Abstract
2′,3′-Didehydro-3′-deoxy-4′-ethynylthymidine (4′-Ed4T), a derivative of stavudine (d4T), has potent activity against human immunodeficiency virus and is much less inhibitory to mitochondrial DNA synthesis and cell growth than its progenitor, d4T. 4′-Ed4T triphosphate was a better reverse transcriptase (RT) inhibitor than d4T triphosphate, due to the additional binding of the 4′-ethynyl group at a presumed hydrophobic pocket in the RT active site. Previous in vitro selection for 4′-Ed4T-resistant viral strains revealed M184V and P119S/T165A/M184V mutations on days 26 and 81, respectively; M184V and P119S/T165A/M184V conferred 3- and 130-fold resistance to 4′-Ed4T, respectively. We investigated the relative contributions of these mutations, engineered into the strain NL4-3 background, to drug resistance, RT activity, and viral growth. Viral variants with single RT mutations (P119S or T165A) did not show resistance to 4′-Ed4T; however, M184V and P119S/T165A/M184V conferred three- and fivefold resistance, respectively, compared with that of the wild-type virus. The P119S/M184V and T165A/M184V variants showed about fourfold resistance to 4′-Ed4T. The differences in the growth kinetics of the variants were not more than threefold. The purified RT of mutants with the P119S/M184V and T165A/M184V mutations were inhibited by 4′-Ed4TTP with 8- to 13-fold less efficiency than wild-type RT. M184V may be the primary resistance-associated mutation of 4′-Ed4T, and P119S and T165A are secondary mutations. On the basis of our findings and the results of structural modeling, a virus with a high degree of resistance to 4′-Ed4T (e.g., more than 50-fold resistance) will be difficult to develop. The previously observed 130-fold resistance of the virus with P119S/T165A/M184V to 4′-Ed4T may be partly due to mutations both in the RT sequence and outside the RT sequence.
Following the fusion of the human immunodeficiency virus type 1 (HIV-1) envelope and host cell membrane, the genetic material (RNA) of HIV-1 is introduced into the cytoplasm; the virus-encoded reverse transcriptase (RT) enzyme converts the single-stranded RNA genome into a double-stranded DNA. Nucleoside RT inhibitors (NRTIs) target the reverse transcription step for their anti-HIV activity. NRTIs were the first therapeutic agents to demonstrate clinical efficacy as antiretroviral therapy for HIV-1 infection and continue to be the backbone of most highly active antiretroviral therapy (HAART) (http://aidsinfo.nih.gov/Guidelines). The effectiveness of HAART may be offset by the increased burden of long-term drug toxicity mediated specifically by NRTI therapy (18, 20, 29, 42). Furthermore, mutations within the RT domain lead to resistance and treatment failure (7, 24, 36). The single M184V mutation in the RT confers high-level phenotypic resistance to lamivudine (3TC) (38, 39). However, for zidovudine (AZT; a thymidine analog), the accumulation of multiple mutations is required to confer high-level phenotypic resistance (4, 22, 24-26, 35). There are two major biochemical mechanisms of resistance to NRTIs: (i) altered NRTI binding or incorporation (i.e., 3TC resistance) and (ii) altered NRTI excision after incorporation (i.e., AZT resistance) (6). Furthermore, mutations can be classified functionally as either a primary mutation (i.e., a mutation that confers resistance to an inhibitor) or a secondary mutation (i.e., a mutation that enhances the resistance conferred by the primary mutation and improves the replicative capacity of the virus without necessarily increasing resistance) (14).
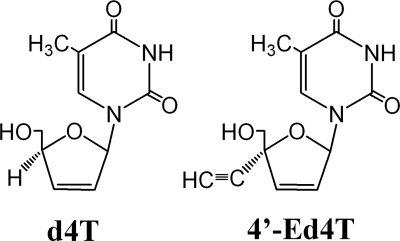
All NRTIs require stepwise intracellular phosphorylation to their triphosphate metabolites, which are preferentially incorporated into HIV DNA by HIV-1 RT and which cause the premature termination of viral DNA chain elongation (20). Among the approved NRTIs, stavudine (d4T) is a highly potent inhibitor of HIV-1 replication in vitro (2, 15, 28). However, the use of d4T in vivo has been limited by delayed toxicity, notably, peripheral neuropathy and myopathy caused by mitochondrial damage (5, 20, 30). The recently discovered drug 2′,3′-didehydro-3′-deoxy-4′-ethynylthymidine (4′-Ed4T) is a derivative of d4T (Fig. 1) (9, 16). It is a more potent inhibitor of HIV-1 replication and is much less inhibitory to mitochondrial DNA synthesis and cell growth than its progenitor, d4T (9). In steady-state enzymatic analyses, 4′-Ed4T triphosphate (4′-Ed4TTP) inhibited the DNA polymerase activity of RT more efficiently than d4T triphosphate (d4TTP), and the inhibition of DNA replication was more effective with an RNA template than with a DNA template (49). Furthermore, pre-steady-state kinetic studies, together with computer modeling, illustrated that 4′-Ed4TTP was a better RT inhibitor than d4TTP due to the additional binding of the 4′-ethynyl group at a presumed hydrophobic pocket in the RT active site (50).
FIG. 1.
Chemical structures of d4T and 4′-Ed4T.
In a selection for 4′-Ed4T drug resistance mutations study by Nitanda et al. (32), the M184V mutation was observed on day 26 and two additional mutations (P119S and T165A) were found on day 81. The M184V mutation and the triple mutation (M184V/P119S/T165A) conferred 3- to 5-fold and 130-fold resistance to 4′-Ed4T, respectively. The M184V mutation, which is in a highly conserved motif (YMDD) of the polymerase active site of RT, is known to confer high-level resistance to 3TC and low-level resistance to zalcitabine, didanosine, and abacavir (13, 44). Moreover, the M184V mutation has been reported to diminish viral fitness, reduce RT processivity, increase RT fidelity, and diminish the rescue of viral DNA synthesis in the presence of RT inhibitors (10, 27, 43). However, the consequence of the combination of the P119S and/or the T165A mutation with M184V remains unknown. These mutations, P119S and/or T165A, are not observed in HIV-1-infected individuals. Moreover, P119 is completely nonpolymorphic and T165A is nonpolymorphic, although position 165 is polymorphic, potentially reflecting a high genetic barrier to resistance (http://hiv.lanl.gov).
In this study, we investigated the relative contributions of the P119S and T165A mutations to wild-type HIV-1 and an M184V variant with regard to the activity of RT, viral growth, and drug resistance.
MATERIALS AND METHODS
Chemicals.
4′-Ed4T was synthesized as described previously (16); d4T was purchased from Sigma-Aldrich (St. Louis, MO). The mono-, di-, and triphosphate forms of 4′-Ed4T and d4T were synthesized and purified by published protocols (23). The purities of these compounds were verified by high-pressure liquid chromatography analysis. [γ-32P]ATP was purchased from NEN Life Sciences Company (Boston, MA). Deoxynucleoside triphosphates (dNTPs) were purchased from Amersham/Pharmacia (Piscataway, NJ). All other chemicals used were of analytical grade.
Cell lines.
The CEM cell line was received from the AIDS Research and Reference Reagent Program of the National Institutes of Health. The TZM-bl indicator cell line (46), obtained from J. Kappes through the AIDS Research and Reference Reagent Program, is a derivative of the HeLa cell line that expresses high levels of CD4 and CCR5, along with endogenously expressed CXCR4. TZM-bl cells contain HIV long terminal repeat-driven β-galactosidase and luciferase reporter cassettes that are activated by HIV Tat expression. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere.
Site-directed mutagenesis.
The P119S, T165A, and M184V mutations were created individually and in combination (for a total of eight clones, including the wild type) by site-directed mutagenesis with a Quickchange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) in plasmid p83-2, which contained 5′ NL4-3 sequences (1). The resultant clones were subjected to fluorescent sequencing (ABI, Foster City, CA) to verify the presence of the desired mutations and the absence of extraneous mutations.
Generation of virus stocks.
Infectious clones were generated as described previously (8). In brief, wild-type p83-2 plasmid DNA or derivative mutant clones was digested with EcoRI (New England Biolabs, Beverly, MA), column purified (Qiagen, Valencia, CA), and combined with an equal amount of similarly prepared EcoRI-digested p83-10 plasmid DNA (containing 3′ NL4-3 sequences). The two plasmids were fused by using T4 DNA ligase (New England Biolabs). The mixture was then introduced into the permissive cell line CEMx174 by DEAE dextran transfection (31). The transfected cells were cultured in RPMI 1640 medium (Gibco BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (Gibco BRL). For the generation of virus stocks, culture supernatants containing virus were harvested 48 h after the first observation of a cytopathic effect. To ensure that the mutation was retained in the HIV-1 strains and that no other mutations were present after transfection and virus production, we sequenced each variant to verify the presence of the desired mutations and the absence of extraneous mutations.
The pTKD chimeric virus, which contains the NL4-3 backbone but which has the RT region swapped from the IIIB-based virus carrying the P119S/T165A/M184V mutation, was generated as follows: primers HIV-IIIB-2096F (5′-CCCATTAGTCCTATTGAAACTGTACC-3′) and HIV-IIIB-3776R (5′-TAGTACTTTCCTGATTCCAGCACTG-3′) were used to amplify by PCR the entire RT region from genomic DNA isolated from MT-4 cells infected by the P119S/T165A/M184V mutant virus (in the strain IIIB background) (32). The resulting 1.7-kb DNA fragment was then used as a megaprimer for domain swapping to replace the RT region (with the strain NL4-3 background) in plasmid p83-2 by using the Quikchange site-directed mutagenesis kit (Stratagene). Plasmid p83-2 containing the P119S/T165A/M184V RT in a IIIB background was fused with p83-10 to generate the pTKD chimeric virus, as described above.
Growth kinetic assay.
In order to assess the growth kinetics of NL4-3 and derivative strains, 2 × 106 CEMx174 cells were infected with virus at a multiplicity of infection (MOI) of 0.01 by using stocks derived from transfected CEMx174 cells. Culture supernatants were collected each day until day 8 for HIV-1 RNA extraction. HIV-1 RNA was extracted with a QIAmp viral RNA extraction kit (Qiagen), according to the manufacturer's instructions. The concentration of HIV-1 RNA at each time point was determined by quantitative real-time RT-PCR, as described previously (8). Primers 5′-GGCCAGGGAATTTTCTTCAGA-3′ and 5′-TCCCCAAACCTGAAGCTC TCT-3′ and probe 5′-6-carboxyfluorescein-AGACCAGAGCCAACAGCCCCACC-6-carboxytetramethylrhodamine-3′ were used. The primers and probe annealed to conserved sequences located near the 3′ end of gag. The cycling parameters used for these experiments were 48°C for 30 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The number of cycles required to reach a threshold fluorescence (the threshold cycle number) was determined, and the quantity of sequences initially present was calculated by extrapolation onto the standard curve.
Drug susceptibility assay.
TZM-bl cells plated at 5 × 104 cells per well in a 96-well microtiter plate and maintained in RPMI 1640 medium supplemented with 10% dialyzed fetal bovine serum were preincubated with various concentrations of inhibitor (4′-Ed4T, d4T, 3TC, or AZT) for 16 h at 37°C. The cells were then infected with 4′-Ed4T-resistant HIV-1 clones (i.e., clones with the P119S, T165A, M184V, P119S/T165A, P119S/M184V, T165A/M184V, or P119S/T165A/M184V mutation or the wild type) at an MOI of 0.01. After 72 h of infection, the cells in each well were lysed, and the luciferase activity of each well was measured with a luciferase assay reagent (Promega, Madison, WI) and a luminometer (FARCyte; Amersham Biosciences Co., Piscataway, NJ). The background luminescence from uninfected cells was determined and was subtracted from the luminescence for all experimental wells. The anti-HIV activity of the drugs was measured as a function of the inhibition of viral growth (inhibition of viral growth as a percentage of the growth of the control not treated with drug), calculated by dividing the mean number of luciferase units at each concentration of a drug by the mean number of luciferase units from the well containing no drug. Inhibition efficiency was expressed as the concentration of inhibitor that produced 50% inhibition of virus growth (the 50% effective concentration [EC50]).
DNA/RNA primer/template and RT protein preparation for kinetic analysis.
A DNA 23-mer (5′-TCAGGTCCCTGTTCGGGCGCCAC-3′) was annealed with an RNA 36-mer (5′-UCUCUAGCAGUGGCGCCCGAACAGGGACCUGAAAGC-3′) and used as the primer/template for the kinetic analysis. The wild-type HIV-1 RT cDNA and mutants with the P119S, T165A, or M184V mutation in the NL4-3 background were amplified from the wild-type or corresponding mutant p83-2 plasmids described above and constructed into the HIV-1 RT expression plasmid provided by Stephen Hughes (National Cancer Institute, Frederick, MD) for RT protein expression in Escherichia coli. The expressed RT proteins were purified by a previously published method (11).
Steady-state single nucleotide incorporation assays.
All enzymatic reactions in this study were carried out as described previously (49) in a standard reaction buffer containing 50 mM Tris-HCl, pH 8.0, 60 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, and 0.1 mg/ml bovine serum albumin. Unless otherwise stated, all the concentrations reported in this study were the final concentrations achieved after mixing of the components. In the steady-state single nucleotide incorporation assays for determination of the Km and kcat values for HIV-1 RT, a mixture containing 250 nM 23-mer/36-mer primer/template and various concentrations of [α-32P]dTTP in the standard reaction buffer was preincubated at 37°C, and the reaction was initiated by adding 2.5 nM wild-type or mutant RT. Aliquots (4 μl each) were removed at different times and mixed with 2 μl 0.5 M EDTA to stop the reaction. Then, 3 μl of each sample was spotted onto DE81 filter paper. The filter paper was subsequently washed three times with 0.5 M Na2HPO4 (pH 7.0), dried, and exposed to a phosphorimager screen. The amount of incorporated dTTP was quantified by phosphorimaging (GE Healthcare Bio-Sciences Corp., Piscataway, NJ).
Steady-state inhibition of RT by 4′-Ed4TTP or d4TTP was determined by an assay similar to the assay used for determination of the Km and kcat values, and 4′-Ed4TTP or d4TTP was included at various concentrations in the reaction mixture as an inhibitor.
Data analysis.
The steady-state kinetic parameters were determined from linear steady-state velocities by using the Prism program (GraphPad Software Inc., San Diego, CA).
RESULTS
Growth profile of HIV variants with mutations at position 119, 165, and/or 184 of RT.
HIV-1 RT variants with the P119S, T165A, M184V, P119S/T165A, P119S/M184V, T165A/M184V, and P119S/T165A/M184V mutations were generated by site-directed mutagenesis in an NL4-3 background. Viral stocks were prepared by transfection, and the viral variants were used to infect CEMx174 cells at an MOI of 0.01. Culture supernatants were harvested at several days postinfection for HIV RNA quantification by quantitative RT-PCR. The concentration of HIV-1 RNA at 2 days postinfection (i.e., after a single cycle of replication) was similar for the variants with single mutations (Table 1). The mutant with the double P119S/T165A mutation produced more virus than the variants with the P119S/M184V and T165A/M184V mutations. In general, the variants with the M184V background tended to produce less virus on postinfection day 2, with the virus with the P119S/T165A/M184V mutation producing the least amount of virus. However, the viral titers for all variants on day 4 were comparable. The peak level of virus production for all the variants occurred on day 6 postinfection (data not shown). In this 8-day growth assay, there was no more than a threefold difference in viral replication between the wild type and the variants harboring the P119S, T165A, or M184V mutation.
TABLE 1.
HIV-1 production by 4′-Ed4T mutant variants in CEM cells
| Virus | HIV-1 RNA (copy no. [103]/ml) |
|
|---|---|---|
| Day 2 | Day 4 | |
| pNL4-3 (wild type) | 61.4 ± 0.8b | 159.2 ± 11.8 |
| P119S | 50.3 ± 4.7 | 140.9 ± 22.8 |
| T165A | 52.2 ± 1.1 | 118.2 ± 2.5 |
| M184V | 52.3 ± 9.1 | 138.4 ± 13.0 |
| P119S/T165A | 76.2 ± 1.2 | 147.0 ± 16.5 |
| P119S/M184V | 49.3 ± 13.2 | 133.4 ± 15.3 |
| T165A/M184V | 47.0 ± 7.9 | 134.8 ± 21.2 |
| P119S/T165A/M184V | 25.3 ± 6.8 | 119.5 ± 16.1 |
| pTKDa | 41.7 ± 2.0 | 127.8 ± 21.6 |
A chimeric NL4-3 clone with the full RT region replaced by the corresponding RT sequence in the IIIB background carrying the P119S/T165A/M184V mutations, which was isolated by Nitanda et al. (32).
The data represent the means ± standard deviations for two separate experiments.
Susceptibilities of viruses harboring P119S, T165A, and/or M184V mutations to HIV RT inhibitors.
The degree of resistance to 4′-Ed4T, d4T, AZT, and 3TC that the mutations conferred when they were present in the variants as either single mutations or combinations of mutations was assessed. TZM-bl cells were infected with the variants at an MOI of 0.01 in the presence of various concentrations of the HIV RT inhibitors (4′-Ed4T, d4T, AZT, or 3TC). The relative luciferase activity was used as a measure of the inhibition of viral growth, and the EC50 for each inhibitor for each variant was calculated (Table 2). The EC50s of 4′-Ed4T were 0.37, 0.51, and 0.58 μM for the wild-type, P119S, and T165A viruses, respectively. The M184V variant had a two- to threefold higher EC50. There was a trend, although it was not statistically significant, of an increased EC50 of 4′-Ed4T when the P119S or T165A mutation was introduced into variants harboring the M184V mutation. The variants harboring the M184V mutation either alone or in combination with P119S and T165A were less susceptible to 4′-Ed4T than the wild-type virus; the change in the EC50 ranged from twofold (M184V) to fivefold (P119S/T165A/M184V). Thus, the high level of resistance of the virus previously isolated by Nitanda et al. could not be due to these three mutations in RT alone (32). The M184V mutation did not have any significant impact on susceptibility to d4T; however, the variant with double mutations (P119S/T165A) but not the variant with triple mutations exhibited about a twofold increase in the EC50 in comparison to that for the wild-type virus. As expected, the EC50 of 3TC was high for all the variants harboring the M184V mutation. All the variants were highly susceptible to treatment with AZT.
TABLE 2.
Anti-HIV-1 activities of 4′-Ed4T, d4T, 3TC, and AZT against 4′-Ed4T-resistant NL4-3 strains in TZM-bl cells
| Virus | EC50 (μM) |
|||
|---|---|---|---|---|
| 4′-Ed4T | d4T | 3TC | AZT | |
| pNL4-3 (wild type) | 0.37 ± 0.09a | 1.25 ± 0.21 | 0.65 ± 0.31 | 0.023 ± 0.02 |
| P119S | 0.51 ± 0.27 | 1.44 ± 0.16 | 0.47 ± 0.16 | 0.017 ± 0.01 |
| T165A | 0.58 ± 0.31 | 1.46 ± 0.27 | 1.08 ± 0.60 | 0.022 ± 0.01 |
| M184V | 0.98 ± 0.55 | 1.40 ± 0.40 | >60 | 0.011 ± 0.01 |
| P119S/T165A | 0.35 ± 0.10 | 2.48 ± 0.74 | 2.10 ± 0.73 | 0.026 ± 0.02 |
| P119S/M184V | 1.38 ± 0.48 | 1.55 ± 0.35 | >60 | 0.011 ± 0.01 |
| T165A/M184V | 1.32 ± 0.20 | 1.50 ± 0.93 | >60 | 0.024 ± 0.02 |
| P119S/T165A/M184V | 1.96 ± 0.27 | 1.28 ± 0.57 | >60 | 0.053 ± 0.09 |
| pTKD | 2.10 ± 0.68 | 1.00 ± 0.18 | >60 | 0.008 ± 0.001 |
The data represent the means ± standard deviations for three separate experiments.
Effect of P119S, T165A, and/or M184V mutation on RT activity.
To further understand the mechanism of resistance to 4′-Ed4T caused by the M184V mutation when it was present either singly or in combination with the P119S and/or T165A mutation, the activities of the wild-type and mutant RTs were compared by using a 23-mer/36-mer DNA/RNA primer/template. The DNA/RNA primer/template was used instead of the DNA/DNA primer/template because the reverse transcription action is more sensitive than the DNA polymerase action to 4′-Ed4T(11). Steady-state kinetic constants (Km and kcat) were determined (Table 3). Most of the RT mutants showed close Km and kcat values, so their efficiencies (kcat/Km) of incorporation of dTTP into the 23-mer/36-mer DNA/RNA primer/template were similar (i.e., within a twofold change). However, the RT of the mutant with the triple mutation (P119S/T165A/M184V) exhibited a fourfold decrease in efficiency compared to that of the wild-type RT.
TABLE 3.
Steady-state kinetic parameters for inhibition of wild-type RT and drug-resistant mutants by 4′-Ed4TTP and d4TTP
| Enzyme | dTTP |
4′-Ed4TTP |
d4TTP |
||||
|---|---|---|---|---|---|---|---|
| Km(dTTP) (μM) | kcat (s−1) | kcat/Km(dTTP) (μM−1 s−1) | Ki (μM) | Ki(4′-Ed4TTP)/Km(dTTP) | Ki (μM) | Ki(d4TTP)/Km(dTTP) | |
| Wild-type RT | 0.12 ± 0.03a | 3.2 × 10−3 ± 1.4 × 10−4 | 2.7 × 10−2 | 0.054 ± 0.016 | 0.45 | 0.63 ± 0.05 | 5.3 |
| P119S | 0.13 ± 0.03 | 3.3 × 10−3 ± 1.7 × 10−4 | 2.5 × 10−2 | 0.11 ± 0.013 | 0.85 | 0.64 ± 0.01 | 2.1 |
| T165A | 0.10 ± 0.008 | 2.7 × 10−3 ± 3.5 × 10−5 | 2.7 × 10−2 | 0.08 ± 0.02 | 0.8 | 0.21 ± 0.03 | 2.1 |
| M184V | 0.12 ± 0.04 | 3.1 × 10−3 ± 2.6 × 10−4 | 2.6 × 10−2 | 0.21 ± 0.03 | 1.7 | 0.79 ± 0.17 | 6.6 |
| P119S/T165A | 0.07 ± 0.01 | 3.6 × 10−3 ± 1.2 × 10−4 | 5.1 × 10−2 | 0.024 ± 0.003 | 0.34 | 0.37 ± 0.02 | 5.3 |
| P119S/M184V | 0.18 ± 0.03 | 4.0 × 10−3 ± 3.6 × 10−5 | 2.2 × 10−2 | 0.64 ± 0.18 | 3.6 | 0.48 ± 0.02 | 2.7 |
| T165A/M184V | 0.11 ± 0.02 | 1.4 × 10−3 ± 4.8 × 10−5 | 1.3 × 10−2 | 0.66 ± 0.12 | 6 | 0.3 ± 0.06 | 2.7 |
| P119S/T165A/M184V | 0.15 ± 0.03 | 9.0 × 10−4 ± 3.6 × 10−5 | 6.0 × 10−3 | 0.73 ± 0.10 | 4.9 | 1.05 ± 0.2 | 7 |
The data represent the means ± standard deviations for three separate experiments.
Inhibition of RT mutants by 4′-Ed4TTP.
We next investigated the ability of 4′-Ed4TTP to inhibit the incorporation of dTTP into the growing DNA chain by either the wild type or the mutant variants. By measuring the apparent Km values of dTTP under various concentrations of 4′-Ed4TTP, the inhibition constants (Ki) for 4′-Ed4TTP inhibition of dTTP incorporation were determined, and the value of the ratio of the Ki of 4′-Ed4TTP to the Km of dTTP [Ki (4′-Ed4TTP)/Km (dTTP)] was used as an index to evaluate the inhibition efficiency. The results are summarized in Table 3. The RTs with the P119S and T165A single mutations as well as the RT with the P119S/T165A double mutation showed similar efficiencies of inhibition of 4′-Ed4TTP compared to that of the wild-type RT; i.e., these mutations did not confer resistance to 4′-Ed4T. On the other hand, the M184V RT mutant exhibited about a fourfold increase in the Ki/Km value compared with that of the wild-type RT, suggesting a fourfold resistance to 4′-Ed4TTP. This observation is consistent with our previous analysis with M184V in the strain IIIB background (49). Interestingly, the RTs with the P119S/M184V and T165A/M184V double mutations showed 8- and 13-fold increases in Ki/Km values, respectively, compared to the Ki/Km value of the wild-type RT. However, the RT with the P119S/T165A/M184V triple mutations did not show further resistance than the RTs with double mutations, with the Ki/Km value being only 10-fold higher than that of the wild-type RT. We also examined the profiles of d4TTP inhibition of these RT mutants. Consistent with the results from drug susceptibility assays, none of these mutants showed resistance to d4T, as judged by their Ki/Km values (Table 3), suggesting specific profiles of resistance to 4′-Ed4T. Interestingly, the RTs of four of the mutant variants, those with the P119S, T165A, P119S/M184V, and T165A/M184V mutations, showed about a twofold higher sensitivity to d4T than the wild-type RT (Table 3).
Computer modeling.
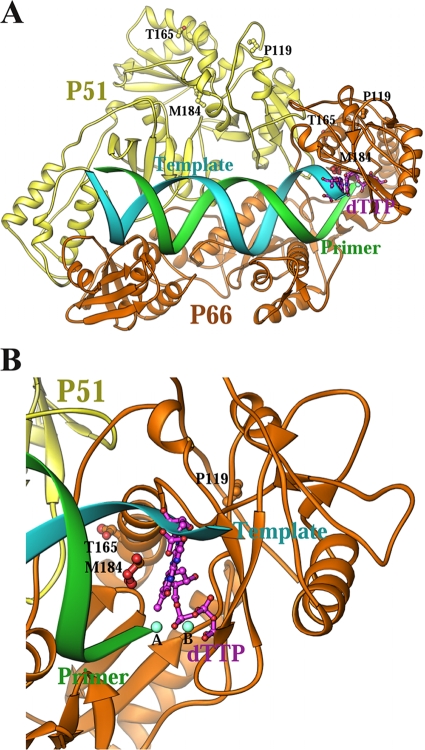
Computer modeling based on the solved crystal structure of the HIV-1 RT-primer/template-dTTP ternary complex (with the DNA/DNA primer/template) with the nucleotide-binding pocket (11) can be used to predict the possible interactions of HIV-1 RT and the RTs with mutations for resistance to 4′-Ed4T with d4TTP and 4′-Ed4TTP. To explain the observed growth and resistance phenotypes of the 4′-Ed4T-resistant variants, we modeled the relationship of dTTP with the side chains of P119, T165, and M184 in the HIV-1 RT ternary complex (Fig. 2). The distance of C-4′ of dTTP, which is the attachment site of 4′-Ed4T, to the carbon of M184 of P66 is 6.7 Å; we previously described a possible role for M184 in interactions with the 4′-Ed4TTP (11). However, the distances of C-4′ to the carbons of P119 and T165 of P66 are 15 and 14 Å, respectively. Also, the distances of C-4′ to the carbons of P199, T165, and M184 of P51 are 40, 56, and 45 Å, respectively. These distances all fall outside the ideal range for van de Waals interactions, making the direct interaction of the mutant variants' side chains with the binding pocket of 4′-Ed4TTP very unlikely.
FIG. 2.
Relationship of dTTP to the side chains of P119, T165, and M184 in the HIV-1 RT ternary complex. (A) Overall structure; (B) close-up view of the dNTP binding site. The catalytic P66 subunit, the noncatalytic P51 subunit, the template, and the primer strands of the DNA duplex are shown in gold, yellow, cyan, and green, respectively. The distance of C-4′ of dTTP, which is the attachment site of 4′-Ed4T, to the carbon of M184 of P66, which we previously described as possibly having a role in interactions with 4′-Ed4T, is 6.7 Å (50).
DISCUSSION
The novel RT mutations P119S and T165A as single mutations had no significant impact on either viral replication or RT activity; viruses with these two mutations have not been identified as clinically relevant in HIV-1-infected individuals. However, in the background of the M184V mutation (P119S/M184V, T165A/M184V, or P119S/T165A/M184V), the viruses exhibited reduced RT activity and decreased susceptibility to 4′-Ed4T in comparison to the RT activity and susceptibility of the wild-type virus. Although variants with the M184V mutation tended to have slow growth within the first 48 h, the overall growth of all the mutant variants was comparable to that of wild-type virus. Our findings are consistent with those of previous studies. Virus with the M184V mutation was found to have replication kinetics similar to that of wild-type virus when the SupT1 T-cell line was infected at different MOIs (3). In an in vitro assay with phytohemagglutinin-stimulated peripheral blood mononuclear cells, Sharma and Crumpacker found that the virus with the M184V mutation had a fitness loss of only 2.4% in comparison to the fitness of the wild-type virus (40). Furthermore, in a single-cycle replication assay, the titer of the variant with the M184V mutation was approximately 90% of that of the wild-type virus (19). Our results from viral growth and RT activity analyses suggest that P119S, T165A, and M184V mutations do not have significant effects on RT. This contradicts the findings of previous studies, which found that the M184V mutation leads to reduced RT activity and a loss of viral fitness (12, 47, 48). The inconsistency of our findings and those of previous studies may partly be due to the sensitivities of the different methods used and viral or cellular factors (3).
The kinetic behaviors of 4′-Ed4T-resistant RT mutants are consistent with drug susceptibility assay results.
When the 4′-Ed4T resistance mutant profiles are compared at both the RT and the viral levels (in the strain NL4-3 background), the results are consistent. M184V showed 4-fold and 2.5-fold resistance to 4′-Ed4T at the RT and viral levels, respectively; M184V, in addition to P119S or T165A, or both, showed about 10-fold resistance at the RT level and 5-fold resistance at the viral level. When the RT domain with the P119S/T165A/M184V mutation (in the strain IIIB background) was swapped into the NL4-3 virus, we observed only fivefold resistance to 4′-Ed4T. This result indicates that the background difference between the IIIB RT and the NL4-3 RT has no impact on 4′-Ed4T resistance. Moreover, our findings are consistent with those of a study conducted by Kawamoto et al. with 2′-deoxy-4′-C-ethynyl-2-halo-adenosine (EFdA), another 4′-ethynyl-substituted nucleoside analog (21). In their study, moderate resistance (22-fold) to EFdA was conferred by a combination of three RT mutations (I142V, T165R, and M184V). The 4′-Ed4T resistance mutation P119S/T165A/M184V conferred 14-fold resistance to EFdA, similar to the resistance to 4′-Ed4T that it conferred.
Mechanism of resistance to 4′-ethynyl analogs.
On the basis of our studies of resistance to 4′-Ed4T, the P119S and/or T165A mutation can confer further resistance, although not statistically significant resistance, to 4′-Ed4T when one or both of those mutations are present in combination with M184V. However, the mutant with the triple mutation did not show more resistance than the mutants with double mutations, indicating a lack of a synergistic or an additive effect between P119S and T165A with respect to resistance to 4′-Ed4T. M184V was the first mutation observed during the selection of 4′-Ed4T-resistant viral strains and conferred about threefold resistance to 4′-Ed4T (32); M184V may be the primary mutation. Since the P119S and/or T165A mutation did not confer any resistance to 4′-Ed4T in the absence of the M184V mutation, they may act as secondary mutations. This is consistent with the observations from studies with EFdA, in which I142V and T165R exhibited resistance to EFdA only in the presence of M184V (21).
The interaction of 4′-ethynyl-substituted nucleoside analog triphosphate and HIV-1 RT remains unclear. On the basis of pre-steady-state kinetic data, we previously proposed a model suggesting that the higher affinity of binding of 4′-Ed4TTP than of d4TTP to RT could be due to the additional binding of the 4′-ethynyl group in a presumed hydrophobic pocket, which is preformed by the side chains of A114, Y115, M184, F160, and D185 (50). We also suggested that M184V could indirectly affect the binding of incoming dNTPs, leading to three- to fivefold resistance to 4′-Ed4T since the M184V mutation can create a gap between the polymerase and the DNA minor groove of the nascent base pair. Notably, residues at the 4′-ethynyl binding pocket are highly conserved, and all mutations at this pocket except M184V and M184A caused a complete loss of RT activity (50). In another model suggested by Siddiqui et al. (41), the mechanism of resistance to 4′-ethynyl analogs conferred by M184V involves steric hindrance between the 4′-ethynyl group of the inhibitors and the side chain of Val184, reminiscent of the mechanism of resistance to 3TC (17, 37). We observed a qualitative correlation but not a quantitative correlation of the susceptibilities of the different variants of HIV-1 to 4′-Ed4T with the sensitivities of their RTs to 4′-Ed4TTP. This could be due to the fact that the recombinant HIV-1 RT does not totally reflect HIV-1 RT activity in vivo, in which there may be interactions with other proteins.
The mechanism of resistance to 4′-ethynyl analogs by P119S, I142V, and T165A/R in the background of M184V is even more difficult to understand, because all these residues are far away from nucleotide-binding site of RT, on the basis of the structure of the RT-primer/template-dTTP ternary complex (17). It has been suggested that the Ala165 or Arg165 mutation in RT would destroy the hydrogen bond from Thr165 to Gln182 and therefore indirectly affect the positioning of Val184 (21). This could lead to the exaggeration of the steric interactions between the side chain of Val184 and the 4′-ethynyl group of EFdA, resulting in greater resistance (21). However, in our modeling, none of these interactions were apparent. Further investigations are needed to test this model and to understand the mechanism of resistance caused by these novel mutations (P119S, I142V, and T165A).
Our findings that P119S and T165A in the background of M184V lead to a decrease in susceptibility to 4′-Ed4T are consistent with those of Nitanda et al. (32); however, our observation of only fivefold resistance of the variant with the P119S/T165A/M18V mutation contradicts the 130-fold resistance observed previously. Recent reevaluation of the activity of 4′-Ed4T against strain IIIB with the P119S/T165A/M18V mutation revealed that it was only 10-fold less susceptible to the compound than the wild-type of strain IIIB (T. Hamasaki et al., unpublished data). We postulated that the difference might have been due to differences in the experimental conditions. We replaced the RT of NL4-3 with the full RT sequence from Nitanda et al. (TKD) and investigated the growth and drug susceptibility of this chimeric clone (pTKD). Although the pTKD variant with all three mutations had about a 1.6-fold higher HIV titer than the variant with the P119S/T165A/M184V mutation engineered into the NL4-3 RT by site-directed mutagenesis (Table 1), there was no significant difference in drug susceptibility between pTKD and the variant with the P119S/T165A/M184V mutation (Table 2). The difference between the findings of the current study and the findings of the study of Nitanda et al. is not clear and is being investigated.
We anticipate that the emergence of significant phenotypic resistance to 4′-Ed4T will be difficult when clinically relevant dosages are used. Resistance to NRTIs as a result of nucleoside-associated mutations develops by at least three pathways: thymidine analog resistance mutations (e.g., 41L, 67N, 70R, 210W, 215Y/F, and 219Q/E); Q151 complex-151M mutation, followed by the mutations 62V, 75I, 77L, and 116Y; and the 69 insertion complex, generally accompanied by several NRTI-associated mutations (34). As a thymidine analog, we expected 4′-Ed4T to at least either select for or have cross-resistance to thymidine analog resistance mutations. The activity of 4′-Ed4T against drug-resistant clinical isolates (nucleoside-associated mutations) has been demonstrated by Nitanda et al. (32). 4′-Ed4T was found to inhibit the replication of four multidrug-resistant clinical HIV isolates in peripheral blood mononuclear cells with EC50s ranging from 0.089 to 0.35 μM. These resistant isolates showed considerable resistance to nonnucleoside RT inhibitor analogs, such as nevirapine and efavirenz, and several protease inhibitors. Interestingly, 4′-Ed4T was found in this study to have the same activity that it has against the wild type against viruses containing the K65R and Q151M mutations. Furthermore, the intracellular concentration of the triphosphate metabolite is critical in the selection for resistance mutations. The half-life of 4′-Ed4TTP is 8.0 to 9.7 h after removal of the drug from culture when CEM cells are used (45). Also, the anti-HIV activity of 4′-Ed4T persisted longer than that of the other thymidine analogs after removal from culture (33). Residues at the 4′-ethynyl binding pocket are highly conserved, and mutations at this pocket could be deleterious to HIV RT activity (50). Taking these together, when an appropriate clinical dose is used, virus with a high degree of resistance to 4′-Ed4T may be difficult to develop.
In conclusion, 4′-Ed4T is a novel anti-HIV NRTI that not only has unique features, including improved potency and reduced cytotoxicity compared to the potency and cytotoxicity of its progenitor (d4T), but also may have a higher genetic barrier to the development of drug resistance mutations. Our findings suggest that P119S and T165A are secondary mutations that make a minimum contribution to drug resistance and do so only in the background of M184V. The M184V mutation may confer intermediate resistance to 4′-Ed4T, similar to the case for didanosine, zalcitabine, and abacavir, but no significant phenotypic resistance in HIV-1-infected patients (3, 15, 44).
Acknowledgments
We are grateful to Louis Alexander for his advice.
This work was supported by Public Health Service grant AI-38204 from the National Institutes of Health to Y.-C.C. E.P. is supported by grants from the National Institutes of Health (grant K08AI074404) and a Yale Child Health Research Center Award (award K12HD001401-08). Y.-C.C. is a fellow of the National Foundation for Cancer Research.
Footnotes
Published ahead of print on 24 August 2009.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, M., R. Pauwels, P. Herdewijn, E. De Clercq, J. Desmyter, and M. Vandeputte. 1987. Both 2′,3′-dideoxythymidine and its 2′,3′-unsaturated derivative (2′,3′-dideoxythymidinene) are potent and selective inhibitors of human immunodeficiency virus replication in vitro. Biochem. Biophys. Res. Commun. 142:128-134. [DOI] [PubMed] [Google Scholar]
- 3.Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, B. O. Oude Essink, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, C., E. O'Sullivan, J. W. Mulder, C. Ramautarsing, P. Kellam, G. Darby, J. M. A. Lange, J. Goudsmit, and B. Larder. 1992. Ordered appearance of zidovudine (AZT) resistance mutations during treatment. J. Infect. Dis. 165:105-110. [DOI] [PubMed] [Google Scholar]
- 5.Brinkman, K., H. J. ter Hofstede, D. M. Burger, J. A. Smeitink, and P. P. Koopmans. 1998. Adverse effects of reverse transcriptase inhibitors: mitochondrial toxicity as common pathway. AIDS 12:1735-1744. [DOI] [PubMed] [Google Scholar]
- 6.Clavel, F., and A. J. Hance. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023-1035. [DOI] [PubMed] [Google Scholar]
- 7.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 8.Collins, J. A., M. G. Thompson, E. Paintsil, M. Ricketts, J. Gedzior, and L. Alexander. 2004. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J. Virol. 78:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutschman, G. E., S. P. Grill, E. A. Gullen, K. Haraguchi, S. Takeda, H. Tanaka, M. Baba, and Y. C. Cheng. 2004. Novel 4′-substituted stavudine analog with improved anti-human immunodeficiency virus activity and decreased cytotoxicity. Antimicrob. Agents Chemother. 48:1640-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng, J. Y., and K. S. Anderson. 1999. Mechanistic studies examining the efficiency and fidelity of DNA synthesis by the 3TC-resistant mutant (184V) of HIV-1 reverse transcriptase. Biochemistry 38:9440-9448. [DOI] [PubMed] [Google Scholar]
- 11.Feng, J. Y., E. Murakami, S. M. Zorca, A. A. Johnson, K. A. Johnson, R. F. Schinazi, P. A. Furman, and K. S. Anderson. 2004. Relationship between antiviral activity and host toxicity: comparison of the incorporation efficiencies of 2′,3′-dideoxy-5-fluoro-3′-thiacytidine-triphosphate analogs by human immunodeficiency virus type 1 reverse transcriptase and human mitochondrial DNA polymerase. Antimicrob. Agents Chemother. 48:1300-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel, F. A., C. F. Invernizzi, M. Oliveira, and M. A. Wainberg. 2007. Diminished efficiency of HIV-1 reverse transcriptase containing the K65R and M184V drug resistance mutations. AIDS 21:665-675. [DOI] [PubMed] [Google Scholar]
- 13.Gao, Q., Z. Gu, M. A. Parniak, J. Cameron, N. Cammack, C. Boucher, and M. A. Wainberg. 1993. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:1390-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Lerma, J. G., H. MacInnes, D. Bennett, H. Weinstock, and W. Heneine. 2004. Transmitted human immunodeficiency virus type 1 carrying the D67N or K219Q/E mutation evolves rapidly to zidovudine resistance in vitro and shows a high replicative fitness in the presence of zidovudine. J. Virol. 78:7545-7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamamoto, Y., H. Nakashima, T. Matsui, A. Matsuda, T. Ueda, and N. Yamamoto. 1987. Inhibitory effect of 2′,3′-didehydro-2′,3′-dideoxynucleosides on infectivity, cytopathic effects, and replication of human immunodeficiency virus. Antimicrob. Agents Chemother. 31:907-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haraguchi, K., S. Takeda, H. Tanaka, T. Nitanda, M. Baba, G. E. Dutschman, and Y. C. Cheng. 2003. Synthesis of a highly active new anti-HIV agent 2′,3′-didehydro-3′-deoxy-4′-ethynylthymidine. Bioorg. Med. Chem. Lett. 13:3775-3777. [DOI] [PubMed] [Google Scholar]
- 17.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, A. A., A. S. Ray, J. Hanes, Z. Suo, J. M. Colacino, K. S. Anderson, and K. A. Johnson. 2001. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J. Biol. Chem. 276:40847-40857. [DOI] [PubMed] [Google Scholar]
- 19.Julias, J. G., P. L. Boyer, M. J. McWilliams, W. G. Alvord, and S. H. Hughes. 2004. Mutations at position 184 of human immunodeficiency virus type-1 reverse transcriptase affect virus titer and viral DNA synthesis. Virology 322:13-21. [DOI] [PubMed] [Google Scholar]
- 20.Kakuda, T. N. 2000. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin. Ther. 22:685-708. [DOI] [PubMed] [Google Scholar]
- 21.Kawamoto, A., E. Kodama, S. G. Sarafianos, Y. Sakagami, S. Kohgo, K. Kitano, N. Ashida, Y. Iwai, H. Hayakawa, H. Nakata, H. Mitsuya, E. Arnold, and M. Matsuoka. 2008. 2′-Deoxy-4′-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Int. J. Biochem. Cell Biol. 40:2410-2420. [DOI] [PubMed] [Google Scholar]
- 22.Kellam, P., C. A. Boucher, and B. A. Larder. 1992. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc. Natl. Acad. Sci. USA 89:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnan, P., J. Y. Liou, and Y. C. Cheng. 2002. Phosphorylation of pyrimidine l-deoxynucleoside analog diphosphates. Kinetics of phosphorylation and dephosphorylation of nucleoside analog diphosphates and triphosphates by 3-phosphoglycerate kinase. J. Biol. Chem. 277:31593-31600. [DOI] [PubMed] [Google Scholar]
- 24.Larder, B., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155-1158. [DOI] [PubMed] [Google Scholar]
- 25.Larder, B. A., K. E. Coates, and S. D. Kemp. 1991. Zidovudine-resistant human immunodeficiency virus selected by passage in cell culture. J. Virol. 65:5232-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larder, B. A., P. Kellam, and S. D. Kemp. 1991. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS 5:137-144. [DOI] [PubMed] [Google Scholar]
- 27.Larder, B. A., S. D. Kemp, and P. R. Harrigan. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696-699. [DOI] [PubMed] [Google Scholar]
- 28.Lin, T. S., R. F. Schinazi, and W. H. Prusoff. 1987. Potent and selective in vitro activity of 3′-deoxythymidin-2′-ene (3′-deoxy-2′,3′-didehydrothymidine) against human immunodeficiency virus. Biochem. Pharmacol. 36:2713-2718. [DOI] [PubMed] [Google Scholar]
- 29.Martin, J. L., C. E. Brown, N. Matthews-Davis, and J. E. Reardon. 1994. Effects of antiviral nucleoside analogs on human DNA polymerases and mitochondrial DNA synthesis. Antimicrob. Agents Chemother. 38:2743-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medina, D. J., C. H. Tsai, G. D. Hsiung, and Y. C. Cheng. 1994. Comparison of mitochondrial morphology, mitochondrial DNA content, and cell viability in cultured cells treated with three anti-human immunodeficiency virus dideoxynucleosides. Antimicrob. Agents Chemother. 38:1824-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naidu, Y. M., H. W. Kestler III, Y. Li, C. V. Butler, D. P. Silva, D. K. Schmidt, C. D. Troup, P. K. Sehgal, P. Sonigo, M. D. Daniel, et al. 1988. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J. Virol. 62:4691-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nitanda, T., X. Wang, H. Kumamoto, K. Haraguchi, H. Tanaka, Y. C. Cheng, and M. Baba. 2005. Anti-human immunodeficiency virus type 1 activity and resistance profile of 2′,3′-didehydro-3′-deoxy-4′-ethynylthymidine in vitro. Antimicrob. Agents Chemother. 49:3355-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paintsil, E., S. P. Grill, G. E. Dutschman, and Y. C. Cheng. 2009. Comparative study of the persistence of anti-HIV activity of deoxynucleoside HIV reverse transcriptase inhibitors after removal from culture. AIDS Res. Ther. 6:5. doi: 10.1186/1742-6405-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinones-Mateu, M. E., Y. Gao, S. C. Ball, A. J. Marozsan, A. Abraha, and E. J. Arts. 2002. In vitro intersubtype recombinants of human immunodeficiency virus type 1: comparison to recent and circulating in vivo recombinant forms. J. Virol. 76:9600-9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richman, D. D., J. C. Guatelli, J. Grimes, A. Tsiatis, and T. Gingeras. 1991. Detection of mutations associated with zidovudine resistance in human immunodeficiency virus by use of the polymerase chain reaction. J. Infect. Dis. 164:1075-1081. [DOI] [PubMed] [Google Scholar]
- 36.Richman, D. D., D. Havlir, J. Corbeil, D. Looney, C. Ignacio, S. A. Spector, J. Sullivan, S. Cheeseman, K. Barringer, D. Pauletti, et al. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarafianos, S. G., K. Das, A. D. Clark, Jr., J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schinazi, R. F., B. I. Hernandez-Santiago, and S. J. Hurwitz. 2006. Pharmacology of current and promising nucleosides for the treatment of human immunodeficiency viruses. Antivir. Res. 71:322-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuurman, R., M. Nijhuis, R. van Leeuwen, P. Schipper, D. de Jong, P. Collis, S. A. Danner, J. Mulder, C. Loveday, C. Christopherson, et al. 1995. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J. Infect. Dis. 171:1411-1419. [DOI] [PubMed] [Google Scholar]
- 40.Sharma, P. L., and C. S. Crumpacker. 1999. Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: a comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J. Virol. 73:8448-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siddiqui, M. A., S. H. Hughes, P. L. Boyer, H. Mitsuya, Q. N. Van, C. George, S. G. Sarafinanos, and V. E. Marquez. 2004. A 4′-C-ethynyl-2′,3′-dideoxynucleoside analogue highlights the role of the 3′-OH in anti-HIV active 4′-C-ethynyl-2′-deoxy nucleosides. J. Med. Chem. 47:5041-5048. [DOI] [PubMed] [Google Scholar]
- 42.Squires, K. E. 2001. An introduction to nucleoside and nucleotide analogues. Antivir. Ther. 6(Suppl. 3):1-14. [PubMed] [Google Scholar]
- 43.Wainberg, M. A., W. C. Drosopoulos, H. Salomon, M. Hsu, G. Borkow, M. Parniak, Z. Gu, Q. Song, J. Manne, S. Islam, G. Castriota, and V. R. Prasad. 1996. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 271:1282-1285. [DOI] [PubMed] [Google Scholar]
- 44.Walter, H., B. Schmidt, M. Werwein, E. Schwingel, and K. Korn. 2002. Prediction of abacavir resistance from genotypic data: impact of zidovudine and lamivudine resistance in vitro and in vivo. Antimicrob. Agents Chemother. 46:89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, X., H. Tanaka, M. Baba, and Y. C. Cheng. 2009. Study of the retention of metabolites of 4′-Ed4T, a novel anti-HIV-1 thymidine analog, in cells. Antimicrob. Agents Chemother. 53:3317-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, X., C. Liang, M. Gotte, and M. A. Wainberg. 2002. The M184V mutation in HIV-1 reverse transcriptase reduces the restoration of wild-type replication by attenuated viruses. AIDS 16:2391-2398. [DOI] [PubMed] [Google Scholar]
- 48.Wei, X., C. Liang, M. Gotte, and M. A. Wainberg. 2003. Negative effect of the M184V mutation in HIV-1 reverse transcriptase on initiation of viral DNA synthesis. Virology 311:202-212. [DOI] [PubMed] [Google Scholar]
- 49.Yang, G., G. E. Dutschman, C. J. Wang, H. Tanaka, M. Baba, K. S. Anderson, and Y. C. Cheng. 2007. Highly selective action of triphosphate metabolite of 4′-ethynyl d4T: a novel anti-HIV compound against HIV-1 RT. Antivir. Res. 73:185-191. [DOI] [PubMed] [Google Scholar]
- 50.Yang, G., J. Wang, Y. Cheng, G. E. Dutschman, H. Tanaka, M. Baba, and Y. C. Cheng. 2008. Mechanism of inhibition of human immunodeficiency virus type 1 reverse transcriptase by a stavudine analogue, 4′-ethynyl stavudine triphosphate. Antimicrob. Agents Chemother. 52:2035-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]