Abstract
A combination of phenotypic and genotypic methods was used to investigate 70 unique Escherichia coli clinical isolates identified as producing extended-spectrum β-lactamases (ESBLs) at a medical center in Pittsburgh, PA, between 2007 and 2008. Fifty-seven isolates (81%) produced CTX-M-type ESBLs, among which CTX-M-15 was predominant (n = 46). Isolates producing CTX-M-2, -9, -14, and -65 were also identified. One CTX-M-producing isolate coproduced CMY-2 cephalosporinase. Ten isolates (14%) produced SHV-type ESBLs, either SHV-5 or SHV-7. Two isolates produced only CMY-2 or -32. Pulsed-field gel electrophoresis revealed the presence of two major clusters of CTX-M-15-producing E. coli isolates, one in phylotype B2 (n = 15) and the other in phylotype A (n = 19). Of four phylotype B2 isolates that were able to transfer the blaCTX-M-15-carrying plasmids, three showed fingerprints related (>60%) to those of plasmids from phylotype A isolates. In phylotype B2, all CTX-M-15-producing isolates, as well as three isolates producing CTX-M-14, two producing SHV-5, and one producing SHV-7, belonged to the international epidemic clone defined by serotype O25:H4 and sequence type 131. The plasmids from eight of nine CTX-M-15-producing E. coli isolates of phylotype A that were examined were highly related to each other and were also found in two isolates belonging to phylotype D, suggesting horizontal transfer of this blaCTX-M-15-carrying plasmid between phylotypes. Our findings underscore the need to further investigate the epidemiology and virulence of CTX-M-producing E. coli in the United States.
The most common cause of resistance to expanded-spectrum cephalosporins in Escherichia coli is the production of extended-spectrum β-lactamases (ESBLs) (29). In the past decade, CTX-M-type ESBLs have replaced TEM- and SHV-type ESBLs in Europe, Canada, and Asia as the most common ESBL type in this species (1, 4, 19, 22, 25, 30). The United States initially appeared immune to this epidemic of CTX-M-type ESBLs. However, an increasing number of reports now indicate that CTX-M-type ESBLs have emerged in E. coli and other species of Enterobacteriaceae in the United States as well (7, 17, 24). A study performed in Texas revealed that CTX-M-producing E. coli strains were already present in small numbers in 2000 and became predominant by 2004 (22).
CTX-M-type ESBLs can be grouped into five clusters according to genetic relatedness: the CTX-M-1, -2, -8, -9, and -25 groups (5a). Of the different CTX-M-type ESBLs, CTX-M-15, which belongs to the CTX-M-1 group, is now the most widely distributed enzyme worldwide. CTX-M-15 was first identified in an isolate from India in 1999 (18) and then became prevalent worldwide (5, 6, 11, 23). CTX-M-15 differs from CTX-M-3 by one amino acid substitution at position 240 (Asp240Gly). This substitution enhances hydrolytic activity against ceftazidime, which otherwise is not a preferred substrate for CTX-M-type ESBLs (31). Of note, a particular clone of CTX-M-15-producing E. coli, characterized by phylogenetic type (phylotype) B2, serotype O25:H4, and sequence type 131 (ST131), appears to be responsible for a large part of the international epidemic of CTX-M-producing E. coli (11, 27). Here we describe detailed molecular-epidemiology and plasmid analyses of CTX-M-producing E. coli isolates identified at our medical center in Western Pennsylvania.
MATERIALS AND METHODS
Bacterial strains.
A total of 70 consecutive and unique E. coli isolates identified from patient specimens between May 2007 and April 2008 at the clinical microbiology laboratory of the University of Pittsburgh Medical Center were included. These isolates had positive phenotypic confirmatory test results for ESBL production based on ≥5-mm increases in zone diameters when a ceftazidime or cefotaxime disk was combined with clavulanic acid, as described in the Clinical and Laboratory Standards Institute (CLSI) guidelines (10). The sources of isolates and patient locations were recorded, and the isolates were kept at −80°C until use.
Susceptibility testing.
The antimicrobial susceptibilities of the study strains to ceftazidime, ceftazidime-clavulanic acid, cefotaxime, cefotaxime-clavulanic acid, cefoxitin, cefepime, imipenem, ciprofloxacin, gentamicin, tetracycline, chloramphenicol, sulfamethoxazole-trimethoprim, and amikacin were determined by using the disk diffusion assay as defined by the CLSI (10). ESBL production was phenotypically reconfirmed by using the same disk method in the research laboratory.
PCR analysis and nucleotide sequencing. (i) PCR for detection of resistance genes and determination of phylotypes.
Crude genomic DNA was extracted from the isolates by heat lysis. The DNA was then subjected to PCR using specific primer pairs to screen for β-lactamase genes (consensus blaCTX-M, blaCTX-M-2, blaCTX-M-9, blaCTX-M-15, blaSHV, blaTEM, blaCMY, and blaOXA-1) (2, 12, 14, 26, 34, 35) and phylotypes (Table 1) (8). PCR analysis was performed using a GeneAmp thermal cycler, model 9700 (Applied Biosystems, Foster City, CA). DNA sequencing of resistance genes was performed using an ABI 3730 instrument with primers that generated the amplified products. Sequences were then analyzed using the BLAST software available from the National Library of Medicine (http://www.ncbi.nlm.nih.gov/blast).
TABLE 1.
Primers used for amplification of resistance genes and replicon typing
| Primer | Target gene/site | Sequence (5′ to 3′) | Annealing temp (°C) | Reference |
|---|---|---|---|---|
| TEMF | blaTEM | ATGAGTATTCAACATTTCCGTG | 55 | 12 |
| TEMR | TTACCAATGCTTAATCAGTGAG | |||
| SHV S1 | blaSHV | ATTTGTCGCTTCTTTACTCGC | 55 | 12 |
| SHV S2 | TTTATGGCGTTACCTTTGACC | |||
| CTX-M/F | blaCTX-M | TTTGCGATGTGCAGTACCAGTAA | 51 | 14 |
| CTX-M/R | CGATATCGTTGGTGGTGCCATA | |||
| CTX-M-2 group F | blaCTX-M-2 | AAATGTGCTGCTCCTTTCGTGAGC | 60 | 12 |
| CTX-M-2 group R | AGGGTTCGTTGCAAGACAAGACTG | |||
| CTX-M-9 group F | blaCTX-M-9 | GTGACAAAGAGAGTGCAACGC | 60 | 34 |
| CTX-M-9 group R | ATGATTCTCGCCGCTGAAGCC | |||
| CTX-M-15-SF | blaCTX-M-15 | CACACGTGGAATTTAGGGACT | 55 | 26 |
| CTX-M-15-SR | GCCGTCTAAGGCGATAAACA | |||
| CMY-F | blaCMY | CCGGACACCTTTTTGCTTTT | 60 | 35 |
| CMY-R | TATCCTGGGCCTCATCGTCAGTTA | |||
| OXA-1 F | blaOXA-1 | TTTTCTGTTTGGGTTTT | 52 | 2 |
| OXA-2 R | TTTCTTGGCTTTTGTGCTTG | |||
| aac(6′)-1b f | aac(6′)-Ib | TTGCGATGCTCTATGAGTGGCTA | 55 | 28 |
| aac(6′)-1b r | CTCGAATGCCTGGCGTGTTT | |||
| rfb.1bis | O typing for O25 | ATACCGACGACGCCGATCTG | 60 | 9 |
| rfb25.r | TGCTATTCATTATGCGCAGC |
(ii) O25 typing, plasmid incompatibility grouping, and MLST.
PCR-based O25 typing was employed for all E. coli isolates using the primers listed in Table 1. Ten CTX-M-15-producing E. coli isolates were O and H typed at the E. coli Reference Center, Pennsylvania State University. Further, to identify the pandemic clone characterized by serotype O25:H4, STs were determined by multilocus sequence typing (MLST) for O25 PCR-positive isolates as well as for CTX-M-15-producing E. coli isolates belonging to phylotypes D (n = 2) and A (n = 5) (37). PCR products from MLST were also sequenced as described above. Allelic profiling and ST determinations were performed through the E. coli MLST website maintained at the Max-Planck Institut fuer Infektionsbiologie (now at the ERI, University College Cork [http://mlst.ucc.ie/mlst/dbs/Ecoli]).
PFGE.
All isolates were evaluated for genomic clonality by pulsed-field gel electrophoresis (PFGE). Genomic DNAs of 70 isolates were prepared as described previously (32). Fingerprints were generated by restriction endonuclease XbaI (New England Biolabs, Ipswich, MA) and subjected to electrophoresis using a CHEF-DR III system (Bio-Rad, Hercules, CA) at 6 V with pulse times of 2.2 to 54.2 s and linear ramping at 14°C for 22 h. A lambda ladder (New England Biolabs) was used as the DNA size marker. The relatedness of PFGE patterns was determined by the unweighted-pair group method using average linkages and cluster analysis with the Dice setting on Bionumerics software, version 4.0 (Applied Maths, Sint-Martens-Latem, Belgium).
Plasmid extraction, transfer, and Southern hybridization.
Plasmids of 16 isolates carrying blaCTX-M-15 were analyzed in detail, as described below, in order to compare them within the collection and also with blaCTX-M-15-carrying plasmids characterized in non-U.S. isolates. In addition, four isolates producing blaCTX-M-2, blaCTX-M-14, or blaCTX-M-65 were also included in order to compare the relatedness among the plasmids. The transferability of blaCTX-M-15-carrying plasmids was assayed by both transformation and conjugation. The transferability of plasmids by conjugation was analyzed by liquid mating in Luria-Bertani (LB) broth with the sodium azide-resistant E. coli strain J53AziR as the recipient. Transconjugants were selected on LB agar containing sodium azide at 100 μg/ml and ampicillin at 50 μg/ml. The transfer of a blaCTX-M-carrying plasmid was confirmed by the resistance profiles of the transconjugants and positive PCR results for blaCTX-M. For the examination of plasmid transferability by transformation and for use in fingerprinting, plasmids were extracted using the alkaline lysis method and were transformed into E. coli DH10B by electroporation as previously described (35). Transformants were selected on LB agar containing 2 μg/ml of cefotaxime (Sigma-Aldrich, St. Louis, MO). The fingerprints of the blaCTX-M-carrying plasmids were generated by digesting plasmids with restriction enzyme HpaI (New England Biolabs). After electrophoresis, plasmid fingerprints were visualized and then transferred to a positively charged nylon membrane (Roche Diagnostics, Indianapolis, IN). DNA probes were prepared from PCR amplicons of blaCTX-M-15 using a digoxigenin nucleic acid labeling and detection system (Roche Diagnostics). The clonality of the plasmid fingerprints was assessed using Bionumerics software. The presence of blaTEM, blaOXA-1, and aac-(6′)-Ib on these plasmids was determined as well. The plasmid profiles were compared to the published characteristics of blaCTX-M-15-carrying plasmids (5, 11).
RESULTS
Antimicrobial susceptibilities of E. coli isolates.
The sources of the 70 study isolates included urine (48 isolates), superficial wounds (9 isolates), bronchoalveolar lavage fluid (7 isolates), sputum (3 isolates), and blood (3 isolates). No cluster of cases according to patient location was observed. Two isolates showed a phenotype consistent with AmpC production and only minimal inhibition by clavulanic acid. They did not carry ESBL genes but instead carried CMY-type cephalosporinase genes. The remaining 68 ESBL producers were resistant to cefotaxime or ceftazidime or to both and showed inhibition by clavulanic acid (zone diameter increase of ≥5 mm with combination disks containing ceftazidime or cefotaxime plus clavulanic acid compared with disks containing ceftazidime or cefotaxime alone). All but six isolates were resistant to ciprofloxacin (92%). All isolates in this study were susceptible to imipenem.
Resistance genes and phylotypes.
Table 2 shows the distribution of ESBL and CMY-type cephalosporinase genes and phylotypes. CTX-M was the predominant ESBL type, produced by 81% of all isolates (n = 57). All isolates carrying blaCTX-M showed resistance to cefotaxime. Additionally, 46 CTX-M-producing E. coli isolates were also resistant or intermediately resistant to ceftazidime. These 46 isolates carried blaCTX-M-15. The other CTX-M-type ESBL genes identified were in the blaCTX-M-2 and blaCTX-M-9 groups (Table 2). The majority of the 10 isolates in the blaCTX-M-9 group possessed blaCTX-M-14 (n = 8). A single isolate possessed both blaCTX-M-65 (blaCTX-M-9 group) and blaCMY-2, which were carried on two separate transferable plasmids. SHV-producing E. coli isolates were detected less frequently (n = 10 [14%]). These isolates carried blaSHV-5 or blaSHV-7 and were resistant to ceftazidime and intermediately resistant to cefotaxime. The two isolates possessing blaCMY-2 or blaCMY-32 but no ESBL gene were considered false-positive ESBL producers. This rare phenomenon has been reported previously for some isolates possessing plasmid-mediated cephalosporinase (33, 35). Only a single isolate had a TEM-type ESBL gene, which was identified as blaTEM-10. This isolate was resistant to ceftazidime and had reduced susceptibility to cefotaxime. The blaTEM-10-carrying plasmid was also transferable, and the transformant showed the same resistance profile as the parent isolate.
TABLE 2.
Resistance genes and phylotypes of ESBL-producing E. coli isolates
| β-Lactamase gene | No. (%) of isolates | No. of isolates of phylotype: |
|||
|---|---|---|---|---|---|
| A | B1 | B2 | D | ||
| CTX-M | 57 (81) | 26 | 0 | 18 | 13 |
| CTX-M-15 | 46 (66) | 25a | 0 | 15b | 6 |
| CTX-M-2 | 1 (1) | 0 | 0 | 0 | 1 |
| CTX-M-9 group | 10 (14) | 1 | 0 | 3 | 6 |
| CTX-M-9 | 1 (1) | 0 | 0 | 0 | 1 |
| CTX-M-14 | 8 (11) | 1 | 0 | 3c | 4 |
| CTX-M-65 | 1 (1) | 0 | 0 | 0 | 1d |
| SHV-5 or -7 | 10 (14) | 1 | 1 | 5e | 3 |
| TEM-10 | 1 (1) | 0 | 0 | 1 | 0 |
| CMY-2 or -32 | 2 (3) | 0 | 0 | 0 | 2 |
| Total | 70 | 27 | 1 | 24 | 18 |
All isolates possessed blaOXA-1 and aac-6′-Ib, and 19 isolates shared ≥75% similarities based on PFGE.
All 15 isolates were O25-ST131. The presence of blaTEM, blaOXA-1, and aac-6′-Ib differed among the isolates.
All isolates were O25-ST131.
This isolate also possessed blaCMY-2.
Three isolates were O25-ST131.
Overall, there was no predominance of a particular phylotype in this study. A total of 27, 1, 24, and 18 isolates belonged to phylotypes A, B1, B2, and D, respectively (Table 2). Of the 24 isolates in phylotype B2, 15 produced CTX-M-15. Most of the phylotype A isolates (25 of 27) were CTX-M-15 producers as well. ESBLs produced by phylotype D isolates were more variable (Table 2).
O25 typing and MLST.
Twenty-one isolates belonging to phylotype B2 (15 producing CTX-M-15, 3 producing CTX-M-14, 2 producing SHV-5, and 1 producing SHV-7) were determined to be serotype O25:H4 and ST131. All six CTX-M-15-producing E. coli isolates belonging to phylotype A that were tested for ST had ST410, which belongs to the ST23 complex. These strains were found to have serotype O20:H9. The two CTX-M-15-producing E. coli isolates belonging to phylotype D were nonserotypeable and were determined to be ST648.
Molecular typing for clonal detection.
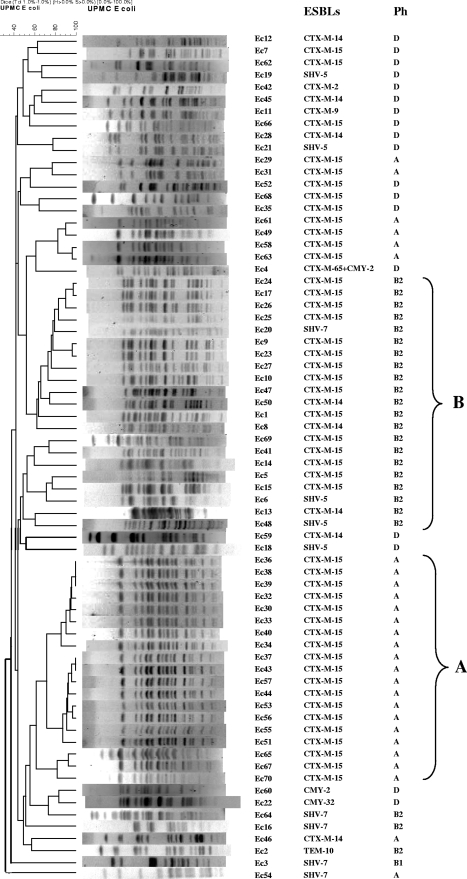
DNA fingerprints resulting from PFGE were assigned to pulsotypes. Each pulsotype comprised genetically related or indistinguishable isolates. PFGE revealed the presence of two predominant pulsotypes, designated pulsotype A (n = 19), with ≥75% similarities, and pulsotype B (n = 18), with ≥50% similarities (Fig. 1). All 19 isolates in pulsotype A had phylotype A and produced CTX-M-15. All 15 CTX-M-15-producing isolates in pulsotype B had phylotype B2. Additionally, three phylotype B2 isolates producing CTX-M-14 and three producing SHV (two SHV-5 isolates and one SHV-7 isolate) belonged to pulsotype B. The rest of the isolates (including 12 CTX-M-15 producers) were clonally diverse, with each pulsotype containing three isolates or fewer.
FIG. 1.
Dendrogram based on PFGE profiles of 70 E. coli isolates. Ph, phylotype; A, pulsotype A; B, pulsotype B.
Plasmid analysis.
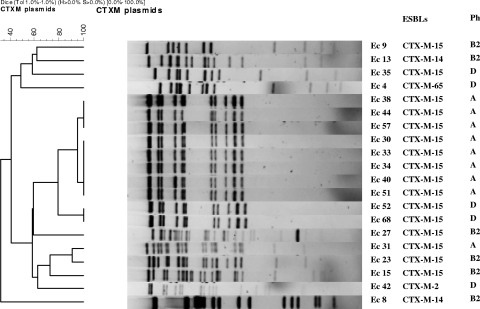
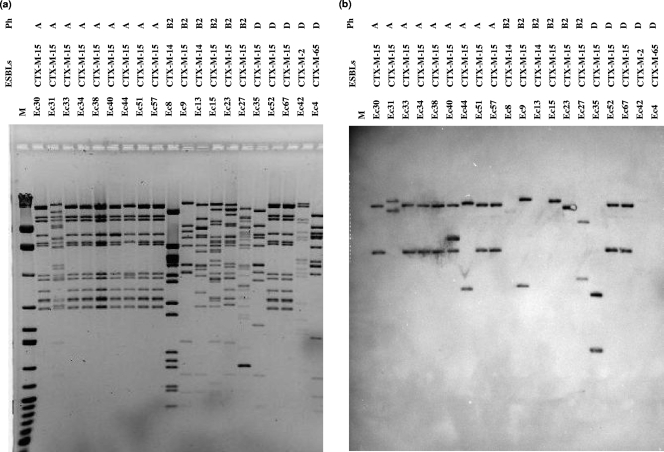
The antimicrobial resistance genes blaOXA-1 and aac-(6′)-Ib were consistently present on the blaCTX-M-15-carrying plasmids of all phylotype A isolates tested. Unlike the international epidemic clone, however, our blaCTX-M-15-carrying plasmids in this clonal group did not possess blaTEM (5, 11). For CTX-M-15-producing E. coli isolates in phylotype B2, after multiple attempts at conjugation and transformation from all parent strains, plasmids from only four isolates were transferable by conjugation or transformation. The other antimicrobial resistance genes located on this blaCTX-M-15-carrying plasmid were variable, with plasmids from two isolates carrying blaOXA-1, a plasmid from one isolate carrying blaTEM, and a plasmid from one isolate carrying both blaTEM and blaOXA-1. The restriction profiles of three of these four plasmids showed 60% to 85% similarities to each other and up to 60% similarity to those of phylotype A isolates (Fig. 2 and 3). However, Southern hybridization analysis showed that the plasmid fragments containing blaCTX-M-15 were all different in size from each other and from those of phylotype A isolates. The profile of a single blaCTX-M-15-carrying plasmid from a phylotype B2 isolate (Ec9) was very similar to that of pC15-1a, previously reported from Canada (5). However, the blaCTX-M-15 probe hybridized to ca. 23-kb and 4.5-kb plasmid fragments, whereas the Canadian plasmids hybridized to ca. 23-kb and 12-kb plasmid fragments (Fig. 3). The other three plasmids in phylotype B2 isolates (Ec15, Ec23, and Ec27) had profiles similar to those of “cluster I” plasmids, reported in isolates from multiple countries (Fig. 2) (11). Plasmid profiles of phylotype A did not match with published plasmid profiles.
FIG. 2.
Dendrogram of plasmids carrying blaCTX-M. Ph, phylotypes.
FIG. 3.
Profiles of HpaI-digested plasmids from CTX-M-producing E. coli isolates and Southern hybridization. (a) Plasmid profiles of transformants; (b) Southern hybridization of plasmids using a digoxigenin-labeled blaCTX-M-15 probe. M, λ/HindIII and 100-bp ladder (MBI Fermentas); Ph, phylotypes.
DISCUSSION
Prior to 2007, CTX-M-producing E. coli isolates were rarely reported in the United States (7, 22). At our institution, TEM- and SHV-type ESBLs were still predominant until late 2006, when CTX-M-15-producing E. coli isolates were identified (13). The number of clinical cases caused by CTX-M-producing E. coli then increased steadily after around August 2007. The proportion of CTX-M production among ESBL-producing E. coli isolates in our study was high (57 of 70 isolates [81%]). This result was consistent with the findings of a nationwide survey, which detected CTX-M-type ESBLs in 83% of ESBL-producing E. coli isolates (7). A recent study conducted in two hospitals in Philadelphia also reported that a significant portion (48%) of their cephalosporin-resistant E. coli isolates produced CTX-M-type ESBLs (24). Our findings add to the growing evidence that CTX-M-type ESBLs, in particular CTX-M-15, are becoming the predominant ESBLs in E. coli strains in the United States.
Our study identified several noteworthy features of the molecular epidemiology of CTX-M-producing E. coli isolates at our institution. First, two major clones of CTX-M-15-producing E. coli were identified. The first was a CTX-M-15-producing E. coli clone with the characteristics of the international epidemic clone, i.e., phylotype B2, serotype O25:H4, and ST131 (9, 11, 27, 38). Some variation in PFGE profiles was observed for isolates within the ST131 clonal group. This phenomenon has been reported previously (20, 27). The second clone, which is unique to this study, was a CTX-M-15-producing E. coli clone belonging to phylotype A and ST410. A recent study of CTX-M-15-producing E. coli isolates in the United States reported that 45% of such isolates were ST131 (17). Our study found a lower percentage of ST131 among our CTX-M-15-producing E. coli isolates (33%). The predominant CTX-M-15-producing clone at our institution was that in phylotype B2 until late 2007. However, the phylotype A clone replaced the phylotype B2 clone thereafter and continued to predominate until the end of the study period. Isolates in phylotype A, along with those in phylotype B1, are conventionally considered commensals, less virulent than isolates in phylotypes B2 and D. The tendency of the CTX-M-15-producing phylotype A clone to spread therefore mandates further investigation, including detailed clinical analysis and determination of virulence factors.
Second, in addition to the clonal spread of phylotype A CTX-M-15-producing E. coli, which was demonstrated by both PFGE and plasmid analysis, horizontal transfer of the blaCTX-M-15-carrying plasmid appears to have occurred between phylotypes A and D. While we cannot determine which phylotype acquired this plasmid first, the fact that this plasmid is able to maintain its stability regardless of different phylogenetic backgrounds is intriguing. On the other hand, plasmid profiles of phylotype B2 isolates were more variable. This is consistent with the findings from Europe, Canada, and Asia documenting the genetic variability of blaCTX-M-15-carrying plasmids in this phylotype (11). It is worth noting that the fingerprints of the blaCTX-M-15-carrying plasmids from phylotype B2 were clonally related (>60% similarity) to that of the plasmid representing the CTX-M-15-producing phylotype A clone, suggesting the possibility of common ancestry of these plasmids across the phylotypes.
We tried to identify the location of blaCTX-M-15 in the rest of the phylotype B2 isolates, whose plasmids were not transferred by conjugation and transformation using several approaches. Southern hybridization analysis using a blaCTX-M-15 probe and a PFGE gel showed that blaCTX-M-15 was located on fragments larger than 200 kb, indicating that these fragments were either on very large plasmids or on the chromosome. Southern hybridization analysis of plasmids from these isolates was negative for blaCTX-M-15 (data not shown). Finally, S1 nuclease analysis revealed the presence of large plasmids (>50 kb) in these isolates, none of which hybridized with the blaCTX-M-15 probe (data not shown) (16). These results suggested that blaCTX-M-15 was likely located on the chromosome for these isolates (11, 15).
Third, while the majority of ST131 isolates in our study produced CTX-M-15 (n = 15), we also observed ST131 for E. coli isolates producing CTX-M-14 (n = 3) and SHV-5 or -7 (n = 3). A recent study from Japan showed an association of serotype O25 with ST73 in addition to ST131 (36). In contrast to other studies, all of these ST131 isolates in this Japanese study produced enzymes of the CTX-M-2 and CTX-M-9 groups. ST131 isolates without blaCTX-M-15 have also been isolated from healthy humans in France and have been found among virulent E. coli isolates in the United States (3, 21). Thus, it is possible that ST131 strains are particularly adept at acquiring plasmids that carry various resistance determinants, not limited to blaCTX-M-15.
In summary, we characterized the emergence of two predominant clones of E. coli producing CTX-M-15 at our institution: one that constitutes part of the worldwide epidemic and one that has not been reported previously and thus appears to have emerged locally. Our findings further demonstrate the versatility of CTX-M-15 and the plasmids encoding this ESBL. Detailed molecular typing of plasmids and genomes of isolates from various locations will help better define the epidemiology of CTX-M-15-producing E. coli in the Unites States. In addition, further work is required to examine the virulence of CTX-M-15-producing isolates in various phylotypes and to evaluate the clinical impact of acquisition of these isolates.
Acknowledgments
We thank the staff of the Microbiology Laboratory of the University of Pittsburgh Medical Center for providing the study isolates. We also thank George A. Jacoby for the kind gift of E. coli J53AziR.
Footnotes
Published ahead of print on 17 August 2009.
REFERENCES
- 1.Apisarnthanarak, A., P. Kiratisin, P. Saifon, R. Kitphati, S. Dejsirilert, and L. M. Mundy. 2007. Clinical and molecular epidemiology of community-onset, extended-spectrum β-lactamase-producing Escherichia coli infections in Thailand: a case-case-control study. Am. J. Infect. Control 35:606-612. [DOI] [PubMed] [Google Scholar]
- 2.Bert, F., C. Branger, and N. Lambert-Zechovsky. 2002. Identification of PSE and OXA β-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J. Antimicrob. Chemother. 50:11-18. [DOI] [PubMed] [Google Scholar]
- 3.Billig, M., R. Lynfield, X. Qin, J. R. Johnson, and S. J. Weissman. 2008. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. G2-1273.
- 4.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, and M. R. Mulvey. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum β-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48:3758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Cantón, R., and T. M. Coque. 2006. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9:466-475. [DOI] [PubMed] [Google Scholar]
- 6.Carattoli, A., A. Garcia-Fernandez, P. Varesi, D. Fortini, S. Gerardi, A. Penni, C. Mancini, and A. Giordano. 2008. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases isolated in Rome, Italy. J. Clin. Microbiol. 46:103-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castanheira, M., R. E. Mendes, P. R. Rhomberg, and R. N. Jones. 2008. Rapid emergence of blaCTX-M among Enterobacteriaceae in U.S. medical centers: molecular evaluation from the MYSTIC Program (2007). Microb. Drug Resist. 14:211-216. [DOI] [PubMed] [Google Scholar]
- 8.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clermont, O., M. Lavollay, S. Vimont, C. Deschamps, C. Forestier, C. Branger, E. Denamur, and G. Arlet. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024-1028. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; seventeenth informational supplement. M100-17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Coque, T. M., A. Novais, A. Carattoli, L. Poirel, J. Pitout, L. Peixe, F. Baquero, R. Canton, and P. Nordmann. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Oliveira Garcia, D., Y. Doi, D. Szabo, J. M. Adams-Haduch, T. M. Vaz, D. Leite, M. C. Padoveze, M. P. Freire, F. P. Silveira, and D. L. Paterson. 2008. Multiclonal outbreak of Klebsiella pneumoniae producing extended-spectrum β-lactamase CTX-M-2 and novel variant CTX-M-59 in a neonatal intensive care unit in Brazil. Antimicrob. Agents Chemother. 52:1790-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doi, Y., J. Adams, A. O'Keefe, Z. Quereshi, L. Ewan, and D. L. Paterson. 2007. Community-acquired extended-spectrum β-lactamase producers, United States. Emerg. Infect. Dis. 13:1121-1123. [DOI] [PubMed] [Google Scholar]
- 14.Edelstein, M., M. Pimkin, I. Palagin, I. Edelstein, and L. Stratchounski. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galani, I., M. Souli, Z. Chryssouli, and H. Giamarellou. 2007. Detection of CTX-M-15 and CTX-M-33, a novel variant of CTX-M-15, in clinical Escherichia coli isolates in Greece. Int. J. Antimicrob. Agents 29:598-600. [DOI] [PubMed] [Google Scholar]
- 16.García, A., F. Navarro, E. Miro, B. Mirelis, S. Campoy, and P. Coll. 2005. Characterization of the highly variable region surrounding the blaCTX-M-9 gene in non-related Escherichia coli from Barcelona. J. Antimicrob. Chemother. 56:819-826. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. R., B. D. Johnston, J. H. Jorgensen, J. S. Lewis II, A. Robisek, M. Menard, C. Clabots, S. J. Weissman, N. D. Hanson, R. Owens, K. Lolans, and J. Quinn. 2008. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. K-3444.
- 18.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 19.Karisik, E., M. J. Ellington, R. Pike, R. E. Warren, D. M. Livermore, and N. Woodford. 2006. Molecular characterization of plasmids encoding CTX-M-15 β-lactamases from Escherichia coli strains in the United Kingdom. J. Antimicrob. Chemother. 58:665-668. [DOI] [PubMed] [Google Scholar]
- 20.Lau, S. H., M. E. Kaufmann, D. M. Livermore, N. Woodford, G. A. Willshaw, T. Cheasty, K. Stamper, S. Reddy, J. Cheesbrough, F. J. Bolton, A. J. Fox, and M. Upton. 2008. UK epidemic Escherichia coli strains A-E, with CTX-M-15 β-lactamase, all belong to the international O25:H4-ST131 clone. J. Antimicrob. Chemother. 62:1241-1244. [DOI] [PubMed] [Google Scholar]
- 21.Leflon-Guibout, V., J. Blanco, K. Amaqdouf, A. Mora, L. Guize, and M. H. Nicolas-Chanoine. 2008. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J. Clin. Microbiol. 46:3900-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis, J. S., II, M. Herrera, B. Wickes, J. E. Patterson, and J. H. Jorgensen. 2007. First report of the emergence of CTX-M-type extended-spectrum β-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob. Agents Chemother. 51:4015-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livermore, D. M., R. Canton, M. Gniadkowski, P. Nordmann, G. M. Rossolini, G. Arlet, J. Ayala, T. M. Coque, I. Kern-Zdanowicz, F. Luzzaro, L. Poirel, and N. Woodford. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165-174. [DOI] [PubMed] [Google Scholar]
- 24.McGettigan, S. E., B. Hu, K. Andreacchio, I. Nachamkin, and P. H. Edelstein. 8 July 2009. Prevalence of CTX-M β-lactamases in Philadelphia. J. Clin. Microbiol. doi: 10.1128/JCM.00319-09. [DOI] [PMC free article] [PubMed]
- 25.Moland, E. S., J. A. Black, A. Hossain, N. D. Hanson, K. S. Thomson, and S. Pottumarthy. 2003. Discovery of CTX-M-like extended-spectrum β-lactamases in Escherichia coli isolates from five U.S. states. Antimicrob. Agents Chemother. 47:2382-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muzaheed, Y. Doi, J. M. Adams-Haduch, A. Endimiani, H. E. Sidjabat, S. M. Gaddad, and D. L. Paterson. 2008. High prevalence of CTX-M-15-producing Klebsiella pneumoniae among inpatients and outpatients with urinary tract infection in Southern India. J. Antimicrob. Chemother. 61:1393-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicolas-Chanoine, M. H., J. Blanco, V. Leflon-Guibout, R. Demarty, M. P. Alonso, M. M. Canica, Y. J. Park, J. P. Lavigne, J. Pitout, and J. R. Johnson. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273-281. [DOI] [PubMed] [Google Scholar]
- 28.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paterson, D. L. 2006. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control 34:S20-S28, S64-S73. [DOI] [PubMed] [Google Scholar]
- 30.Pitout, J. D., D. B. Gregson, D. L. Church, S. Elsayed, and K. B. Laupland. 2005. Community-wide outbreaks of clonally related CTX-M-14 β-lactamase-producing Escherichia coli strains in the Calgary health region. J. Clin. Microbiol. 43:2844-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum β-lactamase CTX-M-15 and of its structurally related β-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 32.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 33.Robberts, F. J., P. C. Kohner, and R. Patel. 2009. Unreliable extended-spectrum β-lactamase detection in the presence of plasmid-mediated AmpC in Escherichia coli clinical isolates. J. Clin. Microbiol. 47:358-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabaté, M., R. Tarrago, F. Navarro, E. Miro, C. Verges, J. Barbe, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sidjabat, H. E., D. L. Paterson, Z. A. Qureshi, J. M. Adams-Haduch, A. O'Keefe, A. Pascual, J. Rodriguez-Bano, and Y. Doi. 2009. Clinical features and molecular epidemiology of CMY-type β-lactamase-producing Escherichia coli. Clin. Infect. Dis. 48:739-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, S., N. Shibata, K. Yamane, J. Wachino, K. Ito, and Y. Arakawa. 2009. Change in the prevalence of extended-spectrum-β-lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 63:72-79. [DOI] [PubMed] [Google Scholar]
- 37.Tartof, S. Y., O. D. Solberg, A. R. Manges, and L. W. Riley. 2005. Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J. Clin. Microbiol. 43:5860-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yumuk, Z., G. Afacan, M. H. Nicolas-Chanoine, A. Sotto, and J. P. Lavigne. 2008. Turkey: a further country concerned by community-acquired Escherichia coli clone O25-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 62:284-288. [DOI] [PubMed] [Google Scholar]