Abstract
Transposon inactivation of ycgE, a gene encoding a putative transcriptional regulator, led to decreased multidrug susceptibility in an Escherichia coli lon mutant. The multidrug susceptibility phenotype (e.g., to tetracycline and β-lactam antibiotics) required the inactivation of both lon and ycgE. In this mutant, a decreased amount of OmpF porin contributes to the lowered drug susceptibility, with a greater effect at 26°C than at 37°C.
Inhibition of MarR, the repressor of the marRAB operon, causes increased expression of MarA, leading to upregulation of transcription of the acrAB operon and of tolC, which together specify a multidrug efflux pump (1, 2). Additionally MarA upregulates the micF gene (8) encoding a small RNA preventing the translation of the porin ompF mRNA (19). However the multidrug susceptibility of M113R, an Escherichia coli K-12 AG100 strain carrying a lon3::IS186 mutation (lon) was not different from the parental strain, AG100 (23), despite a greater level of MarA than that in AG100. In the lon strain, the amount of MarA was about half that in a multidrug-resistant marR mutant strain, yet the levels of expression of the MarA-regulated acrAB transcript were apparently similar in both M113R and AG100 (23). In order to identify a locus, possibly preventing multidrug resistance (MDR) in the lon strain, we performed random mutagenesis of the M113R strain and selected mutants with decreased drug susceptibility.
After electroporation with the EZ-Tn5 <Kan-2> transposon (Epicentre), kanamycin-resistant transformants were replica plated onto Luria-Bertani (LB) agar plates containing 35 μg/ml kanamycin and either 1.5 μg/ml of tetracycline or 3 μg/ml ampicillin (two and three times the wild-type AG100 MIC, respectively) and grown overnight at 37°C. From 1,000 transformants, one with decreased susceptibility to ampicillin and tetracycline was selected and termed lon EZ3. The reduced susceptibility to ampicillin and tetracycline could be cotransduced with Tn5 from the lon EZ3 mutant into another M113R by P1 transduction (data not shown; see reference 23 for P1 transduction method). Interestingly, transduction of the Tn5 insertion from the lon EZ3 into the wild-type AG100 did not lead to decreased antibiotic susceptibility, indicating that the resistance phenotype of the lon EZ3 strain was dependent on a lon background. Inverse PCR, as recommended by Epicentre, identified the Tn5-inactivated gene in lon EZ3 as ycgE, specifying a protein similar to MlrA, the curli regulator of Salmonella and E. coli (3).
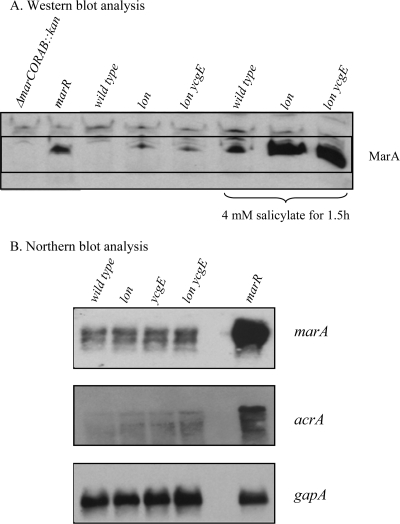
We examined the possible involvement of MarA in the decreased susceptibility of the lon ycgE mutant. With a half-life of <1 min (12), MarA is unstable in the presence of Lon protease. We grew wild-type AG100 and mutants in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.5. After sonication in lysis buffer {20 mM Tris-HCl, 100 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride] [pH 7.5]} and centrifugation to remove unlysed cells, total proteins were quantified by bicinchoninic acid assay (Pierce), separated by a 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and detected with anti-MarA antibodies. MarA was undetectable in AG100, and its stabilized basal level was present in similar amounts in the lon and lon ycgE mutants, although at a level less than that in the marR mutant (Fig. 1A). When MarA production was induced in the presence of 4 mM salicylate (Fig. 1A), lon and lon ycgE mutants showed similar MDR phenotypes to ampicillin, tetracycline, chloramphenicol, and nalidixic acid (data not shown), indicating that MarA was active in all strains. We investigated the expression of the marRAB and acrAB operons in the wild type and mutants by Northern blot analysis using RNA isolated from cells grown in LB medium at 37°C to an OD600 of 0.5. As expected, marRAB and acrAB operons were upregulated in the marR mutant (Fig. 1B), consistent with the increased MarA and MDR of the marR strain. The wild-type, lon, ycgE, and lon ycgE strains all showed the same low level of expression of marRAB. Contrary to previously reported semiquantitative reverse transcription-PCR results (23), the transcription of acrAB in the wild type was similar to that in the lon mutant and was clearly less than that in the marR mutant, consistent with the drug susceptibility of the lon mutant (23).
FIG. 1.
Analysis of MarA amounts and marRAB and acrAB expression. (A) Immunodetection of MarA in whole-cell extracts by Western blot analysis. MarA was detected using anti-MarA antibody (26) diluted 1/2,000 and horseradish peroxidase conjugated to anti-rabbit immunoglobulin G (Cell Signaling Technology) as secondary antibody, with the Western lighting kit (Perkin Elmer). See Table 2 for details about the strains. A total of 25 μg of proteins was loaded in each lane of the gel. (B) Northern blot analysis of total RNA (3 μg) prepared from E. coli AG100 wild type and mutants. Northern blot analyses were performed with the NorthernMax kit (Ambion) using biotinylated RNA probes (see primers listed in Table 1). Detection was carried out using the BrightStar BioDetect kit (Ambion). A concentration of 0.1 nM of each probe was hybridized for 16 h at 68°C. A gapA probe was used as control for hybridization.
The inactivation of ycgE in a lon-positive or lon-negative background had no effect on the transcription of the marRAB and acrAB operons (Fig. 1B). Recently, Martin et al. (16) showed that the genes belonging to the mar regulon require a relatively high level of MarA for activation, especially the acrAB operon. Even though we did not quantify the amount of MarA protein in lon-positive and lon-negative strains because of its different stabilities in these strains, we concluded that the lack of resistance of the lon mutant described previously (23) is because the amount of MarA is insufficient to activate the transcription of acrAB. We also concluded that the decreased susceptibility of a lon ycgE mutant compared to that of a lon mutant to ampicillin and tetracycline was not caused by an increased level or activity of MarA or by an increased expression of AcrAB.
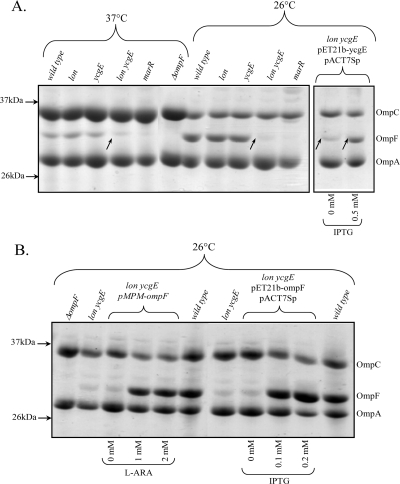
OmpF functions as a diffusion pore, allowing small, hydrophilic molecules into the periplasm of E. coli. The decreased susceptibility to tetracycline and ampicillin observed for the lon ycgE mutant could be associated with a decrease in OmpF in the strain. The outer membrane proteins of each strain were prepared as the Sarkosyl-insoluble fraction of the membrane preparation (8) of cells grown in LB medium to an OD600 of 0.5 and separated by 10% urea-SDS-PAGE. The amount of OmpF in the outer membrane was determined in each strain at 37°C and 26°C, since OmpF expression is regulated by temperature (14) (Fig. 2A). As expected, the marR mutant expressed a reduced amount of OmpF at both temperatures because of the MarA-dependent upregulation of micF (8). At both temperatures, a single mutation of either lon or ycgE had no effect on OmpF expression. The lon ycgE mutant produced slightly less OmpF than did the wild type at 37°C. While the wild-type, lon, and ycgE strains responded to decreased temperature by upregulating OmpF, the lon ycgE strain did not (Fig. 2A). We performed complementation experiments to confirm the effect of YcgE on the OmpF level, using the pET21b plasmid (see Table 2 for details). After amplification using the primers listed in Table 1, ycgE was cloned into pET21b, and the nucleotide sequence was verified. YcgE was overproduced in the presence of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in cells carrying pET21b-ycgE and pACT7Sp (see Table 2 for details). Figure 2A shows that the overexpression of YcgE in the lon ycgE mutant restored OmpF in the outer membrane, confirming that the OmpF level is affected by YcgE.
FIG. 2.
Effect of YcgE and Lon on OmpF levels. (A) Six molar urea-SDS-PAGE (10%) of outer membrane protein extracts from wild-type E. coli AG100 and mutants. To determine the effect of the overexpression of YcgE on the lon ycgE mutant, the outer membrane proteins were isolated from cells carrying plasmids pACT7Sp and pET21b-ycgE (see text and Table 2 for details). Arrows within the gel point to OmpF in the lon ycgE mutant. (B) Six molar urea-SDS-PAGE (10%) of outer membrane protein extracts from lon ycgE mutant expressing OmpF from a plasmid (see text and Table 2 for details). For both panels, the gels were stained with Coomassie blue. A total of 10 μg of protein extracts was loaded in each lane. L-ARA, l-arabinose.
TABLE 2.
Bacterials strains and plasmidsa
| Strain or plasmid | Genotype or characteristic(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| Wild-type AG100 | argE3 thi-1 rpsL xyl mtl supE44 λ lysogen | 11 |
| lon mutant | AG100 lon3::IS186 | 23 |
| lon ycgE mutant | AG100 lon3::IS186 ycgE::Tn5 | This study |
| ycgE mutant | AG100 ycgE::Tn5 by P1 transduction of AG100 × lon ycgE | This study |
| marR mutant | AG100 marR mutant | 24 |
| ΔmarCOAB::kan mutant | AG100 in which the marCORAB divergon is deleted | 15 |
| Plasmids | ||
| pACT7 | T7 RNA polymerase regulated by the lacUV5; ori p15A; Kanr | 18 |
| pACT7Sp | pACT7 Spr; this plasmid is used in conjunction with pET21b | This study |
| pET21b | Expression cloning vector; Ampr; the transcription of the cloned gene is driven by the T7 RNA polymerase and controlled by the LacI repressor | Novagen |
| pET21b-ycgE | pET21b in which ycgE was cloned using the NdeI and XhoI restriction sites | This study |
| pET21b-ompF | pET21b in which ompF was cloned using the NdeI and XhoI restriction sites | This study |
| pMPM | Expression cloning vector; Tetr; ori p15A; the araBAD promoter initiates the transcription of the cloned gene | 17 |
| pMPM-ompF | pMPM in which ompF was cloned using the EcoRI and XhoI restriction sites | This study |
| pKD46 | Red helper plasmid carrying the Red recombinase from the phage λ; Ampr; ori R101 | 9 |
| pKD3 | Plasmid carrying an FRT-flanked chloramphenicol resistance (cat) gene; Cmr; ori Rγ | 9 |
| pCP20 | Flp helper plasmid carrying gene encoding the yeast Flp recombinase; thermal induction of Flp; Ampr Cmr; λpR Repts | 5 |
Amp, ampicillin; Cm, chloramphenicol; Sp, spectinomycin; Tet, tetracycline.
TABLE 1.
Primers used during this study
| Primer | Sequence (5′→3′) | Description |
|---|---|---|
| MarA-UP | ATGTCCAGACGCAATACTGACGCTATTAC | Construction of the marA RNA antisense probe using the Maxiscript kit (Ambion) for Northern blot analysis. The resulting PCR product contains the promoter for the T7 RNA polymerase (shown in boldface type). |
| MarA-T7 | TAATACGACTCACTATAGGCGTTGCGACTCGAAGCCATA | |
| AcrA-UP | ATGAACAAAAACAGAGGGTTTACGCCT | Construction of the acrA RNA antisense probe using the Maxiscript kit (Ambion) for Northern blot analysis. The resulting PCR product contains the promoter for the T7 RNA polymerase (shown in boldface type). |
| AcrA-T7 | TAATACGACTCACTATAGGCGCTTCAGGATAATCCCGCT | |
| GapA-UP | ATGATGAAGCGCAATATTCTGGCAGTG | Construction of the gapA RNA antisense probe using the Maxiscript kit (Ambion) for Northern blot analysis. The resulting PCR product contains the promoter for the T7 RNA polymerase (shown in boldface type). |
| GapA-T7 | TAATACGACTCACTATAGGCCACCAGCGGTGATGTGTTTACGAGCAG | |
| OmpF-A | CTTATTGACGGCAGTGGCAGGTGTCATAAAAAAAACCATGAGGGTAATAAATAATGATGGTGTAGGCTGGAGCTGCTTCGAAG | Amplification of the Flp recombination target (FRT)-flanked chloramphenicol resistance gene (cat) from plasmid pKD3 containing sequences upstream and downstream of ompF (shown in boldface type). cat was inserted into ompF by recombination mediated by the λ Red recombinase. Afterwards, cat was deleted using the pCP20 plasmid (9). |
| OmpF-B | GTATAAAAAAAACAGGACCAAAGTCCTGTTTTTTCGGCATTTAACAAAGAGGTGTGCTATTAATGGGAATTAGCCATGGTCCATATG | |
| OmpF-C | GTTACATATTTTTTCTTTTTGAAACCAAATC | Verification of the insertion of cat into ompF (combination of primers OmpF-C/Cat-1 and OmpF-D/Cat-2). |
| OmpF-D | TAATGTTCTCAAACATGACGAGGTTCC | |
| Cat-1 | CTTCGAAGCAGCTCCAGCCTACAC | |
| Cat-2 | ACGTGCCGATCAACGTCTCATTTTC | |
| OmpF-EcoRI | AAAAAAGAATTCACCATGATGAAGCGCAATATTCTGGCA | Amplification of ompF nucleotide sequence from AG100 genomic DNA. Primers OmpF-EcoRI and OmpF-XhoI contain EcoRI and XhoI restriction sites, respectively, at the 5′ end (shown in boldface type) in order to clone the ompF nucleotide sequence into the pMPM plasmid (17). |
| OmpF-XhoI | TTTTTTCTCGAGTTAGAACTGGTAAACGATACCCACAGC | |
| OmpF-NdeI | AAAAAACCATATGATGAAGCGCAATATTCTGGCAGTG | Amplification of ompF nucleotide sequence from AG100 genomic DNA. Primers OmpF-NdeI and OmpF-XhoI contain NdeI and XhoI restriction sites, respectively, at the 5′ end (shown in boldface type) in order to clone the ompF nucleotide sequence into the pET21b plasmid (Novagen). |
| OmpF-XhoI | TTTTTTCTCGAGTTAGAACTGGTAAACGATACCCACAGC | |
| YcgE-NdeI | CATATGGCTTATAGCATTGGTGATGTTG | Amplification of ycgE nucleotide sequence from AG100 genomic DNA. Primers YcgE-NdeI and YcgE-XhoI contain NdeI and XhoI restriction sites, respectively, at the 5′ end (shown in boldface type) in order to clone the ycgE nucleotide sequence into the pET21b plasmid (Novagen). |
| YcgE-XhoI | CTCGAGTTAGGGGGCATGAAAGATGACTTTATAAC |
To prove that the reduced amount of OmpF was responsible for the lower susceptibility of the lon ycgE mutant, ompF was deleted in the wild-type strain and in the lon ycgE mutant by double recombination (primers listed in Table 1) (9), and the MICs of several antibiotics were measured at 37°C and 26°C (Table 3). The loss of ompF significantly decreased the wild-type susceptibility to ampicillin, cefoxitin, cephalothin (cefalotin), tetracycline, and doxycycline at both temperatures (Table 3). The wild-type strain presented a higher susceptibility at 26°C than at 37°C for β-lactam and tetracycline antibiotics (Table 3). This increased susceptibility observed at 26°C for the wild type presumably results from the large amount of OmpF that allows the rapid influx of these antibiotics into the cell. The ratio of the MIC of the ompF mutant to the MIC of the wild type for β-lactam and tetracycline antibiotics was higher at 26°C than at 37°C, indicating that a decreased OmpF level had a greater impact on susceptibility to these antibiotics at 26°C than at 37°C. The MICs of monoanionic cephalosporins, such as cephalothin and cefoxitin, observed for the ompF mutant were 4.7 and 4 times higher than the wild type at 26°C. This observation confirms that OmpF is important in the accumulation of these hydrophilic drugs into E. coli. The MIC of the ompF mutant to ampicillin, a penicillin, was only two times that of the wild-type strain at 26°C. Cephalosporins diffuse through the porin channel at rates that are two to three times higher than those of ampicillin, presumably related to their greater hydrophilicity (28). The MIC of tetracyclines toward the ompF mutant was four times higher than for the wild type at 26°C. Strains of E. coli K-12 lacking OmpF have been shown to be 1.5- to 3-fold less susceptible to tetracycline, a small hydrophilic compound (6, 7). The lon ycgE, the ΔompF, and the lon ycgE ΔompF mutants showed similar MICs to β-lactams and tetracyclines at both temperatures, indicating that the decreased susceptibility observed in the lon ycgE mutant was associated with a loss of OmpF. Nalidixic acid, a hydrophobic quinolone, penetrates mainly through the phospholipid bilayer (4, 13), explaining why the loss of OmpF in these mutants had no impact on nalidixic acid susceptibility (Table 3) (7). The decreased nalidixic acid susceptibility observed for the marR mutant presumably comes from the MarA-dependent increased expression of the AcrAB/TolC efflux pump.
TABLE 3.
MIC determined by Etests on LB medium at 37°C and 26°Ca
| Strain | MIC (μg/ml) at 37°C/26°C |
||||||
|---|---|---|---|---|---|---|---|
| FX | AM | CE | Cm | TC | DC | NA | |
| Wild-type AG100 | 1.5/0.75 | 1/0.75 | 2/1 | 4/4 | 1/0.5 | 1/0.5 | 4/4 |
| lon mutant | 1.7/0.92 | 1/0.83 | 2/1.2 | 4/4 | 1/0.5 | 1.17/0.75 | 4/3.7 |
| ycgE mutant | 1.7/0.92 | 1/0.75 | 2.3/1.3 | 4/4 | 1/0.5 | 1.17/0.83 | 4/3.7 |
| lon ycgE mutant | 4/4 | 1.7/1.8 | 4.7/6 | 9.3/12 | 2/2 | 2/2 | 4/3.3 |
| marR mutant | 6/7.3 | 3/4 | 9.3/10.7 | 21/24 | 4/6 | 3.3/3.7 | 10.7/6.7 |
| ΔompF mutant | 3/3 | 1.83/1.5 | 4.7/4.7 | 6.7/6 | 1.83/2 | 2/2 | 4/3.3 |
| lon ycgE ΔompF mutant | 3/4.7 | 2.2/2.3 | 4.7/6.7 | 10.7/13.3 | 2/2 | 2/2 | 4/3.3 |
| lon ycgE mutant carrying pMPM | 6 | 2.3 | 5.3 | ND | 3 | ||
| lon ycgE mutant carrying pMPM-ompF | 1 | 1.5 | 2 | ND | 3 | ||
| lon ycgE mutant carrying pET21b and pACT7SP | ND | ND | ND | 2 | 3 | ||
| lon ycgE mutant carrying pET21b-ompF and pACT7Sp | ND | ND | ND | 0.75 | 3 | ||
FX, cefoxitin; AM, ampicillin; CE, cephalothin; Cm, chloramphenicol; TC, tetracycline; DC, doxycycline; NA, nalidixic acid; ND, not able to be determined because of intrinsic resistance of the host. Numbers represent an averages of at least four experimental measurements. The standard errors of the means were <20% for all experiments. For the mutants carrying plasmids, MICs were determined at 26°C.
The relationship between the reduced β-lactam and tetracycline susceptibility of the lon ycgE mutant to the amount of OmpF was examined in cells with OmpF restored via pMPM (see Table 2 for details) and pET21b. ompF was amplified by the PCR using primers listed in Table 1 and was cloned into the pMPM and pET21b, and the nucleotide sequences of the resulting clones were verified. The MICs of ampicillin, cefoxitin, cephalothin, and nalidixic acid were determined in the presence of 20 μg/ml tetracycline and 1.3 mM l-arabinose, a concentration allowing the restoration of OmpF in the outer membrane in the lon ycgE strain carrying the pMPM-ompF plasmid (Fig. 2B). The MICs of doxycycline and nalidixic acid were measured in the presence of 100 μg/ml ampicillin, 50 μg/ml spectinomycin, and 0.1 mM IPTG, a concentration allowing the restoration of OmpF in the outer membrane of the lon ycgE mutant carrying pET21b-ompF and pACT7Sp (Fig. 2B). The restoration of OmpF in the lon ycgE mutant increased its susceptibility to β-lactams and doxycycline but, as expected, not to nalidixic acid (Table 3). Our results indicated that the major cause of the lower susceptibility of the lon ycgE mutant to β-lactams and tetracyclines was the loss of OmpF, with a more noticeable effect at 26°C than at 37°C correlating with the greater reduction of OmpF seen at 26°C (Fig. 2A). The ΔompF mutant was less susceptible than the wild type but more susceptible than the lon ycgE mutant to chloramphenicol. This suggests that although the loss of OmpF in the lon ycgE mutant contributed to the decreased susceptibility of the strain to chloramphenicol, another ompF-independent mechanism was involved in the chloramphenicol resistance of the lon ycgE mutant. It has been shown that the loss of OmpF reduced accumulation of chloramphenicol by only <10% (21).
A network of transcriptional regulators and small noncoding RNAs control the expression of porins in enterobacteria in response to changes in temperature, osmolarity, phosphate concentration, and stress (10, 20, 25, 27). ycgE and lon now join the growing number of genetic loci which affect OmpF expression. The inactivation of YcgE by YcgF results in the induction of eight small proteins (26). YcgE directly represses the ycgZ-ymgABC operon and downregulates yliL and ynaK by as-yet-uncharacterized mechanisms (26). With several target genes, YcgE seems to affect different regulation pathways. We demonstrated the involvement of ycgE and lon together in the regulation of the outer membrane protein OmpF. The absence of lon and ycgE led to decreased susceptibility to different types of drugs with a greater impact at 26°C because the lon ycgE mutant failed to increase the OmpF level at 26°C.
Bacteria face low concentrations of antimicrobial drugs in natural environments, to which they may respond with various mechanisms of resistance, possibly involving lon mutations. lon mutants arose on agar plates containing small amounts of antibiotics (22). These lon mutants had a mutator phenotype allowing the selection of spontaneous drug-resistant mutants at a higher frequency (10 to >100 times) than in the wild type (22). How the Lon protease is involved in the regulation of OmpF through YcgE remains unclear. Protein stability experiments have shown that YcgE is not a substrate of Lon protease (data not shown). We postulate that the regulation of OmpF through YcgE is indirect, involving an intermediate protein which is a substrate for Lon and is negatively regulated by YcgE. This would explain why both lon and ycgE mutations were necessary to observe a decrease in the OmpF level. This intermediate could be one of the YcgE targets pinpointed by Tschowri et al. (26) or an as-yet-undiscovered YcgE regulated protein.
In summary, this study demonstrated that the lower susceptibility of the lon ycgE mutant to β-lactam and tetracycline antibiotics did not result from an overexpression of MarA or AcrAB. Rather, the lower drug susceptibility of the lon ycgE mutant is linked to a decreased level of OmpF.
Acknowledgments
This work was supported by United States Public Health Service grant AI56021 from the National Institutes of Health to S.B.L. V.D. was supported in part by a Fellowship of the Belgian American Educational Foundation as a Université de Liège Fellow.
We thank Mark Silby and Laura McMurry for stimulating discussion.
Footnotes
Published ahead of print on 31 August 2009.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aono, R., N. Tsukagoshi, and M. Yamamoto. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, P. K., C. M. Dozois, C. A. Nickerson, A. Zuppardo, J. Terlonge, and R. Curtiss III. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 41:349-363. [DOI] [PubMed] [Google Scholar]
- 4.Chapman, J. S., and N. H. Georgopapadakou. 1988. Routes of quinolone permeation in Escherichia coli. Antimicrob. Agents Chemother. 32:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 6.Chopra, I., and S. J. Eccles. 1978. Diffusion of tetracycline across the outer membrane of Escherichia coli K-12: involvement of protein Ia. Biochem. Biophys. Res. Commun. 83:550-557. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. P., L. M. McMurry, D. C. Hooper, J. S. Wolfson, and S. B. Levy. 1989. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 33:1318-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, S. P., L. M. McMurry, and S. B. Levy. 1988. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J. Bacteriol. 170:5416-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forst, S., and M. Inouye. 1988. Environmentally regulated gene expression for membrane proteins in Escherichia coli. Annu. Rev. Cell Biol. 4:21-42. [DOI] [PubMed] [Google Scholar]
- 11.George, A. M., and S. B. Levy. 1983. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J. Bacteriol. 155:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffith, K. L., I. M. Shah, and R. E. Wolf, Jr. 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol. Microbiol. 51:1801-1816. [DOI] [PubMed] [Google Scholar]
- 13.Hirai, K., H. Aoyama, T. Irikura, S. Iyobe, and S. Mitsuhashi. 1986. Differences in susceptibility to quinolones of outer membrane mutants of Salmonella typhimurium and Escherichia coli. Antimicrob. Agents Chemother. 29:535-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lugtenberg, B., R. Peters, H. Bernheimer, and W. Berendsen. 1976. Influence of cultural conditions and mutations on the composition of the outer membrane proteins of Escherichia coli. Mol. Gen. Genet. 147:251-262. [DOI] [PubMed] [Google Scholar]
- 15.Maneewannakul, K., and S. B. Levy. 1996. Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin, R. G., E. S. Bartlett, J. L. Rosner, and M. E. Wall. 2008. Activation of the Escherichia coli marA/soxS/rob regulon in response to transcriptional activator concentration. J. Mol. Biol. 380:278-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer, M. P. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163:41-46. [DOI] [PubMed] [Google Scholar]
- 18.McMurry, L. M., and S. B. Levy. 1995. The NH2-terminal half of the Tn10-specified tetracycline efflux protein TetA contains a dimerization domain. J. Biol. Chem. 270:22752-22757. [DOI] [PubMed] [Google Scholar]
- 19.Mizuno, T., M. Y. Chou, and M. Inouye. 1984. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. USA 81:1966-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuno, T., and S. Mizushima. 1990. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol. Microbiol. 4:1077-1082. [DOI] [PubMed] [Google Scholar]
- 21.Mortimer, P. G., and L. J. Piddock. 1993. The accumulation of five antibacterial agents in porin-deficient mutants of Escherichia coli. J. Antimicrob. Chemother. 32:195-213. [DOI] [PubMed] [Google Scholar]
- 22.Nicoloff, H., V. Perreten, and S. B. Levy. 2007. Increased genome instability in Escherichia coli lon mutants: relation to emergence of multiple-antibiotic-resistant (Mar) mutants caused by insertion sequence elements and large tandem genomic amplifications. Antimicrob. Agents Chemother. 51:1293-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicoloff, H., V. Perreten, L. M. McMurry, and S. B. Levy. 2006. Role for tandem duplication and Lon protease in AcrAB-TolC-dependent multiple antibiotic resistance (Mar) in an Escherichia coli mutant without mutations in marRAB or acrRAB. J. Bacteriol. 188:4413-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oethinger, M., I. Podglajen, W. V. Kern, and S. B. Levy. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob. Agents Chemother. 42:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 26.Tschowri, N., S. Busse, and R. Hengge. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 23:522-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel, J., and K. Papenfort. 2006. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9:605-611. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimura, F., and H. Nikaido. 1985. Diffusion of beta-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob. Agents Chemother. 27:84-92. [DOI] [PMC free article] [PubMed] [Google Scholar]