes on plasmids played an important role in both resistance of biofilm cells to antibiotics and in formation of mature biofilms, as they could trigger specific chromosomal resistance mechanisms to confer a high-level resistance during biofilm formation.
Bacteria that adhere to abiotic and biotic surfaces, including medical implants and human tissues, can become the cause of refractory infections (60). The microscopic observations of these refractory infections have revealed bacteria growing as biofilms (49). Recently, it has been observed that resistance of biofilms to antibiotics is greater than what is normally seen with planktonic cells (29, 41). Several mechanisms, including the failure of antibiotic penetration into biofilms, have been proposed for the resistance to antibiotics, with the biofilm acting as a barrier (41, 60). However, the exopolysaccharide matrix does not always act as an impenetrable barrier to the diffusion of antibiotics, especially to β-lactams or tetracyclines, and thus other mechanisms must be sought to explain the biofilm resistance (1, 2, 15, 61). In addition, it has been suggested that high-cell-density stationary-phase characteristics and/or quorum-sensing molecules play an important role in resistance to antibiotics, but again, their exact roles are not clear. Conversely, resistance mechanisms of planktonic cells, such as the production of antibiotic-degrading enzymes (β-lactamase for β-lactams) and the increase in antibiotic efflux pumps (Tet pump for tetracyclines) have been well studied. The TetA(C) efflux pump and the β-lactamase (bla) genes are often encoded on transmissible elements, such as plasmids, transposons, or intergrons, which can spread rapidly among bacteria. Since the efflux pumps can extrude antibiotics from the cell, the induction of the pumps is thought to be one of the key alterations conferring resistance to biofilm cells (66). In Escherichia coli, a putative multidrug resistance pump, YhcQ, was reported to be involved in antibiotic resistance of biofilms (39, 69), and Mar, Sox, and AcrAB-TolC efflux pump-encoded genes have been found to be upregulated under stress conditions, such as a stationary growth condition, growth in biofilms, and exposure to several antimicrobial agents (5, 14, 68). Furthermore, inactivation of efflux pumps by a number of efflux pump inhibitors reduces biofilm formation, indicating the efflux system is required for biofilm formation (32). However, the exact role of efflux pumps in biofilm formation and their importance in biofilm-mediated antibiotic resistance remain under investigation.
The rapid spread of antibiotic resistance among bacteria is mainly due to the localization of antibiotic resistance marker genes on plasmids (46, 58). Recently, it has been reported that there is a connection between the antibiotic marker genes on the plasmid and bacterial biofilm formation. The presence of the β-lactamase gene reduced the amount of biofilm formed by E. coli strains harboring β-lactamase-encoded plasmid, while the presence of either a gentamicin or tetracycline resistance gene did not impair biofilm formation (4, 18). These findings indicate that the resistance to antibiotics is linked to biofilm development and maturation. Therefore, biofilms of bacteria that harbor marker genes on the plasmid may gain resistance to some antibiotics at the expense of other protective phenotypes, and the effect of marker genes on bacterial biofilm phenotypes must be investigated.
In this study, we analyzed the effects of the marker genes, an ampicillin and a tetracycline resistance gene on the pBR322 plasmid, on E. coli biofilm formation in relation to the resistance of biofilms to antimicrobial agents. Furthermore, we investigated the interplay among the genes on pBR322 which effectively promoted E. coli biofilm development and maturation. A global transcriptional approach was also used to analyze the connection between genes on the plasmid and bacterial chromosome regarding the induction of efflux pumps during biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A pBR322 plasmid, which encodes the blaTEM-1 and tetA(C) genes conferring resistance to ampicillin and tetracycline, respectively, was used in this study (7). The laboratory E. coli strain MG1655 was used in all experiments. E. coli OCL62 [genotype 1655sp rpsL polA12 Zih::Tn10 Δ(emrY-ddg)::kan] was obtained from National Institute of Genetics (Japan). OCL62 is a medium deletion strain, with mutations in the emrY, emrK, evgA, and evgS operons, in which the related and neighboring genes were removed counterclockwise with respect to the oriC position (22). In this study, OCL62 was genetically modified by replacing the downstream kan gene with the tetR gene by using the method of Datsenko and Wanner (12). Thus, the modified OCL62 is Δ(emrY-ddg)::tetR and has a counteraction of the constitutive tetA(C) gene by the expression of the tetR repression gene. The modified OCL62 was then used to investigate the counteraction of TetA(C) pump and the loss of function in the EmrY/K and EvgA/S pumps on the bacterial chromosome. Luria-Bertani (LB) broth or agar (Difco) was used for standard cultivation. Appropriate antibiotics (ampicillin, tetracycline, penicillin, erythromycin, chroramphenicol, kanamycin, and gentamicin) and chemicals (NiCl2, NaCl, and MgCl2) were added when required. All antibiotics and chemicals were purchased from Wako Chemical (Japan).
Biofilm formation assay and flow cell experiment.
Biofilms were cultured by using LB broth on polystyrene (TPP, Switzerland) or polyvinylchloride (PVC; Costar) 96 well-plates, which were inoculated with early-stationary-phase growing cultures of E. coli strains diluted to an optical density at 600 nm (OD600) of 0.1 and incubated at 37°C for 24 h without shaking. Antibiotic(s) was diluted and was then added directly to the wells without washing for an additional 24 h. The conditions and concentrations of antibiotics used in this study are shown in Table 1. The biofilm formation assay protocol was the modified Reisner's method (54). Briefly, the culture plates were washed with 0.85% NaCl, and the attached cells were then strained with crystal violet (CV) by semiautomatic staining using a microplate washer (ImmunoWash 1575; Bio-Rad). The OD570 absorbances of CV-binding biofilms were measured with a microplate reader (ARVO 1420 multilabel counter; PerkinElmer). The average values of CV-stained biofilms were obtained from at least 24 independently grown biofilms. The error bars represent the standard deviations from these averages. The bacterial growth activity was determined by subculturing (1:100) the relevant strain into the medium without shaking. The OD600 readings were taken over time using the microplate reader.
TABLE 1.
Conditions and concentrations of antibiotics used in this study
| Exposure group and E. coli strain | Antibiotic(s)a | Concn range (μg/ml)d |
|---|---|---|
| Ampicillin and/or tetracycline (administered separately or in combination) | ||
| MG1655 | Ampicillin, tetracycline | 0.0000005-500 |
| Mixture of ampicillin and tetracyclineb | 0.0000005-500 | |
| MG1655(pBR322) | Ampicillin, tetracycline | 0.0000005-500 |
| Mixture of ampicillin and tetracyclineb | 0.0000005-500 | |
| Ampicillin (administered separately or in combination with tetracycline) | ||
| OCL62 | Ampicillin | 0.0005-50 |
| OCL62(pBR322) | Ampicillin | 0.0005-50 |
| Mixture of ampicillin and tetracyclineb | 0.0005-50 | |
| Penicillin, erythromycin, chloramphenicol, kanamycin, and/or gentamicin (administered separately or in combination with the mixture of ampicillin and tetracycline [5 μg/ml each]) | ||
| MG1655(pBR322) | Penicillin, erythromycin, chloramphenicol, kanamycin, and gentamicin | 0.0005-50 |
| Mixture of ampicillin and tetracyclinec + penicillin | 0.000005-50 | |
| Mixture of ampicillin and tetracyclinec + erythromycin | 0.000005-50 | |
| Mixture of ampicillin and tetracyclinec + chloramphenicol | 0.000005-50 | |
| Mixture of ampicillin and tetracyclinec + kanamycin | 0.000005-50 | |
| Mixture of ampicillin and tetracyclinec + gentamicin | 0.000005-50 | |
| Ni2+, Na+, and Mg2+ salts (administered separately or in combination with the mixture of ampicillin and tetracycline [5 μg/ml each]) | ||
| MG1655(pBR322) | Mixture of ampicillin and tetracyclinec + NiCl2 | 0.001-1000 |
| Mixture of ampicillin and tetracyclinec + NaCl | 0.005-5000 | |
| Mixture of ampicillin and tetracyclinec + MgCl2 | 0.001-1000 |
The antibiotic(s) was administered separately or in combination, depending on each condition.
The mixture of ampicillin and tetracycline was diluted to concentrations of 0.0000005 to 500 μg/ml each.
The mixture of ampicillin and tetracycline was fixed at concentrations of 5 μg/ml each.
With a series of 10-fold dilutions.
For flow cell conditions, biofilms were cultivated in three-channel flow cell reactors (Stovall) by using M9 minimal medium (Difco) supplemented with 20% LB broth to optimize the biofilm biomass of MG1655. Flow cells were inoculated with early-stationary-phase growing cultures of E. coli strains diluted to a 0.1 OD600. After inoculation, the medium flow was arrested for 2 h to allow the injected bacteria to attach to the glass surface. After bacterial cells attached to the glass surface, the medium, which supplemented with appropriate concentration of antibiotic(s), was fed to the flow cell chamber for another 48 h. Medium flow was controlled at a constant rate of 0.20 ml min−1 by an Ismatec IPC8 peristaltic tubing pump (Ismatec, Switzerland).
Determination of MICs.
Comparative MIC determinations were performed according to the standard protocols (34, 52). The conditions and concentrations of antibiotic(s) used in this study are shown in Table 1. Briefly, the planktonic MICs were determined in 96-well polystyrene microtitration plates, which contain an array of 12 by 8 wells with a series of 10-fold dilutions of antibiotic(s), administered separately or in combination. The antibiotic(s), when added into the wells, was placed at room temperature for 1 h. Next, the E. coli cultures were inoculated into the wells, and the plates were then incubated aerobically for 24 h at 37°C. The planktonic MICs were determined as the lowest antibiotic concentration causing the greatest diminution of growth. The biofilm MICs were determined in 96-well PVC microtitration plates. Biofilms were cultured without shaking for 24 h at 37°C, and then the antibiotic(s) was added directly to the wells without washing. The plates were incubated for an additional 24 h before the assay of CV staining of biofilm formation as described above. The biofilm MICs were determined as the lowest antibiotic concentration causing the greatest inhibition of biofilm formation. Statistical analysis was conducted to evaluate the significance of each MIC using a t test with a P value of ≤0.05.
Microscopes and image analyses.
Microscopic observation was performed with a LSM 510 confocal laser scanning microscope (CLSM; Zeiss, Germany). In order to study the spatial localization of the cells in biofilms using the CLSM, the strains were genetically marked by insertion of green fluorescent protein (GFP) into the chromosome according to the Diederich protocol (13). Images were obtained using the IMARIS software package (Bitplane AG, Switzerland). For biofilm quantification, CSLM image stacks were analyzed by using the computer program COMSTAT (24).
For electron microscopy, cells were fixed with 1 M sodium cacodylate buffer (pH 7.2) containing 1% glutaraldehyde. The samples were then washed with the same buffer and postfixed in 1% osmium tetroxide in the cacodylate buffer. The postfixed samples were dehydrated in a graduated ethanol series (30 to 100%) and then treated with an HCP-2 critical point dryer (HITACHI, Japan) and observed with a Hitachi S4000 scanning electron microscope (SEM; Hitachi, Japan).
Determination of plasmid copy number.
The genomic DNA and plasmid DNA were extracted by using a Qiagen DNeasy tissue kit and plasmid minikit (Qiagen), respectively, according to the manufacturer's manuals. The protocol was the modified Lee's method (36). Briefly, DNA was used directly as a template with Power SYBR green PCR master mix (ABI). The blaTEM-1 gene were used for monitoring the copy number of the pBR322 plasmid and normalized with the ftsZ housekeeping gene of the genomic DNA. A list of PCR primers is shown in Table 2. The copy number of plasmids was then calculated as the number of copies per chromosome.
TABLE 2.
PCR/RT-PCR primers used in this study
| Genea | Sequencesb | Tm (°C)b | Product size (bp)b |
|---|---|---|---|
| ftsZ | Forward, 5′-ATGGAACTTACCAATGACGCG-3′ | 57 | 100 |
| Reverse, 5′-TCAACACCTTCAATGCGCTC-3′ | 56 | ||
| blaTEM-1 | Forward, 5′-GCATCTTACGGATGGCATGA-3′ | 56 | 100 |
| Reverse, 5′-GTCCTCCGATCGTTGTCAGAA-3′ | 59 | ||
| tetA(C) | Forward, 5′-TCTAACAATGCGCTCATCGTCATCC-3′ | 61 | 109 |
| Reverse, 5′-GGAATGGACGATATCCCGCA-3′ | 57 | ||
| rop | Forward, 5′-CAGGAAAAAACCGCCCTTAACATG-3′ | 59 | 101 |
| Reverse, 5′-ATGTCTGCCTGTTCATCCGC-3′ | 54 | ||
| cpsE | Forward, 5′-CTATTCTCGACGCCATCAACG-3′ | 59 | 104 |
| Reverse, 5′-CCTGCAAAGAAATGCGCTCT-3′ | 59 |
Primers were used for determinations of pBR322 plasmid copy number and mRNA gene expression levels. CpsE, colonic acids biosysthesis gene; blaTEM-1, TEM-1 β-lactamase gene; tetA(C), tetracycline efflux pump gene; rop, pBR322 plasmid stabilization gene; ftsZ, cell division FtsZ protein (E. coli essential gene).
Primers were designed by using Primer Express software (Applied Biosystems). The primer conditions were determined according to the dye manufacturer.
Gene expression analysis and reverse transcription (RT)-quantitative PCR analysis.
Samples were taken at an OD600 of 0.5 from a suspended culture (planktonic growth) and/or directly from a flow cell (biofilm growth). The E. coli antisense GeneChip array (Affymetrix) was used to study the global gene expression pattern, according to the Affymetrix expresion analysis technical manual. For each experiment, three biological replicates were analyzed. Statistical analysis was carried out by using the computer program DNA-Chip Analyzer (37). The data were normalized using the invariant set method. The model-based expression value was calculated. After the statistical filtering process, the transcripts were subjected to cluster analysis.
For the RT-quantitative PCR experiment, RNA was converted to cDNA by using a PrimeScript RT reagent kit (Takara Bio, Japan) as described by the manufacturer. The remaining RNA was digested with RNase H, and first-strand cDNA was used directly as a template with SYBR Premix Ex Taq (Takara Bio, Japan) using an Applied Biosystems 7000 sequence detection system. A list of RT-PCR primers is shown in Table 2. The ftsZ housekeeping gene on the host cell chromosome was used as a standard curve, equal in quantity to the test article RNA. The mRNA expression level was calculated as the number of relative cDNA copies per host genome copy detected for each of eight independent dilutions of the RNA samples.
Microarray data accession number.
The data reported in this paper have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession no. E-MEXP-953.
RESULTS AND DISCUSSION
Subinhibitory concentrations of the antibiotic mixture promoted E. coli biofilm formation.
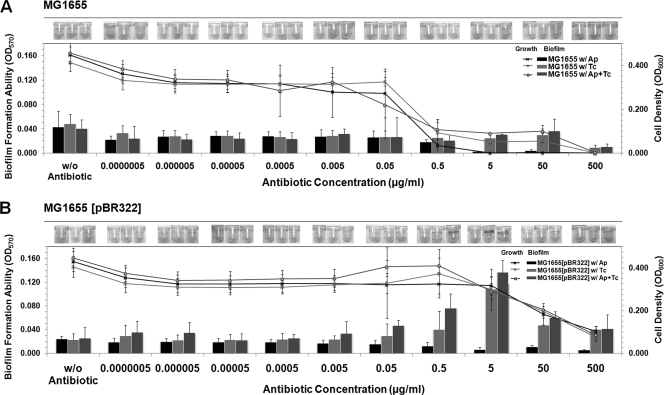
The tetracycline resistance gene on pBR322 vector, a source of tetA(C) in this study, provides for its constitutive expression of the TetA(C) efflux pump in most E. coli strains due to a lack of the tetR repressor on the plasmid (45, 66). It carries along with an ampicillin resistance gene (blaTEM-1) that encodes an ampicillin-degrading enzyme, β-lactamase. The purified β-lactamase of pBR322 is a ubiquitous TEM-1 enzyme (62) which has been reported to decrease the ability of E. coli to form biofilms (18). However, the effects of both the tetracycline resistance TetA(C) efflux pump and the β-lactamase-encoded gene on E. coli biofilm formation in the presence of the mixture of ampicillin and tetracycline have never been elucidated. Therefore, the antibiotic susceptibility and the biofilm formation ability of E. coli harboring pBR322 were investigated and compared with those of a plasmid-free E. coli strain (Fig. 1 and Table 3).
FIG. 1.
Subinhibitory concentrations of ampicillin and tetracycline promoted biofilm formation by E. coli harboring pBR322. Results show quantification of biofilm biomass of MG1655 (A) and MG1655(pBR322) (B) in the absence of antibiotic, in the presence of ampicillin (Ap), tetracycline (Tc), and ampicillin and tetracycline (Ap+Tc). Biofilms were treated with the antibiotic(s) at different concentrations from 0.0000005 to 500 μg/ml for 24 h after the initial attachment, and then the growth activity of the total bacterial cells, which included both the suspended cells and biofilm cells (line graph; “growth”) and the biofilm biomass, which attached to the wells (bar graph; “biofilm”) were quantified. Results are averages of eight replicates ± standard deviations and are representative of four independent experiments. Photographs of biofilms formed on PVC wells, which were stained with crystal violet, under each condition were captured and are represented in the top panels. w/, with; w/o, without. The statistically comparative MICs under each condition are shown in Table 3.
TABLE 3.
Comparative MICs (in planktonic cells and in biofilm cultures) of all antibiotics against all E. coli strains used in this study
| Exposure group and E. coli strain | Antibiotic(s) | MIC (μg/ml)c when grown: |
|
|---|---|---|---|
| Planktonic | As biofilm | ||
| Ampicillin and/or tetracycline (administered separately or in combination) | |||
| MG1655 | Ampicillin | 0.5 | 5 |
| Tetracycline | 0.5 | 500 | |
| Mixture of ampicillin and tetracycline | 0.5d | 500d | |
| MG1655(pBR322) | Ampicillin | 500 | 5 |
| Tetracycline | 500 | >500 (0.5-50 promoted biofilm)e | |
| Mixture of ampicillin and tetracycline | 500d | >500d (0.05-50 promoted biofilm)e | |
| Ampicillin (separately or in combination with tetracycline) | |||
| OCL62 | Ampicillin | 5 | 5 |
| OCL62(pBR322) | Ampicillin | 5 | 50 |
| Mixture of ampicillin and tetracycline | 5d | 50d | |
| Penicillin, erythromycin, chloramphenicol, kanamycin, and/or gentamicin (administered separately or in combination with the mixture of ampicillin and tetracycline [5 μg/ml each]) | |||
| MG1655(pBR322) | Penicillin | 5 | 0.5 |
| Erythromycin | 0.5 | 5 | |
| Chloramphenicol | 0.5 | 5 | |
| Kanamycin | 0.50.005 | ||
| Gentamicin | 0.5 | 0.005 | |
| Mixture of ampicillin and tetracyclinea + penicillinb | >50 | >50 (0.005-5 promoted biofilm)e | |
| Mixture of ampicillin and tetracyclinea + erythromycinb | 50 | 50 (0.05-0.5 promoted biofilm)e | |
| Mixture of ampicillin and tetracyclinea + chloramphenicolb | 5 | 0.5 (0.0005-0.05 promoted biofilm)e | |
| Mixture of ampicillin and tetracyclinea + kanamycinb | 5 | 0.00005 | |
| Mixture of ampicillin and tetracyclinea + gentamicinb | 5 | 0.00005 | |
The mixture of ampicillin and tetracycline was fixed at a concentration of 5 μg/ml each.
When penicillin, erythromycin, chloramphenicol, kanamycin, or gentamicin was supplemented in combination with the mixture of ampicillin and tetracycline, the representative MICs were determined for penicillin, erythromycin, chloramphenicol, kanamycin, or gentamicin.
Statistical analysis was applied to evaluate the clear cut of each MIC using the t test with a P value of ≤0.05.
The representative MICs were determined at the same concentration of ampicillin and tetracycline each.
The subinhibitory concentration of antibiotic(s) that promoted biofilm formation.
The biofilm formation ability of plasmid-free MG1655 in the absence of antibiotic was defined as a baseline and so was called the wild-type biofilm phenotype (Fig. 1A). The planktonic MICs for ampicillin and tetracycline were 0.5 μg/ml. When supplemented with ampicillin, we found the concentration range of 0.0000005 to 0.5 μg/ml slightly reduced biofilm formation, but the concentration range from 5 to 500 μg/ml inhibited biofilm formation, indicating a biofilm MIC of 5 μg/ml. Similar MIC trends were obtained for planktonic cells when supplemented with either tetracycline alone or the antibiotic mixture (both ampicillin and tetracycline). However, the biofilm formation was inhibited at much higher concentrations (the biofilm MICs were 500 μg/ml for either tetracycline or the mixture). Since the antibiotics were supplemented after biofilms were formed in this study, our results indicate that the biofilms were 1,000 times more tolerable to tetracycline and the antibiotic mixture treatment than planktonic cells. In contrast, for ampicillin, the resistance of biofilms over planktonic cells was only 10 times higher. These results suggested that blaTEM-1 does not have much effect, and the response to the exposure to the mixture is dominated by the response to TetA(C).
For MG1655 harboring the plasmid, MG1655(pBR322) (Fig. 1B), in the absence of antibiotic, the adherence to the surface was reduced compared with a plasmid-free strain, showing that the presence of the constitutive TetA(C) efflux pump did not counteract the effect of TEM-1 β-lactamase. In the presence of antibiotic, the planktonic MICs for both antibiotics were 500 μg/ml. The presence of ampicillin inhibited biofilm formation because the addition of ampicillin induces β-lactamase production. β-Lactams, such as ampicillin, inhibit bacterial growth by inactivating penicillin-binding proteins that are involved in synthesis of peptidoglycan (65). Peptidoglycan is thought to play an important role in biofilm formation and is required for optimal assembly of surface molecules. The biofilm MICs for either tetracycline or the mixture were at a concentration of more than 500 μg/ml. Surprisingly, the tetracycline concentration from 0.5 to 50 μg/ml promoted biofilm formation. When treated with the antibiotic mixture, the biofilm formation was further promoted at concentration ranges from 0.05 to 50 μg/ml, and so these were called the subinhibitory concentration levels (Table 3). Thus, the mixture of ampicillin and tetracycline had the strongest biofilm promotion effect versus tetracycline alone. In addition, we also tested that the E. coli strain harboring the β-lactamase-encoding gene alone clearly delayed biofilm formation compared with the wild type. An E. coli strain harboring the TetA(C)-encoding gene alone had no effect on biofilm formation (data not shown). In contrast, only E. coli strains harboring both the genes, like pBR322, promoted biofilm formation. Therefore, the subinhibitory concentration of the antibiotic mixture at 5 μg/ml each (which gave the highest relative biofilm formation ability) was used for the following flow cell experiments.
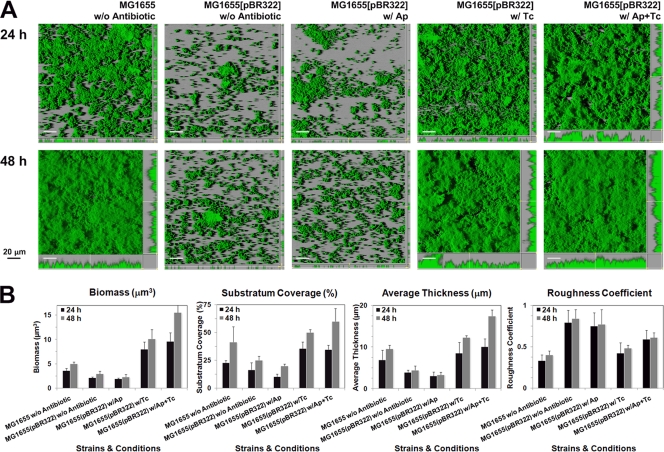
Biofilm structures of E. coli harboring pBR322 in the presence of the antibiotic mixture.
To confirm that the subinhibitory level of the antibiotic mixture promotes cell attachment and subsequent biofilm formation, we cultured biofilms by using flow cell systems and monitored biofilm formation at 24 h and 48 h after addition of antibiotic(s) (Fig. 2A). The biofilm architecture was quantitatively analyzed using COMSTAT software (Fig. 2B). In the absence of antibiotic, E. coli strain MG1655 formed a rather uniform biofilm at 48 h, which is defined as a flow cell wild-type biofilm phenotype. On the other hand, the same strain harboring pBR322 formed less biofilm due to the presence of the blaTEM-1 gene and subsequent β-lactamase expression. In addition, the amount of biofilm was obviously reduced when supplemented with ampicillin, which induces a high level of β-lactamase production and consequently inhibits biofilm development under the flow cell conditions. The presence of β-lactamase makes bacteria resistant to ampicillin by enzymatic activity under mainly planktonic conditions. However, the expression of β-lactamase harms the peptidoglycan remodeling and assembly on the bacterial cell wall, resulting in the lack of a coherence ability to form biofilm (56). In contrast, addition of tetracycline could restore a wild-type biofilm phenotype, showing similar substratum coverage and average thickness to MG1655 in the absence of antibiotics. Notably, tetracycline is a broad-spectrum bacteriostatic antibiotic that inhibits bacterial protein synthesis by preventing aminoacyl-tRNA from binding to the bacterial ribosome (10, 55). From these results, the activation of the TetA(C) pump by addition of tetracycline did not interrupt construction of bacterial cell surface structures because they still could form mature biofilms. As we expected, the antibiotic mixture enhanced biofilm formation of E. coli harboring pBR322. Its mature biofilm at 48 h had a higher roughness coefficient and biofilm thickness than a wild-type biofilm (Fig. 2B). The expression of TetA(C) seemed to interfere with the β-lactamase activity. Therefore, the interplay between marker genes in the presence of a subinhibitory level of the antibiotic mixture might contribute to the development of mature biofilms.
FIG. 2.
The morphology of early (24-h) and late (48-h) biofilms formed by the GFP-tagged strains of a plasmid-free MG1655 (a wild-type biofilm) and MG1655(pBR322) in the absence of antibiotic or in the presence of the mixture of ampicillin and tetracycline at 5 μg/ml each under flow cell conditions (A). Antibiotics were supplemented directly to the medium after the bacterial cells were allowed to attach to the glass surface for 12 h, and the biofilm photographs were taken at 24 h and 48 h, respectively, after the addition of antibiotic(s). All experiments were repeated three times, and representative images are shown. The substratum coverage, average thickness, and roughness coefficient of the biofilms were quantified using the program COMSTAT (B). Values are means of data from 20 image stacks (10 image stacks from two independent channels). The error bars in the individual columns represent the standard deviations. w/, with; w/o, without.
The copy number of pBR322 was stably maintained in biofilms.
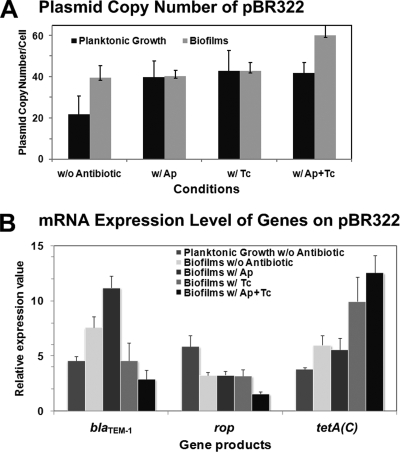
Although the promotion of biofilm formation due to the presence of plasmids has been investigated in several studies (38, 44, 53, 54), it is still not known whether the copy number of plasmids per cell in biofilms is different from those in planktonic cells. The effect of antibiotics on the plasmid copy number and its contribution to biofilm formation are also unclear. Therefore, we determined the copy numbers of pBR322 in biofilms (Fig. 3A). Normally, the copy number of pBR322 is about 40 copies per cell in both minimal and complex media (23, 35, 36). In this study, the plasmid was stably maintained around 40 copies per cell in biofilms in the absence of antibiotic, while it was only about 20 copies per cell in the planktonic culture. The presence of each antibiotic could sustain the copy number of pBR322 at around 40 copies per cell under either the biofilm or planktonic conditions. Surprisingly, when supplemented with the antibiotic mixture, the copy number significantly increased up to 60.2 ± 7.6 copies per cell in biofilms, but not in planktonic cultures. This indicates that the biofilm is able to increase the relative fitness of the plasmid and enhance the propagation of the resistant plasmid. Thus, the presence of antibiotic markers in association with an addition of subinhibitory concentrations of the antibiotic mixture is a great selective pressure to keep the resistant genes in the biofilm community.
FIG. 3.
The interplay between antibiotic marker genes on pBR322 during E. coli biofilm formation resulted in the increase of plasmid copy number, high-level expression of the TetA(C) pump, and the interruption of β-lactamase expression. (A) The plasmid copy numbers of pBR322 in the suspended cultures and in biofilms were determined. (B) The mRNA expression levels of the ampicillin resistance gene (blaTEM-1), the tetracycline resistance gene [tetA(C)], and the copy number regulator (rop) on pBR322 were analyzed using quantitative RT-PCR. All experiments were repeated three times, and the average expression values with their standard derivations are shown. w/, with; w/o, without.
Interplay between the marker genes: high-level expression of TetA(C) reduced the expression of β-lactamase in the biofilm.
There are two marker genes on pBR322, the ampicillin (blaTEM-1) and tetracycline [tetA(C)] resistance genes, and one plasmid control (rop) gene (11). Rop protein is the plasmid copy number regulator and serves to decrease the copy number (3). To understand the interplay mechanism between the marker genes, we quantified mRNA expression levels of those genes (Fig. 3B). The rop gene was more highly expressed under planktonic conditions than in biofilms, supporting the notion of the stable maintenance of plasmid during biofilm formation. In contrast, both tetA(C) and blaTEM-1 genes were more expressed in biofilms. This indicates that E. coli harboring pBR322 produced more β-lactamase inside biofilm cells than in planktonic culture, even in the absence of antibiotic. It might be due to the mature biofilms maintaining more copies of plasmid than in the planktonic cells. Addition of ampicillin further increased the expression of the blaTEM-1 gene. Therefore, E. coli biofilm formation is impaired by β-lactamase that could damage peptidoglycan. These results could confirm that the horizontal acquisition of ampicillin resistance enzymes damages the bacterial cell surface and leads to the reduction of biofilm formation. However, blaTEM-1 could protect planktonic cells, but reduces biofilm formation, which is a clear-cut distinction between a biofilm and planktonic phenotype. Interestingly, the antibiotic mixture further increased the expression of the tetA(C) gene but repressed the expression of the blaTEM-1 gene, indicating the reduction of β-lactamase production. Notably, addition of tetracycline resulted in high-level expression of the TetA(C) efflux pump in biofilms. In addition, the antibiotic mixture also reduced the expression of the rop gene, which could explain the increased copy number of pBR322 inside mature biofilms (Fig. 3A). However, this interplay mechanism only occurred when ampicillin was added together with tetracycline at subinhibitory levels. Therefore, the interplay between marker genes on pBR322 led to the expression of a high level of the TetA(C) efflux pump, the interruption of the ampicillin-degrading enzymatic activity, and the increase in the plasmid copy number and its stability in biofilms.
High-level expression of the tetracycline efflux pump stimulated several efflux systems on the E. coli chromosome.
Since the TetA(C) efflux pump is mainly responsible for the tetracycline resistance mechanism (19), we analyzed whether the biofilms can be resistant to ampicillin without the production of β-lactamase. Recently, the possibility of cooperation between the β-lactam efflux pump and β-lactamase was suggested by Masuda and Church (42). This cooperation may effectively decrease the concentration of β-lactams in the periplasmic spaces of gram-negative bacteria to the point where penicillin-binding proteins are no longer saturated. These bacteria might then display the high-level resistance without producing a high level of β-lactamase (33, 43). Thus, we hypothesized that the interplay between marker genes on the plasmid was stimulated by the presence of subinhibitory concentrations of the antibiotic mixture, which consequently promotes other resistance mechanisms on E. coli bacterial chromosome. To verify this hypothesis, we analyzed global gene expression with DNA microarray technology. We first analyzed the relationship between the interruption of β-lactamase and the expression of E. coli biofilm-specific genes. Second, we investigated the chromosomal efflux mechanisms, which were activated when the high-level TetA(C) pump was present during the biofilm development of E. coli harboring pBR322.
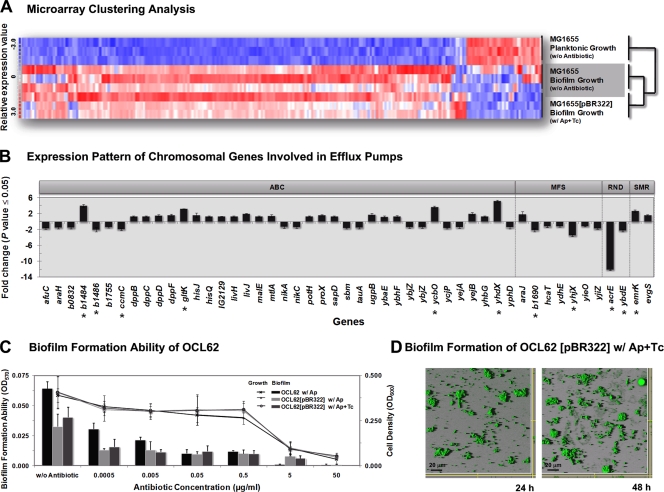
First, since the morphology of MG1655(pBR322) biofilms is similar to the wild-type biofilm (MG1655) when the antibiotic mixture is supplemented (Fig. 2A), we assumed that the presence of both the plasmid and subinhibitory level of the antibiotic mixture must have impacts on E. coli gene expression. Based on the clustering analysis, we found gene expression profiles similar to those of a wild-type biofilm (Fig. 4A). We found that E. coli harboring pBR322 in biofilms also induced the genes involved in several bacterial surface structures, such as flagella, fimbriae, autotransporter proteins, colanic acid, and exopolysaccharide poly-N-acetyl-glucosamine (see Table S1 in the supplemental material). Since β-lactamase affects assembly and function of large macromolecular bacterial surface structures (18, 56), the interplay between marker genes could remodify the cell wall structures and reproduce necessary bacterial cell surface adhesins that are important for biofilm formation.
FIG. 4.
The interplay between the marker genes on the plasmid-induced chromosomal SMR efflux pumps during biofilm development and maturation. (A) Dynamic hierarchical clustering analysis of E. coli gene expression during biofilm formation. Each column represents one gene. Red, upregulated; white, no change; blue, downregulated. The sample profiles were grouped into two clusters: MG1655 planktonic growth versus MG1655 (plasmid free) and MG1655(pBR322) biofilms. For each experiment, three biological replicates were analyzed. (B) Pattern of 49 regulated genes, which encode chromosomal antibiotic efflux pumps, during the biofilm formation of MG1655(pBR322) in the presence of antibiotics but not in MG1655 in neither suspended cultures nor biofilms. Eleven significantly regulated genes (change of ≥2-fold) are labeled with an asterisk. The classification of efflux systems in this study was based on the gene information reported in Affymetrix's NetAffx database. (C) Quantification of the total bacterial growth activity (line graph) and biofilm formation (bar graph) by the modified OCL62 strain, in which emrY, emrK, evgA, and evgS were removed and counteracted with TetA(C), with or without pBR322 plasmid, in the absence or in the presence of antibiotics. Results are averages of eight replicates ± standard deviations and are representative of four independent experiments. (D) CSLM photographs of the modified OCL62(pBR322) biofilms in the presence of antibiotic mixture at 24 h and 48 h, respectively. The experiment was duplicated and representative images are shown.
Second, we screened the regulated genes encoding chromosomal efflux pumps in the MG1655(pBR322) biofilm in the presence of the antibiotic mixture, but not in MG1655 under the planktonic conditions or biofilms. Generally, the antibiotic efflux pumps in bacteria are classified into four superfamilies according to their energy source, sequence alignment, and substrate specificity: the ABC superfamily (ATP-binding cassette), the MFS superfamily (major facilitator), the RND superfamily (resistance nodulation division), and the SMP superfamily (small multidrug resistance) (48, 66). The TetA(C) pump on pBR322 belongs to the MFS superfamily (55). In this study, we found 49 bacterial chromosomal efflux pump-related genes. Most of the genes belonging to the MFS and RND superfamilies were repressed (except for araJ), whereas the genes belonging to the SMR superfamily were upregulated. Eleven out of 49 genes were overexpressed more than twofold (in which b1484, gltK, ycbO, yhdX, and emrK were upregulated while b1486, b1690, acrE, ccmC, yhjX, and ybdE were downregulated) (Fig. 4B; see also Table S2 in the supplemental material). This indicates that the interplay between the marker genes on pBR322 stimulated several chromosomal efflux pumps, resulting in multidrug resistance of biofilms. For this reason, the production of β-lactamase on pBR322 was repressed by the high-level expression of the TetA(C) pump during biofilm development (Fig. 3B), and then other effective resistance mechanisms on the E. coli chromosome were stimulated instead.
Since the genes belonging to the SMR superfamily were clearly upregulated, we believed that the EmrY/K pumps were involved in the antibiotic resistance. Recently, it was reported that subinhibitory concentrations of tetracycline effectively induced the EmrY/K operon (64), and the tetracycline-dependent promoter of EmrY/K is located between the emrKY and evgAS operons (16, 40). The response of the EvgA/S system to environmental signals is likely to be an important factor in EmrK/Y drug resistance (30). It has been reported that the EmrK/Y system could efflux various antibiotic(s) and chemical(s) when the EvgA/S two-component system was initiated (16, 25). Therefore, we examined the contribution of the EmrY/K and EvgA/S systems to the antibiotic resistance in biofilms (Fig. 4C and D) using the modified strain OCL62, in which emrY, emrK, evgA, and evgS genes have been deleted (31), counteracting the TetA(C) pump. The disruption of these genes abolished biofilm formation (Fig. 4C and Table 3). The modified OCL62 harboring pBR322 formed patchy biofilms when the antibiotic mixture was added, similar to MG1655 harboring pBR322 when only ampicillin, but not the antibiotic mixture, was supplemented (Fig. 3D and 2A). These results indicate that the EmrY/K and EvgA/S efflux systems also play an important role in the antibiotic resistance during biofilm formation, and these systems are induced by the high-level TetA(C) expression. Thus, E. coli harboring the pBR322 plasmid-encoding antibiotic resistance genes triggered other chromosomal resistance mechanisms to confer the high-level resistance within biofilms.
Induction of multidrug resistance in biofilms.
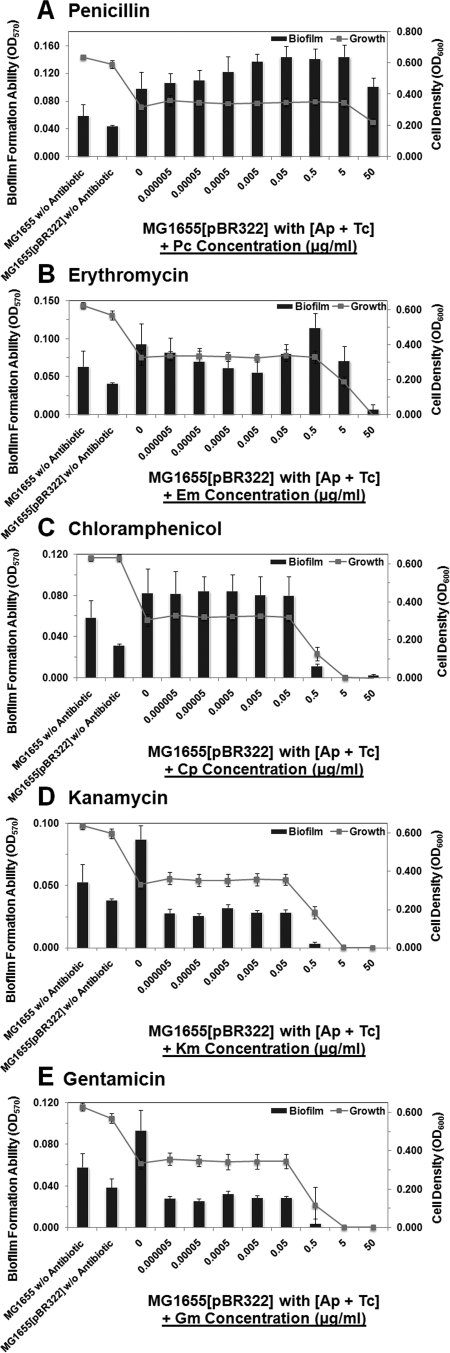
The EmrY/K and EvgA/S efflux systems played an important role in the antibiotic resistance during the biofilm formation, in association with the high-level TetA(C) efflux pump on the plasmid. Both the EmrY/K and EvgA/S pumps are drug/metabolite transporters or proton-drug antiporters coupling substrate removal to electrochemical proton gradients in the opposite direction (17, 20). The overexpression of these genes could make bacteria become resistant to a wide variety of cationic hydrophobic compounds such as tetracycline, erythromycin, β-lactam, macrolide, surfamide, as well as other antiseptics and intercalating dyes (26, 50, 66). Therefore, we investigated whether the activated efflux pumps confer resistance to other antibiotics on the biofilm. We assayed other 5 antibiotics, penicillin, erythromycin, chroramphenicol, kanamycin and gentamicin (Fig. 5A to E and Table 3). The antibiotic was separately added after the biofilms were cultured for 12 h and treated with 5 μg/ml of the mixture of ampicillin and tetracycline for 12 h, and then the plate(s) was incubated for another 24 h. We found that the β-lactam antibiotic penicillin, when given in combination with ampicillin and tetracycline, promoted biofilm formation (Fig. 1B). Similarly, the biofilms could resist the protein synthesis inhibitor, erythromycin, and bacteriostatic antibiotic, chloramphenicol. In contrast, two aminoglycoside antibiotics, kanamycin and gentamicin, inhibited biofilm formation. Therefore, the resistance mechanisms by TetA(C) in combination with EmrY/K and EvgA/S pumps in biofilm cells are likely to be solely due to inhibition of specific protein synthesis or interaction with cell wall. However, they were sensitive to the aminoglycoside antibiotics because these drugs might accumulate inside the cell (19, 21).
FIG. 5.
The high-level resistance phenomenon was found in biofilms of E. coli harboring pBR322 in the presence of subinhibitory concentrations of ampicillin and tetracycline, which confer resistance to penicillin, erythromycin, and chloramphenicol but not to kanamycin and gentamicin. Results are from a quantification assay for the total bacterial growth activity (line graph) and biofilm formation (bar graph) of MG1655(pBR322) in the presence of the antibiotic mixture, in association with separately supplemented of the following antibiotics: (A) penicillin (Pc); (B) erythromycin (Em); (C) chroramphenicol (Cp); (D) kanamycin (Km); (E) gentamicin (Gm). Results are averages of eight replicates ± standard deviations and are representative of four independent experiments. The statistically comparative MICs under each condition are shown in Table 3.
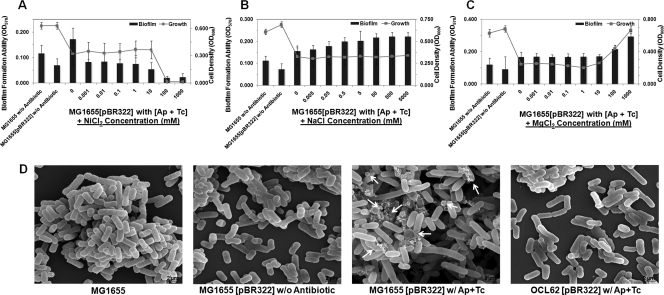
Efflux pumps responded to osmotic stress, which is responsible for colanic acid production and biofilm maturation.
We investigated how the high-level TetA(C) and additional EmrY/K-EvgA/S pumps promote the development and maturation of E. coli biofilms. Both the pumps mediated energy-dependent efflux of the antibiotic from the bacterial cell (47, 67). It has been reported that TetA(C) protein facilitates the uptake of nickel ions and aminoglycoside antibiotics into the cell (51), therefore the increasing copy number of the tetA(C) gene might make cells more susceptible to NiCl2 and aminoglycoside antibiotics. The results shown in Fig. 6A and Fig. 5D-E indicate that the influx and accumulation of toxic metal salts and aminoglycoside antibiotics increased inside bacterial cells. In addition, when present at high levels, the tetA(C) gene product significantly alters the structure of the inner membrane and this confers an additional phenotype of osmotic sensitivity (59). This was investigated by determining the growth sensitivity and the biofilm formation ability of the cultures to increasing concentrations of NaCl and MgCl2. As shown in Fig. 6B-C, biofilm formation was promoted by NaCl (at concentration 0.005 to 5,000 mM) and MgCl2 (at concentration 100 to 1000 mM). Thus, there was a positive correlation between the enhanced efflux system and the increased osmotic pressure in biofilms.
FIG. 6.
Characteristics of the induced efflux pumps in E. coli harboring pBR322 contribute to the increase in the osmotic stress response and the production of capsular colanic acid. Results show quantification of the total bacterial growth activity (line graph) and biofilm formation (bar graph) by MG1655(pBR322) at various concentrations of NiCl2 (A), NaCl (B), and MgCl2 (C) in the presence of antibiotic mixture. Results are averages of eight replicates ± standard deviations and are representative of four independent experiments. (D) SEM micrographs of the structure of biofilms that developed in the flow cells. The presence of capsular exopolysaccharide colanic acid (indicated by white arrows) was found only in MG1655(pBR322) biofilms. The experiment was duplicated and representative images are shown.
We, therefore, hypothesized that the induced efflux pumps in response to osmotic stress contribute to development of a three-dimensional mature biofilms. Accordingly, it has been reported that osmotic stress induced the production of exopolysaccharide, such as capsular colanic acid and/or poly-N-acetyl-glucosamine, which generally acts as an important cell-to-cell adhesin during biofilm development and maturation (9, 28, 44, 57). To confirm this possibility, we observed the morphology and compositions of the biofilms using SEM (Fig. 6D and 7B). As we expected, the MG1655(pBR322) biofilms had depositions of colanic acid in association with other ultramicroscopic bacterial adhesins when supplemented with the antibiotic mixture, while there were no such depositions under any other tested conditions. The identification of the bacterial cell surface structures was based on our previous study conducted with the specific strains (44). Therefore, changes in bacterial surface properties, including the production of essential exopolysaccharide such as colanic acid, promoted the formation of thick mature biofilms. We also found the upregulation of fimbriae (mot, yad), poly-N-acetyl-glucosamine (aga), and colanic acid (wca) biosynthesis genes but none of the colanic acid transporters, such as csp genes in E. coli harboring pBR322 (see Table S1 in the supplemental material).
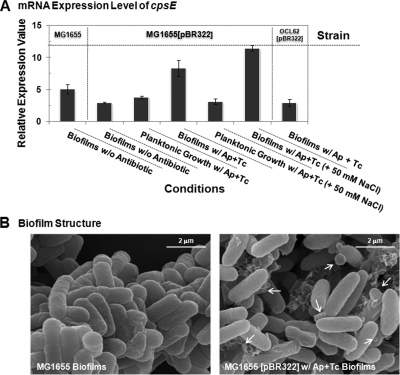
FIG. 7.
The induced efflux pumps cause a change in bacterial surface properties during biofilm formation. (A) The mRNA expression levels of cpsE show overproduction of colanic acid by E. coli harboring pBR322 in response to osmotic stress. All experiments were repeated three times, and the average expression values with their standard derivations are shown. (B) SEM micrographs of biofilm of E. coli harboring pBR322 compared with a wild-type biofilm. Arrows point at the deposition of colanic acid. The results are the means of 20 SEM images from two independent experiments, and representative images are shown.
We further analyzed the mRNA expression levels of the cspE gene in the MG1655 biofilm and found that its level was higher than that in the MG1655(pBR322) biofilm in the absence of antibiotic (Fig. 7A). As we expected, the highest cspE gene expression was obtained in the biofilms when 50 mM NaCl was added. This indicates that the high-level TetA(C) pumps and the induced SMR pumps responded to osmotic stress, which subsequently promoted production of colanic acid and biofilm maturation. In addition, the expression level of the cspE gene in the modified OCL62 biofilm was low, supporting that the development of biofilm with E. coli harboring pBR322 is specifically attributed to the presence of the high-level tetracycline efflux pump and the induced SMR pumps. The further maturation process is related to the secondary characteristics of these pumps, which induce effective bacterial adhesins such as colanic acid (Fig. 7B). Overall, these results indicate that subinhibitory concentrations of antibiotics could result in biofilm maturation, leading to high-level antibiotic resistance.
In summary, there is growing evidence that bacteria respond specifically and defensively to subinhibitory concentrations of antibiotics (6, 8, 27, 63). The evidence presented in this study clearly demonstrates that E. coli harboring marker genes on the plasmid which encodes an efflux pump gene could gain high-level resistance to various antimicrobial agents by stimulating other efflux pump systems on the host cell chromosome and by forming a biofilm when supplemented with a subinhibitory level of the antibiotic mixture. Additionally, the interplay between the marker genes also stimulated high osmotic pressure and then directly promoted the progressive development and maturation of biofilms, which would make the biofilms more difficult to treat. Therefore, care should be taken with regard to use of antibiotics and their spread in the environment.
Supplementary Material
Acknowledgments
We thank Andreas Reisner of the University of Graz in Austria and Maarten Leyssen of the Glaxosmithkline Biologicals S.A. in Belgium for critical comments on the original manuscript, Søren Molin from Biocentrum-DTU in Denmark for supplying the GFP plasmids, and Keiko Ito of the National Institute of Genetics in Japan for kindly providing E. coli strains and the pBR322 plasmid.
Footnotes
Published ahead of print on 31 August 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderl, J. N., J. Zahller, F. Roe, and P. S. Stewart. 2003. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 47:1251-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin, S., S. Friedman, and D. Ludtke. 1986. Partition functions of unit-copy plasmids can stabilize the maintenance of plasmid pBR322 at low copy number. J. Bacteriol. 168:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagge, N., M. Hentzer, J. B. Andersen, O. Ciofu, M. Givskov, and N. Hoiby. 2004. Dynamics and spatial distribution of β-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 48:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey, A. M., M. A. Webber, and L. J. V. Piddock. 2006. Medium plays a role in determining expression of acrB, marA, and soxS in Escherichia coli. Antimicrob. Agents Chemother. 50:1071-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baquero, F., and J. Blázquez. 1997. Evolution of antibiotic resistance. Trends Ecol. Evol. 12:482-487. [DOI] [PubMed] [Google Scholar]
- 7.Bolivar, F. 1978. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene 4:121-136. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, A., N. Shapir, and M. J. Sadowsky. 2004. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70:2503-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., S. M. Lee, and Y. Mao. 2004. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157:H7 to osmotic and oxidative stress. Int. J. Food Microbiol. 93:281-286. [DOI] [PubMed] [Google Scholar]
- 10.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covarrubias, L., L. Cervantes, A. Covarrubias, X. Soberón, I. Vichido, A. Blanco, Y. M. Kupersztoch-Portnoy, and F. Bolivar. 1981. Construction and characterization of new cloning vehicles. V. Mobilization and coding properties of pBR322 and several deletion derivatives including pBR327 and pBR328. Gene 13:25-35. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for integration into the λ attachment site attB of the Escherichia coli chromosome. Plasmid 28:14-24. [DOI] [PubMed] [Google Scholar]
- 14.Domain, F., X. R. Bina, and S. B. Levy. 2007. Transketolase A, an enzyme in central metabolism, derepresses the marRAB multiple antibiotic resistance operon of Escherichia coli by interaction with MarR. Mol. Microbiol. 66:383-394. [DOI] [PubMed] [Google Scholar]
- 15.Dunne, W. M., Jr., E. O. Mason, Jr., and S. L. Kaplan. 1993. Diffusion of rifampin and vancomycin through a Staphylococcus epidermidis biofilm. Antimicrob. Agents Chemother. 37:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eguchi, Y., T. Oshima, H. Mori, R. Aono, K. Yamamoto, A. Ishihama, and R. Utsumi. 2003. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149:2819-2828. [DOI] [PubMed] [Google Scholar]
- 17.Elbaz, Y., T. Salomon, and S. Schuldiner. 2008. Identification of a glycine motif required for packing in EmrE, a multidrug transporter from Escherichia coli. J. Biol. Chem. 283:12276-12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallant, C. V., C. Daniels, J. M. Leung, A. S. Ghosh, K. D. Young, L. P. Kotra, and L. L. Burrows. 2005. Common β-lactamases inhibit bacterial biofilm formation. Mol. Microbiol. 58:1012-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith, J. K., D. H. Cuellar, C. A. Fordyce, K. G. Hutchings, and A. A. Mondragon. 1994. Structure and function of the class C tetracycline/H+ antiporter: three independent groups of phenotypes are conferred by TetA (C). Mol. Membr. Biol. 11:271-277. [DOI] [PubMed] [Google Scholar]
- 20.Grinius, L. L., and E. B. Goldberg. 1994. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 269:29998-30004. [PubMed] [Google Scholar]
- 21.Guay, G. G., and D. M. Rothstein. 1993. Expression of the tet(K) gene from Staphylococcus aureus in Escherichia coli: comparison of substrate specificities of TetA(B), TetA(C), and Tet(K) efflux proteins. Antimicrob. Agents Chemother. 37:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto, M., T. Ichimura, H. Mizoguchi, K. Tanaka, K. Fujimitsu, K. Keyamura, T. Ote, T. Yamakawa, Y. Yamazaki, H. Mori, T. Katayama, and J. Kato. 2005. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 55:137-149. [DOI] [PubMed] [Google Scholar]
- 23.Herman, A., A. Wegrzyn, and G. Wegrzyn. 1994. Regulation of replication of plasmid pBR322 in amino acid-starved Escherichia coli strains. Mol. Gen. Genet. 243:374-378. [DOI] [PubMed] [Google Scholar]
- 24.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 25.Hirakawa, H., Y. Inazumi, T. Masaki, T. Hirata, and A. Yamaguchi. 2005. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 55:1113-1126. [DOI] [PubMed] [Google Scholar]
- 26.Hirakawa, H., K. Nishino, J. Yamada, T. Hirata, and A. Yamaguchi. 2003. β-Lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 52:576-582. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman, L. R., D. A. D'Argenio, M. J. Maccoss, Z. Zhang, R. A. Jones, and S. I. Miller. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171-1175. [DOI] [PubMed] [Google Scholar]
- 28.Ionescu, M., and S. Belkin. 2009. Overproduction of exopolysaccharides by an Escherichia coli K-12 rpoS mutant in response to osmotic stress. Appl. Environ. Microbiol. 75:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito, A., A. Taniuchi, T. May, K. Kawata, and S. Okabe. 2009. An increase in antibiotic resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbiol. 75:4093-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato, A., H. Ohnishi, K. Yamamoto, E. Furuta, H. Tanabe, and R. Utsumi. 2000. Transcription of emrKY is regulated by the EvgA-EvgS two-component system in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 64:1203-1209. [DOI] [PubMed] [Google Scholar]
- 31.Kato, J., and M. Hashimoto. 2007. Construction of consecutive deletions of the Escherichia coli chromosome. Mol. Syst. Biol. 3:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kvist, M., V. Hancock, and P. Klemm. 2008. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 74:7376-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakaye, B., A. Dubus, S. Lepage, S. Groslambert, and J.-M. Frere. 1999. When drug inactivation renders the target irrelevant to antibiotic resistance: a case story with β-lactams. Mol. Microbiol. 31:89-101. [DOI] [PubMed] [Google Scholar]
- 34.Lampe, M. F., C. L. Aitken, P. G. Dennis, P. S. Forsythe, K. E. Patrick, F. D. Schoenknecht, and J. C. Sherris. 1975. Relationship of early readings of minimal inhibitory concentrations to the results of overnight tests. Antimicrob. Agents Chemother. 8:429-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, C., J. Kim, S. G. Shin, and S. Hwang. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123:273-280. [DOI] [PubMed] [Google Scholar]
- 36.Lee, C. L., D. S. W. Ow, and S. K. W. Oh. 2006. Quantitative real-time polymerase chain reaction for determination of plasmid copy number in bacteria. J. Microbiol. Methods 65:258-267. [DOI] [PubMed] [Google Scholar]
- 37.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch, S. V., L. Dixon, M. R. Benoit, E. L. Brodie, M. Keyhan, P. Hu, D. F. Ackerley, G. L. Andersen, and A. Matin. 2007. Role of the rapA gene in controlling antibiotic resistance of Escherichia coli biofilms. Antimicrob. Agents Chemother. 51:3650-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma, C., and G. Chang. 2004. Structure of the multidrug resistance efflux transporter EmrE from Escherichia coli. Proc. Natl. Acad. Sci. USA 101:2852-2857. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Mah, T.-F. C., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 42.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 43.Masuda, N., N. Gotoh, C. Ishii, E. Sakagawa, S. Ohya, and T. Nishino. 1999. Interplay between chromosomal β-lactamase and the MexAB-OprM efflux system in intrinsic resistance to β-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.May, T., and S. Okabe. 2008. Escherichia coli harboring a natural IncF conjugative F plasmid develops complex mature biofilms by stimulating synthesis of colanic acid and curli. J. Bacteriol. 190:7479-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNicholas, P., I. Chopra, and D. M. Rothstein. 1992. Genetic analysis of the tetA(C) gene on plasmid pBR322. J. Bacteriol. 174:7926-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255-261. [DOI] [PubMed] [Google Scholar]
- 47.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27:S32-S41. [DOI] [PubMed] [Google Scholar]
- 48.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1:516-523. [DOI] [PubMed] [Google Scholar]
- 49.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 50.Paulsen, I. T., R. A. Skurray, R. Tam, M. H. Saier, R. J. Turner, J. H. Weiner, E. B. Goldberg, and L. L. Grinius. 1996. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol. 19:1167-1175. [DOI] [PubMed] [Google Scholar]
- 51.Podolsky, T., S.-T. Fong, and B. T. O. Lee. 1996. Direct selection of tetracycline-sensitive Escherichia coli cells using nickel salts. Plasmid 36:112-115. [DOI] [PubMed] [Google Scholar]
- 52.Reeves, D. S., A. Holt, M. J. Bywater, R. Wise, M. N. Logan, J. M. Andrews, and J. M. Broughall. 1980. Comparison of Sensititre dried microtitration trays with a standard agar method for determination of minimum inhibitory concentrations of antimicrobial agents. Antimicrob. Agents Chemother. 18:844-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reisner, A., J. A. J. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 54.Reisner, A., B. M. Höller, S. Molin, and E. L. Zechner. 2006. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 188:3582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts, M. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 56.Sauvage, E., F. Kerff, M. Terrak, J. A. Ayala, and P. Charlier. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234-258. [DOI] [PubMed] [Google Scholar]
- 57.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 59.Stavropoulos, T. A., and C. A. Strathdee. 2000. Expression of the tetA(C) tetracycline efflux pump in Escherichia coli confers osmotic sensitivity. FEMS Microbiol. Lett. 190:147-150. [DOI] [PubMed] [Google Scholar]
- 60.Stewart, P. S., and C. J. William. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 61.Stone, G., P. Wood, L. Dixon, M. Keyhan, and A. Matin. 2002. Tetracycline rapidly reaches all the constituent cells of uropathogenic Escherichia coli biofilms. Antimicrob. Agents Chemother. 46:2458-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takahashi, N., K. Ishihara, R. Kimizuka, K. Okuda, and T. Kato. 2006. The effects of tetracycline, minocycline, doxycycline and ofloxacin on Prevotella intermedia biofilm. Oral Microbiol. Immunol. 21:366-371. [DOI] [PubMed] [Google Scholar]
- 64.Tanabe, H., K. Yamasak, M. Furue, K. Yamamoto, A. Katoh, M. Yamamoto, S. Yoshioka, H. Tagami, H. A. Aiba, and R. Utsumi. 1997. Growth phase-dependent transcription of emrKY, a homolog of multidrug efflux emrAB genes of Escherichia coli, is induced by tetracycline. J. Gen. Appl. Microbiol. 43:257. [DOI] [PubMed] [Google Scholar]
- 65.Thomson, J. M., and R. A. Bonomo. 2005. The threat of antibiotic resistance in gram-negative pathogenic bacteria: β-lactams in peril! Curr. Opin. Microbiol. 8:518-524. [DOI] [PubMed] [Google Scholar]
- 66.Van Bambeke, F., E. Balzi, and P. M. Tulkens. 2000. Antibiotic efflux pumps. Biochem. Pharmacol. 60:457-470. [DOI] [PubMed] [Google Scholar]
- 67.Viveiros, M., M. Dupont, L. Rodrigues, I. Couto, A. Davin-Regli, M. Martins, J.-M. Pagès, and L. Amaral. 2007. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS ONE 2:e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, L., and T.-F. Mah. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 190:4447-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.