Abstract
Three biologically active β-defensins were purified by chromatography from chicken bone marrow extract: avian β-defensin 1 (AvBD1), AvBD2, and the newly isolated β-defensin AvBD7. Mass spectrometry analyses showed that bone marrow-derived AvBD1, -2, and -7 peptides were present as mature peptides and revealed posttranslational modifications for AvBD1 and AvBD7 in comparison to their in silico-predicted amino acid sequences. Tandem mass spectrometry analysis using the nanoelectrospray-quadrupole time of flight method showed N-terminal glutaminyl cyclization of mature AvBD7 and C-terminal amidation of mature AvBD1 peptide, while posttranslational modifications were absent in bone marrow-derived mature AvBD2 peptide. Furthermore, mass spectrometry analysis performed on intact cells confirmed the presence of these three peptides in mature heterophils. In addition, the antibacterial activities of the three β-defensins against a large panel of gram-positive and -negative bacteria were assessed. While the three defensins displayed similar antibacterial spectra of activity against gram-positive strains, AvBD1 and AvBD7 exhibited the strongest activity against gram-negative strains in comparison to AvBD2.
Defensins belong to the large family of antimicrobial peptides which are important components of innate and adaptive immunity (23). They are 30 to 50 amino acids long and rich in cationic residues, and their tertiary structure is maintained by three disulfide bridges. Depending on their disulfide motifs and the positions of conserved cysteines, defensins are divided into three major groups in vertebrates: α-, β-, and θ-defensins. The largest families comprise α- and β-defensins, which are produced by leukocytes and epithelial cells. The roles of both defensin families are not restricted to antimicrobial effectors at host-pathogen interfaces. In mammals, they have been shown to modulate adaptive immunity by attracting antigen-presenting cells and lymphocytes (45).
Interest in defensins as therapeutic drugs is growing because defensins may constitute an alternative to the controversial use of antibiotics. Moreover, defensins may be used as an additive in food preservation. Bird defensins have a double economic interest: besides their interesting antimicrobial properties, the poultry industry may represent an important source of production. The only known bird defensins belong to the β-defensin family (see reference 40 for a review). The three-dimensional structure and broad antimicrobial activity spectrum have been studied only for king penguin avian β-defensin 103b (AvBD103b) (spheniscin-2), which gave new insights into the structure-activity relationship of AvBDs (11, 36). β-Defensins have also been isolated from ostrich heterophils (AvBD1, -2, -4, -7, and -8), the so-called ostricacins, which are highly similar to chicken defensins. The antimicrobial activity of these ostrich peptides is sensitive to cations (35), and their targets seem to be intracellular (34). In chicken, the β-defensin family encompasses 14 genes that have mostly been identified in nucleic acid sequence data (40). The mature peptide of AvBD1 and AvBD2, formerly called gallinacin 1 and gallinacin 2, were the only ones isolated from heterophil granulocytes (7, 8). In these previous studies, the resolution of the mass spectrometry (MS) techniques used allowed for amino acid sequence identification from the average masses of these two peptides but not posttranslational modification characterization. Therefore, in order to further characterize the sequences and the antimicrobial activities of AvBD1 and AvBD2, larger amounts of defensins were required. Bone marrow, in which defensin genes have been shown to be highly expressed (15), should be a relevant source of natural peptides. It is the active site of leukopoiesis and contains the heterophil precursors, which are AvBD1- and AvBD2-producing cells. These cell precursors are present in larger numbers in bone marrow than mature heterophils in blood (25).
In the present work, we describe the extraction of AvBD1 and AvBD2 from bone marrow and their fine characterization. With modern proteomic analyses, the primary structure of AvBDs has been refined as well as the N- and C-terminal cleavage sites of mature defensin. Besides AvBD1 and AvBD2, we characterize for the first time AvBD7, whose mature primary sequence was known only as bioinformatic prediction (15, 43). The specific expression of AvBD1, AvBD2, and AvBD7 in mature effector cells has been confirmed using intact-cell matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS (ICM-MS). Finally, the antimicrobial activity of the three bone marrow-extracted defensins has been characterized and compared against a large panel of bacterial strains, including zoonotic agents.
MATERIALS AND METHODS
Sample preparation from bone marrow and extraction.
Bone marrow was obtained from femurs and tibias of 10 2-month-old PA12 chickens. The bones were cut longitudinally, and the marrow was collected by several phosphate-buffered saline (pH 7.4) rinses. The phosphate-buffered saline solution containing bone marrow was centrifuged at 1,500 × g for 10 min. The pellet was then resuspended in 5% acetic acid. The mixture obtained was sonicated on ice for 5 min (6 s at a power of 40 W [Vibra-Cell 75455; Bioblock] separated by 1 s of resting) and stirred for 16 h at 4°C. Finally, acid-insoluble material was removed by ultracentrifugation at 70,000 × g for 30 min at 4°C, and the supernatant was lyophilized.
Purification of bone marrow-extracted peptides.
Bone marrow-lyophilized extract was dissolved in 5% acetic acid and fractionated on a 2- by 100-cm Bio-Gel P10 column (Bio-Rad) equilibrated in 5% acetic acid. The column effluent at a flow rate of 0.5 ml/min was monitored at 280 nm. The fractions (6 ml) corresponding to the major gel filtration peaks were screened by MALDI-TOF MS. The peptides characterized on the basis of their intact mass were further purified by semipreparative C18 reverse-phase high-performance liquid chromatography (RP-HPLC) (Nucleosil C18, 300 Å, 5 μm, 250 by 10 mm; Merck) using a linear acetonitrile gradient with 0.1% trifluoroacetic acid (TFA) at a constant flow rate of 3 ml/min. The TFA counter anions of the purified peptides were finally exchanged by evaporation of acetonitrile and loading on a Sep-Pak C18 6-ml column (Waters), followed by washings with 5% acetic acid, and eluted by 10% acetic acid in 50% acetonitrile and lyophilized. The powder was dissolved in ultrapure water at a 500-μg/ml concentration.
Heterophil collection from peritoneal exudates.
A suspension (20 ml) of Sephadex G75 (Sigma) (3% in 0.15 M NaCl) was injected in the peritoneal cavities of three 5-week-old PA12 chickens. The animals were sacrificed 12 hours after the injection. Peritoneal cavities were washed with 50 ml of Hank's buffered salt solution (Sigma) to collect the cells. Each cell suspension was centrifuged at 300 × g for 10 min at 4°C, and the cell pellets were suspended in fresh Hank's buffered salt solution. Peritoneal exudates gave approximately 108 cells (95% heterophils) per chicken. Cells were finally centrifuged in 1.5-ml tubes, and the cell pellets were frozen until ICM-MS analysis was carried out.
MALDI-TOF MS.
All mass spectra were generated on a M@LDI LR (Waters, Micromass, Ltd., Manchester, United Kingdom) MALDI-TOF mass spectrometer, operating in positive linear mode. External mass calibration was performed with a mixture containing Glu-fibrinopeptide B, adrenocorticotropin (18-39 fragment), insulin, and ubiquitin, all at 1 pmol/μl; cytochrome c at 2 pmol/μl; myoglobin at 4 pmol/μl; and trypsinogen at 8 pmol/μl. Mass spectra were recorded in the m/z range of 2,000 to 20,000, acquiring 10 shots per spectrum at a laser firing rate of 10 Hz. Data processing was performed using MassLynx 4.0 software. The background of the spectrum result for each sample well was subtracted using a polynomial order of 10% below the curve and smoothed with the minimum peak width at half height set to 15 channels. Smoothing was performed twice using the Savitzky Golay algorithm. All the spectra were processed using the same parameters.
For gel filtration eluted-peptide mass fingerprint analysis, 1 μl of each fraction and the matrix (1:1, vol/vol) were loaded on the target by using the dried droplet method. The matrix used was sinapinic acid at 20 mg/ml dissolved in 50% acetonitrile-50% H2O-0.1% TFA.
For ICM-MS analysis, the frozen whole-cell pellets were resuspended with a 10-μl aliquot of a sinapinic acid matrix solution prepared by dissolving 20 mg of the solid crystals in 1 ml of 50% acetonitrile-50% H2O-1% TFA reagent. The matrix/sample was spotted onto the MALDI sample probe and dried at room temperature. To obtain a mass accuracy of around 0.06%, internal calibration was accomplished using the major identified peak of AvBD2 as the lock mass (average mass, 3,316.66 Da) for all spectra.
Nano-LC-MS-MS: bottom-up analysis.
Identification by bottom-up proteomic analysis was conducted on peptides from gel filtration fractions. Aliquots were dried and resuspended in 20 μl of 50 mM NH4HCO3, then reduced with 5 mM dithiothreitol, alkylated with 12.5 mM iodoacetamide, and incubated overnight at 37°C with 0.3 μg of bovine trypsin (sequencing grade; Roche, Paris, France). The tryptic digests were acidified with 5 μl of 5% formic acid and sonicated for 10 min. Nanoscale capillary liquid chromatography-tandem MS (nano-LC-MS-MS) analysis of the digested peptides was performed using a CapLC system coupled to a hybrid quadrupole TOF (Q-TOF) Ultima Global mass spectrometer (Waters, Manchester, United Kingdom) fitted with a Z-spray ion source. The capillary voltage was set at 3,200 V, the cone voltage at 100 V, and the microchannel plate voltage at 2,300 V. Samples were desalted and concentrated using an on-line precolumn (Monolithic Trap column, 200 m [inside diameter] by 5 mm; LC Packings, Dionex). Peptide separations were conducted on a C18 column (Atlantis dC18, 75 μm [inside diameter] by 150 mm; NanoEase, Waters) running with a 200-nl/min flow. The gradient profile consisted of a gradient from 100% A (0.1% formic acid-2% acetonitrile-98% H2O, vol/vol) to 15% B (0.1% formic acid-20% H2O-80% acetonitrile, vol/vol) in 2 min and 15% to 55% B in 60 min. Mass data were acquired using automatic switching between MS and MS-MS (fragmentation) modes: one MS survey scan was followed by MS-MS scans on the three most intense peptide ions detected. Only doubly and triply charged ions were allowed to be selected as precursors over an m/z range of 400 to 1,300. The collision energy was selected depending on the precursor ion mass and charge. The mass spectrometer was calibrated using the fragmentation spectrum of Glu-fibrinopeptide B at 500 fmol/μl in 50% water-50% acetonitrile-0.1% formic acid. Data were processed using ProteinLynx Global server 2.2 (Waters). The peptide and fragment masses obtained were matched automatically to proteins in a nonredundant database (Swiss-Prot 56.5) and a decoy database, using the MS-MS ion search option in the MASCOT software program (Matrix Science, United Kingdom) against the all-taxa section (as of 15 December 2008, with 402,482 sequences). Enzyme specificity was set to semitrypsin with two missed cleavages, using carbamidomethylcysteine and methionine oxidation as variable modifications. The tolerance of the ions was set to 0.3 Da for both parent and fragment ion matches. All hits with P values of <0.05 were manually verified, and proteins detected by one peptide were considered positively identified with five consecutive fragment ions and with a false-discovery rate of 0% (peptide matches above the identity threshold).
Nanoelectrospray MS: top-down proteomic analysis.
HPLC fractions were diluted with a mixture of water-formic acid-acetonitrile (45:5:50) and were analyzed by MS and MS-MS on a nanoelectrospray ionization (nano-ESI)-Q-TOF Ultima Global mass spectrometer (Waters, Manchester, United Kingdom). The samples were loaded into nanoelectrospray capillaries (Proxeon, Odense, Denmark). The capillary voltage was set at 1,200 V, the cone voltage at 100 V, the microchannel plate voltage at 2,300 V, and collision energies on the order of 20 to 50 eV. Argon was used as the collision gas. Data acquisition and analysis were performed using MassLynx version 4.0 software (Waters, Manchester, United Kingdom). Multicharged precursor ions with m/z values of 900.99 (+5), 979.23 (+4), and 892.27 (+6) were selected for AvBD1, AvBD2, and AvB7, respectively, for the fragmentation of intact molecular species.
Bacterial strains.
To test the antimicrobial activity, the following strains were used: Bacillus subtilis, Bacillus cereus ATCC 14579, Staphylococcus aureus ATCC 29740, Staphylococcus saprophyticus ATCC 19701, Staphylococcus haemolyticus DSM 20263, Listeria monocytogenes strain EGD, Enterobacter cloacae ATCC 13047, Klebsiella pneumoniae ATCC 13882, Pseudomonas aeruginosa ATCC 25010, Salmonella enterica serovar Enteritidis ATCC 13076, Salmonella enterica serovar Enteritidis LA5, and Salmonella enterica serovar Typhimurium ATCC 14028. Salmonella enterica serovar Enteritidis LA5 is a wild-type strain (nalidixic acid resistant at 20 μg ml−1) isolated from natural chicken infections (1). Enterobacter cloacae ATCC 13047, Klebsiella pneumoniae ATCC, 13882 and Pseudomonas aeruginosa ATCC 25010 were purchased from the Centre International de Ressources Microbiennes-Bactéries Pathogènes (CIRM-BP, INRA, Nouzilly, France). Salmonella enterica serovar Enteritidis ATCC 13076 and Salmonella enterica serovar Typhimurium ATCC 14028 were purchased from the Centre de Ressources Biologiques de l'Institut Pasteur (CRBIP, Institut Pasteur, Paris, France). Staphylococcus strains were kindly provided by Pascal Rainard (INRA, UR1282, Nouzilly, France). Bacillus strains, Listeria monocytogenes strain EGD, and Salmonella strain LA5 were kindly provided by Philippe Velge (INRA, UR1282, Nouzilly, France).
Antimicrobial activity test.
The antibacterial activities of peptides were measured by a radial diffusion assay according to the method described by Lehrer and colleagues (12, 31). Bacteria incubated overnight were diluted in Trypticase soy broth (TSB) or brain heart infusion at an absorbance of 0.02 and were incubated 2.5 to 4 h according to the bacterial strain at 37°C to obtain a mid-logarithmic-phase culture for which bacterial concentration was determined by plating serial 10-fold dilutions of bacterial suspension on Trypticase soy agar plates and by counting CFU after 24 h of incubation at 37°C. A volume containing bacteria at 1.107 CFU was centrifuged at 900 × g for 5 min at 4°C, and bacteria were washed once with cold 10 mM sodium phosphate buffer (pH 7.4), resuspended in a small volume of cold sodium phosphate buffer, and mixed to 25 ml of previously autoclaved, warm (42°C) 10 mM sodium phosphate buffer containing 7.5 mg of TSB medium, 1% (wt/vol) low-electroendosmosis-type agarose (Sigma), and 0.02% Tween 20. The agarose solution containing bacteria was poured into a square petri dish (120 by 120 by 15 mm) to form a 1-mm-deep uniform layer. A 2.5-mm-diameter gel punch was used to make 36 evenly spaced wells. In each well, 5 μl of peptide dilutions or control solutions was added. MSI-94 (a broad-antimicrobial-spectrum linear amphipathic magainin variant) was used as a positive control and was a kind gift from Philippe Bulet (CNRS UMR 5525, Archamps, France). Peptide and control solutions were allowed to diffuse in the gel containing bacteria by incubating the plates for 3 h at 37°C. The gel was then overlaid with 25 ml of agar consisting of a double-strength (6% [wt/vol]) solution of TSB containing 1% (wt/vol) agarose. After overnight incubation at 37°C, the diameter of the clear zone surrounding each well was measured. For each bacterial strain, three identical independent measurements of antibacterial activity were performed. The MIC of each peptide was determined according to the method of Lehrer and colleagues (31) from a graph constructed by plotting the log10 peptide concentration against the diameter of the clear zone on the plate minus the diameter of the well as a function of the log peptide concentration. The best-fit straight line was determined using linear regression with GraphPad Prism 5 software (GraphPad Software). The MIC was calculated by finding the intersection of the line with the x axis, indicating the lowest peptide concentration from which no clear zone is obtained.
RESULTS
AvBD purification and identification from bone marrow extract.
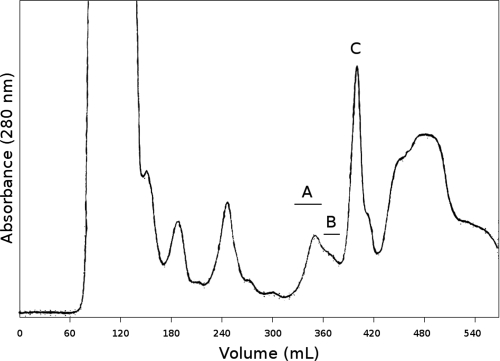
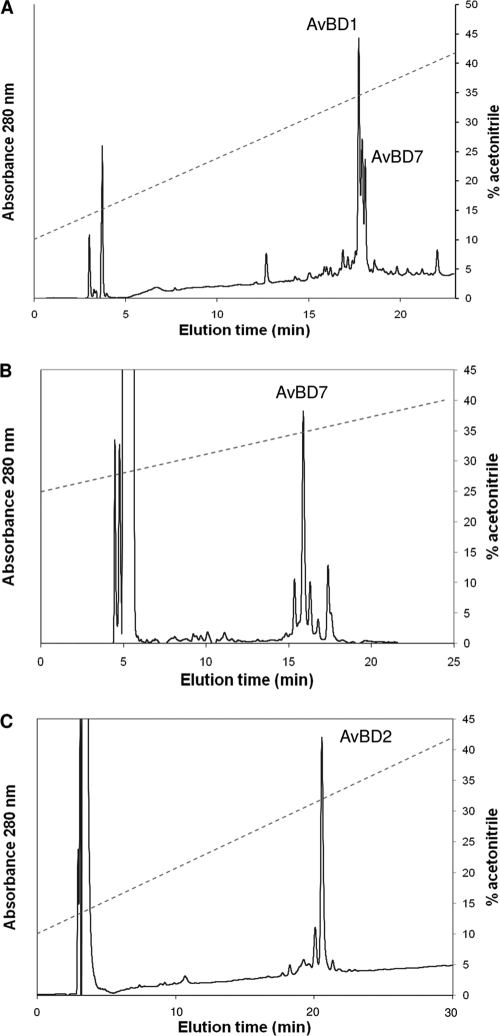
AvBD1, AvBD2, and AvBD7 were identified in the bone marrow extract. As a first purification step, a size exclusion chromatography was run on P10 gel (Fig. 1). MALDI-TOF MS analysis of fractions revealed three main peptides corresponding to average molecular masses ([M+H]+) of 4,504, 5,353, and 3,916 Da at peaks A, B, and C, respectively (Fig. 1). The gel filtration fractions containing the three main peptides were subjected to trypsin digestion, and the proteolyzed peptides were analyzed by nano-LC-MS-MS. AvBD1, AvBD2, and AvBD7 were identified (Table 1). The fractions containing each AvBD, selected by MALDI-TOF MS measurements, were pooled. An additional step of RP-HPLC was necessary to purify AvBD1 in fractions from peak A (Fig. 2A) and AvBD7 in fractions from peak B (Fig. 2B). The AvBD2 RP-HPLC chromatogram showed that gel filtration-pooled elution fractions corresponding to AvBD2 peak were quite pure (Fig. 2C). After the final step of HPLC purification, the amount of recovered peptides was between 1 and 2 milligrams of defensin from 10 chickens.
FIG. 1.
Gel filtration chromatography profile. Bone marrow extract was eluted by a 5% acetic acid aqueous solution on a P10 column with a flow rate of 0.5 ml/min. Peaks A, B, and C encompassed AvBD1, AvBD7, and AvBD2 peptides, respectively.
TABLE 1.
Peptide list for all nano-LC-Q-TOF-identified gel filtration fractionsa
| Fraction | m/z | z | Mobs | Mtheo | Δmass | No. of missed cleavages | Peptide score | Sequence | Positions | Observed modification | Uniprot accession no. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peak A | 465.2037 | 2 | 928.3928 | 928.4477 | −0.0548 | 0 | 22 | SGFCAFLK | 10-17 | CAM | AMP1_CHICK |
| 529.2473 | 2 | 1,056.48 | 1,056.5426 | −0.0626 | 1 | 41 | KSGFCAFLK | 9-17 | CAM | AMP1_CHICK | |
| 538.2618 | 2 | 1,074.509 | 1,074.5743 | −0.0653 | 0 | 79 | CPSLTLISGK | 18-28 | CAM | AMP1_CHICK | |
| Peak B | 597.241 | 2 | 1,192.4674 | 1,192.5481 | −0.0807 | 0 | 35 | NGICFPGICR | 35-44 | 2 CAM | GLL7_CHICK |
| Peak C | 538.55 | 3 | 1,612.6282 | 1,612.7239 | −0.0957 | 0 | 49 | GGSCHFGGCPSHLIK | 33-47 | 2 CAM | GLL2_CHICK |
m/z, ratio of mass/charge fragmented; z, charge; Mobs, experimental molecular weight of peptide; Mtheo, theoretical molecular weight of peptide; CAM, carbamidomethylation of cysteine; 2 CAM, carbamidomethylation of two cysteines. “Peptide score” represents the individual MASCOT score for peptide, and “positions” represents the positions in the full sequence.
FIG. 2.
RP-HPLC profile of pooled gel filtration fractions corresponding to peak A, peak B, and peak C. The fractions were chromatographed on a C18 column. The broken line represents an acetonitrile gradient.
Structural characterization of AvBDs by use of top-down analysis.
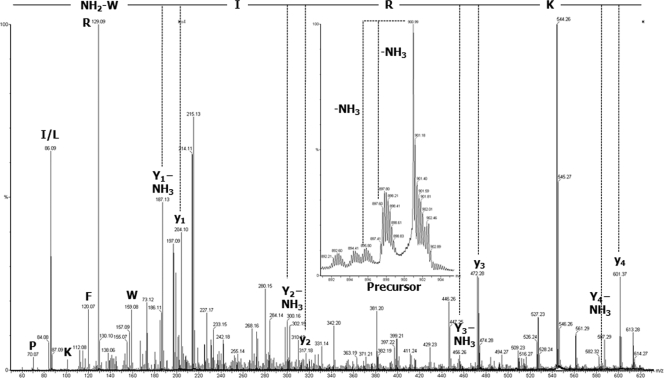
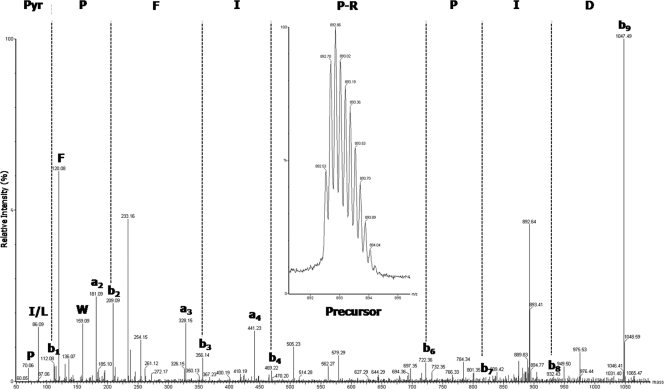
To complete the characterization of the primary structure sequence and posttranslational modifications and to sequence the N and C termini, a top-down MS strategy was applied on intact proteins. An aliquot of HPLC-purified intact AvBDs was subjected to top-down MS and MS-MS analyses using nano-ESI-Q-TOF MS without any prior reduction or alkylation steps. The m/z values of the precursor multiple-charged monoisotopic ions were measured with a mass accuracy of 100 ppm, affording an m/z of 901.04 (z = 5) for AvBD1 ([M+H]+, 4,501.2 Da), an m/z of 979.23 (z = 4) for AvBD2 ([M+H]+, 3,913.9 Da), and an m/z of 1,070.84 (z = 5) for AvBD7 ([M+H]+, 5,350.2 Da). Bone marrow AvBD2 exhibited the expected monoisotopic mass in comparison to AvBD2 extracted from heterophils (8) with three disulfide bonds (Δmass, −6 Da) (Table 2). Bone marrow AvBD1 presented a monoisotopic mass corresponding to an amidated C terminus with three disulfide bonds (Table 2). Fragment ions y1 and y4 in the MS-MS spectrum of the fivefold-charged precursor ion with an m/z of 901 (Fig. 3) confirmed this posttranslational modification. Moreover, the double loss of NH3 from the precursor with low collision energy confirmed the presence of an amidated C-terminal tryptophan (16). The monoisotopic mass of AvBD7 corresponded to the mass of the AvBD7 propeptide predicted by Xiao et al., with three disulfide bonds (43) and with the N-terminal Gln being cyclized as a pyroglutamic acid residue (see Table 2 for comparison). The presence of this modified amino acid at the N terminus explained the failure of the AvBD7 sequence determination using Edman degradation (data not shown). Fragmentation of the intact AvBD7 sixfold-charged precursor ion (m/z = 892.53) corresponded to the N-terminal sequence of the propeptide AvBD7 predicted by Xiao et al., with the pyroglutamic acid at the first position (Fig. 4).
TABLE 2.
AvBD primary structures and corresponding theoretical massesa
| β-Defensin | Sequence | Cys reduced |
Cys oxidized |
||
|---|---|---|---|---|---|
| Avg mass | Monoisotopic mass | Avg mass | Monoisotopic mass | ||
| AvBD1 | H-GRKSDCFRKSGFCAFLKCPSLTLISGKCSRFYLCCKRIW-OHb | 4,511.495 | 4,508.253 | 4,505.447 | 4,502.206 |
| H-GRKSDCFRKSGFCAFLKCPSLTLISGKCSRFYLCCKRIWG-OHc | 4,568.547 | 4,565.274 | 4,562.499 | 4,559.227 | |
| H-GRKSDCFRKSGFCAFLKCPSLTLISGKCSRFYLCCKRIW-NH2d | 4,510.510 | 4,507.268 | 4,504.462 | 4,501.221 | |
| AvBD2 | H-LFCKGGSCHFGGCPSHLIKVGSCFGFRSCCKWPWNA-OHb | 3,922.662 | 3,919.763 | 3,916.614 | 3,913.716 |
| AvBD7 | H-QPFIPRPIDTCRLRNGICFPGICRRPYYWIGTCNNGIGSCCARGWRS-OHe | 5,377.318 | 5,373.585 | 5,371.270 | 5,367.538 |
| PyrQ-PFIPRPIDTCRLRNGICFPGICRRPYYWIGTCNNGIGSCCARGWRS-OHf | 5,360.292 | 5,356.559 | 5,354.244 | 5,350.512 | |
| H-RPIDTCRLRNGICFPGICRRPYYWIGTCNNGIGSCCARGWRS-OHc | 4,794.618 | 4,791.269 | 4,788.570 | 4,785.222 | |
The masses were calculated with PROWL (http://prowl.rockefeller.edu/prowl-cgi/sequence.exe) and are shown as [M+H]+.
Mature AvBDs extracted from heterophils (8).
Mature AvBD sequence determined by bioinformatic prediction (15).
Putative mature AvBD1 with the C-terminal Trp amidated from predictive terminal sequence WG.
Propeptide AvBD7 sequence determined by bioinformatic prediction (43).
Propeptide AvBD7 sequence determined by bioinformatic prediction (43), completed with N-terminal Gln cyclized as a pyroglutamic acid (PyrQ).
FIG. 3.
C terminus bone marrow-extracted AvBD1 sequence with posttranslational modification. Shown is an MS-MS spectrum (×4 zoom on baseline) obtained by collision-induced dissociation with nano-ESI-Q-TOF MS from intact AvBD1 peptide without prior reduction or alkylation. The precursor ion had an m/z of 901 in charge state 5. The C-terminal sequence KRIW, determined by manual interpretation, was essentially obtained by four consecutive y ions (y1 to y4) and y ions with loss of NH3 confirming an amidation in the C terminus. Moreover, the double loss of NH3 from the protonated precursor indicated the presence of a peptide amide. Single letters indicate immonium ions. The amidated tryptophan (W) is shown as CONH2-W.
FIG. 4.
N terminus bone marrow-extracted AvBD7 sequence with posttranslational modification. Shown is an MS-MS spectrum obtained by collision-induced dissociation with nano-ESI-Q-TOF MS from intact AvBD7 peptide without reduction or alkylation. The precursor ion had an m/z of 892 in charge state 6. The N-terminal sequence PyrQ-PFIPRPID, determined by manual interpretation, was essentially obtained by eight b ions (b1 to b9). Despite the discontinuous fragmentation pattern (b5 ion not observed), we were able to reveal the sequence information. Few a ions were noted on the spectrum. Single letters indicate immonium ions. The pyroglutamic acid residue formed from Gln (Q) is shown as Pyr.
Characterization of mature AvBDs in heterophils.
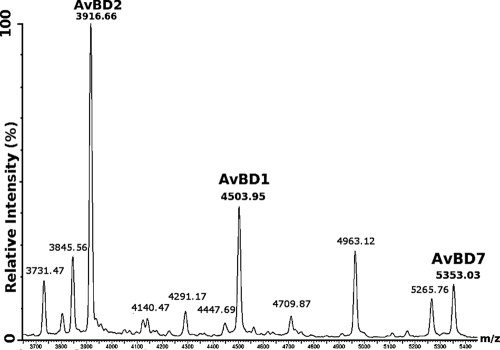
In order to strengthen the finding that the three AvBD sequences determined as described above correspond to the mature peptide sequences exported from the bone marrow, spectral profiling by MALDI-TOF MS was performed on intact heterophils isolated from peritoneal exudates (Fig. 5). This ICM-MS experiment demonstrated that the observed average masses in whole heterophils correspond to the masses observed by nano-ESI-MS for AvBD1, AvBD2, and AvBD7 in bone marrow.
FIG. 5.
ICM-MS spectrum of heterophils from peritoneal exudates. The spectrum was processed by a subtracted baseline and smoothed with an m/z range of 3,650 to 5,450. The ICM-MS approach shows specific expression of AvBD2, AvBD1, and AvBD7.
Antimicrobial activity and MIC.
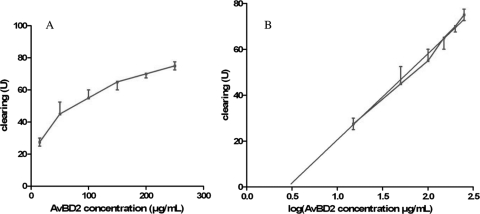
The antimicrobial activity of each purified AvBD against a large panel of bacterial strains was evaluated in a radial diffusion assay. Clearing units (U) were calculated by the diameter in mm of the clear zone surrounding the well minus the diameter in mm of the well and were plotted as a function of log (peptide concentration) for each bacterial strain, as illustrated in Fig. 6 for AvBD2 against Escherichia coli. The various MICs obtained are shown in μM in Table 3. In comparison to control antimicrobial peptide MSI-94, similar MICs were observed for AvBD1, AvBD2, and AvBD7 against gram-positive bacterial strains. However, AvBD1 displayed a particularly low MIC of 80 nM for Staphylococcus aureus. Indicative of a high-level antibacterial effect, the MICs of AvBD1 and AvBD7 against the gram-negative bacteria tested as well as against gram-positive ones remained low. In contrast, the antimicrobial activity of AvBD2 was 2 to more than 100 times less effective against gram-negative bacteria than against gram-positive ones. Indeed, AvBD2 antibacterial activity is significantly lower than those of AvBD1 and AvBD7 toward gram-negative bacteria. The MIC of AvBD2 against Enterobacter cloacae was above the measurement range under our conditions, indicative of a weak activity of AvBD2 against this strain. Interestingly, it could be noticed that AvBD2 was far less efficient against the Salmonella enterica serovar Enteritidis LA5 strain, corresponding to a field isolate, than against Salmonella enterica serovar Enteritidis ATCC 13076.
FIG. 6.
Antimicrobial activity measurement for AvBD2 on E. coli. (A) Dose-effect curve between the AvBD2 concentration and the inhibitory effect on E. coli ATTC 25922, reflected by clearing (the diameter in mm of the clear zone on the plate minus the diameter in mm of the well). (B) The best-fit straight line (in gray) was determined using linear regression with GraphPad Prism 5 software (GraphPad Software). The AvBD2 MIC for E. coli ATTC 25922 was calculated by finding the intersection of the line with the x axis, indicating the lowest peptide concentration from which no clear zone is obtained.
TABLE 3.
Antimicrobial activities of purified AvBD1, AvBD2, and AvBD7 and control MSI-94
| Bacterial group | MIC (μM) (95% CI)a |
|||
|---|---|---|---|---|
| MSI-94 | AvBD1 | AvBD2 | AvBD7 | |
| Gram positive | ||||
| B. subtilis | 0.73 (0.37-1.23) | 0.19 (0.13-0.27) | 0.28 (0.19-0.39) | 0.21 (0.15-0.28) |
| B. cereus | 0.32 (0.18-0.50) | 0.21 (0.14-0.31) | 0.47 (0.29-0.70) | 0.20 (0.13-0.31) |
| S. aureus | 0.34 (0.22-0.50) | 0.08 (0.02-0.18) | 0.42 (0.14-0.86) | 0.11 (0.04-0.23) |
| S. haemolyticus | 0.56 (0.37-0.78) | 0.14 (0.07-0.24) | 0.25 (0.12-0.43) | 0.21 (0.14-0.29) |
| S. saprophyticus | 0.37 (0.25-0.52) | 0.16 (0.08-0.27) | 0.22 (0.09-0.42) | 0.16 (0.08-0.27) |
| L. monocytogenes | 0.41 (0.20-0.69) | 0.26 (0.13-0.43) | 0.22 (0.08-0.43) | 0.31 (0.18-0.48) |
| Gram negative | ||||
| S. Enteritidis ATCC 13076 | 0.32 (0.22-0.44) | 0.17 (0.11-0.24) | 0.80 (0.47-1.22) | 0.16 (0.11-0.22) |
| S. Enteritidis LA5 | 0.31 (0.23-0.41) | 0.16 (0.09-0.24) | 6.05 (3.45-8.88) | 0.21 (0.16-0.27) |
| S. Typhimurium | 0.37 (0.24-0.54) | 0.15 (0.08-0.24) | 2.39 (0.75-4.66) | 0.21 (0.14-0.31) |
| E. cloacae | 0.44 (0.30-0.62) | 0.20 (0.09-0.35) | >64 | 0.32 (0.12-0.61) |
| K. pneumoniae | 0.26 (0.12-0.46) | 0.10 (0.04-0.19) | 0.71 (0.26-1.40) | 0.32 (0.18-0.50) |
| E. coli | 0.59 (0.31-0.97) | 0.27 (0.16-0.41) | 0.72 (0.50-1.00) | 0.28 (0.20-0.37) |
| P. aeruginosa | 0.42 (0.31-0.56) | 0.27 (0.13-0.46) | 1.86 (0.75-3.36) | 0.33 (0.25-0.42) |
The MIC was determined by a radial diffusion assay for every bacterial strain. MSI-94 was used as a control antibiotic.
DISCUSSION
The aim of our work was to improve knowledge of the primary structure and the antibacterial activity of chicken bone marrow-extracted β-defensins and to demonstrate that chicken bone marrow can be a source of bioactive defensins. In this study, a hybrid strategy combining “bottom-up” and “top-down” proteomic approaches enabled characterization of AvBD1, AvBD2, and AvBD7. A strategy based on multiple MS techniques was employed to characterize the peptides, including mass fingerprinting by MALDI-TOF MS, de novo sequencing of tryptic digested molecular species by nano-LC-Q-TOF MS, intact peptide fragmentation by nano-ESI-Q-TOF MS, and direct cell analysis by MALDI-TOF MS. The use of complementary MS technologies allowed us to obtain structural information (sequence and posttranslational modifications) and to visualize these structures in their biological context in cells.
Intact-molecular-species analysis demonstrated that three peptides (AvBD1, AvBD2, and AvBD7) were efficiently purified by chromatography. AvBD1 and AvBD2, formerly known as gallinacins 1 and 2, respectively, have been isolated from chicken heterophils (7, 8). Our results show that these peptides are abundantly present in bone marrow, which is consistent with the recent data from Kannan et al. (9). Furthermore, our results demonstrate the presence of AvBD7 peptide in bone marrow cells whose sequence corresponds to the putative one predicted from in silico studies (15, 43). Taken together, the presence of AvBD7 in heterophils (as judged by ICM-MS analysis) and its activity against several bacterial strains lead us to conclude that the isolated form of AvBD7 was the mature one. This sequence revealed a longer N terminus than the predicted mature sequence of Gal7 (43). In spite of sequence variability among chicken defensins, AvBD6 displays a signal peptide sequence and N-terminal cleavage site sequence (VAGQP) identical to those of AvBD7. We can thus hypothesize that AvBD6 should possess the same cleavage site as AvBD7 and a pyroglutamic acid at the first position (Fig. 6).
The monoisotopic molecular masses ([M+H]+) of mature AvBD1, AvBD2, and AvBD7 were shown to be 4,501.2 Da, 3,913.9 Da, and 5,350.2 Da, respectively. Upon comparison of measured mass with theoretical mass, a difference of 6 Da indicated the presence of three disulfide bonds, which is consistent with the sequence conservation of six cysteines in β-defensins. Posttranslational modifications were not determined for AvBD1 when this peptide was initially purified from heterophils and its sequence characterized by MS and Edman sequencing (7, 8). However, Brockus et al. (3) suspected a cleavage after the C-terminal tryptophan of AvBD1 by comparison between the mRNA sequence and the amino acid sequence isolated by Harwig et al. (8). Here, higher-resolution MS analyses allow us to demonstrate the amidation of the C-terminal tryptophan residue of AvBD1, a posttranslational modification resulting from the elimination of a C-terminal glycine residue. Elsewhere, the presence of pyroglutamic acid as the N-terminal residue, another kind of posttranslational modification, was found in the newly characterized AvBD7 sequence. Amidation and glutaminyl cyclization are commonly found in bioactive peptides and short polypeptides, such as hormones, neuropeptides, antimicrobial peptides, and venom toxins (6, 21, 24, 27, 29, 32, 41, 42). The C-terminal amide has been shown to increase antibacterial potency by inducing α-helix formation or stabilizing α-helix (13, 28, 32). This property leads to a greater level of stability, associated with resistance to proteolytic degradation (32) and with better abilities to bind receptor (10) and to penetrate the bacterial membrane (32). Pyroglutamic acid at the first position offers similar properties, such as implication in receptor binding and resistance to protease (21). Unlike AvBD1 and AvBD7, AvBD2 peptide does not contain any modified N- or C-terminal amino acid and displays the weakest activity against gram-negative bacteria. Although the β-sheet structure stabilized through disulfide bonds commonly found in mammalian β-defensins contributes to the resistance to protease (17), this lack of N- and C-terminal end protection could be related to a greater susceptibility to bacterial defense mechanisms (22).
The mechanism of action of cationic antimicrobial peptides such as β-defensin is also mainly dependent on electrostatic interactions between positive charges from the peptide sequence and the anionic surface of the outer membrane of gram-negative bacteria (4, 18). Comparison of the activities of the three chicken β-defensins against gram-negative bacteria shows that AvBD2, which is less cationic (+4) than AvBD1 (+8) or AvBD7 (+6) (Fig. 7), exhibits the weakest activity toward gram-negative bacteria. Interestingly, the N-terminal part is less charged. This extremity, as proposed for highly similar ostrich defensins, could be important in antimicrobial activity against gram-negative bacteria (8, 33). However, there is no direct correlation between the net charge of the protein and its potency against gram-negative bacteria, since AvBD7 (+6) and AvBD1 (+8) exhibit similar levels of activity. Moreover, the potencies of the three chicken β-defensins against gram-positive bacteria are comparable for most of the strains tested. Once more, even if cationicity is essential for the activity, no direct correlation between the net charge of the protein and its potency against gram-positive bacteria can be established.
FIG. 7.
Sequence comparison of 14 AvBDs. The predicted signal peptide of each AvBD is highlighted in orange, and the mature peptide sequences of isolated AvBD1, AvBD2, and AvBD7 are highlighted in blue. “*” indicates positions which have a single, fully conserved residue. “:” indicates conserved substitutions. “.” indicates semiconserved substitutions.
Mammalian α-defensins are produced as inactive propeptides stored in vesicles of granulocyte progenitor cells (37, 39). The role of the propiece has been demonstrated to counteract the positive charges in mature peptide and to limit the defensin activity in producing cells of mammals (14, 37). In chicken defensins, the putative propiece is shorter, and negative charges are barely present in its sequence (Fig. 7), which is not consistent with a role of neutralization attributable to chicken defensin propiece. The number of residues between the predicted signal peptide and the mature sequence of AvBD7 is reduced to a glycine residue and may not constitute a functional propiece (Fig. 7). The only β-defensin which has been isolated in proforms is human hBD1, and all the propeptides exhibit an antibacterial activity contrary to α-defensins (38, 39). In addition, in silico predictions suggest that maturation of β-defensins occurs by signal peptidase cleavage (2). Furthermore, the fully active, mature forms of AvBD1, AvBD2, and AvBD7 appear as available in the bone marrow as they are in the granulocyte effector cells, as shown by the ICM-MS experiment. This indicates that the mature form of chicken β-defensins is already stored in granules of heterophils and precursor cells. It seems that no proregion processing occurs either in mature heterophils or in bone marrow precursor cells. The β-defensin family seems to be at the origin of defensin gene diversification, as α- and θ-defensins are not present in older vertebrates (26). Therefore, the negatively charged mammalian propiece may have appeared later in evolution to replace costly mechanisms, such as chaperone proteins, for processing active defensin in older vertebrates.
On the other hand, the primary structure and the difference in activity between the three defensins may indicate that they are implicated in functions other than antimicrobial activity. The antimicrobial activity spectra of our AvBDs showed that the Salmonella LA5 strain, which is a field isolate (1), was less susceptible to AvBD2 antimicrobial activity than to those of other AvBDs. Avian defensins have been linked to host resistance to Salmonella intestinal carriage (19, 20). AvBD1 and AvBD2 have recently been shown to be expressed by chicken intestinal epithelial cells (5). Surprisingly, live Salmonella strain LA5 was shown to inhibit AvBD2 expression in this cell population, in spite of having relatively low susceptibility to this avian defensin. Taken together, these data could indicate that Salmonella has developed a strategy to escape from another function of AvBD2 peptide. This function could be the chemotactic ability, as shown by comparison with homologous duck AvBD2 and some mammalian defensins which have been shown to attract immune cells (30, 44). The shorter N-terminal extremity of AvBD2 may reduce steric effects and thus facilitate coupling with a receptor. Future work will be performed to identify attracted cells and characterize targeted receptors.
In conclusion, new insights have been gained into the sequence characteristics, antimicrobial activity spectra, and maturation of chicken β-defensins, revealing posttranslational modifications. Future investigations of multiple biological activities, such as chemotactism, will be helpful for exploiting chicken β-defensins as therapeutic or preservative agents.
Acknowledgments
We thank Philippe Marceau for technical assistance with gel filtration chromatography, Robert Lehrer for helpful discussions and useful advice, and Jocelyn Spragg for careful reading of the manuscript. We also thank Laurence Merat and the staff of the Plateforme d'Infectiologie Expérimentale of the Centre INRA, Nouzilly, France, for providing animals.
Chrystelle Derache was supported by a doctoral fellowship from INRA and Région Centre. The Plateforme de Protéomique Analytique et Fonctionnelle was financially supported by the Région Centre, CNRS, and INRA. The work was supported by a Biotechnocentre grant (no. 3200068) from the Région Centre and by a European Union grant (FP6-2002-FOOD1 no. 505523).
Footnotes
Published ahead of print on 8 September 2009.
REFERENCES
- 1.Allen-Vercoe, E., M. Dibb-Fuller, C. J. Thorns, and M. J. Woodward. 1997. SEF17 fimbriae are essential for the convoluted colonial morphology of Salmonella Enteritidis. FEMS Microbiol. Lett. 153:33-42. [DOI] [PubMed] [Google Scholar]
- 2.Beckloff, N., and G. Diamond. 2008. Computational analysis suggests beta-defensins are processed to mature peptides by signal peptidase. Protein Pept. Lett. 15:536-540. [DOI] [PubMed] [Google Scholar]
- 3.Brockus, C. W., M. W. Jackwood, and B. G. Harmon. 1998. Characterization of beta-defensin prepropeptide mRNA from chicken and turkey bone marrow. Anim. Genet. 29:283-289. [DOI] [PubMed] [Google Scholar]
- 4.Chan, D. I., E. J. Prenner, and H. J. Vogel. 2006. Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim. Biophys. Acta 1758:1184-1202. [DOI] [PubMed] [Google Scholar]
- 5.Derache, C., E. Esnault, C. Bonsergent, Y. Le Vern, P. Quéré, and A. Lalmanach. 2009. Differential modulation of beta-defensin gene expression by Salmonella Enteritidis in intestinal epithelial cells from resistant and susceptible chicken inbred lines. Dev. Comp. Immunol. 33:959-966. [DOI] [PubMed] [Google Scholar]
- 6.Destoumieux, D., P. Bulet, D. Loew, A. Van Dorsselaer, J. Rodriguez, and E. Bachère. 1997. Penaeidins, a new family of antimicrobial peptides isolated from the shrimp penaeus vannamei (decapoda). J. Biol. Chem. 272:28398-28406. [DOI] [PubMed] [Google Scholar]
- 7.Evans, E. W., G. G. Beach, J. Wunderlich, and B. G. Harmon. 1994. Isolation of antimicrobial peptides from avian heterophils. J. Leukoc. Biol. 56:661-665. [DOI] [PubMed] [Google Scholar]
- 8.Harwig, S. S., K. M. Swiderek, V. N. Kokryakov, L. Tan, T. D. Lee, E. A. Panyutich, G. M. Aleshina, O. V. Shamova, and R. I. Lehrer. 1994. Gallinacins: cysteine-rich antimicrobial peptides of chicken leukocytes. FEBS Lett. 342:281-285. [DOI] [PubMed] [Google Scholar]
- 9.Kannan, L., N. C. Rath, R. Liyanage, and J. O. J. Lay. 2009. Direct screening identifies mature {beta}-defensin 2 in avian heterophils. Poult. Sci. 88:372-379. [DOI] [PubMed] [Google Scholar]
- 10.Kotani, M., M. Detheux, A. Vandenbogaerde, D. Communi, J. M. Vanderwinden, E. Le Poul, S. Brézillon, R. Tyldesley, N. Suarez-Huerta, F. Vandeput, C. Blanpain, S. N. Schiffmann, G. Vassart, and M. Parmentier. 2001. The metastasis suppressor gene KISS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J. Biol. Chem. 276:34631-34636. [DOI] [PubMed] [Google Scholar]
- 11.Landon, C., C. Thouzeau, H. Labbe, P. Bulet, and F. Vovelle. 2004. Solution structure of spheniscin, a beta-defensin from the penguin stomach. J. Biol. Chem. 279:30433-30439. [DOI] [PubMed] [Google Scholar]
- 12.Lehrer, R. I., M. Rosenman, S. S. Harwig, R. Jackson, and P. Eisenhauer. 1991. Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137:167-173. [DOI] [PubMed] [Google Scholar]
- 13.Li, Z. Q., R. B. Merrifield, I. A. Boman, and H. G. Boman. 1988. Effects on electrophoretic mobility and antibacterial spectrum of removal of two residues from synthetic sarcotoxin IA and addition of the same residues to cecropin B. FEBS Lett. 231:299-302. [DOI] [PubMed] [Google Scholar]
- 14.Liu, L., and T. Ganz. 1995. The pro region of human neutrophil defensin contains a motif that is essential for normal subcellular sorting. Blood 85:1095-1103. [PubMed] [Google Scholar]
- 15.Lynn, D. J., R. Higgs, S. Gaines, J. Tierney, T. James, A. T. Lloyd, M. A. Fares, G. Mulcahy, and C. O'Farrelly. 2004. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics 56:170-177. [DOI] [PubMed] [Google Scholar]
- 16.Mouls, L., G. Subra, J. Aubagnac, J. Martinez, and C. Enjalbal. 2006. Tandem mass spectrometry of amidated peptides. J. Mass Spectrom. 41:1470-1483. [DOI] [PubMed] [Google Scholar]
- 17.Nicolas, P., and A. Mor. 1995. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu. Rev. Microbiol. 49:277-304. [DOI] [PubMed] [Google Scholar]
- 18.Powers, J. S., and R. E. W. Hancock. 2003. The relationship between peptide structure and antibacterial activity. Peptides 24:1681-1691. [DOI] [PubMed] [Google Scholar]
- 19.Sadeyen, J., J. Trotereau, J. Protais, C. Beaumont, N. Sellier, G. Salvat, P. Velge, and A. Lalmanach. 2006. Salmonella carrier-state in hens: study of host resistance by a gene expression approach. Microbes Infect. 8:1308-1314. [DOI] [PubMed] [Google Scholar]
- 20.Sadeyen, J., J. Trotereau, P. Velge, J. Marly, C. Beaumont, P. A. Barrow, N. Bumstead, and A. Lalmanach. 2004. Salmonella carrier state in chicken: comparison of expression of immune response genes between susceptible and resistant animals. Microbes Infect. 6:1278-1286. [DOI] [PubMed] [Google Scholar]
- 21.Schilling, S., C. Wasternack, and H. Demuth. 2008. Glutaminyl cyclases from animals and plants: a case of functionally convergent protein evolution. Biol. Chem. 389:983-991. [DOI] [PubMed] [Google Scholar]
- 22.Schmidtchen, A., I. Frick, E. Andersson, H. Tapper, and L. Björck. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46:157-168. [DOI] [PubMed] [Google Scholar]
- 23.Selsted, M. E., and A. J. Ouellette. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551-557. [DOI] [PubMed] [Google Scholar]
- 24.Selsted, M. E., Y. Q. Tang, W. L. Morris, P. A. McGuire, M. J. Novotny, W. Smith, A. H. Henschen, and J. S. Cullor. 1996. Purification, primary structures, and antibacterial activities of beta-defensins, a new family of antimicrobial peptides from bovine neutrophils. J. Biol. Chem. 271:16430. [DOI] [PubMed] [Google Scholar]
- 25.Semerad, C. L., F. Liu, A. D. Gregory, K. Stumpf, and D. C. Link. 2002. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity 17:413-423. [DOI] [PubMed] [Google Scholar]
- 26.Semple, C. A., P. Gautier, K. Taylor, and J. R. Dorin. 2006. The changing of the guard: molecular diversity and rapid evolution of beta-defensins. Mol. Divers. 10:575-584. [DOI] [PubMed] [Google Scholar]
- 27.Sforça, M. L., S. J. Oyama, F. Canduri, C. C. B. Lorenzi, T. A. Pertinhez, K. Konno, B. M. Souza, M. S. Palma, J. Ruggiero Neto, W. F. J. Azevedo, and A. Spisni. 2004. How C-terminal carboxyamidation alters the biological activity of peptides from the venom of the eumenine solitary wasp. Biochemistry 43:5608-5617. [DOI] [PubMed] [Google Scholar]
- 28.Shalev, D. E., A. Mor, and I. Kustanovich. 2002. Structural consequences of carboxyamidation of dermaseptin S3. Biochemistry 41:7312-7317. [DOI] [PubMed] [Google Scholar]
- 29.Silva, P. I. J., S. Daffre, and P. Bulet. 2000. Isolation and characterization of gomesin, an 18-residue cysteine-rich defense peptide from the spider acanthoscurria gomesiana hemocytes with sequence similarities to horseshoe crab antimicrobial peptides of the tachyplesin family. J. Biol. Chem. 275:33464-33470. [DOI] [PubMed] [Google Scholar]
- 30.Soman, S. S., D. S. Arathy, and E. Sreekumar. 2009. Discovery of anas platyrhynchos avian beta-defensin 2 (apl_avbd2) with antibacterial and chemotactic functions. Mol. Immunol. 46:2029-2038. [DOI] [PubMed] [Google Scholar]
- 31.Steinberg, D. A., and R. I. Lehrer. 1997. Designer assays for antimicrobial peptides. Disputing the “one-size-fits-all” theory. Methods Mol. Biol. 78:169-186. [DOI] [PubMed] [Google Scholar]
- 32.Strömstedt, A. A., M. Pasupuleti, A. Schmidtchen, and M. Malmsten. 2009. Evaluation of strategies for improving proteolytic resistance of antimicrobial peptides by using variants of EFK17, an internal segment of LL-37. Antimicrob. Agents Chemother. 53:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiarto, H., and P. Yu. 2006. Identification of three novel ostricacins: an update on the phylogenetic perspective of beta-defensins. Int. J. Antimicrob. Agents 27:229-235. [DOI] [PubMed] [Google Scholar]
- 34.Sugiarto, H., and P. Yu. 2007. Mechanisms of action of ostrich beta-defensins against Escherichia coli. FEMS Microbiol. Lett. 270:195-200. [DOI] [PubMed] [Google Scholar]
- 35.Sugiarto, H., and P. Yu. 2007. Effects of cations on antimicrobial activity of ostricacins-1 and 2 on E. coli O157:H7 and S. aureus 1056MRSA. Curr. Microbiol. 55:36-41. [DOI] [PubMed] [Google Scholar]
- 36.Thouzeau, C., Y. Le Maho, G. Froget, L. Sabatier, C. Le Bohec, J. A. Hoffmann, and P. Bulet. 2003. Spheniscins, avian beta-defensins in preserved stomach contents of the king penguin, Aptenodytes patagonicus. J. Biol. Chem. 278:51053-51058. [DOI] [PubMed] [Google Scholar]
- 37.Valore, E. V., and T. Ganz. 1992. Posttranslational processing of defensins in immature human myeloid cells. Blood 79:1538-1544. [PubMed] [Google Scholar]
- 38.Valore, E. V., C. H. Park, A. J. Quayle, K. R. Wiles, P. B. J. McCray, and T. Ganz. 1998. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J. Clin. Investig. 101:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valore, E. V., E. Martin, S. S. Harwig, and T. Ganz. 1996. Intramolecular inhibition of human defensin HNP-1 by its propiece. J. Clin. Investig. 97:1624-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Dijk, A., E. J. A. Veldhuizen, and H. P. Haagsman. 2008. Avian defensins. Vet. Immunol. Immunopathol. 124:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verleyen, P., G. Baggerman, W. D'Hertog, E. Vierstraete, S. J. Husson, and L. Schoofs. 2006. Identification of new immune induced molecules in the haemolymph of drosophila melanogaster by 2D-nanoLC MS/MS. J. Insect Physiol. 52:379-388. [DOI] [PubMed] [Google Scholar]
- 42.Walsh, G., and R. Jefferis. 2006. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 24:1241-1252. [DOI] [PubMed] [Google Scholar]
- 43.Xiao, Y., A. L. Hughes, J. Ando, Y. Matsuda, J. Cheng, D. Skinner-Noble, and G. Zhang. 2004. A genome-wide screen identifies a single beta-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genomics 5:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, D., A. Biragyn, L. W. Kwak, and J. J. Oppenheim. 2002. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 23:291-296. [DOI] [PubMed] [Google Scholar]
- 45.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schröder, J. M. Wang, O. M. Howard, and J. J. Oppenheim. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]