Abstract
A cell culture system for the production of hepatitis C virus (HCV) whole virions has greatly accelerated studies of the virus life cycle and the discovery of anti-HCV agents. However, the quantification of the HCV titers in a whole-virus infection/replication system currently relies mostly on reverse transcription-PCR or immunofluorescence assay, which would be cumbersome for high-throughput drug screening. To overcome this problem, this study has generated a novel cell line, Huh7.5-EG(Δ4B5A)SEAP, that carries a dual reporter, EG(Δ4B5A)SEAP. The EG(Δ4B5A)SEAP reporter is a viral protease-cleavable fusion protein in which the enhanced green fluorescence protein is linked to secreted alkaline phosphatase (SEAP) in frame via Δ4B5A, a short peptide cleavage substrate for NS3/4A viral protease. This study demonstrates that virus replication/infection in the Huh7.5-EG(Δ4B5A)SEAP cells can be quantitatively indicated by measuring the SEAP activity in cell culture medium. The levels of SEAP released from HCV-infected Huh7.5-EG(Δ4B5A)SEAP cells correlated closely with the amounts of HCV in the inocula. The Huh7.5-EG(Δ4B5A)SEAP cells were also shown to be a suitable host for the discovery of anti-HCV inhibitors by using known compounds that target multiple stages of the HCV life cycle. The Z′-factor of this assay ranged from 0.64 to 0.74 in 96-well plates, indicating that this reporter system is suitable for high-throughput screening of prospective anti-HCV agents.
Infection with hepatitis C virus (HCV) is a major global health problem. More than 170 million individuals are infected worldwide. The infection is strongly associated with risks of liver cirrhosis and hepatocellular carcinoma (42, 43). HCV has a single, positive-stranded RNA genome of approximately 9.6 kb which encodes a polyprotein of about 3,000 amino acids (38). Cellular and viral proteases are involved in processing viral polyprotein into at least 10 proteins (core, E1, E2, p7, nonstructure protein 2 [NS2], NS3, NS4A, NS4B, NS5A, and NS5B), and the cleavages at the NS3-4A, NS4A-4B, NS4B-5A, and NS5A-5B junctions are mediated by NS3/4A protease (13, 24, 26). In the past few years, subgenomic HCV replicons have been employed extensively for studying viral replication, protein processing, and virus-host interactions and for discovering anti-HCV agents (4, 15, 16, 29, 34, 52). However, such subgenomic systems are not useful for studying the entry, assembly, or release of viral particles, because they lack structure proteins. In 2005, an infectious clone was isolated from a Japanese patient with fulminant hepatitis (JFH1) (44). The advent of the JFH1 isolate represented an important breakthrough for the investigation of the full viral life cycle as well as HCV interactions with host cells. Notably, this system is also useful for the testing of antiviral agents targeting viral attachment, entry, trafficking, assembly, and release. Recently, JFH1-based intergenotypic HCV virions were generated by substituting the JFH1 structure protein region for that of other strains (12, 40, 46, 47, 50). These chimeras may be important to the development of effective antivirals and vaccines against all genotypes. Several robust virus-producing systems have also led to important advances in studies of HCV (5, 50, 51). In particular, a stably JFH1 cDNA-transfected human hepatoma cell line, Huh7-GL, is a valuable tool for the robust and durative production of infectious virus. The JFH1 virion generated from the Huh7-GL cells was utilized in this work.
To elucidate the viral life cycle and the inhibitory effects of anti-HCV agents, several methods have been employed to measure the degree of HCV infection/replication. They include quantitative reverse transcription-PCR (qRT-PCR) to determine RNA levels or Western blot analysis to determine the expression levels of proteins (2, 12). These methods are time-consuming and labor-intensive because of the need to prepare cell lysates and to purify RNA. Therefore, a high-throughput screening system for accelerating the discovery and development of anti-HCV therapeutics is urgently needed. Recombinant JFH1 viruses that contain a luciferase or green fluorescent protein (GFP) reporter gene have recently been generated to investigate viral infection (1, 15, 25, 50). However, the associated common problem for these constructions is the low yield of viral progeny, possibly due to the insertion of undesirable elements that impair RNA replication. We have developed an efficient reporter-based assay for monitoring HCV protease activity and the degree of subgenomic replication (29-31). These studies involved the construction of a substrate vector, pEG(Δ4AB)SEAP, which contained a dual reporter gene. The enhanced green fluorescent protein (EGFP) gene was separated from a secreted alkaline phosphatase (SEAP) reporter gene by a fragment encoding a cleavable peptide substrate of the viral NS3/4A protease. These systems exhibited not only a high signal-to-noise ratio but also a close relationship between SEAP activity and HCV subgenomic replication.
In this study, we established a stable reporter cell line, Huh7.5-EG(Δ4B5A)SEAP, that can be used to determine JFH1 infection via the quantification of SEAP activity in culture medium. This system has allowed us to study the detailed kinetics of viral infection in real time without the preparation of cell lysates, isolation of RNA, and the subsequent qRT-PCR experiments. Several known inhibitors that target various stages in the viral life cycle were used to validate that this novel Huh7.5 cell-based system can indeed be utilized to screen anti-HCV agents of different modes of action. Statistical analyses were performed with anti-HCV inhibitors after treatment in a 96-well format, and the results indicated that both the accuracy and sensitivity of this assay system are suitable for high-throughput screening of anti-HCV drugs.
MATERIALS AND METHODS
Cells and virus.
Huh7.5 cells were generously provided by C. Rice (Rockefeller University, NY) and were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Life Technologies) plus penicillin (50 U/ml), streptomycin (50 mg/ml), and nonessential amino acids as a basal medium. Huh7.5 cells are a mutant line of Huh7 cells and are highly permissive for HCV RNA replication (3). Huh7-GL cells were kindly provided by Takaji Wakita (Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan) and were maintained in basal medium plus sodium pyruvate and 5 μg/ml blasticidine (Invitrogen). Huh7-GL cells contain a chromosomally integrated genotype 2a HCV cDNA and continuously generate infectious virus (JFH1) (5). In this study, stably transfected Huh7.5 cells expressing EG(Δ4B5A)SEAP fusion protein, Huh7.5-EG(Δ4B5A)SEAP cells, were maintained in basal medium plus 0.5 mg/ml G418.
Plasmid constructions.
In this study, we used pEGFP-C1 plasmid (Clontech) as a backbone vector to construct pEG(Δ4A4B)SEAP, pEG(Δ4B5A)SEAP, and pEG(Δ5A5B)SEAP. The SEAP gene was amplified by PCR using pSEAP2-control (Clontech) as a template. The forward primers 5′-CGG GGT ACC GAT GAG ATG GAG GAA TGC GCC TCT AGG GCG ATG CTG CTG CTG CTG CTG CTG CTG GGC CTG-3′, 5′-CGG GGT ACC ACT GAG GAC TGC CCC ATC CCA TGC TCC GGA TCC TGG CTC CGC GAC GTG ATG CTG CTG CTG CTG CTG CTG CTG GGC CTG-3′, and 5′-CGG GGT ACC GAG GAC GAT ACC ACC GTG TGC TGC TCC ATG TCA TAC TCC TGG ACC GGG ATG CTG CTG CTG CTG CTG CTG CTG GGC CTG-3′ contain the junction sites (genotype 2a HCV cDNA sequences) of NS4A/NS4B, NS4B/NS5A, and NS5A/NS5B, respectively, encoding in-frame amino acid sequences Δ4A4B (DEMEECASRA), Δ4B5A (TEDCPIPCSGSWLRDV), and Δ5A5B (EDDTTVCCSMSYSWTG) as substrates for HCV NS3/4A protease. The reverse primer sequence was 5′-GTG ATT GGT ACC TTC ATG TCT GCT CG-3′. The PCR product was digested with KpnI and inserted into the KpnI site of pEGFP-C1.
Transient transfection analysis of plasmid.
Huh7.5 cell lines were seeded at a density of 1 × 105 cells per well in 24-well plates and were incubated at 37°C overnight. Subsequently, cells were transfected with 0.2 μg of individual reporter plasmids by using FuGENE 6 reagent (Roche). The transfection procedure was performed according to the manufacturer's instructions. The plasmid-transfected Huh7.5 cells were infected with JFH1 virus at a multiplicity of infection (MOI) of 0.02 in each well for 6 h. At the end of infection, unbound viruses were removed by medium replacement and infected cells were further cultured in phenol red-free basal medium. At 6 days postinfection, SEAP activity was measured following one more medium replacement for 24 h. The expression of EGFP within reporter genes served as an internal control, and each experiment was normalized by the average number of EGFP-positive cells detected in multiple foci.
Stable cell line selection.
Huh7.5 cells were seeded at a density of 5 × 104 cells per well in a 24-well plate. After incubation at 37°C overnight, cells were transfected with 0.2 μg of pEG(Δ4B5A)SEAP plasmid in single well. At 2 days posttransfection, cells were maintained in basal medium plus 0.5 mg/ml G418 for selection of neomycin-resistant clones. After approximately 1 week of selection, expended clones were examined for expression of the EG(Δ4B5A)SEAP fusion protein and the extracellular SEAP activity under the proteolytic digestion of NS3 protease by using Western blot analysis and a Phospha-Light assay kit (Applied Biosystems), respectively. The stable cell line was designated Huh7.5-EG(Δ4B5A)SEAP.
Western blot analysis.
In 24-well plates, Huh7.5-EG(Δ4B5A)SEAP cells were seeded at a density of 1 × 105 cells per well and infected with JFH1 virus at an MOI of 0.02 in each well for 6 h. Huh7.5-EG(Δ4B5A)SEAP cells infected or noninfected by JFH1 were incubated for 3 days and washed with phosphate-buffered saline. Cells were lysed in lysis buffer (50 mM Tris-HCl, 150 mM sodium chloride, 1% Nonidet P-40, and 0.5% sodium deoxycholate), and the cell lysates were collected and clarified by centrifugation at 15,000 × g for 10 min at 4°C. The protein concentration of cell extracts was measured by using a protein assay buffer (Bio-Rad). Equal amounts of protein samples were resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and then proteins in gel were transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was blocked with NET buffer (0.25 g/liter gelatin, 150 mM sodium chloride, 5 mM disodium EDTA dihydrate, 50 mM Tris, and 0.05% Tween 20) for 1 h at room temperature. The expression levels of the EG(Δ4B5A)SEAP fusion protein and free-form EGFP were detected using mouse polyclonal anti-EGFP antibody. The intracellular NS3 protease of HCV JFH1-infected cells was detected using mouse monoclonal anti-NS3 antibody (Virostat). The membrane was also probed with mouse monoclonal antiactin antibody (Santa Cruz Biotechnology). The amount of β-actin served as a loading control. After primary antibody incubation, the membrane was washed with TBST buffer (150 mM sodium chloride, 0.1% Tween 20, 50 mM Tris-HCl) and probed with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G antibody. Finally, the membrane was treated with an enhanced chemiluminescence reagent and exposed to X-ray film.
Detection of SEAP activity released from HCV-infected Huh7.5-EG(Δ4B5A)SEAP cells.
HCV-infected Huh7.5-EG(Δ4B5A)SEAP cells were continually maintained in phenol red-free basal medium containing 0.5 mg/ml G418. At 5 days postinfection, culture medium was replaced and cells were incubated one more day. Subsequently, SEAP activity in the culture medium was detected using a Phospha-Light assay kit (Applied Biosystems), and cell viability was measured with the MTS assay as previously described (8).
Quantitative reverse transcription-PCR.
Total cellular RNA and virus RNA were extracted using TRIzol reagent (Invitrogen) and the QIAamp viral RNA minikit (Qiagen), respectively. One microgram of total RNA was used in the reverse transcription reaction performed with the Moloney murine leukemia virus reverse transcriptase kit (Promega). Briefly, an RNA sample was mixed with 1 μM random hexamer primer and the secondary structure of the RNA was allowed to unfold by heating for 8 min at 65°C. The reverse transcription reaction was carried out under the following conditions: annealing at 25°C for 10 min, extension at 42°C for 90 min, and denaturation at 95°C for 5 min. A 20-μl aliquot of the reaction mixture contained 200 U of Moloney murine leukemia virus RNase H(-) reverse transcriptase, 1 mM each deoxynucleotide triphosphate, and 20 U of RNase OUT reagent (Invitrogen).
Two microliters of reverse transcription reaction mixture was used in the qPCR performed on a LightCycler (Roche) with a LightCycler DNA master SYBR green I kit, according to the manufacturer's instructions. Primers for HCV (genotype 2a) cDNA were as follows: forward, 5′-CCC GTT GAG ACA CTC G-3′; reverse primer, 5′-CCT AAT GTT GGG ATT GAT GC-3′. The primers for glyceraldehyde 3-phosphate dehydrogenase were as follows: forward, 5′-GAA GGT GAA GGT CGG AGT C-3′; reverse, 5′-GAA GAT GGT GAT GGG ATT TC-3′. The qPCR conditions were as follows: denaturation at 95°C for 600 s and 45 cycles of amplification at 95°C for 0 s, 64°C for 5 s, and 72°C for 15 s. At the end of the qPCR procedure, the PCR cycle numbers of each sample were converted to the relative copy number of HCV RNA according to the calibration curve. The calibration curve was made from serial 10-fold dilutions of reference DNA that consisted of a full-length HCV (genotype 2a) cDNA. The copy number of intracellular HCV RNA in each sample was normalized by dividing the corresponding glyceraldehyde 3-phosphate dehydrogenase copy numbers. All experiments were performed in triplicate to determine statistical parameters.
Enzymatic assays for the HCV NS3/4A protease.
At 6 days postinfection, HCV-infected Huh7.5-EG(Δ4B5A)SEAP cells were washed with phosphate-buffered saline and lysed in lysis buffer without protease inhibitor. The cell lysates were collected and clarified by centrifugation at 15,000 × g for 10 min at 4°C. The protein concentration of cell extract was measured and 50 μg of total protein was analyzed in the HCV NS3/4A protease assay. The HCV NS3/4A protease activity was determined by using an internally quenched fluorogenic depsipeptide (HCV protease fluorescence resonance energy transfer [FRET] substrate [RET S1]), Ac-DED(EDANS)EEαAbuψ[COO]ASK(DABCYL)-NH2 (AnaSpec Inc.), in a black F96 MicroWell plate (Nunc) (11). The cell extract was incubated with reaction buffer (50 mM Tris-HCl [pH 7.5], 50% glycerol, and 30 mM dithiothreitol). The reaction was started by the addition of 1.5 μM RET-S1 substrate and incubated at 25°C. The reaction volume was 100 μl. The assay was run in a kinetics model for 10 min, and product release was detected using a Fluoroskan Ascent FL plate reader (Thermo Scientific) with excitation at 355 nm and emission at 510 nm. The protease activity was determined by the linear slope of product release with time.
Preparation of HCV JFH1 stock.
Approximately 30 to 40% confluence of Huh7-GL cells in a T150 flask was maintained in basal medium without 0.5 μg/ml blasticidine. After incubation for 4 days, culture medium was collected and cleared by using low-speed centrifugation (1,000 × g). The supernatant was passed through a 0.45-μm low-protein-binding filter. The filtrate was concentrated 1/50 using an Amicin Ultra-15 centrifugal filter (100 kDa; Millipore) (44), and the remaining filtrate was aliquoted for storage at −80°C in 15% glycerol. The amount of infectious HCV JFH1 in virus stock was measured by immunofluorescence assay.
Titration of infectious HCV JFH1.
In this study, we estimated the amount of infectious HCV JFH1 in viral stock by immunofluorescence assay (51). Briefly, viral stock was serially diluted 10-fold in Dulbecco's modified Eagle's medium and then used to infect 8 × 103 Huh7.5 cells per well in 96-well plates. After 6 h, the supernatants were removed and culture medium was supplemented. After 4 days, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Subsequently, fixed cells were blocked with 5% bovine serum albumin for 1 h at room temperature. The intracellular HCV NS3 proteins in infected cells were detected by incubation with mouse monoclonal anti-NS3 antibody (1:20 dilution; Virostat) for 1 h at room temperature. Tetramethyl rhodamine isothiocyanate-conjugated AffiniPure donkey anti-mouse immunoglobulin G (H+L; 1:200 dilution; obtained from the optical biology core lab in the core facilities of the National Health Research Institutes) was applied to visualize the positive cells. The number of HCV foci in each dilution was counted, and the viral titer is expressed as the number of focus-forming units per milliliter.
Effects of anti-CD81 antibody and H-89 on HCV JFH1 infection.
Huh7.5-EG(Δ4B5A)SEAP cells were seeded at a density of 8 × 103 cells per well in 96-well plates. After incubation overnight, cells were treated with increasing concentrations of anti-CD81 antibody (Santa Cruz Biotechnology, Inc.) or H-89 (Calbiochem) in basal medium for 1 h and infected with JFH1 virus at an MOI of 0.02 per well for 6 h in the presence of inhibitor. At the end of infection, unbound virus/inhibitors were removed by medium replacement, and HCV-infected cells were continually cultured. At 5 days postinfection, the culture medium in each well was replaced with phenol red-free basal medium, and cells were cultured for one more day. Subsequently, SEAP activity in culture medium was measured, and cell viability was estimated by MTS assay. Each experiment was performed in triplicate for statistical analysis.
Effects of naringenin, BILN2061, and IFN-α on intracellular activity of HCV.
To estimate the levels of secreted HCV JFH1 from various compound-treated Huh7-GL cells, cells at a density of 1.5 × 104 cells per well in 96-well plates were treated with increasing concentrations of reference compounds, naringenin (Sigma), BILN2061 (Acme Bioscience, Inc.), and alpha interferon (IFN-α; Sigma) for 3 days. At the end of treatment, the supernatants containing secreted JFH1 cells were collected and cell viability was estimated in an MTS assay. The level of secreted JFH1 was detected by infecting Huh7.5-EG(Δ4B5A)SEAP cells, and HCV-infected cells were cultured further. At 5 days postinfection, the culture medium in each well was replaced. Subsequently, SEAP activity and cell viability in each well were measured at 6 days postinfection.
Calculation of S/B, S/N, and Z′-factor.
To evaluate the performance of this developed assay for high-throughput screening, the signal-to-noise ratio (S/N), signal-to-background ratio (S/B), and Z′-factor values were calculated using the methods of Zhang et al. (49). In HCV JFH1 entry inhibition, Huh7.5-EG(Δ4B5A)SEAP cells were seeded at a density of 8 ×103 cells per well in a single 96-well plate overnight. H-89 was added to the 30 wells for 1 h at a volume of 100 μl and a final concentration of 10 μM in 1% (vol/vol) dimethyl sulfoxide (DMSO) as a background set. A 1% (vol/vol) DMSO solution was added to the other 30 wells as a signal set. Subsequently, cells were infected with JFH1 virus at an MOI of 0.02 per well for 6 h in the presence of H-89 or 1% DMSO. At the end of infection, unbound virus and compounds were removed by medium replacement and HCV-infected cells were further cultured. At 6 days postinfection, medium was collected from each well for SEAP activity analysis and cell viability was measured. For assessment of intracellular HCV JFH1 activity, Huh7-GL cells were seeded at a density of 1.5 × 104 cells per well in a single 96-well plate. Cells in the 30 wells were treated with 100 nM BILN2061 in 1% (vol/vol) DMSO, as a background set, for 3 days while 1% (vol/vol) DMSO was added to cells in the other 30 wells, as a signal set, for 3 days. Three days later, supernatant media were collected and cell viability was measured. Subsequently, Huh7.5-EG(Δ4B5A)SEAP cells at a density of 8 × 103 cells per well in a 96-well plate were treated with collected supernatant media for 6 h by the plate-to-plate method. At the end of treatment, reporter cells were further cultured for 6 days followed by detection of SEAP activity and cell viability. Finally, the data were collected and S/B, S/N, and Z′-factor values were determined.
Data analysis and statistics.
Data analyses for SEAP activity and cell viability were performed with the SigmaPlot software (SPSS Inc.). The SEAP signal data are presented as means ± standard deviations. Statistically significant effects between groups were examined using the paired Student t test. Statistical significance was defined as a P level of <0.01.
RESULTS
Characterization of HCV JFH1 infection in Huh7.5-EG(Δ4B5A)SEAP cells.
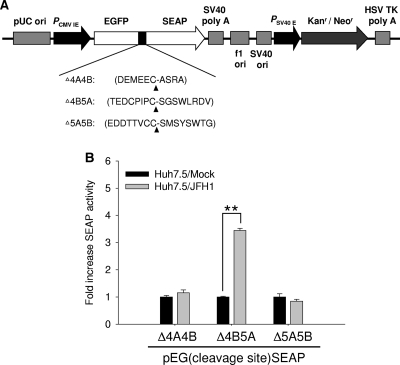
To develop a convenient tool for monitoring HCV JFH1 infection, several reporter plasmids were constructed based on our earlier reporter-based assay system, which involves the HCV Con1 strain of the genotype 1b subgenomic replicon (30), with EGFP fused to SEAP via three prospective peptide linkers corresponding to the JFH1 genotype 2a NS3 protease cleavage sites at various junctions. The amount of SEAP that is secreted into the medium depends on the expression of NS3 protease following HCV infection. The evaluated cleavage peptides of JFH genotype 2a included DEMEEC-ASRA (designated Δ4A4B), TEDCPIPC-SGSWLRDV (designated Δ4B5A), and EDDTTVCC-SMSYSWTG (designated Δ5A5B) (Fig. 1A). Initially, these reporter constructs were transiently transfected into Huh7.5 cells to examine whether they could be efficiently cleaved by NS3 protease following HCV infection. At 6 days postinfection, cell culture media were harvested and analyzed for SEAP activity. Surprisingly, elevated SEAP levels were observed only in media from cells that were transfected with the EG(Δ4B5A)SEAP construct but not in media from the other two constructs (Fig. 1B). This result differed substantially from that in a previous investigation in which the linked Δ4A4B peptide sequence (DEMEEC-ASHL) of the HCV genotype 1b was cleavable by NS3 protease in HCV replicon cells harboring the HCV subgenome of genotype 1b (29). Indeed, we observed that the SEAP activity was also significantly detectable with the EG(Δ4A4B)SEAP construct containing the peptide sequence DEMEEC-ASHL following HCV JFH1 infection (data not shown), suggesting that interference with cleavage may be caused by different protein conformations when the various peptide sequences are inserted into the chimera fusion protein. The JFH1 viruses were collected from Huh7-GL cells that contained a chromosomally integrated full-length JFH1 HCV cDNA (5).
FIG. 1.
Diagram of dual-function reporter vectors used in the HCV NS3/4A protease activity assay. (A) The various junction regions, NS4A/NS4B, NS4B/NS5A, and NS5A/NS5B, as substrates of HCV NS3/4A protease were inserted between the egfp gene and the seap gene by an in-frame fusion. The amino acid sequences of the junction regions Δ4A4B, Δ4B5A, and Δ5A5B are shown. Expression of the reporter genes is controlled by the human cytomegalovirus immediate-early (CMV IE) promoter, and the selection marker for the generation of the stable cell line is neomycin phosphotransferase (neo). SV40, simian virus 40. (B) Huh7.5 cells were seeded at a density of 1 × 105 cells per well in 24-well plates and transfected with 0.2 μg of individual plasmids. The expression of EGFP within reporter genes served as an internal control for the normalization of transfection efficiency. Transiently transfected Huh7.5 cells were infected with or without JFH1 virus at an MOI of 0.02 per well. At 6 days postinfection, the medium was harvested and SEAP activity was measured. Each experiment was performed in triplicate, and error bars reflecting standard deviations are shown. **, P < 0.01.
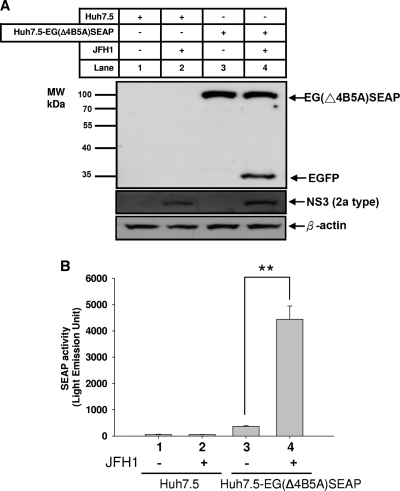
To confirm that the EG(Δ4B5A)SEAP fusion protein was indeed cleaved by NS3 protease, an EG(Δ4B5A)SEAP stably transfected Huh7.5 cell line, designated Huh7.5-EG(Δ4B5A)SEAP, was infected with JFH1 virus (each well of cells in a 24-well plate received a viral inoculum at an MOI of 0.02) for 6 days. Cell lysates were analyzed by Western blotting with anti-EGFP antibody and anti-NS3 antibody, in which the levels of β-actin served as the loading controls. As shown in Fig. 2A, high levels of intact EG(Δ4B5A)SEAP fusion protein were observed in Huh7.5-EG(Δ4B5A)SEAP reporter cells (lanes 3 and 4, upper panel) but not in the parental Huh7.5 cells (lanes 1 and 2). Upon infection of the Huh7.5 control cells (lane 2) and the Huh7.5-EG(Δ4B5A)SEAP reporter cells by JFH1 (lane 4), viral replication was revealed by the expression of NS3 viral protease, as determined by Western blot analysis. A distinct band that corresponds to free EGFP was clearly present in lane 4, indicating that JFH1-derived NS3 protease effectively cleaved the EG(Δ4B5A)SEAP into EGFP and SEAP.
FIG. 2.
Characterization of Huh7.5-EG(Δ4B5A)SEAP cells stably expressing EG(Δ4B5A)SEAP fusion protein in Huh7.5. (A) Cells were seeded at a density of 1 × 105 cells per well in 24-well plates and infected with JFH1 virus at an MOI of 0.02 per well. The lysates of HCV JFH1-infected and noninfected cells were harvested at 6 days postinfection. Expression levels of EG(Δ4B5A)SEAP and NS3 protease were analyzed using Western blotting with anti-EGFP (upper panel) and anti-NS3 antibody (middle panel), respectively. Lane 1, Huh7.5 cells; lane 2, JFH1-infected Huh7.5 cells; lane 3, Huh7.5-EG(Δ4B5A)SEAP cells; lane 4, JFH1-infected Huh7.5-EG(Δ4B5A)SEAP cells. β-Actin was the loading control (lower panel). (B) SEAP activity in culture medium was measured at 6 days postinfection. Column 1, Huh7.5 cells; column 2, JFH1-infected Huh7.5 cells; column 3, Huh7.5-EG(Δ4B5A)SEAP cells; column 4, JFH1-infected Huh7.5-EG(Δ4B5A)SEAP cells. Each experiment was performed in triplicate, and error bars reflecting standard deviations are shown. **, P < 0.01.
The SEAP levels in the culture media were also measured. In Huh7.5 cells without the EG(Δ4B5A)SEAP reporter, only basal levels of SEAP activity were detected in the culture medium (Fig. 2B, columns 1 and 2). The SEAP level in the culture medium of the Huh7.5-EG(Δ4B5A)SEAP cells was slightly higher (column 3). The SEAP level in the culture medium of Huh7.5-EG(Δ4B5A)SEAP cells infected with JFH1 virus was many times higher than that of noninfected cells (column 4), presumably because of proteolytic processing of EG(Δ4B5A)SEAP by the NS3 protease.
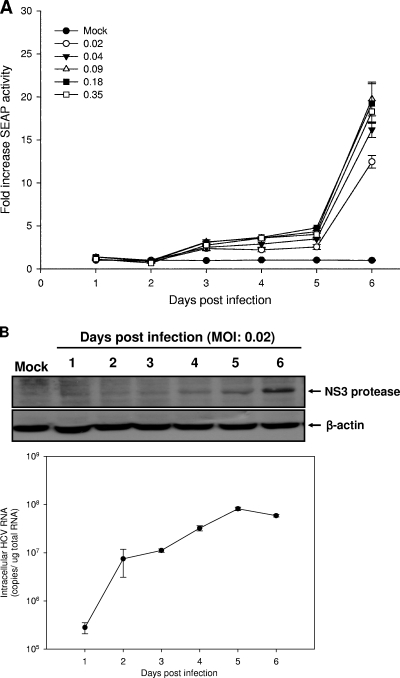
To optimize the assay conditions, Huh7.5-EG(Δ4B5A)SEAP cells were infected with JFH1 at increasing MOIs and SEAP activity in each culture medium was analyzed at various time points postinfection. After inoculation with the JFH1 virus, culture media were harvested daily to measure the activities of SEAP. As shown in Fig. 3A, increasing MOIs in the inoculum progressively increased the SEAP signals in a time-dependent manner. The SEAP signals became detectable at 3 days postinfection. When Huh7.5-EG(Δ4B5A)SEAP cells were infected with JFH1 virus at an MOI of 0.02, at 6 days postinfection, the SEAP activity was approximately 14.9-fold ± 0.7-fold higher than that at 2 days. With a low MOI (0.04) or higher MOI (>0.04), at 6 days postinfection, the SEAP activity also was approximately 15-fold higher than that at 2 days. Such kinetics of SEAP signals is consistent with the infection kinetics observed by Zhong et al. using immunofluorescence staining on naïve JFH1-infected Huh7.5.1 cells (51). Notably, the time course of SEAP detection was similar to the results recently described by Yu et al., who used an NS3-based FRET assay to monitor HCV infection (48). Furthermore, we examined the relationship between intracellular HCV RNA levels and NS3 protease expression during virus infection. It was interesting to find that the induction of SEAP correlated directly with increased NS3 protein levels but not with HCV RNA (Fig. 3B). We suggest that the slow onset of the SEAP signal may be caused by the lag time for HCV to overcome the innate infection in the reporter cells before HCV starts to express protein efficiently.
FIG. 3.
Kinetics of HCV JFH1 infection in Huh7.5-EG(Δ4B5A)SEAP cells. (A) A virus stock generated from Huh7-GL was diluted to infect Huh7.5-EG(Δ4B5A)SEAP cells (1 × 105 cells per well in 24-well plates) with various MOIs (0, 0.02, 0.04, 0.09, 0.18, and 0.35). The supernatant medium was collected, and SEAP activity was measured at the indicated time points postinfection. The SEAP activity in HCV JFH1 infection with various MOIs increased in a time-dependent manner. (B) Huh7.5-EG(Δ4B5A)SEAP cells inoculated with HCV JFH1 at an MOI of 0.02 were analyzed for the intracellular HCV NS3 protein and viral RNA levels at the indicated time points postinfection. The HCV NS3 and β-actin protein levels were analyzed by Western blotting (upper panel). The intracellular HCV RNA level was quantified by qRT-PCR (lower panel). Each infection experiment was performed in triplicate, and error bars reflecting standard deviations are shown.
Correlation between levels of secreted SEAP activity and HCV JFH1 replication inside cells.
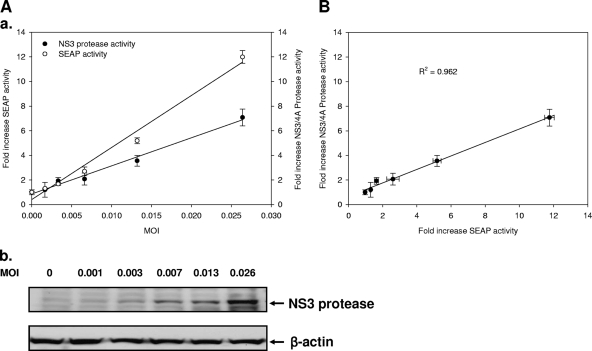
Whether the levels of SEAP activity in culture supernatants correlated with the extent of HCV replication inside infected cells was examined. The Huh7.5-EG(Δ4B5A)SEAP cells were inoculated with increasing MOIs and incubated for 6 days. Subsequently, the culture supernatants were analyzed for SEAP activity. The cell lysates were analyzed for both NS3 expression levels and protease activity using Western blotting with anti-NS3 antibody and a FRET assay with an internally quenched fluorogenic depsipeptide of NS3/4A protease substrate, as described in Materials and Methods, respectively. As shown in Fig. 4A, a linear relationship exists between the SEAP levels and the amount of the HCV inoculums (Fig. 4A, panel a). A similar linear relationship was also observed between the NS3/4A protease activity and the titers of virus inoculums (panel a). In panel b of Fig. 4A, the viral NS3/4A protease expression levels clearly increased with the amount of the HCV inoculum. The square of the correlation constant (R2) was 0.962, as calculated by linear regression (Fig. 4B). This value indicates that the secretion of SEAP was closely correlated with the abundance of the viral proteins, as reflected by the NS3 expression levels and relative activity. Altogether, in a manner similar to the representation of viral replication using the replicon-based reporter system that we established previously (29, 30), the secreted SEAP reporter level suitably represented the amount of the virus inoculum.
FIG. 4.
Correlation between intracellular NS3/4A protease activity and extracellular SEAP activity in HCV JFH1-infected Huh7.5-EG(Δ4B5A)SEAP cells. In 24-well plates, 1 × 105 Huh7.5-EG(Δ4B5A)SEAP cells per well were infected with HCV JFH1 virus at various MOIs (0, 0.001, 0.003, 0.006, 0.013, and 0.026) for 6 h. Six days postinfection, the cell lysates and supernatant media were harvested to measure NS3/4A protease activity and SEAP activity, respectively. (A) Linear regression analysis of the relationship between the various numbers of HCV JFH1 copies and intracellular NS3/4A protease activities or extracellular SEAP activities (panel a). Cell lysates were analyzed by Western blotting with anti-NS3 antibody (panel b). β-Actin was the loading control. (B) Linear regression analysis of SEAP activities and NS3/4A protease activities in HCV JFH1-infected Huh7.5-EG(Δ4B5A)SEAP cells. Each point was determined in triplicate, and error bars reflecting standard deviations are shown.
Evaluations of various kinds of HCV inhibitors in Huh7.5-EG(Δ4B5A)SEAP cells infected with JFH1 virus.
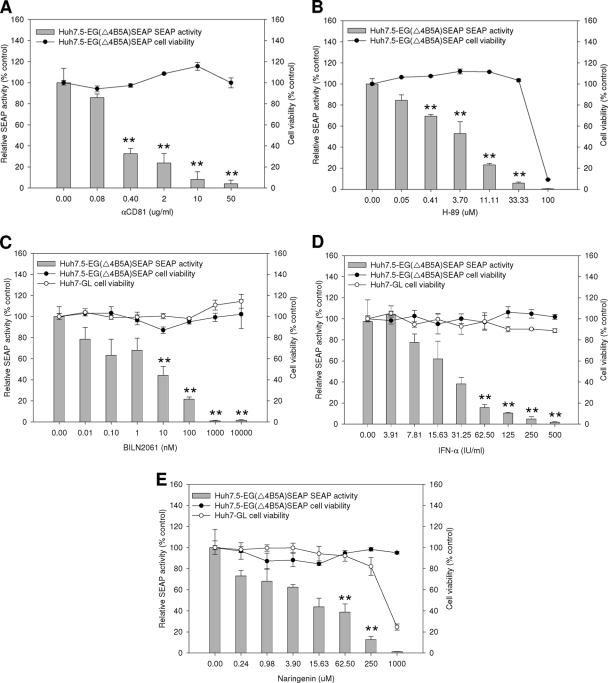
To verify the utility of the JFH1-infected Huh7.5-EG(Δ4B5A)SEAP cells as a quantitative antiviral assay system, several known HCV inhibitors that target multiple stages of the HCV life cycle were examined. These HCV inhibitors included (i) anti-CD81 antibody, an entry blocker that interferes with the CD81-E2 interaction (37); (ii) H-89, a protein kinase A inhibitor that reduces viral receptor activity by inducing the redistribution of CLDN1 at the plasma membrane (23); (iii) BILN2061, an NS3 protease inhibitor (28); (iv) IFN-α, a clinical drug against viral replication (41); and (v) naringenin, a blocker of apolipoprotein B-dependent hepatitis C virus secretion (36). These inhibitors were added to JFH1-infected Huh7.5-EG(Δ4B5A)SEAP cells at concentrations that did not cause cytotoxicity. Then, the levels of SEAP activity in culture supernatants were measured to reflect the anti-HCV activities of the compounds.
In the virus-neutralizing test, Huh7.5-EG(Δ4B5A)SEAP cells were incubated with the neutralizing anti-CD81 antibody at concentrations of 0, 0.08, 0.4, 2, 10, and 50 μg/ml for 1 h before the addition of JFH1 virus. SEAP activity was measured 6 days later. As shown in Fig. 5A, the anti-CD81 antibody showed anti-HCV activity with a 50% effective concentration (EC50) of 0.25 ± 0.03 μg/ml, which was comparable to the results recently reported by Iro et al. with a similar NS3-based reporter assay (21). Furthermore, the treatment of Huh7.5-EG(Δ4B5A)SEAP cells with another entry inhibitor, H-89, also resulted in the dose-dependent reduction of SEAP activity, with an EC50 at 4.8 ± 0.2 μM (Fig. 5B) which was similar to the result of Farquhar et al. (10).
FIG. 5.
Attenuation of HCV JFH1 infectivity by reference compounds. (A and B) To analyze inhibition of HCV JFH1 entry, Huh7.5-EG(Δ4B5A)SEAP cells at a density of 8 × 103 cells per well in 96-well plates were preincubated with increasing concentrations of anti-CD81 antibody (αCD81) or H-89 for 1 h and then infected with HCV JFH1 at an MOI of 0.02 per well in the presence of the inhibitor. SEAP activity and cell viability in each well were measured at 6 days postinfection. The 50% effective concentrations of αCD81 and H-89 were 0.34 μg/ml and 2.26 μM, respectively. (C to E) For inhibition of HCV JFH1 intracellular activity, Huh7-GL cells at a density of 1.5 × 104 cells per well in 96-well plates were treated with BILN2061, IFN-α, or naringenin for 3 days. Following the treatment, the supernatant medium containing secreted JFH1 was collected and used to infect Huh7.5-EG(Δ4B5A)SEAP cells. Cell viability was also estimated. Huh7.5-EG(Δ4B5A)SEAP cells were seeded at a density of 8 × 103 cells per well in 96-well plates and infected with the virus. Six days postinfection, SEAP activity and cell viability were measured. The 50% effective concentrations of BILN2061, IFN-α, and naringenin were 6.98 nM, 21.91 IU/ml, and 8.33 μM, respectively. Each infection experiment was performed in triplicate, and error bars reflecting standard deviations are shown. **, P < 0.01.
To clarify whether this reporter system is useful for analyzing inhibitors against HCV replication in the later stages of the viral life cycle, Huh7-GL cells, a cell line that constitutively produces the infectious virus, were incubated with various concentrations of BILN2061 and IFN-α for 72 h. Subsequently, the culture supernatant containing secreted JFH1 virions was employed to infect Huh7.5-EG(Δ4B5A)SEAP reporter cells. As revealed by SEAP activity, BILN2061 showed an EC50 of 6.9 ± 0.8 nM (Fig. 5C). IFN-α also significantly reduced SEAP activity in a dose-dependent manner (Fig. 5D).
To verify if an HCV secretion inhibitor could also be discovered using this reporter system, Huh7-GL cells were cultured in the presence of naringenin, with increasing concentrations for 72 h. Then, the secreted JFH1 viruses were collected to infect Huh7.5-EG(Δ4B5A)SEAP reporter cells for 6 h. At 6 days postinfection, the culture media were assayed for SEAP activity after they had been replaced. As shown in Fig. 5E, naringenin was facilely shown to reduce HCV secretion by 87.2% ± 3.2% at a concentration of 250 μM. The results were similar to those obtained by Nahmias et al., who used the rather cumbersome immunofluorescence staining of HCV core protein (36). Taken together, the EC50s are comparable to previous reports, demonstrating that SEAP activity really reflects the activity of the anti-HCV agents of different modes of action.
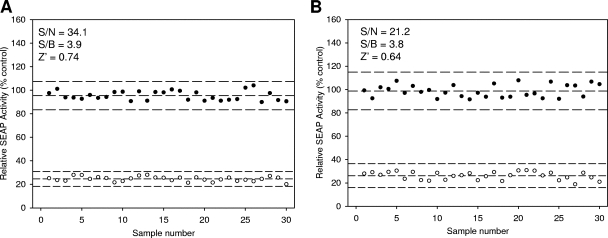
Statistical evaluation of reporter-based HCV JFH1 infection assay for robust analysis.
To assess whether these protease-based Huh7.5-EG(Δ4B5A)SEAP reporter cells can be adopted for a high-throughput screening assay for novel anti-HCV agents, assay parameters, including the S/N, S/B, and Z′-factor, were measured in the presence and absence of H-89 or BILN-2061. As a control for screening of viral entry inhibitors, Huh7.5-EG(Δ4B5A)SEAP cells were treated with 10 μM H-89 for 1 h. Six days after JFH1 virus inoculation, the levels of SEAP activity following replacement of medium are shown graphically in Fig. 6A. The S/N ratio and S/B ratio were 34.1 and 3.9, respectively. Using statistical calculations as defined by Zhang's formula (49), a Z′ value of 0.74 was obtained. Similarly, the Huh7.5-EG(Δ4B5A)SEAP cells were evaluated for the screening of inhibitors targeting the viral life cycle after the entry and uncoating of vision. Toward this end, Huh7-GL cells were first treated with 100 nM BILN2061 for 3 days. Then, the viruses produced in the supernatants were harvested to inoculate Huh7.5-EG(Δ4B5A)SEAP cells. The SEAP activity was measured at 6 days postinfection. As shown in Fig. 6B, the S/N and S/B ratios were 21.2 and 3.8, respectively. The Z′ factor was 0.64. Collectively, both of the different models of virus infection exhibited higher S/B ratios, lower S/N ratios, and a Z′-factor of more than 5, which indicated that the signal-to-noise scatter in the assay plates was highly reproducible with little variation for robust determination of inhibitors against viral infection.
FIG. 6.
Estimates of statistical parameters of the screening system. (A) HCV JFH1 entry system. Huh7.5-EG(Δ4B5A)SEAP cells at a density of 8 × 103 cells per well in a 96-well plate were utilized to determine the statistical parameters. Cells were pretreated with 10 μM H-89 (open circles), as a background set, and 1% DMSO (closed circles), as a signal set, for 1 h before being infected with HCV JFH1 at an MOI of 0.02 per well for 6 h. Six days postinfection, SEAP activity and cell viability were measured. The S/N, S/B, and Z′ factor values were 34.1, 3.9, and 0.74, respectively. (B) In the HCV JFH1 secretion system. Huh7-GL cells at a density of 1.5 × 104 cells per well in a 96-well plate were treated with 100 nM BILN2061 (open circles), as a background set, and 1% DMSO (closed circles), as a signal set, for 3 days. Subsequently, supernatant media were inoculated in Huh7.5-EG(Δ4B5A)SEAP cells (8 × 103 cells per well) for 6 h in a plate-to-plate manner. Six days postinfection, SEAP activity and cell viability were measured. S/N, S/B, and the Z′ factor were 21.2, 3.8, and 0.64, respectively.
DISCUSSION
Currently, the quantification of the levels of HCV RNA or viral proteins relies mainly on real-time qPCR or immunofluorescence assay. However, these methods are rather time-consuming and labor-intensive. In recent years, several protease-based approaches have been successfully established for developing inhibitors for a number of different diseases, including hypertension, diabetes, thrombosis, osteoporosis, and infectious diseases (7, 14, 35, 45). To discover novel HCV inhibitors, we previously developed a rapid and reliable assay system in HCV subgenomic replicon cells based on the principle employed in this study (17-19, 29, 32). The utility of such an assay system has been validated by the fact that we have indeed discovered several bioactive compounds against HCV replication by using the reporter system.
The study of HCV in cell culture has been greatly facilitated by the advent of the JFH1 clone (33, 44, 51). In this study, a facile reporter system was established for the evaluation of HCV infectivity in cells. Several engineered reporter fusion proteins were made with several NS3/4A-cleavable peptides encoded in the polyprotein of the JFH1 clone. One important finding was that, out of the three prospective constructs examined, only the EG(Δ4B5A)SEAP reporter was functional (Fig. 1). Notably, the EG(Δ4B5A)SEAP reporter described in this study is also a functional reporter for monitoring HCV 1b replication in a subgenomic replicon cell line (results not shown). Whether the EG(Δ4B5A)SEAP fusion protein can be employed as a reporter for other subtypes of HCV is currently being evaluated in our laboratory.
The use of EG(Δ4B5A)SEAP for HCV replication is advantageous because this reporter does not need to be engineered into the viral genome. Several chimeric JFH1 reporter viruses have been developed for analyzing virus infection (1, 15, 22, 25, 39, 50). One common strategy is to modify HCV cDNA by inserting GFP or a luciferase gene into the viral genome in a monocistronic or bicistronic reporter construction. However, genetic modification of a viral genome may be cumbersome. Furthermore, the engineered viral genome rendered more deviation from the authentic RNA structure (27, 39), and the enlarged genome size might slow RNA synthesis and compromise the packaging of viral progeny (9, 20, 25, 27). Schaller et al. also found that expression of the GFP reporter gene rapidly disappeared during prolonged passage of monocistronic HCV reporter virus (39). Accordingly, the higher risk of detrimental mutation associated with the high mutation rate of virus-encoded RNA-dependent RNA polymerase is another caveat for use of the monocistronic HCV reporter genome. In the case of the bicistronic reporter genome, Schaller et al. and Koutsoudakis et al. also observed that the kinetics of RNA replication and the release of infectious particles were lower than those of the parental genome (27, 39). The addition of a controlling element, such as the internal ribosome entry site of encephalomyocarditis virus or the foot-and-mouse disease virus 2A protease, also reduces the specificity of anti-HCV drug screening, as the prospective hits may interfere with targets and cause the emergence of false positives (27, 39, 50).
In this report, we also showed that a 6-day infection assay is the optimal assay condition for achieving detectable SEAP activity in culture medium (Fig. 3). Such a time course of SEAP induction in our assay system is consistent with a very recent study (48). In a protease-based HCV infection assay, Yu et al. demonstrated that the kinetics of HCV NS3/4A protease protein accumulation correlated well with FRET signal detection in Huh7 cells infected with HCV at various MOIs, and a greater FRET signal was detectable up to day 6 postinfection, which is similar to our observations (Fig. 3 and 4). However, by use of a similar dual reporter, EGFP-SEAP, linked by an octapeptide, DEDEDEDE followed by the HCV genotype 1b NS4A/4B recognition sequence (DEMEEC-ASHL) of NS3/4A protease, Iro et al. defined a significant increase of SEAP at day 4 postinfection. In addition, the levels of induction of SEAP were greater than in our system (21). This may be due to the inclusion of the DEDEDEDE peptide in Iro's study. Chou et al. demonstrated that the DEDEDEDE residues prior to the natural 4A/4B cleavage markedly enhanced the cleavage efficiency (6). However, we cannot exclude the possibility that there are different copy numbers of chromosomally integrated fusion reporter genes in selected reporter cell clones.
Another interesting observation from this study is that, upon JFH1 infection of Huh7.5-EG(Δ4B5A)SEAP cells, the expression levels of HCV NS3 viral proteins continued to increase up to day 6 postinfection while the viral RNA level seemed to reach a plateau at day 5 (Fig. 3B). This indicates that cellular anti-HCV immunity may exist in the Huh7.5-based reporter cells to mollify HCV replication during the earlier days postinfection. It will be important to further engineer the host cells so that they can become more permissive to HCV infection, that is, efficient expression of HCV viral proteins would take place immediately upon viral infection. Alternatively, mutant JFH1 clones with various degrees of replication competence in host cells could be established to provide a cell culture system that can be more reflective of HCV infection in humans.
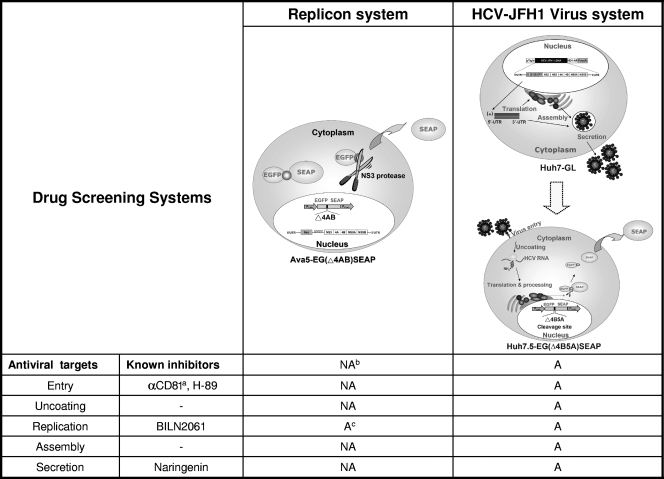
Finally, as shown in Fig. 7, compared to the Ava5-EG(Δ4AB)SEAP system that we established previously (29), Huh7.5-EG(Δ4B5A)SEAP reporter cells can be facilely utilized for investigating all stages of the HCV life cycle, including viral entry, uncoating, replication, virion assembly, and secretion. This novel reporter-based assay system has great potential to be used for screening novel anti-HCV agents. The SEAP activity in culture medium may reflect the level of HCV replication, and the viability of cells can be analyzed simultaneously.
FIG. 7.
Comparison of a reporter-based replicon system and HCV JFH1 infection model for screening novel anti-HCV agents. The replicon system was applied as described by Lee et al. (29). αCD8, anti-CD81 antibody; NA, not applicable; A, applicable.
Acknowledgments
We thank Charles Rice at Rockefeller University and Apath, LLC, for providing Huh7.5. We also thank Takaji Wakita at Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan, for providing Huh7-GL cells.
This work was supported by the National Health Research Institutes (BP-093-CP-04) and National Science Council (NSC 95-2745-B-400-MY3) in Taiwan.
Footnotes
Published ahead of print on 31 August 2009.
REFERENCES
- 1.Ahmed, M. M., T. H. Huang, and Q. D. Xie. 2008. A sensitive and rapid assay for investigating vertical transmission of hepatitis B virus via male germ line using EGFP vector as reporter. J. Biomed. Biotechnol. 2008:495436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne, N., R. B. Pyles, M. Yi, R. L. Veselenak, M. M. Davis, and S. M. Lemon. 2005. Screening for hepatitis C virus antiviral activity with a cell-based secreted alkaline phosphatase reporter replicon system. Antivir. Res. 67:76-82. [DOI] [PubMed] [Google Scholar]
- 5.Cai, Z., C. Zhang, K. S. Chang, J. Jiang, B. C. Ahn, T. Wakita, T. J. Liang, and G. Luo. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 79:13963-13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou, C. F., S. Shen, G. Mahadevappa, S. G. Lim, W. Hong, and Y. J. Tan. 2007. The use of hepatitis C virus NS3/4A and secreted alkaline phosphatase to quantitate cell-cell membrane fusion mediated by severe acute respiratory syndrome coronavirus S protein and the receptor angiotensin-converting enzyme 2. Anal. Biochem. 366:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow, W. A., C. Jiang, and M. Guan. 2009. Anti-HIV drugs for cancer therapeutics: back to the future? Lancet Oncol. 10:61-71. [DOI] [PubMed] [Google Scholar]
- 8.Cory, A. H., T. C. Owen, J. A. Barltrop, and J. G. Cory. 1991. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 3:207-212. [DOI] [PubMed] [Google Scholar]
- 9.Dansako, H., M. Ikeda, K. Abe, K. Mori, K. Takemoto, Y. Ariumi, and N. Kato. 2008. A new living cell-based assay system for monitoring genome-length hepatitis C virus RNA replication. Virus Res. 137:72-79. [DOI] [PubMed] [Google Scholar]
- 10.Farquhar, M. J., H. J. Harris, M. Diskar, S. Jones, C. J. Mee, S. U. Nielsen, C. L. Brimacombe, S. Molina, G. L. Toms, P. Maurel, J. Howl, F. W. Herberg, S. C. van Ijzendoorn, P. Balfe, and J. A. McKeating. 2008. Protein kinase A-dependent step(s) in hepatitis C virus entry and infectivity. J. Virol. 82:8797-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallinari, P., D. Brennan, C. Nardi, M. Brunetti, L. Tomei, C. Steinkuhler, and R. De Francesco. 1998. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J. Virol. 72:6758-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottwein, J. M., T. K. Scheel, A. M. Hoegh, J. B. Lademann, J. Eugen-Olsen, G. Lisby, and J. Bukh. 2007. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 133:1614-1626. [DOI] [PubMed] [Google Scholar]
- 13.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grum-Tokars, V., K. Ratia, A. Begaye, S. C. Baker, and A. D. Mesecar. 2008. Evaluating the 3C-like protease activity of SARS-coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 133:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao, W., and R. Duggal. 2009. High-throughput screening of HCV RNA replication inhibitors by means of a reporter replicon system. Methods Mol. Biol. 510:243-250. [DOI] [PubMed] [Google Scholar]
- 16.Hao, W., K. J. Herlihy, N. J. Zhang, S. A. Fuhrman, C. Doan, A. K. Patick, and R. Duggal. 2007. Development of a novel dicistronic reporter-selectable hepatitis C virus replicon suitable for high-throughput inhibitor screening. Antimicrob. Agents Chemother. 51:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang, D. R., R. K. Lin, G. Z. Leu, T. Y. Lin, T. W. Lien, M. C. Yu, C. T. Yeh, and J. T. Hsu. 2005. Inhibition of hepatitis C virus replication by antimonial compounds. Antimicrob. Agents Chemother. 49:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang, D. R., Y. C. Tsai, J. C. Lee, K. K. Huang, R. K. Lin, C. H. Ho, J. M. Chiou, Y. T. Lin, J. T. Hsu, and C. T. Yeh. 2004. Inhibition of hepatitis C virus replication by arsenic trioxide. Antimicrob. Agents Chemother. 48:2876-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang, D. R., Y. S. Wu, C. W. Chang, T. W. Lien, W. C. Chen, U. K. Tan, J. T. Hsu, and H. P. Hsieh. 2006. Synthesis and anti-viral activity of a series of sesquiterpene lactones and analogues in the subgenomic HCV replicon system. Bioorg. Med. Chem. 14:83-91. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda, M., K. Abe, H. Dansako, T. Nakamura, K. Naka, and N. Kato. 2005. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem. Biophys. Res. Commun. 329:1350-1359. [DOI] [PubMed] [Google Scholar]
- 21.Iro, M., J. Witteveldt, A. G. Angus, I. Woerz, A. Kaul, R. Bartenschlager, and A. H. Patel. 2009. A reporter cell line for rapid and sensitive evaluation of hepatitis C virus infectivity and replication. Antivir. Res. 83:148-155. [DOI] [PubMed] [Google Scholar]
- 22.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung, E. Y., M. N. Lee, H. Y. Yang, D. Yu, and K. L. Jang. 2001. The repressive activity of hepatitis C virus core protein on the transcription of p21waf1 is regulated by protein kinase A-mediated phosphorylation. Virus Res. 79:109-115. [DOI] [PubMed] [Google Scholar]
- 24.Kato, N. 2001. Molecular virology of hepatitis C virus. Acta Med. Okayama 55:133-159. [DOI] [PubMed] [Google Scholar]
- 25.Kim, C. S., J. H. Jung, T. Wakita, S. K. Yoon, and S. K. Jang. 2007. Monitoring the antiviral effect of alpha interferon on individual cells. J. Virol. 81:8814-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J. C., C. F. Chang, Y. H. Chi, D. R. Hwang, and J. T. Hsu. 2004. A reporter-based assay for identifying hepatitis C virus inhibitors based on subgenomic replicon cells. J. Virol. Methods 116:27-33. [DOI] [PubMed] [Google Scholar]
- 30.Lee, J. C., Y. F. Shih, S. P. Hsu, T. Y. Chang, L. H. Chen, and J. T. Hsu. 2003. Development of a cell-based assay for monitoring specific hepatitis C virus NS3/4A protease activity in mammalian cells. Anal. Biochem. 316:162-170. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. C., M. C. Yu, T. W. Lien, C. F. Chang, and J. T. Hsu. 2005. High-throughput cell-based screening for hepatitis C virus NS3/4A protease inhibitors. Assay Drug Dev. Technol 3:385-392. [DOI] [PubMed] [Google Scholar]
- 32.Leu, G. Z., T. Y. Lin, and J. T. Hsu. 2004. Anti-HCV activities of selective polyunsaturated fatty acids. Biochem. Biophys. Res. Commun. 318:275-280. [DOI] [PubMed] [Google Scholar]
- 33.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 34.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 35.McKerrow, J. H., P. J. Rosenthal, R. Swenerton, and P. Doyle. 2008. Development of protease inhibitors for protozoan infections. Curr. Opin. Infect. Dis. 21:668-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nahmias, Y., J. Goldwasser, M. Casali, D. van Poll, T. Wakita, R. T. Chung, and M. L. Yarmush. 2008. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology 47:1437-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owsianka, A. M., J. M. Timms, A. W. Tarr, R. J. Brown, T. P. Hickling, A. Szwejk, K. Bienkowska-Szewczyk, B. J. Thomson, A. H. Patel, and J. K. Ball. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Penin, F., J. Dubuisson, F. A. Rey, D. Moradpour, and J. M. Pawlotsky. 2004. Structural biology of hepatitis C virus. Hepatology 39:5-19. [DOI] [PubMed] [Google Scholar]
- 39.Schaller, T., N. Appel, G. Koutsoudakis, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81:4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheel, T. K., J. M. Gottwein, T. B. Jensen, J. C. Prentoe, A. M. Hoegh, H. J. Alter, J. Eugen-Olsen, and J. Bukh. 2008. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc. Natl. Acad. Sci. USA 105:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soriano, V., M. G. Peters, and S. Zeuzem. 2009. New therapies for hepatitis C virus infection. Clin. Infect. Dis. 48:313-320. [DOI] [PubMed] [Google Scholar]
- 42.Tan, S. L., A. Pause, Y. Shi, and N. Sonenberg. 2002. Hepatitis C therapeutics: current status and emerging strategies. Nat. Rev. Drug Discov. 1:867-881. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, D. L. 2000. Hepatitis C epidemiology. Curr. Top. Microbiol. Immunol. 242:25-41. [DOI] [PubMed] [Google Scholar]
- 44.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, Y., J. Kitagaki, H. Wang, D. X. Hou, and A. O. Perantoni. 2009. Targeting the ubiquitin-proteasome system for cancer therapy. Cancer Sci. 100:24-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, X., B. Sainz, Jr., and S. L. Uprichard. 20 July 2009. Development of a cell-based hepatitis C virus (HCV) infection FRET assay for high-throughput antiviral compound screening. Antimicrob. Agents Chemother. doi: 10.1128/AAC.00495-09. [DOI] [PMC free article] [PubMed]
- 49.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Y., P. Weady, R. Duggal, and W. Hao. 2008. Novel chimeric genotype 1b/2a hepatitis C virus suitable for high-throughput screening. Antimicrob. Agents Chemother. 52:666-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuck, P., E. M. Murray, E. Stec, J. A. Grobler, A. J. Simon, B. Strulovici, J. Inglese, O. A. Flores, and M. Ferrer. 2004. A cell-based beta-lactamase reporter gene assay for the identification of inhibitors of hepatitis C virus replication. Anal. Biochem. 334:344-355. [DOI] [PubMed] [Google Scholar]