Abstract
The population structure of the opportunistic yeast pathogen Candida dubliniensis is composed of three main multilocus sequence typing clades (clades C1 to C3), and clade C3 predominantly consists of isolates from the Middle East that exhibit high-level resistance (MIC50 ≥ 128 μg/ml) to the fungicidal agent flucytosine (5FC). The close relative of C. dubliniensis, C. albicans, also exhibits clade-specific resistance to 5FC, and resistance is most commonly mediated by an Arg101Cys substitution in the FUR1 gene encoding uracil phosphoribosyltransferase. Broth microdilution assays with fluorouracil (5FU), the toxic deaminated form of 5FC, showed that both 5FC-resistant and 5FC-susceptible C. dubliniensis isolates exhibited similar 5FU MICs, suggesting that the C. dubliniensis cytosine deaminase (Fca1p) encoded by C. dubliniensis FCA1 (CdFCA1) may play a role in mediating C. dubliniensis clade-specific 5FC resistance. Amino acid sequence analysis of the CdFCA1 open reading frame (ORF) identified a homozygous Ser29Leu substitution in all 12 5FC-resistant isolates investigated which was not present in any of the 9 5FC-susceptible isolates examined. The tetracycline-inducible expression of the CdFCA1 ORF from a 5FC-susceptible C. dubliniensis isolate in two separate 5FC-resistant clade C3 isolates restored susceptibility to 5FC, demonstrating that the Ser29Leu substitution was responsible for the clade-specific 5FC resistance and that the 5FC resistance encoded by FCA1 genes with the Ser29Leu transition is recessive. Quantitative real-time PCR analysis showed no significant difference in CdFCA1 expression between 5FC-susceptible and 5FC-resistant isolates in either the presence or the absence of subinhibitory concentrations of 5FC, suggesting that the Ser29Leu substitution in the CdFCA1 ORF is the sole cause of 5FC resistance in clade C3 C. dubliniensis isolates.
Candida dubliniensis is an opportunistic yeast pathogen that was first described in 1995 in human immunodeficiency virus-infected patients in Ireland (39). Since then the organism has been shown to have a worldwide distribution and has been recovered from other groups of immunocompromised individuals and from patients with severe underlying disease (2-4, 11, 29, 30, 36-38, 44). The population structure of C. dubliniensis has previously been investigated by using the species-specific complex DNA fingerprinting probe Cd25 and multilocus sequence typing (MLST) (4, 11, 15, 18). Early Cd25 fingerprinting analyses demonstrated that C. dubliniensis consists of two fingerprinting groups, termed Cd25 group I and Cd25 group II (15). Group I isolates comprise the majority of isolates investigated from many countries around the world and are very closely related, with an average similarity coefficient value (SAB) of 0.8. Group II isolates are less closely related and have an average SAB value of 0.47 (15). These results were later confirmed with a larger collection of isolates by Gee et al. (11), who also showed that Cd25 group I isolates comprised a single genotype (genotype 1) on the basis of sequence analysis of the internal transcribed spacer (ITS) region of the ribosomal DNA operon. Furthermore, Cd25 group II isolates were found to belong to three ITS genotypes (genotypes 2 to 4). In 2005, a study by Al Mosaid et al. (4) identified a third Cd25 fingerprinting group, termed Cd25 group III, which exhibited an average SAB value of 0.35, among C. dubliniensis isolates recovered exclusively in Egypt, Saudi Arabia, and Israel, all of which belonged to ITS genotypes 3 or 4. All isolates belonging to Cd25 group III examined exhibited high-level intrinsic resistance to the antifungal drug flucytosine (5FC), apart from one Israeli isolate that was 5FC susceptible. This phenotype did not occur in isolates belonging to either Cd25 group I or Cd25 group II, including isolates from Cd25 groups I and II recovered from Egypt, Saudi Arabia, and Israel (4). Recent studies that have used MLST analysis to investigate the population structure of C. dubliniensis revealed the presence of three distinct clades, termed clades C1 to C3 (18). All 5FC-resistant isolates belonging to Cd25 fingerprint group III were found to cluster exclusively in MLST clade C3 (18). More recently, MLST was used to show that clade C1 C. dubliniensis isolates recovered from avian excrement-associated samples were genetically distinct from other clade C1 isolates that were recovered from humans (19).
The closest relative of C. dubliniensis, Candida albicans, also exhibits clade-specific resistance to 5FC, with 72.7% of isolates in MLST clade C1 (Ca3 fingerprinting clade I) exhibiting reduced susceptibility to this antifungal agent (23, 32). In C. albicans, the 5FC resistance patterns vary among isolates and range from reduced susceptibility (MICs, 0.5 to 2 μg/ml) to intermediate resistance (MICs, 2 to 8 μg/ml) or high-level resistance (MICs, ≥8 μg/ml); and a wide range of 5FC MICs for this drug have been reported among isolates (range, 0.06 μg/ml to ≥128 μg/ml) (7, 13). In C. dubliniensis, the resistance patterns are more clearly defined, with 5FC-susceptible isolates exhibiting 5FC MICs of ≤0.125 μg/ml and 5FC-resistant isolates exhibiting 5FC MICs of ≥128 μg/ml (4). To date, 5FC resistance in C. dubliniensis has been reported only in isolates from the Middle East, all of which that have been tested belong to MLST clade C3 (1, 4, 18, 29).
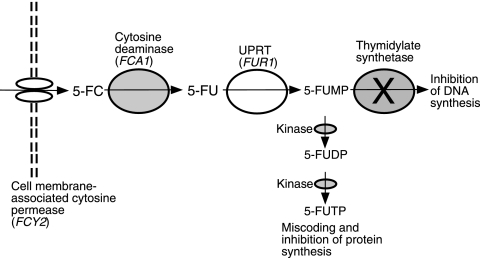
The antifungal action of 5FC relies on the intracellular conversion of 5FC to fluorouracil (5FU) by cytosine deaminases upon entry into fungal cells (Fig. 1). Cytosine deaminase (Fca1p) is encoded by FCA1 in C. albicans and C. dubliniensis (CdFCA1) (4, 9), and the FCA1 genes in these two species are homologues of the FCY1 gene in Saccharomyces cerevisiae (9) and in other Candida species, such as Candida lusitaniae (26). The absence of cytosine deaminases in mammalian cells prevents 5FC toxicity in humans, as the 5FC prodrug itself is nontoxic. After the conversion of 5FC to 5FU, the FUR1-encoded uracil phosphoribosyltransferase (UPRT) catalyzes the phosphorylation of 5FU to fluorouridine monophosphate (5FUMP) (Fig. 1). Two specific kinases catalyze the further phosphorylation of 5FUMP, eventually converting it to fluorouridine triphosphate. Fluorouridine triphosphate in turn gets incorporated into RNA, which causes miscoding, leading to the inhibition of fungal protein synthesis (Fig. 1). As a secondary method of inhibition, 5FUMP inhibits thymidylate synthetase (Fig. 1), leading to the depletion of dTTP and the misincorporation of dUTP into newly synthesized DNA, causing irreversible DNA damage and cell cycle arrest (14, 31, 41).
FIG. 1.
Metabolic pathway and mode of action of 5FC in Candida yeasts. 5FC and 5FU are transported into the cell by cell membrane-associated cytosine-purine permeases. In Candida spp., these are encoded by two genes that display amino acid sequence homology with the FCY2 gene of S. cerevisiae (13). Upon entry into the cell, 5FC is then deaminated to 5FU by Fca1p, encoded by FCA1. 5FU is then phosphorylated by UPRT, encoded by FUR1, yielding 5FUMP. 5FUMP inhibits thymidylate synthetase, which leads to thymidine depletion in the cell and which ultimately interrupts DNA synthesis. 5FUMP is also metabolized by two kinases, yielding fluorouridine diphosphate (5FUDP) and, subsequently, fluorouridine triphosphate (5FUTP), the latter of which is incorporated into RNA in the place of UTP, which leads to miscoding and the inhibition of protein synthesis.
In haploid C. lusitaniae isolates, a missense T26C nucleotide mutation in the FCY1 gene has been reported in four clinical isolates demonstrating 5FC resistance, although 5FC and 5FC-fluconazole cross-resistance has more commonly been attributed to defects in the purine cytosine permease-encoded FCY2 gene in this species (10). In C. albicans, resistance to 5FC is mediated by a reduction in the activity of either the Fca1p encoded by FCA1 or the UPRT encoded by FUR1 (13, 31, 43). Two different research groups reported that in the majority of 5FC-resistant C. albicans isolates, resistance is associated with a homozygous single amino acid substitution, Arg101Cys, in UPRT (7, 13). However, other 5FC-resistant C. albicans isolates lack this substitution (13). One such isolate (5FC MIC, >64 μg/ml) was reported to contain a homozygous Gly28Asp substitution in the cytosine deaminase gene, and a Ser29Leu amino acid substitution was also observed in the same gene of another C. albicans isolate displaying intermediate 5FC resistance (5FC MIC, 4 μg/ml) (13). In C. dubliniensis, the DNA sequences of the FUR1 genes encoding the UPRTs of four 5FC-resistant and five 5FC-susceptible isolates from the Middle East were determined previously, and while several single nucleotide polymorphisms (SNPs) were identified, no amino acid substitutions were observed between the isolates (4).
The purpose of the present study was to investigate the role of Fca1p in C. dubliniensis clade-specific 5FC resistance by the use of broth microdilution assays with 5FC and 5FU, analysis of DNA and amino acid sequences, and analysis of CdFCA1 expression. A tetracycline-inducible expression plasmid was used to incorporate the CdFCA1 gene from a 5FC-susceptible isolate (hereafter called CdFCA1s) into the ADH1 locus of a 5FC-resistant isolate and the CdFCA1 gene from a 5FC-resistant isolate (hereafter called CdFCA1r) into the ADH1 locus of a 5FC-susceptible isolate. These strains were used to determine if 5FC susceptibility or resistance could be induced in isolates upon the acquisition and expression of the respective CdFCA1 gene.
MATERIALS AND METHODS
Isolates and culture conditions.
Twenty-one epidemiologically unrelated human C. dubliniensis isolates were included in the present study, including 9 5FC-susceptible isolates and 12 5FC-resistant isolates (Table 1), as reported previously (2, 4, 11, 30, 39). Previously, 5FC resistance in C. dubliniensis has ever been reported only in isolates from Saudi Arabia, Egypt, Israel, and Kuwait (1, 4, 29); and all but two of these have previously been investigated by MLST analysis and/or Cd25 fingerprint analysis and were shown to belong to C. dubliniensis MLST clade C3 and Cd25 fingerprint group III (4, 18). For these reasons, 20 of the 21 isolates chosen for study (12 5FC-resistant isolates and 8 5FC-susceptible isolates) were originally recovered in Egypt, Saudi Arabia, or Israel (Table 1). The 5FC-susceptible isolates belonged to MLST clade C1 or C2 and Cd25 fingerprint group I or II (4, 18). The C. dubliniensis type strain CD36, originally isolated from the oral cavity of a human immunodeficiency virus-infected individual in Ireland, was also included as a reference isolate because the complete genome sequence of this organism has been determined (http://www.sanger.ac.uk/sequencing/Candida/dubliniensis/). The C. dubliniensis isolates were routinely cultured on yeast extract-peptone-dextrose (YPD) agar, pH 5.6, at 37°C. For liquid culturing, isolates were grown in YPD broth (with the following per liter: 10 g yeast extract [Sigma-Aldrich Ireland Ltd., Wicklow, Ireland], 20 g peptone [Oxoid, Basingstoke, Hampshire, England], and 20 g d-glucose, pH 5.5) at 37°C in an orbital incubator (Gallenkamp, Leicester, United Kingdom) at 200 rpm. Escherichia coli strain DH5α (12) [F− φ80dlacZΔm15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 d− gyrA96 relA1] was routinely grown on Luria-Bertani agar, pH 7.4, at 37°C or in Luria-Bertani broth, pH 7.4, at 37°C with shaking at 200 rpm for liquid culture.
TABLE 1.
Candida dubliniensis isolates used to investigate the molecular mechanism of 5FC resistance and their susceptibilities to 5FC and 5FUa
| Isolate | Country of origin | Yr of isolation | Sample | Cd25 fingerprint group | ITS genotype | MIC50 (μg/ml) |
Reference | |
|---|---|---|---|---|---|---|---|---|
| 5FC | 5FU | |||||||
| CD36b | Ireland | 1988 | Oral | I | 1 | ≤0.25 | 32 | 4, 11, 39 |
| SA101 | S. Arabia | 2002 | Oral | I | 1 | ≤0.25 | 16 | 4 |
| SA105 | S. Arabia | 2002 | Oral | I | 1 | ≤0.25 | 32 | 4 |
| SA115 | S. Arabia | 2002 | Oral | I | 1 | ≤0.25 | 32 | 4 |
| Eg203 | Egypt | 2002 | Oral | I | 1 | ≤0.25 | 16 | 4 |
| Eg206 | Egypt | 2002 | Oral | I | 1 | ≤0.25 | 32 | 4 |
| p7276 | Israel | 1999 | RT | II | 3 | ≤0.25 | 8 | 4, 11 |
| p6785 | Israel | 1999 | Urine | II | 3 | ≤0.25 | 16 | 4, 11, 30 |
| p7718 | Israel | 1999 | Wound | III | 4 | ≤0.25 | 16 | 4, 11 |
| Eg200 | Egypt | 2002 | Oral | III | 4 | ≥128 | 8 | 4 |
| Eg201 | Egypt | 2002 | Oral | III | 4 | ≥128 | 32 | 4 |
| Eg202 | Egypt | 2002 | Oral | III | 4 | ≥128 | 32 | 4 |
| Eg207 | Egypt | 2002 | Oral | III | 4 | ≥128 | 32 | 4 |
| SA100 | S. Arabia | 2002 | Oral | III | 3 | ≥128 | 32 | 4 |
| SA103 | S. Arabia | 2002 | BAL | III | 3 | ≥128 | 32 | 4 |
| SA107 | S. Arabia | 2002 | Oral | III | 3 | ≥128 | 32 | 4 |
| SA108 | S. Arabia | 2002 | Oral | III | 3 | ≥128 | 32 | 4 |
| SA109 | S. Arabia | 2002 | Oral | III | 3 | ≥128 | 8 | 4 |
| SA113 | S. Arabia | 2002 | Oral | III | 4 | ≥128 | 32 | 4 |
| SA118 | S. Arabia | 2002 | Oral | III | 3 | ≥128 | 32 | 4 |
| SA121 | S. Arabia | 2002 | Oral | III | 4 | ≥128 | 32 | 4 |
Abbreviations: S. Arabia, Saudi Arabia; RT, respiratory tract; BAL, broncheoalveolar lavage fluid.
C. dubliniensis type strain.
Susceptibility testing.
The MICs for 5FC and 5FU (Sigma-Aldrich) were determined by the method described in document M27-A2 of the Clinical and Laboratory Standards Institute (CLSI; formerly the National Committee for Clinical Laboratory Standards) by using RPMI 1640 medium (6). Both drugs were titrated from a concentration of 128 μg/ml to one of 0.25 μg/ml for the preliminary analysis of all 21 clinical isolates, as well as for analysis of the recipient isolates and transformant derivatives from cloning experiments. These were determined as the lowest concentrations of the drug that reduced the turbidity by 50% relative to the turbidity of the growth of the drug-free controls. All isolates were tested in duplicate and on two separate occasions.
DNA extraction.
Isolates were grown overnight in 5 ml of YPD broth as described above. Cells were harvested from 1.5 ml of culture by centrifugation at 14,000 × g, and the DNA was extracted from the resulting pellet by using a DNeasy blood and tissue kit, according to the manufacturer's instructions (Qiagen Science, Crawley, West Sussex, United Kingdom) and resuspended in a final volume of 200 μl. Nucleic acids were ethanol precipitated and resuspended in 50 μl of molecular-grade Milli-Q Biocel-purified water (resistivity, 18.2 MΩ/cm; Millipore, Carrigtwohill, Cork, Ireland).
PCR amplification, sequencing, and sequence analysis of CdFCA1.
The complete open reading frame (ORF) of the C. dubliniensis CdFCA1 gene was amplified from 12 5FC-resistant isolates (Table 1) and 9 5FC-susceptible isolates (Table 1) by using oligonucleotide primers FCA1F and FCA1R, which incorporated SalI and BglII restriction endonuclease recognition sites, respectively (Table 2). The reaction mixtures contained 100 ng of purified template DNA, 1× Expand high-fidelity buffer, 2.5 U of Expand high-fidelity PCR system enzyme mixture (Roche Applied Science, Mannheim, Germany), 0.2 mM concentrations of each deoxynucleoside triphosphate (Promega Corporation, Madison, WI), and 0.2 μM concentrations of each oligonucleotide (Sigma Genosys Biotechnologies Europe Ltd., Pampisford, Cambridgeshire, United Kingdom). The reaction mixtures underwent an initial denaturation step of 94°C for 10 min, followed by 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min and a final elongation step of 72°C for 7 min. The 550-bp products were purified with a GenElute PCR cleanup kit (Sigma-Aldrich), and both strands were sequenced by using the same primers that had been used for amplification. DNA sequencing reactions were performed commercially by Cogenics (Essex, United Kingdom) with an ABI 3730xl DNA analyzer. Multiple DNA and amino acid sequence alignments of CdFCA1 genes and their encoded proteins from 5FC-susceptible and 5FC-resistant C. dubliniensis isolates were carried out by using the CLUSTAL W sequence alignment computer program (40), available at the EMBL-EBI website (http://www.ebi.ac.uk/).
TABLE 2.
Oligonucleotide primers used in this study
| Oligonucleotide | Sequence (5′-3′)b | Function |
|---|---|---|
| ADH1F | ATGCAAGCAAGCTTATTCA | PCR screening of transformantsa |
| cartTA | CGGCATACTATCAGTAGTAG | |
| SAT | CAATGCCGCCGAGAGTAAAG | |
| ADH1R | CCCAAGATCTTACCTTCTTCCATT | |
| FCA1F | GACGCGTCGACGATATCAACGATGACATTT | CdFCA1 cloning and PCR screening |
| FCA1R | CGGGATCCAGATCTTTATTCTCCAATATCTTC | |
| RTFCA1F | AAACGCAGGAAGATTGCCAG | Gene expression analysis |
| RTFCA1R | TGGCCCCTGTACACATACTACATG | |
| RTACT1F | AGCTCCAGAAGCTTTGTTCAGACC | |
| RTACT1R | TGCATACGTTCAGCAATACCTGGG |
The regions of the pNIM1-CdFCA1 cassette and background ADH1 locus amplified during PCR screening of transformants are displayed in Fig. 2.
The SalI and BglII restriction endonuclease recognition sites incorporated into the FCA1F and FCA1R primer sequences, respectively, are underlined.
Tetracycline-inducible CdFCA1 expression in C. dubliniensis.
The tetracycline-inducible gene expression plasmid pNIM1, developed by Park and Morschhauser (27), was adapted to investigate the inducible expression of the C. dubliniensis CdFCA1s and CdFCA1r genes in both 5FC-susceptible and 5FC-resistant C. dubliniensis isolates. The pNIM1 cassette was originally designed to integrate into the C. albicans alcohol dehydrogenase encoding gene ADH1. The DNA sequence for the C. albicans ADH1 gene (GenBank accession no. CaO19.3997) was used in a BLAST search against the C. dubliniensis genome sequence database (http://www.sanger.ac.uk/sequencing/Candida/dubliniensis/) in order to identify a homologue in C. dubliniensis. A high level of sequence homology (94%) is shared by the ADH1 ORFs of C. albicans and C. dubliniensis. The tetracycline-inducible promoter Ptet, included in pNIM1 (27), was used to drive expression of the CdFCA1s and CdFCA1r genes individually in the ADH1 locus of a C. dubliniensis isolate with the opposite 5FC phenotype. The complete coding regions of the CdFCA1 gene from 5FC-susceptible C. dubliniensis isolate p7276 and 5FC-resistant C. dubliniensis isolates SA113 and SA109 were amplified from genomic DNA with the FCA1F-FCA1R primer pair (Table 2). The amplimers were ligated into pGEM T-Easy vector I (Promega) vector DNA and transformed into E. coli strain DH5α. Plasmids were recovered from the transformants by using a GenElute plasmid miniprep kit (Sigma-Aldrich), and the cloned DNA was sequenced. The complete CdFCA1 ORF was digested from the pGEM T-Easy plasmids by using the SalI and BglII restriction endonuclease recognition sites, which were introduced upstream and downstream of the ORF, respectively, and the FCA1F-FCA1R primer pair. The CdFCA1 fragments were then gel purified with a Wizard SV Gel and PCR cleanup system (Promega) and separately cloned between Ptet and TACT1 in SalI-BglII-digested pNIM1, incorporating the CdFCA1 ORFs into the pNIM1 cassette in the place of the GFP1 gene (27), (Fig. 2). The resulting plasmids, pNIM1-CdFCA1s and pNIM1-CdFCA1r, were transformed into E. coli strain DH5α for replication of the plasmid, prior to purification with the GenElute plasmid miniprep kit (Sigma-Aldrich), SacII-ApaI linearization of the cassette, gel purification, and transformation into C. dubliniensis as described previously (35). 5FC-resistant isolates SA113 and SA109 were transformed with pNIM1-CdFCA1s, and 5FC-susceptible isolate p7276 was transformed with pNIM1-CdFCA1r (Table 1). All three of these C. dubliniensis isolates were also transformed with the pNIM1 cassette containing the GFP1 gene instead of the CdFCA1 gene as a control for the disruption of the ADH1 locus. In order to confirm the correct integration of the complete pNIM1-CdFCA1 cassette into the ADH1 locus in transformant derivatives, a number of PCR amplifications were carried out. The primer pairs used in these PCR amplifications were ADH1F and cartTA, SAT and FCA1R, FCA1F and ADH1R, and SAT and ADH1R (Table 2). These stepwise amplifications revealed the presence and the correct integration of the full pNIM1-CdFCA1 cassette in transformant derivatives. This was further confirmed by Southern hybridization analysis with two separate probes: a CdFCA1-directed probe and a pNIM1-directed probe which was directed toward the cartTA transactivator region and the background ADH1 locus. The probes were labeled with digoxigenin (DIG) by using a DIG DNA labeling and detecting kit (Roche). The prehybridization, hybridization, wash, and detection steps were carried out according to the manufacturer's instructions.
FIG. 2.
Structure and ADH1 integration site of the pNIM1-CdFCA1 cassette used in the tetracycline-inducible expression transformation studies. The restriction sites used for the excision of the C. albicans GFP1 gene (replaced by the CdFCA1 gene) and the excision of the entire pNIM1-CdFCA1 cassette from the pNIM1 plasmid (27) are indicated. Transcription start sites and the directions of transcription are displayed by right-angled arrows. Terminator sequences are displayed as black hairpin loops and function in the termination of transcription of the Candida-adapted reverse tetracycline-dependent transactivator (cartTA) and the CdFCA1 target gene. The primers used in the PCR screening of the pNIM1-CdFCA1 transformants are indicated by labeled arrows and are listed in Table 2.
CdFCA1 expression analysis.
To monitor the relative gene expression of the CdFCA1 gene in 5FC-resistant wild-type C. dubliniensis isolates (isolates SA113, SA109, and Eg202) and 5FC-susceptible wild-type C. dubliniensis isolates (isolates Eg204 and p7276), as well as in the doxycycline (DOX)-inducible transformant derivatives (strains SA113T1, SA109T1, and SA109T2), quantitative real-time PCR was carried out according to standard protocols. In brief, RNA was extracted from isolates and transformant derivatives that were grown in YPD broth in the presence of DOX (15 μg/ml) only or YPD broth in the presence of DOX (15 μg/ml) and subinhibitory concentrations of 5FC (6.4 ng/ml). The RNAs were extracted with an RNeasy minikit (Qiagen) and were treated with DNeasy (Ambion; Applied Biosystems, Warrington, United Kingdom), according to the manufacturer's instructions. The RNA samples were then reverse transcribed to cDNA by using a Superscript II reverse transcriptase kit (Invitrogen, Biosciences Ltd., Dun Laoghaire, Dublin, Ireland). Quantitative real-time PCR was carried out with two pairs of reverse transcription-PCR primers; one pair amplified the CdFCA1 gene, and the second pair amplified the ACT1 gene as an internal expression control (Table 2). The comparative amplification efficiencies of these primers were assessed, prior to reverse transcription-PCR, by using primer amplification efficiency plot analysis, as described previously (28). Quantitative real-time PCRs were carried out with 0.3 μM of each primer and SYBR green master mixture in an ABI 7500 real-time PCR system (Applied Biosystems), according to the manufacturer's recommended protocols. Data analysis was carried out as described by Schmittgen and Livak (34), and the 2−ΔCT values were calculated from the average threshold cycle (CT) values acquired from three replicates for both the CdFCA1 and the ACT1 genes.
RESULTS
In vitro susceptibility testing.
Nine C. dubliniensis isolates previously reported to be 5FC susceptible and 12 C. dubliniensis isolates previously reported to be 5FC resistant by Al Mosaid et al. (4) (Table 1) were tested for their susceptibilities to 5FC by broth microdilution assays. All nine isolates previously reported to be 5FC susceptible were confirmed as such and exhibited 5FC MICs of ≤0.25 μg/ml. Similarly, the 12 previously reported 5FC-resistant isolates exhibited 5FC MICs of ≥128 μg/ml. In an attempt to localize potential blocks or lesions in the 5FC metabolic pathway in 5FC-resistant isolates which may contribute to resistance, broth microdilution assays were also carried out with 5FU instead of 5FC (Table 1). We hypothesized that if a block in the 5FC metabolic pathway occurred at the level of Fca1p, then 5FC-resistant isolates should be susceptible to 5FU (Fig. 1). All 9 5FC-susceptible isolates and 12 5FC-resistant isolates used in the study (Table 1) were tested for their susceptibilities to 5FU by using a range of concentrations (0.25 to 128 μg/ml). All 21 isolates exhibited 5FU MICs in the range of 8 to 32 μg/ml (Table 1). There was no correlation between the 5FU MIC and susceptibility to 5FC. These findings indicated that a block(s) in the 5FC metabolic pathway occurred in resistant isolates at the level of cytosine deaminase or upstream of this enzyme (Fig. 1).
Sequence analysis of CdFCA1 in C. dubliniensis.
The DNA sequence of the C. albicans FCA1 gene, which encodes Fca1p (GenBank accession no. U55194), was used in a BLAST search against the C. dubliniensis genome sequence (http://www.sanger.ac.uk/sequencing/Candida/dubliniensis/) in order to identify its homologue in C. dubliniensis. In C. dubliniensis, Fca1p is encoded by the CdFCA1 gene, which shares 89% sequence identity with FCA1 from C. albicans, and both contain an internal intron of 81 bp. In order to investigate whether a mutation(s) or a deletion(s) was present in the CdFCA1 gene encoding Fca1p, the DNA sequences of the CdFCA1 ORF were determined for all 12 5FC-resistant isolates (Table 1) and all 9 5FC-susceptible isolates (Table 1) investigated in the study. For each isolate, the CdFCA1 sequence was compared with that of the CdFCA1 consensus sequence of 5FC-susceptible C. dubliniensis type strain CD36 (http://www.sanger.ac.uk/sequencing/Candida/dubliniensis/). Three separate SNPs were identified in the CdFCA1-coding sequences of the 21 C. dubliniensis isolates investigated. Two synonymous SNPs were observed; one of these (position 264, A → T transition) occurred in all isolates sequenced, with the exception of six of the nine 5FC-susceptible isolates (isolates CD36, Eg203, Eg206, SA101, SA105, and SA115), and the second (position 390, T → C transition) occurred in only one of the 5FC-susceptible isolates (isolate p7276). The third SNP was nonsynonymous (position 86, C → T transition), resulting in an amino acid substitution (Ser29Leu) in all 12 of the 5FC-resistant isolates tested, but was not present in the CdFCA1 gene of the 9 5FC-susceptible isolates sequenced.
Tetracycline-inducible expression of CdFCA1 in C. dubliniensis.
Transformation of the pNIM1-CdFCA1r cassette into 5FC-susceptible isolate p7276 yielded several transformants with the correct integration of the pNIM1-CdFCA1r cassette into the ADH1 locus, as determined by Southern hybridization and PCR analysis. Broth microdilution assays were carried out with these transformants by using serial dilutions of 5FC from a concentration of 0.25 to one of 128 μg/ml. All of the transformants were 5FC susceptible (5FC MICs ≤ 0.25 μg/ml). This experiment was replicated with the addition of the tetracycline derivative DOX to the RPMI 1640 broth microdilution medium at a final concentration of 15 μg/ml in order to induce the expression of the pNIM1-CdFCA1r cassette. All of the DOX-induced transformants remained 5FC susceptible (5FC MICs ≤ 0.25 μg/ml). Transformation of pNIM1-CdFCA1s DNA into 5FC-resistant isolates SA113 and SA109 yielded several transformants with the correct integration of the pNIM1-CdFCA1s cassette into the ADH1 locus, as determined by Southern hybridization and PCR analysis (Table 3). All of these transformants were 5FC resistant (5FC MICs ≥ 128 μg/ml). In contrast, in a parallel series of experiments, replicate broth microdilution assays were carried out with the addition of DOX (15 μg/ml) to the RPMI assay medium. This resulted in a dramatic change in the 5FC resistance phenotype of these transformants (isolates SA109T1, SA109T2, and SA113T1) from being 5FC resistant (5FC MICs ≥ 128 μg/ml) to being 5FC susceptible (5FC MICs ≤ 0.25 μg/ml) (Table 3). Similar results were obtained in separate broth microdilution experiments with DOX at a final concentration of 30 μg/ml (Table 3). As transformation controls, 5FC-resistant parental isolates SA109 and SA113 and 5FC-susceptible parental isolate p7276 were also transformed with the pNIM1 cassette containing the GFP1 gene instead of the CdFCA1 gene (Table 3). These control pNIM1-GFP1 transformants were also examined by broth microdilution assays in the presence and absence of DOX (15 μg/ml). The presence or absence of DOX in the broth microdilution medium had no effect on the 5FC MICs, and all the transformant derivatives tested exhibited 5FC MICs similar to those of their respective parental isolates (Table 3). These results strongly suggest that expression of the CdFCA1s gene by the transformants harboring the entire pNIM1-CdFCA1s cassette (i.e., isolates SA109T1, SA109T2, and SA113T1; Table 3) is responsible for the DOX-inducible 5FC susceptibility exhibited by these transformants.
TABLE 3.
Susceptibilities of C. dubliniensis isolates and pNIM1-FCA1/GFP1 transformant derivatives in the presence or absence of DOX
| Isolate or transformant | 5FC MIC50 (μg/ml) with DOX at: |
||
|---|---|---|---|
| None | 15 μg/ml | 30 μg/ml | |
| C. dubliniensis clinical isolates | |||
| SA113 | ≥128 | ≥128 | ≥128 |
| SA109 | ≥128 | ≥128 | ≥128 |
| p7276 | ≤0.25 | ≤0.25 | ≤0.25 |
| C. dubliniensis pNIM1-CdFCA1r transformants | |||
| p7276T1 | ≤0.25 | ≤0.25 | ≤0.25 |
| p7276T2 | ≤0.25 | ≤0.25 | ≤0.25 |
| p7276T3 | ≤0.25 | ≤0.25 | ≤0.25 |
| C. dubliniensis pNIM1-CdFCA1s transformants | |||
| SA109T1 | ≥128 | ≤0.25 | ≤0.25 |
| SA109T2 | ≥128 | ≤0.25 | ≤0.25 |
| SA113T1 | ≥128 | ≤0.25 | ≤0.25 |
| C. dubliniensis pNIM1-GFP1 transformants | |||
| SA113-GFP1T1 | ≥128 | ≥128 | ≥128 |
| SA109-GFP1T1 | ≥128 | ≥128 | ≥128 |
| SA109-GFP1T2 | ≥128 | ≥128 | ≥128 |
| p7276-GFP1T1 | ≤0.25 | ≤0.25 | ≤0.25 |
CdFCA1 expression analysis.
A comparison of CdFCA1 gene expression by the 5FC-susceptible isolates (isolates Eg204 and p7276) and 5FC-resistant isolates (isolates SA113, SA109, and Eg202) and the pNIM1-CdFCA1s transformant derivatives (isolates SA113T1, SA109T1, and SA109T2) was undertaken following exposure to DOX (15 μg/ml) or following exposure to DOX (15 μg/ml) and a subinhibitory concentration (6.4 ng/ml) of 5FC. The expression of CdFCA1 was analyzed by quantitative real-time PCR, and the data were normalized to the level of ACT1 expression, which was used as an internal control. Two-tailed Student's t tests were carried out on the CdFCA1 expression values obtained from 5FC-susceptible and 5FC-resistant clinical isolates, and no significant differences in CdFCA1 expression were observed between the two groups of isolates in either the presence (P = 0.47) or the absence (P = 0.16) of 5FC in the growth medium. This suggests that alterations in CdFCA1 expression do not play a significant role in mediating 5FC resistance. Upon exposure to 5FC (6.4 ng/ml), the level of CdFCA1 expression increased in both the 5FC-susceptible isolates (range, 14.5- to 25-fold) and the 5FC-resistant isolates (range, 4- to 18-fold). Two-tailed Student's t tests confirmed the significance of these CdFCA1 expression increases upon addition of 5FC to the growth medium (P < 0.001). On exposure to DOX, transformant derivatives SA109T1, SA109T2, and SA113T1 all showed significant (P < 0.001) increases in their levels of CdFCA1 expression (range, 5- to 26-fold) in comparison to those of parental isolates SA109 and SA113. These transformant derivatives also showed significant (P < 0.001) increases in their levels of CdFCA1 expression (range, 5- to 22-fold) in comparison to those of their parental isolates, isolates SA109 and SA113, in the presence of a subinhibitory concentration of 5FC as well as DOX.
DISCUSSION
Studies of pyrimidine salvage pathways (Fig. 1) and 5FC resistance mechanisms in yeast species have previously been undertaken with S. cerevisiae (9, 17, 25, 42), C. albicans (9, 13, 43), and C. lusitaniae (5, 22, 26). Investigations with S. cerevisiae have shown that the disruption of the FCY2 or the FUR1 gene can play a role in 5FC resistance, but only the FCA1 gene is absolutely required for the mediation of 5FC susceptibility (24, 25). In C. lusitaniae, Papon et al. (26) reported that inactivation of either the FCA1 or the FCY2 gene mediates 5FC resistance (5FC MICs, 128 μg/ml and 64 μg/ml, respectively) and promotes cross-resistance to 5FC and fluconazole (MICs, 4 μg/ml to 32 μg/ml), and the authors suggested that this was due to the competitive inhibition of fluconazole uptake by extracellular 5FC (5, 22, 26). Further analysis has identified a nonsense mutation in the FCY2 gene that resulted in a truncated purine cytosine permease in seven such isolates. In addition to this finding, a missense mutation (Met9Thr transition) has been identified in the FCY1 genes of four clinical C. lusitaniae isolates that also exhibited 5FC and 5FC-fluconazole cross-resistance (10). In C. albicans, two different research groups (7, 13) identified a homozygous Arg101Cys amino acid substitution in the FUR1-encoded UPRT to be the most common cause of high-level 5FC resistance (5FC MICs, 8 to >64 μg/ml). Isolates that were heterozygous for this transition exhibited reduced 5FC susceptibility (5FC MICs, 0.5 to 1 μg/ml) (13). Furthermore, a homozygous Gly28Asp substitution in Fca1p, encoded by FCA1, was suggested by Hope et al. to be an alternative method of resistance in a C. albicans isolate that did not harbor the UPRT-associated Arg101Cys substitution (13). Finally, Hope et al. (13) also described a C. albicans isolate with a Ser29Leu substitution in Fca1p which exhibited an intermediate level of 5FC resistance (5FC MIC, 4 μg/ml). In the light of the findings of the previous studies, we hypothesized that the C. dubliniensis pyrimidine salvage pathway very likely retains structural and functional homology with the pyrimidine salvage pathways in other Candida species, as C. dubliniensis is the closest relative to C. albicans in the genus Candida. Therefore, we investigated the CdFCA1-encoded Fca1p (Fig. 1) as a possible cause of C. dubliniensis clade-specific 5FC resistance. Initially, broth microdilution assays were carried out with both 5FC and 5FU to determine whether the deamination step in the 5FC metabolic pathway was responsible for 5FC resistance. If the deamination step was responsible for 5FC resistance, bypassing its requirement in the metabolic pathway by exposing 5FC-resistant cells to 5FU should result in susceptibility to 5FU (see Fig. 1). Both 5FC-susceptible and 5FC-resistant isolates exhibited similar 5FU MICs (Table 1), indicating that Fca1p is very likely responsible for the clade-specific 5FC resistance in C. dubliniensis. In order to investigate this possibility further, DNA sequence analysis of the CdFCA1 genes from 12 5FC-resistant and 9 5FC-susceptible C. dubliniensis isolates was undertaken. This identified a homozygous Ser29Leu substitution that occurred exclusively among 5FC-resistant isolates. This radical substitution results in the replacement of a hydrophilic polar amino acid (serine) with a hydrophobic nonpolar residue (leucine) in the β1 strand of the cytosine deaminase enzyme and is closely linked to an active-site residue, according to the yeast cytosine deaminase structure defined by Ko et al. (16). This amino acid substitution may disrupt the quaternary structure of the enzyme, distorting the active site and inhibiting the conversion of the 5FC prodrug to its toxic form, 5FU. As mentioned above, a similar amino acid substitution was reported by Hope et al. in a C. albicans isolate; however, that isolate exhibited intermediate resistance (5FC MIC, 4 μg/ml) to 5FC (13), in comparison to the high levels of 5FC resistance observed in the C. dubliniensis isolates displaying the Ser29Leu substitution in the present study. The differences in the levels of resistance to 5FC exhibited by the C. albicans isolate reported by Hope et al. and the 5FC-resistant C. dubliniensis isolates reported here, all of which harbored the same Ser29Leu substitution in Fca1p, may be due to the fact that different 5FC MIC determination methods were used in the two studies: the EUCAST method (33) was used in the previous study for the C. albicans isolates, and the CLSI method (6) was used in the present study for the C. dubliniensis isolates. Alternatively, differences in CdFCA1 and FCA1 gene expression or posttranscriptional or posttranslational modifications may be responsible for the differences between the two species in the levels of resistance to 5FC exhibited by isolates harboring the Fca1p Ser29Leu substitution.
In order to obtain direct evidence that the Ser29Leu substitution present in Fca1p from 5FC-resistant C. dubliniensis isolates was responsible for the 5FC-resistant phenotype in these isolates, the gene encoding cytosine deaminase from 5FC-susceptible C. dubliniensis isolate p7276 (CdFCA1s), which was originally recovered in Israel and which lacked the Ser29Leu substitution, was introduced into the ADH1 locus of two separate 5FC-resistant Saudi Arabian isolates, SA109 (ITS genotype 3) and SA113 (ITS genotype 4), by the tetracycline-inducible cassette pNIM1 (27). Three transformant derivatives (isolates SA109T1, SA109T2, and SA113T1) harboring the complete pNIM1-CdFCA1s cassette integrated into the ADH1 locus were tested and exhibited DOX-inducible 5FC susceptibility on the acquisition and expression of the CdFCA1s gene (Table 3). In contrast, transformant derivatives of 5FC-susceptible C. dubliniensis isolate p7276 harboring the complete pNIM1-CdFCA1r cassette encoding the FCA1 gene with the Ser29Leu substitution from 5FC-resistant C. dubliniensis isolate SA113 integrated into the ADH1 locus remained 5FC susceptible following DOX induction (Table 3). These findings provided convincing evidence that the Ser29Leu substitution in Fca1p from 5FC-resistant isolates was responsible for 5FC resistance in these isolates, but they also showed that the 5FC resistance mutation is recessive and, thus, that 5FC resistance is not expressed in the presence of a wild-type, functional FCA1 allele.
No significant difference in CdFCA1 expression was detected between the 5FC-susceptible and the 5FC-resistant C. dubliniensis isolates tested by quantitative real-time PCR analysis in either the presence or the absence of subinhibitory concentrations of 5FC in the growth medium. These results indicate that the lack of expression or the reduced expression of CdFCA1 in 5FC-resistant C. dubliniensis isolates following exposure to subinhibitory concentrations of 5FC was not responsible for the 5FC resistance in 5FC-resistant isolates and that the Ser29Leu substitution is very likely the sole method of CdFCA1-mediated 5FC resistance in the C. dubliniensis isolates investigated. Previous studies have shown that free pyrimidines often present in peptones present in some brands of culture media can antagonize the activity of 5FC (8). Antagonism was not observed in the present study with the YPD-grown cultures used in the expression studies. Following the addition of a subinhibitory concentration of 5FC (i.e., 6.4 ng/ml) to YPD-grown cultures, the quantitative real-time PCR experiments consistently showed that both 5FC-susceptible and 5FC-resistant C. dubliniensis isolates exhibited significant upregulation of FCA1 expression: 14.5- to 20-fold and 4- to 18-fold, respectively.
All Cd25 fingerprint group III C. dubliniensis isolates, apart from one (isolate p7718; Table 1), tested so far exhibit high-level 5FC resistance; all were originally recovered in Saudi Arabia or Egypt; and all belong to MLST clade C3 (4, 18). The close genetic relationship shared by these isolates is reflected by their identical CdFCA1 DNA sequences and high-level resistance to 5FC. It is highly likely that an identical mechanism is used to mediate 5FC resistance in all of these isolates. MLST C3 clade (Cd25 fingerprint group III) C. dubliniensis isolates can be subdivided into ITS genotypes 3 and 4 on the basis of the nucleotides sequence of the ITS region of the ribosomal DNA operon (4), although clade C3 isolates of both ITS genotypes exhibit high-level 5FC resistance. In the present study, clade C3 C. dubliniensis isolates SA109 (ITS genotype 3) and SA113 (genotype 4) were both transformed with the pNIM1-CdFCA1s cassette, and both yielded transformant derivatives (isolates SA109T1, SA109T2, and SA113T1) that exhibited DOX-inducible 5FC susceptibility. These findings support our view that clade-specific 5FC resistance in C. dubliniensis is mediated by a common molecular mechanism, i.e., the presence of the Ser29Leu substitution in Fca1p.
This is not the first report of a clade-specific SNP that has resulted in the alteration of a protein involved in antifungal drug resistance in C. dubliniensis. In 2002, Moran et al. (20) reported that 58% of ITS genotype 1 C. dubliniensis isolates (Cd25 group I, MLST clade C1) harbored a TAG nonsense mutation in the CDR1 gene encoding an ABC transporter. In C. albicans, the upregulation of CDR1 is the most common mechanism of fluconazole resistance, whereas in C. dubliniensis, the most common mechanism of fluconazole resistance involves the overexpression of the MDR1 gene encoding a multidrug transporter (21). These studies highlight the fact that despite the close phylogenetic relationship between C. dubliniensis and C. albicans, resistance to particular antifungal drugs can be due to different mechanisms in the two species.
In conclusion, the results of this study demonstrate that the presence of a Ser29Leu substitution in Fca1p in C. dubliniensis isolates is responsible for clade-specific resistance to 5FC. Isolates belonging to C. dubliniensis clade C3 have been recovered only from individuals of Arab ethnicity in Saudi Arabia, Egypt, and Israel (4, 18). Resistance to 5FC has not yet been reported in C. dubliniensis isolates from other countries around the world, apart from Kuwait. In 2004, Ahmad et al. reported the recovery of two 5FC-resistant isolates of C. dubliniensis from Kuwait (1). Because of Kuwait's close proximity to Saudi Arabia, it is likely that these isolates also belong to C. dubliniensis MLST clade C3.
Acknowledgments
This study was supported by the Microbiology Research Unit, Dublin Dental School and Hospital.
Footnotes
Published ahead of print on 24 August 2009.
REFERENCES
- 1.Ahmad, S., Z. Khan, E. Mokaddas, and Z. U. Khan. 2004. Isolation and molecular identification of Candida dubliniensis from non-human immunodeficiency virus-infected patients in Kuwait. J. Med. Microbiol. 53:633-637. [DOI] [PubMed] [Google Scholar]
- 2.Al Mosaid, A., D. Sullivan, I. F. Salkin, D. Shanley, and D. C. Coleman. 2001. Differentiation of Candida dubliniensis from Candida albicans on Staib agar and caffeic acid-ferric citrate agar. J. Clin. Microbiol. 39:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al Mosaid, A., D. J. Sullivan, and D. C. Coleman. 2003. Differentiation of Candida dubliniensis from Candida albicans on Pal's agar. J. Clin. Microbiol. 41:4787-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Mosaid, A., D. J. Sullivan, I. Polacheck, F. A. Shaheen, O. Soliman, S. Al Hedaithy, S. Al Thawad, M. Kabadaya, and D. C. Coleman. 2005. Novel 5-flucytosine-resistant clade of Candida dubliniensis from Saudi Arabia and Egypt identified by Cd25 fingerprinting. J. Clin. Microbiol. 43:4026-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapeland-Leclerc, F., J. Bouchoux, A. Goumar, C. Chastin, J. Villard, and T. Noel. 2005. Inactivation of the FCY2 gene encoding purine-cytosine permease promotes cross-resistance to flucytosine and fluconazole in Candida lusitaniae. Antimicrob. Agents Chemother. 49:3101-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard; M27-A2. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Dodgson, A. R., K. J. Dodgson, C. Pujol, M. A. Pfaller, and D. R. Soll. 2004. Clade-specific flucytosine resistance is due to a single nucleotide change in the FUR1 gene of Candida albicans. Antimicrob. Agents Chemother. 48:2223-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doern, G. V., T. A. Tubert, K. Chapin, and M. G. Rinaldi. 1986. Effect of medium composition on results of macrobroth dilution antifungal susceptibility testing of yeasts. J. Clin. Microbiol. 24:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erbs, P., F. Exinger, and R. Jund. 1997. Characterization of the Saccharomyces cerevisiae FCY1 gene encoding cytosine deaminase and its homologue FCA1 of Candida albicans. Curr. Genet. 31:1-6. [DOI] [PubMed] [Google Scholar]
- 10.Florent, M., T. Noel, G. Ruprich-Robert, B. Da Silva, V. Fitton-Ouhabi, C. Chastin, N. Papon, and F. Chapeland-Leclerc. 2009. Nonsense and missense mutations in FCY2 and FCY1 genes are responsible for flucytosine resistance and flucytosine-fluconazole cross-resistance in clinical isolates of Candida lusitaniae. Antimicrob. Agents Chemother. 53:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gee, S. F., S. Joly, D. R. Soll, J. F. Meis, P. E. Verweij, I. Polacheck, D. J. Sullivan, and D. C. Coleman. 2002. Identification of four distinct genotypes of Candida dubliniensis and detection of microevolution in vitro and in vivo. J. Clin. Microbiol. 40:556-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Hope, W. W., L. Tabernero, D. W. Denning, and M. J. Anderson. 2004. Molecular mechanisms of primary resistance to flucytosine in Candida albicans. Antimicrob. Agents Chemother. 48:4377-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoskins, J., and J. S. Butler. 2008. RNA-based 5-fluorouracil toxicity requires the pseudouridylation activity of Cbf5p. Genetics 179:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joly, S., C. Pujol, M. Rysz, K. Vargas, and D. R. Soll. 1999. Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis. J. Clin. Microbiol. 37:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko, T. P., J. J. Lin, C. Y. Hu, Y. H. Hsu, A. H. Wang, and S. H. Liaw. 2003. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J. Biol. Chem. 278:19111-19117. [DOI] [PubMed] [Google Scholar]
- 17.Kurtz, J. E., F. Exinger, P. Erbs, and R. Jund. 1999. New insights into the pyrimidine salvage pathway of Saccharomyces cerevisiae: requirement of six genes for cytidine metabolism. Curr. Genet. 36:130-136. [DOI] [PubMed] [Google Scholar]
- 18.McManus, B. A., D. C. Coleman, G. Moran, E. Pinjon, D. Diogo, M. E. Bougnoux, S. Borecka-Melkusova, H. Bujdakova, P. Murphy, C. d'Enfert, and D. J. Sullivan. 2008. Multilocus sequence typing reveals that the population structure of Candida dubliniensis is significantly less divergent than that of Candida albicans. J. Clin. Microbiol. 46:652-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McManus, B. A., D. J. Sullivan, G. P. Moran, C. D'Enfert, M. E. Bougnoux, M. A. Nunn, and D. C. Coleman. Genetic differences between avian and human isolates of Candida dubliniensis. Emerg. Infect. Dis. 15:1467-1470. [DOI] [PMC free article] [PubMed]
- 20.Moran, G., D. Sullivan, J. Morschhauser, and D. Coleman. 2002. The Candida dubliniensis CdCDR1 gene is not essential for fluconazole resistance. Antimicrob. Agents Chemother. 46:2829-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran, G. P., D. Sanglard, S. M. Donnelly, D. B. Shanley, D. J. Sullivan, and D. C. Coleman. 1998. Identification and expression of multidrug transporters responsible for fluconazole resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 42:1819-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noel, T., F. Francois, P. Paumard, C. Chastin, D. Brethes, and J. Villard. 2003. Flucytosine-fluconazole cross-resistance in purine-cytosine permease-deficient Candida lusitaniae clinical isolates: indirect evidence of a fluconazole uptake transporter. Antimicrob. Agents Chemother. 47:1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odds, F. C., M. E. Bougnoux, D. J. Shaw, J. M. Bain, A. D. Davidson, D. Diogo, M. D. Jacobsen, M. Lecomte, S. Y. Li, A. Tavanti, M. C. Maiden, N. A. Gow, and C. d'Enfert. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6:1041-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paluszynski, J. P., R. Klassen, and F. Meinhardt. 2008. Genetic prerequisites for additive or synergistic actions of 5-fluorocytosine and fluconazole in baker's yeast. Microbiology 154:3154-3164. [DOI] [PubMed] [Google Scholar]
- 25.Paluszynski, J. P., R. Klassen, M. Rohe, and F. Meinhardt. 2006. Various cytosine/adenine permease homologues are involved in the toxicity of 5-fluorocytosine in Saccharomyces cerevisiae. Yeast 23:707-715. [DOI] [PubMed] [Google Scholar]
- 26.Papon, N., T. Noel, M. Florent, S. Gibot-Leclerc, D. Jean, C. Chastin, J. Villard, and F. Chapeland-Leclerc. 2007. Molecular mechanism of flucytosine resistance in Candida lusitaniae: contribution of the FCY2, FCY1, and FUR1 genes to 5-fluorouracil and fluconazole cross-resistance. Antimicrob. Agents Chemother. 51:369-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, Y. N., and J. Morschhauser. 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 4:1328-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaller, M. A., S. A. Messer, S. Gee, S. Joly, C. Pujol, D. J. Sullivan, D. C. Coleman, and D. R. Soll. 1999. In vitro susceptibilities of Candida dubliniensis isolates tested against the new triazole and echinocandin antifungal agents. J. Clin. Microbiol. 37:870-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polacheck, I., J. Strahilevitz, D. Sullivan, S. Donnelly, I. F. Salkin, and D. C. Coleman. 2000. Recovery of Candida dubliniensis from non-human immunodeficiency virus-infected patients in Israel. J. Clin. Microbiol. 38:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polak, A., and H. J. Scholer. 1975. Mode of action of 5-fluorocytosine and mechanisms of resistance. Chemotherapy 21:113-130. [DOI] [PubMed] [Google Scholar]
- 32.Pujol, C., M. A. Pfaller, and D. R. Soll. 2004. Flucytosine resistance is restricted to a single genetic clade of Candida albicans. Antimicrob. Agents Chemother. 48:262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodríguez-Tudela, J. L., F. Barchiesi, F. Bille, E. Chryssanthou, M. Cuenca-Estrella, D. Denning, J. P. Donnelly, B. Dupont, W. Fegeler, C. Moore, M. Richardson, and P. E. Verweij. 2003. Method for the determination of minimum inhibitory concentration (MIC) by broth dilution of fermentative yeasts. Clin. Microbiol. Infect. 9:i-viii. [Google Scholar]
- 34.Schmittgen, T. D., and K. J. Livak. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101-1108. [DOI] [PubMed] [Google Scholar]
- 35.Staib, P., G. P. Moran, D. J. Sullivan, D. C. Coleman, and J. Morschhauser. 2001. Isogenic strain construction and gene targeting in Candida dubliniensis. J. Bacteriol. 183:2859-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan, D., K. Haynes, J. Bille, P. Boerlin, L. Rodero, S. Lloyd, M. Henman, and D. Coleman. 1997. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J. Clin. Microbiol. 35:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan, D. J., G. P. Moran, and D. C. Coleman. 2005. Candida dubliniensis: ten years on. FEMS Microbiol. Lett. 253:9-17. [DOI] [PubMed] [Google Scholar]
- 38.Sullivan, D. J., G. P. Moran, E. Pinjon, A. Al-Mosaid, C. Stokes, C. Vaughan, and D. C. Coleman. 2004. Comparison of the epidemiology, drug resistance mechanisms, and virulence of Candida dubliniensis and Candida albicans. FEMS Yeast Res. 4:369-376. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan, D. J., T. J. Westerneng, K. A. Haynes, D. E. Bennett, and D. C. Coleman. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507-1521. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waldorf, A. R., and A. Polak. 1983. Mechanisms of action of 5-fluorocytosine. Antimicrob. Agents Chemother. 23:79-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber, E., C. Rodriguez, M. R. Chevallier, and R. Jund. 1990. The purine-cytosine permease gene of Saccharomyces cerevisiae: primary structure and deduced protein sequence of the FCY2 gene product. Mol. Microbiol. 4:585-596. [DOI] [PubMed] [Google Scholar]
- 43.Whelan, W. L., and D. Kerridge. 1984. Decreased activity of UMP pyrophosphorylase associated with resistance to 5-fluorocytosine in Candida albicans. Antimicrob. Agents Chemother. 26:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willis, A. M., W. A. Coulter, D. J. Sullivan, D. C. Coleman, J. R. Hayes, P. M. Bell, and P. J. Lamey. 2000. Isolation of C. dubliniensis from insulin-using diabetes mellitus patients. J. Oral Pathol. Med. 29:86-90. [DOI] [PubMed] [Google Scholar]