Abstract
Since 2002, nine methicillin (meticillin)-resistant Staphylococcus aureus (MRSA) strains that are also resistant to vancomycin (VRSA) have been reported in the United States, including seven clinical isolates from Michigan. The strains harbor a plasmid-borne Tn1546 element following conjugation from a glycopeptide-resistant Enterococcus strain. In the second step, Tn1546 transposed to a resident plasmid in five strains; the acquired plasmid behaved as a suicide gene delivery vector, and the incoming DNA had been rescued by illegitimate recombination. Surprisingly, combination of a glycopeptide with a β-lactam has a strong synergistic effect against VRSA, both in vitro and in an animal model, despite resistance of the strains to both drug classes when administered separately. This results from the fact that the late peptidoglycan precursors ending in d-alanine-d-lactate (d-Ala-d-Lac) that are mainly synthesized in the presence of glycopeptide inducers are not substrates for PBP2′, which is the only transpeptidase that remains active in the presence of oxacillin. One VRSA strain is partially dependent on vancomycin for growth due to a mutation in the host d-Ala:d-Ala ligase, thus having to rely on the inducible resistance pathway for cell wall synthesis. Competition growth experiments in the absence of inducer between the MRSA recipient and isogenic VRSA transconjugant revealed a disadvantage for the transconjugant, accounting, in part, for the low level of dissemination of the VRSA clinical isolates. The association of multiple molecular and environmental factors has been implicated in the regional emergence of VRSA in Michigan.
Staphylococcus aureus is one of the most common causes of hospital- and community-acquired infections, and treatment of staphylococcal infections is complicated by the ability of this bacterial species to become resistant to antibiotics. Vancomycin is the drug of choice for therapy of infections due to methicillin (meticillin)-resistant S. aureus (MRSA), but increase in vancomycin use has led to the emergence of two types of glycopeptide-resistant S. aureus. The first one, designated vancomycin intermediate-resistant S. aureus, is associated with a thickened and poorly cross-linked cell wall, resulting in accumulation of acyl-d-alanyl-d-alanine (X-d-Ala-d-Ala) targets in the periphery that sequester glycopeptides (8). The second type, vancomycin-resistant S. aureus (VRSA), is due to acquisition from Enterococcus spp. of the vanA operon, carried by transposon Tn1546, resulting in high-level resistance (4, 5).
This review is devoted to the genetics of acquisition and to the phenotypic expression of the vanA operon in S. aureus.
VanA-type glycopeptide resistance.
Glycopeptides inhibit cell wall synthesis in gram-positive bacteria by binding to the C-terminal d-Ala-d-Ala of the pentapeptide precursors of peptidoglycan, thus blocking the transglycosylation and transpeptidation reactions (28).
High-level resistance to glycopeptides was first reported in enterococci in 1988 (19, 39), approximately 30 years after the introduction of this antibiotic into clinical practice. Since then, glycopeptide-resistant enterococci have disseminated throughout the world. To date, seven types of resistance (VanA, -B, -C, -D, -E, -G, and -L) in enterococci have been described; these correspond to specific operons (vanA, -B, -C, -D, -E, -G, and -L) responsible for (i) synthesis of a new target (peptidoglycan precursors ending in d-Ala-d-lactate [d-Ala-d-Lac] in VanA, -B, and -D type or d-Ala-d-serine [d-Ala-d-Ser] in VanC, -E, -G, and -L type) having a reduced affinity for glycopeptides and (ii) elimination of the normal d-Ala-d-Ala-terminating precursors (26).
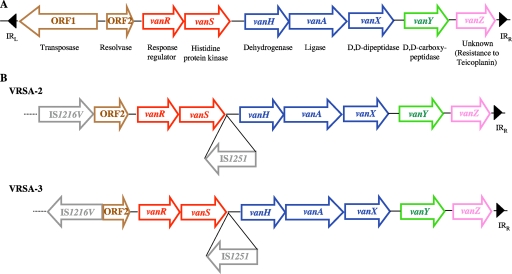
VanA-type resistance, which was the first to be elucidated and which is the most common, is characterized by high levels of resistance to glycopeptides, vancomycin, and teicoplanin and is mediated by transposon Tn1546 or closely related elements that are chromosomally or plasmid located. This 11-kb mobile genetic element, which belongs to the Tn3 family of transposons, codes for nine polypeptides, responsible for transposition (products of ORF1 and ORF2), regulation of expression of resistance (VanR and VanS), synthesis of modified peptidoglycan precursors ending in d-Lac (VanH and VanA), hydrolysis of normal precursors (VanX and VanY), and an unknown function (VanZ) (Fig. 1A). ORF1 and ORF2 encode, respectively, a transposase and a resolvase, responsible for the movements of the transposon. VanH is a dehydrogenase that reduces pyruvate to d-Lac, and VanA is a ligase allowing the formation of the d-Ala-d-Lac depsipeptide that replaces the d-Ala-d-Ala dipeptide in peptidoglycan synthesis. This substitution dramatically decreases the affinity of late peptidoglycan precursors for glycopeptides. The VanX d,d-dipeptidase and the VanY d,d-carboxypeptidase are implicated in the removal of the susceptibility pathway: the former hydrolyzes the dipeptide d-Ala-d-Ala formed by the host chromosomal d-Ala:d-Ala ligase, allowing reduction of the level of peptidoglycan precursors ending in d-Ala-d-Ala, and the latter hydrolyzes the terminal residue of late peptidoglycan precursors, notably the terminal d-Ala of pentapeptide precursors that are produced if elimination of d-Ala-d-Ala by VanX is not complete (5). Inducible expression of VanA-type resistance by glycopeptides is regulated by the VanR/VanS two-component system. The two proteins control the level of expression of the resistance genes in response to the presence of glycopeptides in the culture medium. VanS, a membrane-associated sensor, contains a histidine residue in the cytoplasmic domain which is phosphorylated when glycopeptides are present in the medium. VanR is a transcriptional activator which accepts the phosphoryl group on an aspartate residue from activated VanS. Thus, VanS controls the level of phosphorylation of VanR. The phosphorylated form of VanR activates the cotranscription of the vanH, vanA, vanX, and vanY genes by binding to the PRES promoter (3) and of the vanR and vanS genes by binding to the PREG promoter (10).
FIG. 1.
Comparison of Tn1546 elements. (A) Schematic representation of Tn1546. IRL and IRR, inverted left and right repeats, respectively. Open arrows represent coding sequences and indicate direction of transcription. Brown arrows, genes required for transposition; red arrows, regulatory genes; blue arrows, genes required for resistance; green arrow, accessory gene; pink arrow, gene of unknown function. (B) Organization of the Tn1546 element in strains VRSA-2 and VRSA-3; gray arrows, insertion sequences.
Transfer of the vanA gene cluster from Enterococcus species to S. aureus was demonstrated in vitro and in an in vivo model 15 years ago (22), suggesting that such a phenomenon could occur in humans. Indeed, in 2002, the first MRSA clinical isolate (VRSA-1) exhibiting high-level resistance to glycopeptides (vancomycin MIC > 256 μg/ml; teicoplanin MIC = 128 μg/ml) due to acquisition of the vanA operon was detected in Michigan (34). Since then, 11 VanA-type MRSA strains have been isolated: 9 in the United States (7 from Michigan, 1 from Pennsylvania, and 1 from New York) (17, 21, 43), 1 in India (Kolkata) (31), and 1 in Iran (Tehran) (1).
Nomenclature.
In this review, we concentrate on the clinical isolates from the United States since these strains have been the most extensively studied. We have therefore adopted the nomenclature developed by the Network on Antimicrobial Resistance in S. aureus (NARSA) (http://www.narsa.net) (Table 1): VRSA followed by a number.
TABLE 1.
VRSA strains isolated in the United States
| NARSA designation | Patient | Date of isolation (mo/yr) | State | Age (yr) | Sex | Reference or source |
|---|---|---|---|---|---|---|
| VRSA-1 | 1 | 06/2002 | Michigan | 40 | Female | 34 |
| VRSA-2 | 2 | 09/2002 | Pennsylvania | 70 | Male | 21 |
| VRSA-3 | 3 | 03/2004 | New York | 63 | Female | 17 |
| VRSA-5 | 4 | 02/2005 | Michigan | 78 | Male | 35 |
| VRSA-6 | 5 | 10/2005 | Michigan | 58 | Female | 35 |
| VRSA-7 | 6 | 12/2005 | Michigan | 48 | Male | 35 |
| VRSA-8 | 7 | 10/2006 | Michigan | 43 | Female | 35 |
| VRSA-9 | 8 | 11/2007 | Michigan | NAa | Female | NARSA |
| VRSA-10 | 9 | 12/2007 | Michigan | NA | Female | NARSA |
NA, not available.
Transfer of glycopeptide resistance from Enterococcus to MRSA.
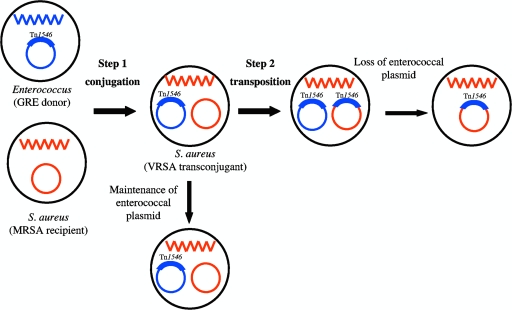
Among the 11 VRSA strains detected so far, the vanA gene cluster has been assigned to a plasmid in at least 10 strains. For the strain isolated in Iran, no data on the location of the operon are yet available. Except for VRSA-8, a glycopeptide-resistant Enterococcus sp. was coisolated with the VRSA strain from the same patient, strongly suggesting that MRSA strains have acquired Tn1546 from glycopeptide-resistant enterococci. Acquisition of vancomycin resistance can result from one or two genetic events (Fig. 2). The first step is plasmid transfer by conjugation from the Enterococcus donor to the S. aureus recipient. Certain enterococcal plasmids can replicate efficiently in staphylococci and be stably maintained in the new host. Others are less efficient in replication: they are lost during cell division and diluted in the progeny. However, in a second step, Tn1546 could transpose from the incoming plasmid to a resident replicon (plasmid or chromosome) in the recipient; the acquired plasmid behaves as a suicide gene delivery vector, and the incoming DNA is rescued by illegitimate recombination. Transposition was demonstrated in VRSA-1 by studying the insertion site of the transposon. Tn1546 belongs to the Tn3 family of transposons, and insertion of these elements generates a 5-bp direct duplication of target DNA. In VRSA-1, a 5-bp AAGTT duplication was found at the target site in the endogenous resident plasmid (24, 42). Among the nine VRSA strains isolated in the United States, three (VRSA-3, -5, and -6) maintained the enterococcal vanA plasmid (one-step transfer) whereas in VRSA-1, -7, -8, -9, and -10, Tn1546 moved from the enterococcal plasmid to a staphylococcal plasmid (two-step transfer) (45). The vanA plasmid in VRSA-2, ca. 120 kb, has not been characterized (38).
FIG. 2.
Schematic representation of Tn1546 transfer from Enterococcus spp. to S. aureus. Blue and red wavy lines represent chromosomal DNA of Enterococcus and S. aureus, respectively. Blue circle, enterococcal plasmid with a broad host range of transfer; red circle, resident staphylococcal plasmid. Acquisition of Tn1546 was obtained in one step by VRSA-3, -5, and -6 and in two steps by VRSA-1, -7, -8, -9, and -10. GRE, glycopeptide-resistant enterococci.
Resistance phenotypes.
Two strains, VRSA-2 and VRSA-3, differ from the others in their levels of resistance to glycopeptides (17, 21). They exhibit moderate resistance to vancomycin (MICs, 32 μg/ml and 64 μg/ml, respectively) and low resistance to teicoplanin (MICs, 4 μg/ml and 16 μg/ml, respectively). These isolates are designated low-level-resistant VRSA (LLR-VRSA). Analysis by PCR mapping and DNA sequencing of the vanA gene clusters in these strains indicated that all the structural genes involved in resistance are present and in the same order as in Tn1546. However, two insertion sequences, IS1216V and IS1251, are inserted in place of ORF1 and into the vanSH intergenic region, respectively, leaving intact the structural genes for the resistance enzymes (Fig. 1B) (7, 25).
In contrast, the remaining VRSA strains exhibited high-level resistance to both glycopeptides (vancomycin MIC > 256 μg/ml; teicoplanin MIC > 32 μg/ml) and are named high-level-resistant VRSA (HLR-VRSA). All the seven HLR-VRSA strains harbored a prototype Tn1546 element (Fig. 1A). For isolates from Kolkata, India (31), and Tehran, Iran (1), there is no clear evidence that the operon responsible for resistance is a typical vanA operon.
Analysis of expression of the resistance genes revealed that the activities of the VanX and VanY d,d-peptidases, as well as the relative amounts of late peptidoglycan precursors, in the LLR- and HLR-VRSA isolates were similar to each other and to those in VanA-type enterococci (24, 25). Furthermore, in the absence of glycopeptides in the culture medium, UDP-N-acetylmuramyl-pentapeptide was the main late peptidoglycan precursor synthesized by all strains whereas, after growth in the presence of inducer, UDP-N-acetylmuramyl-pentadepsipeptide was the predominant precursor. Similar results were obtained by analysis of cell wall precursor accumulation, in the presence and in the absence of vancomycin, for a transconjugant obtained in vitro (32). Similarly, VanX and VanY activities were significantly higher after induction by glycopeptides (24, 25). Taken together, these data confirm inducible expression of the vanA gene cluster and efficient heterologous expression of the vanA operon in S. aureus.
The two LLR-VRSA strains had a copy of an insertion sequence that disrupted the gene for the transposase of Tn1546. In the absence of this protein, the genetic element can no longer transpose. Study by replica plating showed that glycopeptide resistance was lost at a high frequency (ca. 50%) in LLR-VRSA strains after overnight culture in the absence of selective pressure, whereas, in the same conditions, resistance was fully stable in the HLR-VRSA isolates (24, 25). Loss of resistance by the LLR-VRSA strains probably results from inefficient replication of the enterococcal plasmid bearing the resistance determinant in the new host (24, 25).
As already mentioned, VanA-type resistance is inducible by glycopeptides. In the absence of an antibiotic in the medium the resistance pathway is not expressed, whereas, in the presence of an inducer (vancomycin or teicoplanin), the resistance mechanism is activated. The delay before phenotypic expression of resistance corresponds to the time necessary for (i) synthesis of peptidoglycan precursors ending in d-Lac and (ii) elimination of the susceptibility pathway by the sequential action of VanX and VanY. Induction of resistance by vancomycin is greatly delayed (>8 h) in LLR-VRSA compared to HLR-VRSA (ca. 3 h) (24, 25). This long lag phase before growth resumes in the presence of vancomycin could be responsible for lack of detection of resistance by automated in vitro susceptibility testing methods (38). The combination of delay of the growth of the host with instability of the plasmid carrying the vanA operon is responsible for the LLR-VRSA phenotype.
Synergistic activity of glycopeptides and β-lactams against VRSA.
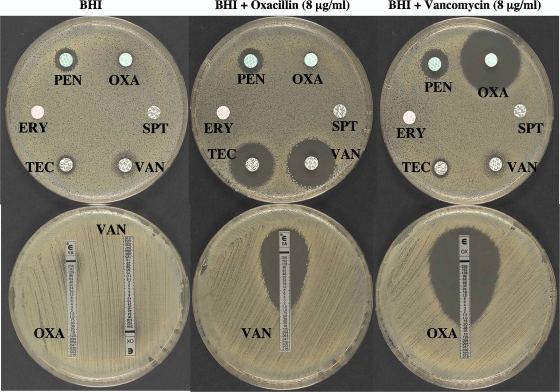
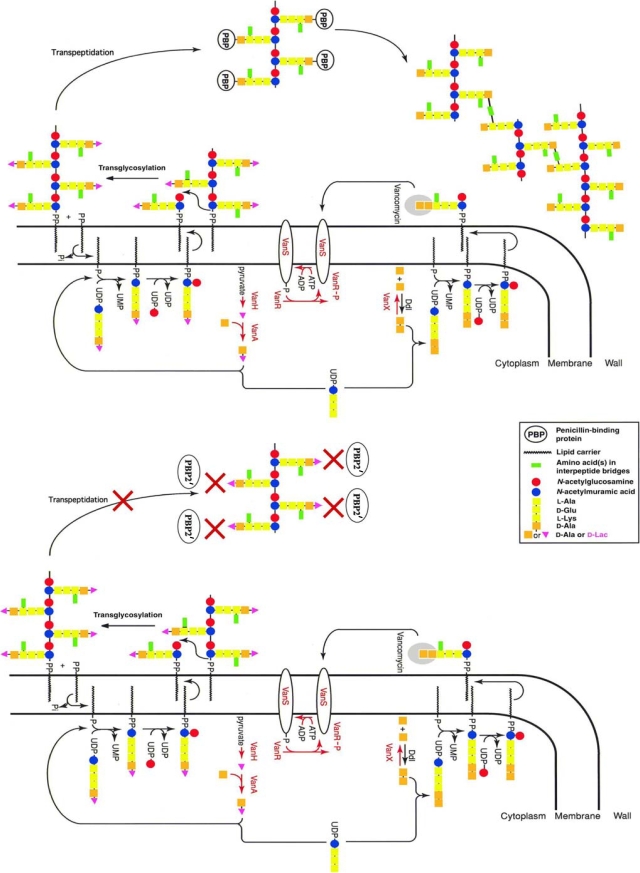
All S. aureus strains that have acquired the vanA operon are resistant to methicillin when grown in the absence of the glycopeptide. It has been demonstrated that vancomycin (or teicoplanin) and oxacillin, at concentrations that are achievable in therapy, have a powerful synergistic effect against VRSA both in vitro (Fig. 3) and in an animal model (15, 25). Methicillin resistance is due to synthesis of an additional penicillin-binding protein, PBP2′, encoded by the mecA gene, that exhibits low-affinity binding for β-lactams (30). PBP2′ is a transpeptidase (TPase) that remains active in the presence of β-lactams, whereas this class of antibiotics inactivates all the native penicillin-binding proteins of S. aureus (9, 29). The cooperativeness of the bifunctional PBP2, having transglycosylase (TGase) and TPase activity, with PBP2′ is required to achieve high-level resistance to methicillin (33). In the presence of β-lactams, the penicillin-insensitive TGase domain of PBP2 remains functional and cooperates with the TPase activity of PBP2′ for synthesis of staphylococcal peptidoglycan (27). It has also been demonstrated that the TGase and TPase activities of PBP2 are essential for VRSA growth in the presence of vancomycin (33). In the presence of representatives of both drug classes, such as vancomycin and oxacillin, bacteria synthesize modified peptidoglycan precursors ending in d-Ala-d-Lac which are not substrates for PBP2′ in the transpeptidation step (Fig. 4) . Because the TPase activity of PBP2 is inactivated by oxacillin, synthesis of peptidoglycan cannot proceed, resulting in death of the bacteria.
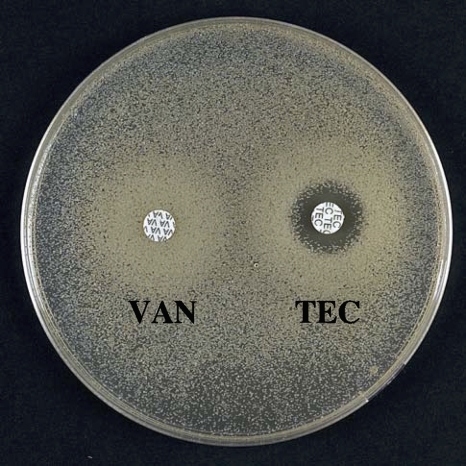
FIG. 3.
Oxacillin-glycopeptide synergism against HLR-VRSA as tested by (top) disk diffusion and (bottom) Etest. BHI, brain heart infusion agar. ERY, erythromycin (15 μg); OXA, oxacillin (5 μg); PEN, penicillin (6 μg); SPT, spectinomycin (100 μg); TEC, teicoplanin (30 μg); VAN, vancomycin (30 μg).
FIG. 4.
Schematic representation of peptidoglycan synthesis by a VRSA strain in the presence of vancomycin (top) or of vancomycin and methicillin (bottom). Top, in the presence of vancomycin, the resistance pathway is activated, leading to synthesis of peptidoglycan precursors ending in d-Ala-d-Lac and elimination of the d-Ala-d-Ala ending precursors. The depsipeptide late precursors are substrates for the transpeptidases (PBP) but are not a target for the glycopeptides. Bottom, in the presence of vancomycin and methicillin, the d-Ala-d-Lac terminating precursors, which are synthesized following induction by vancomycin, are not substrates of PBP2′, which is the only transpeptidase that remains active in the presence of methicillin. Thus, biosynthesis of the cell wall cannot proceed despite the fact that the strain is highly resistant to both drug classes when used separately.
Combinations of vancomycin and β-lactams could therefore be a successful treatment for patients infected with VRSA despite high-level resistance of the strain to both drug classes. This was tested in the rabbit model of endocarditis (15). Treatment of VRSA-1-infected rabbits with nafcillin (a β-lactam antibiotic) alone, vancomycin alone, or the combination showed that therapy with nafcillin plus vancomycin was responsible for a significant reduction in bacterial loading and organ sterilization compared to monotherapy with either antibiotic.
Glycopeptide dependence among VRSA strains.
Vancomycin-dependent enterococci are able to grow only in the presence of vancomycin in the culture medium. Such strains have been isolated in vitro, in animal models, and from patients treated for prolonged periods of time with vancomycin (11, 13, 18, 36, 40). These bacteria possess an inactive chromosomally encoded d-Ala:d-Ala ligase and thus rely on the activity of the d-Ala:d-Lac ligase, encoded by vanA, to synthesize the cell wall (6, 16, 36, 40). Strain VRSA-7 demonstrated in vitro partial dependence on glycopeptides for growth (Fig. 5). Sequence determination of the ddl gene in that strain revealed a point mutation leading to an asparagine-to-lysine (N308K) substitution, resulting in a 1,000-fold decrease in d-Ala:d-Ala ligase activity (21a). Two factors are responsible for weak growth of this strain in the absence of inducer: low-level expression of the resistance genes due to loose regulation of the vanA operon by the VanR/VanS system (2) combined with a gene dosage effect due to location of the vanA gene cluster on a multicopy plasmid. Even in the absence of vancomycin in the culture medium, VRSA-7 synthesizes mainly precursors ending in d-Ala-d-Lac that are, as already discussed, not substrates for PBP2′. Thus, inactivation of the VRSA-7 d-Ala:d-Ala ligase and acquisition of the vanA gene cluster on a multicopy plasmid result in high susceptibility of the host bacterium to methicillin despite the presence of a functional mecA gene.
FIG. 5.
Glycopeptide susceptibility of VRSA-7 as tested by disk diffusion. VAN, vancomycin (30 μg); TEC, teicoplanin (30 μg).
Biological cost of VanA-type resistance in MRSA.
Biological cost is one of the factors that determines the stability and dissemination of antibiotic resistance. Worldwide dissemination of MRSA clones has been associated with their ability to compensate for the cost of harboring the staphylococcal chromosomal cassette mec (SCCmec) element that confers methicillin resistance (12). Deletion of the mecA gene in certain vancomycin intermediate-resistant S. aureus isolates suggests that simultaneous resistance to β-lactams and glycopeptides is too costly for S. aureus (23). Quantification of the exponential growth rates of VRSA-1, VRSA-5, VRSA-6, and a recipient MRSA strain (HIP11713) isogenic to VRSA-1 revealed that (i) in the absence of vancomycin the growth rates of the VRSA strains were similar to that of susceptible MRSA, indicating that the reduction in the fitness of the resistant strains due to acquisition of Tn1546 was minimal in the absence of induction, and (ii) when resistance was induced by vancomycin, there was an important reduction of the growth rate of the VRSA strains relative to those of their noninduced counterparts and to the susceptible MRSA strain (14). The reduction in fitness following induction was evaluated as ca. 20 to 38%, indicating that VanA-type resistance is associated with an important biological cost for the host. However, competition experiments between an isogenic VRSA-1 transconjugant and a MRSA recipient revealed a competitive disadvantage of 0.4 to 3% per 10 generations of the transconjugant versus the recipient when the two strains were mixed in the same environment without an inducer. This slight fitness burden can be attributed to the basal level of expression of the van operon combined with a gene dosage effect due to the presence of the cluster on a multicopy plasmid. The observation that, in competition experiments that mimic best natural conditions, the transconjugant is more rapidly eliminated than the MRSA recipient could explain, in part, the low level of dissemination of the VRSA clinical isolates. The minimal fitness cost in the absence of induction could lead to selection of compensatory mutations that restore the fitness of the host while retaining resistance.
Dissemination of VanA-type vancomycin resistance.
Among the nine VRSA strains from the United States, seven were isolated in Michigan. Analysis of the pulsed-field gel electrophoresis patterns of these strains indicated that five were of USA100 lineage (45), which is the most common in the United States, in particular in the health care setting (20). Strains of this lineage are usually multiresistant, and additional acquisition of glycopeptide resistance could thus have a dramatic effect on therapy.
A broad-host-range conjugative Inc18-like enterococcal vanA plasmid was present at least in five strains, suggesting a possible role for this plasmid group as resistance donors (23a, 45). It has been shown that Inc18-like plasmids are statistically more frequent in enterococci isolated in Michigan (3.6%) than in those from other states representative of all geographic regions of the United States (0.1%). Furthermore, these plasmids occur more commonly in Enterococcus faecalis than in other species of Enterococcus. This could account for the fact that seven of the nine U.S. VRSA strains were isolated in Michigan and that five of the seven Michigan VRSA strains were associated with VanA-type E. faecalis. Consistent with these observations, conjugative transfer of an Inc18-like vanA plasmid from Enterococcus spp. to S. aureus was obtained in vitro at a frequency of 10−7 and the plasmid was maintained in the new host (45a). When conjugation was successful, a large majority (75%) of the transconjugants lost resistance to vancomycin after several subcultures, confirming the relative instability of the plasmid.
Restriction modification is probably the major mechanism that controls uptake of foreign DNA by S. aureus (41). This system protects the host genome from restriction by adding a methyl group to residues within specific target DNA sequences and restricts unmodified foreign DNA that has entered the cell (for a review, see reference 44). The SauI type I system, which recognizes two specific DNA sequences separated by a nonspecific spacer, is composed of genes coding for three subunits: HsdM, implicated in DNA methylation; HsdS, which determines DNA specificity; and HsdR, responsible for restriction. It has been shown that this restriction modification system blocks transfer of mobile genetic elements from other species to S. aureus and limits the spread of resistance genes between isolates of different S. aureus lineages (41). In addition, certain strains appear to be better recipients than others (37). Of five S. aureus strains (three VRSA clinical isolates and strains COL and RN4220) tested as recipients in conjugation experiments, only one (VRSA-1) was a successful recipient (45a). Taken together, these findings, combined with those from the biological cost experiments, could account for the limited dissemination of vancomycin resistance in S. aureus.
Acquisition of high-level vancomycin resistance by S. aureus strains already multiresistant to antibiotics is a major public health problem. Although it is of serious concern for patients infected with such bacteria, it seems that, due to several biological constraints, dissemination of VRSA has so far been limited. However, due to the minimal biological cost in the absence of induction, the potential for spread of such clinical isolates should not be underestimated.
Acknowledgments
We thank P. E. Reynolds for reading the manuscript and NARSA for supplying the S. aureus strains.
The NARSA program is supported under NIAID/NIH contract N01-AI-95359.
Footnotes
Published ahead of print on 8 June 2009.
REFERENCES
- 1.Aligholi, M., M. Emaneini, F. Jabalameli, S. Shahsavan, H. Dabiri, and H. Sedaght. 2008. Emergence of high-level vancomycin-resistant Staphylococcus aureus in the Imam Khomeini Hospital in Tehran. Med. Princ. Pract. 17:432-434. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, M., F. Depardieu, P. Reynolds, and P. Courvalin. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21:33-44. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, M., C. Molinas, and P. Courvalin. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arthur, M., P. E. Reynolds, and P. Courvalin. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401-407. [DOI] [PubMed] [Google Scholar]
- 6.Baptista, M., F. Depardieu, P. Reynolds, P. Courvalin, and M. Arthur. 1997. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol. Microbiol. 25:93-105. [DOI] [PubMed] [Google Scholar]
- 7.Clark, N. C., L. M. Weigel, J. B. Patel, and F. C. Tenover. 2005. Comparison of Tn1546-like elements in vancomycin-resistant Staphylococcus aureus isolates from Michigan and Pennsylvania. Antimicrob. Agents Chemother. 49:470-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El-Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jonge, B. L. M., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. J. Biol. Chem. 267:11248-11254. [PubMed] [Google Scholar]
- 10.Depardieu, F., I. Podglajen, R. Leclercq, E. Collatz, and P. Courvalin. 2007. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20:79-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dever, L. L., S. M. Smith, S. Handwerger, and R. H. K. Eng. 1995. Vancomycin-dependent Enterococcus faecium isolated from stool following oral vancomycin therapy. J. Clin. Microbiol. 33:2770-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ender, M., N. McCallum, R. Adhikari, and B. Berger-Bachi. 2004. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:2295-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrag, N., I. Eltringham, and H. Liddy. 1996. Vancomycin-dependent Enterococcus faecalis. Lancet 348:1581-1582. [DOI] [PubMed] [Google Scholar]
- 14.Foucault, M. L., P. Courvalin, and C. Grillot-Courvalin. 2009. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox, P. M., R. J. Lampert, K. S. Stumpf, G. L. Archer, and M. W. Climo. 2006. Successful therapy of experimental endocarditis caused by vancomycin-resistant Staphylococcus aureus with a combination of vancomycin and beta-lactam antibiotics. Antimicrob. Agents Chemother. 50:2951-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gholizadeh, Y., M. Prevost, F. Van Bambeke, B. Casadewall, P. M. Tulkens, and P. Courvalin. 2001. Sequencing of the ddl gene and modeling of the mutated D-alanine:D-alanine ligase in glycopeptide-dependent strains of Enterococcus faecium. Protein Sci. 10:836-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kacica, M., and L. C. McDonald. 2004. Vancomycin-resistant Staphylococcus aureus—New York, 2004. MMWR Morb. Mortal. Wkly. Rep. 53:322-323. [PubMed] [Google Scholar]
- 18.Kirkpatrick, B. D., S. M. Harrington, D. Smith, D. Marcellus, C. Miller, J. Dick, L. Karanfil, and T. M. Perl. 1999. An outbreak of vancomycin-dependent Enterococcus faecium in a bone marrow transplant unit. Clin. Infect. Dis. 29:1268-1273. [DOI] [PubMed] [Google Scholar]
- 19.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 20.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, D., V. Urdaneta, A. Weltman, and S. Park. 2002. Vancomycin-resistant Staphylococcus aureus. MMWR Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 21a.Moubarek, C., D. Meziane-Cherif, P. Courvalin, and B. Périchon. 2008. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-4176.
- 22.Noble, W. C., Z. Virani, and R. G. A. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC-12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 93:195-198. [DOI] [PubMed] [Google Scholar]
- 23.Noto, M. J., P. M. Fox, and G. L. Archer. 2008. Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 52:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Patel, J. B., W. C. Huskins, W. Zhu, J. A. Jernigan, N. C. Clark, K. F. Anderson, L. K. McDougal, C. Chenoweth, G. J. Alangaden, and P. R. Murray. 2008. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. K-668.
- 24.Périchon, B., and P. Courvalin. 2004. Heterologous expression of the enterococcal vanA operon in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:4281-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Périchon, B., and P. Courvalin. 2006. Synergism between beta-lactams and glycopeptides against VanA-type methicillin-resistant Staphylococcus aureus and heterologous expression of the vanA operon. Antimicrob. Agents Chemother. 50:3622-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Périchon, B., and P. Courvalin. 2000. Update on vancomycin resistance. Int. J. Clin. Pract. 54:250-254. [PubMed] [Google Scholar]
- 27.Pinho, M. G., H. De Lencastre, and A. Tomasz. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant and drug-susceptible staphylococci. Proc. Natl. Acad. Sci. USA 98:10886-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds, P. E. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8:943-950. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds, P. E., and D. F. J. Brown. 1985. Penicillin-binding proteins of beta-lactam-resistant strains of Staphylococcus aureus. FEBS Lett. 192:28-32. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds, P. E., and C. Fuller. 1986. Methicillin-resistant strains of Staphylococcus aureus; presence of identical additional penicillin-binding protein in all strains examined. FEMS Microbiol. Lett. 33:251-254. [Google Scholar]
- 31.Saha, B., A. K. Singh, A. Ghosh, and M. Bal. 2008. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (south Asia). J. Med. Microbiol. 57:72-79. [DOI] [PubMed] [Google Scholar]
- 32.Severin, A., K. Tabei, F. Tenover, M. Chung, N. Clarke, and A. Tomasz. 2004. High level oxacillin and vancomycin resistance and altered cell wall composition in Staphylococcus aureus carrying the staphylococcal mecA and the enterococcal vanA gene complex. J. Biol. Chem. 279:3398-3407. [DOI] [PubMed] [Google Scholar]
- 33.Severin, A., S. W. Wu, K. Tabei, and A. Tomasz. 2004. Penicillin-binding protein 2 is essential for expression of high-level vancomycin resistance and cell wall synthesis in vancomycin-resistant Staphylococcus aureus carrying the enterococcal vanA gene complex. Antimicrob. Agents Chemother. 48:4566-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sievert, D. M., M. L. Boulton, G. Stolman, D. Johnson, M. G. Stobierski, F. P. Downes, P. A. Somsel, J. T. Rudrik, W. Brown, W. Hafeez, T. Lundstrom, E. Flanagan, R. Johnson, J. Mitchell, and S. Chang. 2002. Staphylococcus aureus resistant to vancomycin. MMWR Morb. Mortal. Wkly. Rep. 51:565-567.12139181 [Google Scholar]
- 35.Sievert, D. M., J. T. Rudrik, J. B. Patel, L. C. McDonald, M. J. Wilkins, and J. C. Hageman. 2008. Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. Clin. Infect. Dis. 46:668-674. [DOI] [PubMed] [Google Scholar]
- 36.Sifaoui, F., and L. Gutmann. 1997. Vancomycin dependence in a VanA-producing Enterococcus avium strain with a nonsense mutation in the natural d-Ala-d-Ala ligase gene. Antimicrob. Agents Chemother. 41:1409. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sung, J. M., and J. A. Lindsay. 2007. Staphylococcus aureus strains that are hypersusceptible to resistance gene transfer from enterococci. Antimicrob. Agents Chemother. 51:2189-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uttley, A. H. C., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet i:57-58. [DOI] [PubMed] [Google Scholar]
- 40.Van Bambeke, F., M. Chauvel, P. E. Reynolds, H. S. Fraimow, and P. Courvalin. 1999. Vancomycin-dependent Enterococcus faecalis clinical isolates and revertant mutants. Antimicrob. Agents Chemother. 43:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waldron, D. E., and J. A. Lindsay. 2006. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 43.Weigel, L. M., R. M. Donlan, D. H. Shin, B. Jensen, N. C. Clark, L. K. McDougal, W. Zhu, K. A. Musser, J. Thompson, D. Kohlerschmidt, N. Dumas, R. J. Limberger, and J. B. Patel. 2007. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob. Agents Chemother. 51:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan, R. 1981. Structure and mechanism of multifunctional restriction endonucleases. Annu. Rev. Biochem. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, W., N. C. Clark, L. K. McDougal, J. Hageman, L. C. McDonald, and J. B. Patel. 2008. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Zhu, W., N. C. Clark, and J. B. Patel. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-269.