Abstract
The expanded-spectrum β-lactamase (ESBL) gene blaVEB-1, identified worldwide in Enterobacteriaceae and Pseudomonas aeruginosa, is associated with either class 1 integrons or repeated elements. We report here the first association of blaVEB-1a with the insertion sequence ISCR2 in six Acinetobacter species isolates recovered from Argentina. That genetic structure was likely at the origin of the mobilization of this ESBL gene.
Among the different Ambler class A expanded-spectrum β-lactamase (ESBL) genes, the blaVEB-1 gene is considered to be an emerging one since its presence has been detected in many gram-negative organisms from different parts of the world during the last decade (14, 25). It has been identified in Enterobacteriaceae and Pseudomonas aeruginosa in many countries, such as France, Spain, Algeria, Turkey, Canada, Korea, and Thailand (1, 2, 8, 23, 29). In addition, P. aeruginosa isolates producing the VEB-1a variant (differing from VEB-1 by just a single amino acid located in the leader peptide of the premature protein) have been found in Kuwait and India (3, 24). VEB-1-producing Acinetobacter baumannii strains have been identified in France, having disseminated on a nationwide scale (13, 20).
The blaVEB-1 gene is often part of a gene cassette located in class 1 integrons. In A. baumannii strain AYE from France, the corresponding class 1 integron was itself part of an 86-kb resistance island, the largest identified so far (7). However, in some cases, the blaVEB-1 and blaVEB-1a genes have not been identified in the form of gene cassettes, being associated with peculiar genetic structures called repeated elements (Re) in P. aeruginosa and Enterobacteriaceae (3, 12, 22).
From 2002 to 2004, several Acinetobacter isolates producing the ESBL VEB-1a were recovered from different cities in Argentina (16). Six VEB-1a-positive isolates were retained: four A. baumannii isolates, one Acinetobacter johnsonii isolate, and one Acinetobacter genomospecies 10 isolate (17). Five of them were shown previously to produce the narrow-spectrum β-lactamase SCO-1 (17). All six isolates are resistant to all β-lactams except carbapenems, with the exception of Acinetobacter genomospecies 10, which coproduces the carbapenem-hydrolyzing oxacillinase OXA-58 and is resistant to imipenem (17). Preliminary experiments performed by PCR mapping using corresponding primers (3) showed that the blaVEB-1a gene is neither part of a class 1 integron nor associated with Re elements in those isolates. Accordingly, we have examined the genetic context of the blaVEB-1a gene in those six isolates in order to predict its acquisition mechanism.
First, cloning experiments were performed using DNA from a whole-cell preparation of one of the isolates, A. johnsonii isolate 7037, and pBK-CMV as a cloning vector as described previously (17). BamHI-restricted DNA fragments of A. johnsonii 7037 were ligated into a BamHI-digested plasmid and transferred into Escherichia coli DH10B by electroporation as described previously (18). Selection of recombinant clones was performed using Trypticase soy agar plates containing 100 μg of amoxicillin (amoxicilline) and 30 μg of kanamycin per ml. Recombinant plasmid p7037B1, expressing an ESBL phenotype, was retained for further analysis. Sequencing of the 3,630-bp insert in p7037B1 identified the blaVEB-1a gene (Fig. 1). Upstream of blaVEB-1a, sequences corresponding to insertion sequence ISCR2 (formerly designated the CR2 element) were identified. In fact, the ISCR2 right-end extremity defined as oriIS was located 223 bp upstream of blaVEB-1a, those 223 bp including the GTTAGCG sequence usually defining the core site of the blaVEB-1a gene cassette when blaVEB-1a is present in class 1 integrons. Downstream of blaVEB-1a, a truncated copy of ISCR2 (ΔISCR2) was present, the corresponding tnpA transposase gene being truncated at its 5′ extremity, resulting in a protein lacking 119 of its 497 amino acids at the N-terminal extremity. ISCR2 belongs to the ISCR family, which currently includes 20 members, all presenting some similarities to IS91- like insertion sequences (http://www.cardiff.ac.uk/medic/aboutus/departments/medicalmicrobiology/genetics/iscr/iscr_elements.html). The ISCR2 transposase shares less than 57% amino acid identity with all other ISCR transposases. ISCRs are peculiar since they do not possess inverted repeats, do not generate target site duplications upon transposition, and transpose through a process called rolling-circle transposition (28, 29).
FIG. 1.
Nucleotide sequence of a 3,630-bp BamHI fragment of recombinant plasmid p7037B1 containing the blaVEB-1a gene. The deduced amino acid sequence is designated in single-letter code below the nucleotide sequence. The transcription orientations of the ORFs are indicated by horizontal arrows (orf is the unknown gene, and xre is the gene encoding a putative transcriptional regulator). The right-end boundary of ISCR2, defined as oriIS, is shaded. The putative promoter sequences of the blaVEB-1a gene, consisting of the −35 and −10 regions, are boxed. The ΔtnpA truncated transposase gene of ISCR2 is indicated. The stars indicate stop codons. The vertical arrows indicate the boundaries of the blaVEB-1a gene cassette as found in class 1 integrons (with the core site sequence in bold) and also the location where ΔISCR2 is identified. The BamHI sites of the cloned fragment are underlined.
The downstream 1,170-bp fragment separating the blaVEB-1a gene from the ΔISCR2 element contained two open reading frames (ORFs), in addition to the entire 59-bp sequence defining the blaVEB-1a gene cassette when blaVEB-1a is integron borne. One ORF corresponded to a 92-amino-acid putative protein of unknown function, and the second corresponded to a 103-amino-acid protein showing homology to helix-turn-helix XRE family transcriptional regulators (Fig. 1). The latter protein shared 84% amino acid identity with a protein identified by analysis of the genome of Psychroflexus torquis ATCC 700755 (GenBank accession number ZP_01253739).
We subsequently investigated the five other isolates of our collection for the ISCR2-blaVEB-1a association. PCR assays performed using blaVEB-1-specific primers (27) in combination with ISCR2-specific primer ISCR2A (5′-AAGAATTTCTCCAATGCGGG-3′) or ISCR2B (5′-GCGGCTCCTTTTCCGACAAC-3′) showed that in all isolates the blaVEB-1a gene was bracketed by the ISCR2 elements, as found in isolate 7037. Attempts to identify plasmids by using the Kieser technique (10) permitted visualization of several plasmids in all the blaVEB-1a-positive isolates. However, subsequent Southern hybridization performed with a probe specific for blaVEB-1a indicated that this ESBL gene was very likely chromosomally located in all the isolates tested since a single hybridization signal corresponding only to the chromosomal band was obtained (data not shown). This result is in accordance with the results of electrotransformation assays, performed as described previously (17), that did not allow the transfer of any β-lactam resistance marker into an A. baumannii recipient strain.
The finding of two copies of the ISCR2 element (including one truncated copy) at the extremities of blaVEB-1a strongly suggests that ISCR2 was at the origin of the gene's mobilization. According to the hypotheses raised by Toleman et al. (28), it is very likely that an intact ISCR2 copy originally mobilized blaVEB-1a by a rolling-circle transposition process and that a secondary process of homologous recombination between two ISCR2 copies led to the observed structure. To confirm the hypothesis that such recombination events may occur, we performed a PCR assay using blaVEB-1a-specific outward primers VEB-inv1 (5′-CAGTTTGAGCATTTGAATACAC-3′) and VEB-inv2 (5′-AGCGTATTTGTTGCAGAGTCC-3′). Using DNA samples from all VEB-1a-positive isolates as templates, a ca. 2,900-bp amplicon from each isolate was obtained. Sequencing identified a structure encompassing the blaVEB-1a gene and the corresponding downstream sequences, together with the downstream ΔISCR2 truncated element. However, the sequence of the upstream, intact ISCR2 copy was not included in that amplicon. Therefore, homologous recombination may have been at the source of the mobilization of the blaVEB-1a gene in its present genetic context. Further studies will be conducted to explore the possible role of the ISCR2 transposase in that recombination process, with regard to the fact that the ISCR transposases were initially thought to be recombinases (15).
The genetic structures involved in the mobilization process for resistance genes often play an additional role in the genes’ expression by providing promoter sequences, as demonstrated previously for many IS elements and class 1 integrons (11, 21) and in particular for ISCR1 and ISCR4 (19, 27). In the ISCR2-blaVEB-1a configuration in evidence here, no obvious feature indicates a possible role of ISCR2 in blaVEB-1a expression. Indeed, the distance separating the oriIS extremity of ISCR2 from the blaVEB-1a start codon is quite long and no putative promoter is found in silico inside the ISCR2 sequence. In contrast, a precise analysis of the sequences separating ISCR2 from the blaVEB-1a gene revealed putative promoter sequences made of a −35 motif (TTGTAA) and a −10 motif (TGGTTT) separated by the optimal 17-bp distance (Fig. 1), thus suggesting that blaVEB-1a gene expression is driven by its original promoter in those Acinetobacter isolates.
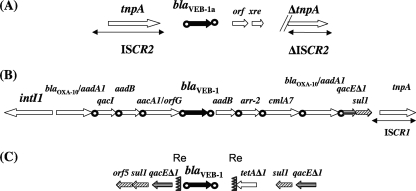
This study demonstrates further that blaVEB-like gene acquisition may be linked to a variety of genetic elements (Fig. 2). This is an uncommon observation since the widespread ESBL genes of the blaTEM, blaSHV, and blaCTX-M types are usually associated with very conserved genetic structures (28). In addition, this finding is the first evidence of ISCR2-mediated acquisition of a β-lactamase gene, while ISCR2 has been identified previously in association with the sul2 sulfonamide resistance gene (26), the dfr18 trimethoprim resistance gene (4, 9), and the floR florfenicol resistance gene (5, 6). Interestingly, in the isolates described herein, a fragment larger than the known blaVEB-1 gene cassette had been mobilized by ISCR2, likely directly from the bacterial progenitor.
FIG. 2.
Comparison of the different genetic structures in which the blaVEB-1-like genes have been identified. The diagram in panel A corresponds to the present description of an ISCR2-associated blaVEB-1a gene, that in panel B shows the blaVEB-1a gene in the form of a gene cassette inside a class 1 integron as reported previously (21), and the diagram in panel C depicts the blaVEB-1a gene bracketed by the Re1 and Re2 elements as described elsewhere (3, 12). The core and inverse core sites bracketing the blaVEB-1a gene and defining its corresponding cassette are indicated by black circles.
Finally, the origin of this ISCR2 element remains be determined to better understand where and how the capture of blaVEB-1a from its natural reservoir has occurred. Although the blaVEB-1a gene has a GC content of 31.3%, that of ISCR2 is 59.5%, clearly not consistent with the elements’ having the same origin. ISCR2 has been so far identified in diverse bacterial species, including A. baumannii (1).
Nucleotide sequence accession number.
The sequences determined in this study have been assigned GenBank accession number FJ808975.
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France, and mostly by grants from the European Community (DRESP2 project contract no. LSHM-CT-2005-018705 and TROCAR project contract no. HEALTH-F3-2008-223031) and by the INSERM.
We are grateful to S. Corvec, who has participated in this project.
Footnotes
Published ahead of print on 24 August 2009.
REFERENCES
- 1.Adams, M. D., K. Goglin, N. Molyneaux, K. M. Hujer, H. Lavender, J. J. Jamison, I. J. MacDonald, K. M. Martin, T. Russo, A. A. Campagnari, A. M. Hujer, R. A. Bonomo, and S. R. Gill. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aragón, L. M., B. Mirelis, E. Miró, C. Mata, L. Gómez, A. Rivera, P. Coll, and F. Navarro. 2008. Increase in β-lactam-resistant Proteus mirabilis strains due to CTX-M- and CMY-type as well as new VEB- and inhibitor-resistant TEM-type β-lactamases. J. Antimicrob. Chemother. 61:1029-1032. [DOI] [PubMed] [Google Scholar]
- 3.Aubert, D., D. Girlich, T. Naas, S. Nagarajan, and P. Nordmann. 2004. Functional and structural characterization of the genetic environment of an extended-spectrum β-lactamase blaVEB gene from a Pseudomonas aeruginosa isolate obtained in India. Antimicrob. Agents Chemother. 48:3284-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blickwede, M., and S. Schwarz. 2004. Molecular analysis of florfenicol-resistant Escherichia coli isolates from pigs. J. Antimicrob. Chemother. 53:58-64. [DOI] [PubMed] [Google Scholar]
- 6.Doublet, B., S. Schwarz, C. Kehrenberg, and A. Cloeckaert. 2005. Florfenicol resistance gene floR is part of a novel transposon. Antimicrob. Agents Chemother. 49:2106-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fournier, P. E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 11.Lévesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 12.Naas, T., D. Aubert, T. Lambert, and P. Nordmann. 2006. Complex genetic structures with repeated elements, a sul-type class 1 integron, and the blaVEB extended-spectrum β-lactamase gene. Antimicrob. Agents Chemother. 50:1745-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naas, T., B. Coignard, A. Carbonne, K. Blanckaert, O. Bajolet, C. Bernet, X. Verdeil, P. Astagneau, J. C. Desenclos, P. Nordmann, and the French Nosocomial Infection Early Warning Investigation and Surveillance Network. 2006. VEB-1 extended-spectrum β-lactamase-producing Acinetobacter baumannii, France. Emerg. Infect. Dis. 12:1214-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naas, T., L. Poirel, and P. Nordmann. 2008. Minor extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1):42-52. [DOI] [PubMed] [Google Scholar]
- 15.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasterán, F., M. Rapoport, A. Petroni, D. Faccone, A. Corso, M. Galas, M. Vázquez, A. Procopio, M. Tokumoto, and V. Cagnoni. 2006. Emergence of PER-2 and VEB-1a in Acinetobacter baumannii strains in the Americas. Antimicrob. Agents Chemother. 50:3222-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel, L., S. Corvec, M. Rapoport, P. Mugnier, A. Petroni, F. Pasteran, D. Faccone, M. Galas, H. Drugeon, V. Cattoir, and P. Nordmann. 2007. Identification of the novel narrow-spectrum β-lactamase SCO-1 in Acinetobacter spp. from Argentina. Antimicrob. Agents Chemother. 51:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel, L., D. Girlich, T. Naas, and P. Nordmann. 2001. OXA-28, an extended-spectrum variant of OXA-10 β-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., M. Magalhaes, M. Lopes, and P. Nordmann. 2004. Molecular analysis of metallo-β-lactamase gene blaSPM-1-surrounding sequences from disseminated Pseudomonas aeruginosa isolates in Recife, Brazil. Antimicrob. Agents Chemother. 48:1406-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel, L., O. Menuteau, N. Agoli, C. Cattoen, and P. Nordmann. 2003. Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 41:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel, L., T. Naas, and P. Nordmann. 2008. Genetic support of extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1):75-81. [DOI] [PubMed] [Google Scholar]
- 23.Poirel, L., J. D. Pitout, L. Calvo, J.-M. Rodriguez-Martinez, D. Church, and P. Nordmann. 2006. In vivo selection of fluoroquinolone-resistant Escherichia coli isolates expressing plasmid-mediated quinolone resistance and expanded-spectrum β-lactamase. Antimicrob. Agents Chemother. 50:1525-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel, L., V. O. Rotimi, E. M. Mokaddas, A. Karim, and P. Nordmann. 2001. VEB-1-like extended-spectrum β-lactamases in Pseudomonas aeruginosa, Kuwait. Emerg. Infect. Dis. 7:468-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel, L., L. Villa, A. Bertini, J. D. Pitout, P. Nordmann, and A. Carattoli. 2007. Expanded-spectrum β-lactamase and plasmid-mediated quinolone resistance. Emerg. Infect. Dis. 13:803-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rådström, P., and G. Swedberg. 1988. RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob. Agents Chemother. 32:1684-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Martinez, J.-M., L. Poirel, R. Canton, and P. Nordmann. 2006. Common region CR1 for expression of antibiotic resistance genes. Antimicrob. Agents Chemother. 50:2544-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 58:1-6. [DOI] [PubMed] [Google Scholar]