Abstract
The Trojan horse antibiotic albomycin, produced by Streptomyces sp. strain ATCC 700974, contains a thioribosyl nucleoside moiety linked to a hydroxamate siderophore through a serine residue. The seryl nucleoside structure (SB-217452) is a potent inhibitor of seryl-tRNA synthetase (SerRS) in the pathogenic bacterium Staphylococcus aureus, with a 50% inhibitory concentration (IC50) of ∼8 nM. In the albomycin-producing Streptomyces sp., a bacterial SerRS homolog (Alb10) was found to be encoded in a biosynthetic gene cluster in addition to another serRS gene (serS1) at a different genetic locus. Alb10, named SerRS2 herein, is significantly divergent from SerRS1, which shows high homology to the housekeeping SerRS found in other Streptomyces species. We genetically and biochemically characterized the two genes and the proteins encoded. Both genes were able to complement a temperature-sensitive serS mutant of Escherichia coli and allowed growth at a nonpermissive temperature. serS2 was shown to confer albomycin resistance, with specific amino acid residues in the motif 2 signature sequences of SerRS2 playing key roles. SerRS1 and SerRS2 are comparably efficient in vitro, but the Km of serine for SerRS2 measured during tRNA aminoacylation is more than 20-fold higher than that for SerRS1. SB-217452 was also enzymatically generated and purified by two-step chromatography. Its IC50 against SerRS1 was estimated to be 10-fold lower than that against SerRS2. In contrast, both SerRSs displayed comparable inhibition kinetics for serine hydroxamate, indicating that SerRS2 was specifically resistant to SB-217452. These data suggest that mining Streptomyces genomes for duplicated aminoacyl-tRNA synthetase genes could provide a novel approach for the identification of natural products targeting aminoacyl-tRNA synthetases.
Aminoacyl-tRNA synthetases (aaRSs) are enzymes that catalyze the ligation of specific amino acids to their cognate tRNAs that are subsequently used in protein synthesis (13). While this process is essential and the reaction mechanisms are universal in all three domains of life, aaRS genes are remarkably evolutionarily divergent, leading to a wide range of aaRS proteins with subtle structural differences in various organisms (19). In most pathogenic bacteria or fungi, aaRSs are encoded by single-copy genes and are attractive antimicrobial drug targets for screening and designing enzyme inhibitors of high specificity (18, 22). For example, the antibiotic pseudomonic acid (mupirocin), a potent inhibitor of bacterial isoleucyl-tRNA synthetase, has been used clinically as a topical agent to prevent Staphylococcus aureus infection (6) and another novel class of aaRS inhibitors has also recently emerged (26). For many environmental bacteria, genomic analyses have revealed two or three homologous genes encoding aaRSs of the same amino acid specificity in one microorganism (21, 37). In several cases, one of these isozymes is demonstrably resistant to an enzyme inhibitor of another (15, 20, 37). This phenomenon is quite common in bacteria such as soil-dwelling actinomycetes that are capable of producing diverse secondary metabolites and antibacterial natural products. A mupirocin-producing Pseudomonas fluorescens uses such a self-resistance mechanism (6, 37), and a similar scenario has been postulated for Streptomyces parvulus producing borrelidin (a threonyl-tRNA synthetase inhibitor) (20). All known Streptomyces species and related bacterial genera host a large array of multiple aaRS gene homologs in their genomes (21). The aaRS genes can either stand alone or associate with biosynthetic gene clusters. Only one of these aaRSs has been biochemically characterized to be drug resistant (15). However, more recent evidence points to an additional physiological role of aaRSs, especially when the second aaRS gene is dedicated to a biosynthetic pathway (9, 10). Because aminoacyl-tRNAs have been shown to serve as substrates in other cellular processes, including phospholipid modification, peptidoglycan biosynthesis, and porphyrin biosynthesis (reviewed in reference 25), a pathway-specific aaRS would obviously meet the need for increasing local concentrations of the charged aminoacyl-tRNA. In Streptomyces viridifaciens, which produces the antibiotic valanimycin, the pathway-specific seryl-tRNA synthetase (SerRS) VlmL is believed to produce l-seryl-tRNA, from which the seryl residue is transferred directly to another reactant in the valanimycin biosynthetic pathway (10). However, vlmL does not encode a valanimycin-resistant SerRS.
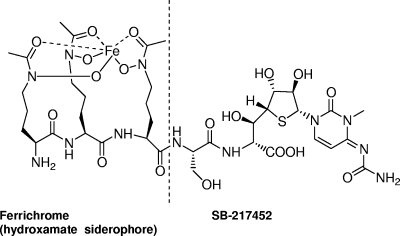
The Trojan horse antibiotic albomycin, produced by Streptomyces sp. strain ATCC 700974, has a thioribosyl nucleoside moiety linked to an iron-chelating siderophore through a serine residue (Fig. 1) (3). The seryl-glycine dipeptide-linked 4′-thioribosyl-N4-carbamyl-5-methyl-4-imino-cytidine structure, named SB-217452, has been isolated from a Streptomyces fermentation culture and identified as a potent inhibitor against Staphylococcus aureus SerRS (50% inhibitory concentration [IC50], ∼8 nM) (31). It was speculated that SB-217452 could mimic seryl-adenylate, the intermediate of the SerRS-catalyzed reaction, thus blocking the aminoacylation activity of the enzyme. In fact, albomycin as a whole can be actively taken up by a bacterium through its siderophore-dependent iron acquisition system, which results in an extremely low MIC (11, 23). SB-217452 is released and activated from the prodrug albomycin inside cells by peptidases reported to be PepN in Escherichia coli and PepN and PepA in Salmonella enterica serovar Typhimurium (4). Several natural antimicrobial agents, such as microcin C (17) and agrocin 84 (14), employ similar Trojan horse strategies to transport hydrophilic aaRS inhibitors across the membrane into target cells. To further investigate this important class of emerging antibiotics, we first cloned and sequenced the albomycin biosynthetic gene cluster in Streptomyces sp. strain ATCC 700974 (38). A bacterial SerRS gene homolog (alb10) was identified in the gene cluster. Given several unusual aspects of the serylation systems in bacteria, yeasts, archaea, and mammalian mitochondria (reviewed in reference 34), it was of interest to study the proposed drug-resistant properties of Alb10 and use it as an example to explore the evolutionary divergence of Streptomyces aaRSs. Here we report the in vitro and in vivo characterization of two SerRSs, one pathway specific and the other housekeeping, in the albomycin-producing Streptomyces strain. Our findings now set the stage to elucidate the albomycin biosynthetic pathway, improve the production level of albomycin, and expand the discovery of microbial natural products that are aaRS inhibitors.
FIG. 1.
Structure of the Trojan horse antibiotic albomycin and the SerRS inhibitor SB-217452 linked to the hydroxamate siderophore.
MATERIALS AND METHODS
Strains, plasmids, and reagents.
Streptomyces sp. strain ATCC 700974 was cultivated in production medium consisting of 20 g starch, 5 g l-ornithine-HCl, 1.8 g KH2PO4, 10.2 g Na2HPO4·2H2O, 2 g (NH4)2SO4, 2 g NaCl, 2 g MgSO4·7H2O, 0.8 g CaCl2·2H2O, 0.28 g FeSO4·7H2O, 0.02 g ZnSO4·7H2O, and 1 liter of deionized water. E. coli JM109 was used as a general cloning host. E. coli K28 was obtained from the Yale E. coli Genetic Stock Center. The vectors used for protein production were pET30a(+) and pET15b (both from Novagen) and pUC19 (NEB). High-fidelity PCR enzyme was used. All reagents were from Sigma, NEB, and Fisher Scientific. Molecular biology procedures followed standard methods.
DNA manipulations.
We created a cosmid library from Streptomyces sp. strain ATCC 700974 genomic DNA. An ∼38-kb region was identified as the albomycin biosynthetic gene cluster (38). serS2 was amplified from cosmid 8F8 using an N-terminal primer containing an NdeI site, serS2-NdeI, and a C-terminal primer with a XhoI site, serS2-XhoI (Table 1) . The PCR product was cloned into the NdeI and XhoI sites of pET30a(+) to produce pET30-SerRS2. serS2 was also amplified with the SerRS2 N-terminal primer and a reverse primer containing a PstI site, serS2-PstI. The PCR product was cloned into pSUP (27) to yield pSUP-SerRS2. An ∼1.3-kb fragment including the glnS′ promoter was excised from pSUP-SerRS2 by HindIII and PstI and cloned into pUC19 to yield pUC-SerRS2. A SerRS2 H270G mutant was created by site-directed mutagenesis of the serS2 gene, using primers serS2mut2-F, serS2mut2-R, serS2mut-Nco-F, and serS2mut-BamH-R. The mutations were introduced in the PCR primers. The product was cloned into the NcoI and BamHI sites of pET15b to produce pET15b-SerRS2mut. The serS1 gene was amplified from genomic DNA. Based on the conserved sequences of the homologous housekeeping gene serS and the adjacent genes in Streptomyces coelicolor and Streptomyces avermitilis, degenerate primer pairs were designed to amplify the two end sequences of serS1 in Streptomyces sp. strain ATCC 700974. For the N-terminal region, primers serS1-N-F and serS1-N-R gave an ∼850-bp product. For the C-terminal region, primers serS1-C-F and serS1-C-R gave an ∼1,000-bp product. Sequencing results confirmed that they both contained either the N or C terminus of a SerRS homolog. The full-length serS1 gene was amplified with an N-terminal primer containing an NdeI site, serS1-NdeI, and a C-terminal primer with a PstI site, serS1-PstI. The SerRS1 PCR product was cloned into pUC-SerRS2 cut by NdeI and PstI to replace the SerRS2 DNA. This resulted in plasmid pUC-SerRS1. The SerRS1 N-terminal primer and a C-terminal primer containing an XhoI site, serS1-XhoI, were used to amplify the serS1 gene that was cloned into the NdeI and XhoI sites of pET30a(+) to produce pET30-SerRS1. The E. coli serS gene, including its native promoter and terminator, was amplified from E. coli JM109 genomic DNA using primers EcserS-F and EcserS-R. The product was cloned into the XbaI and EcoRI sites of pUC19 to produce pUC-EcSerRS. DNA sequencing was performed by the Ohio University Genomics Facility. Sequence analysis and alignments were performed with Vector NTI Advance 10.3 (Invitrogen).
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| serS1-N-F | GTAAAACGACGGCCAGGASGAGCGSCGCMGGTC |
| serS1-N-R | CAGGAAACAGCTATGACCGGATSGWCRGCCGGCGGGMCTGGAA |
| serS1-C-F | CGCTGCCAGGACGGCCGGTTCT |
| serS1-C-R | GACGAAGTCCTCCTCGCCGCCGAC |
| serS1-NdeI | AGGGCATATGATTGACCTTCGCCTGCT |
| serS1-XhoI | AGTCTCGAGCTTGGCTACCGGTTCCAGGA |
| serS1-PstI | GGAAGTCTGCAGTCACTTGGCTACCGGTTCCAGGA |
| serS2-NdeI | CGCCATATGCTTGATCTGGACCTGATTCG |
| serS2-XhoI | TCCCTCGAGACGCTTGGGGCGGACCAG |
| serS2-PstI | TCCCTGCAGCTAACGCTTGGGGCGGACCAG |
| serS2mut2-F | GCCGGAGCCGGCTCCGACGAGCGCGGCACGGTCCGGGGCCA |
| serS2mut2-R | CGCTCGTCGGAGCCGGCTCCGGCCCGCTCCCGCCGGTA |
| serS2mut-Nco-F | CACGCCCATGGTTGATCTGGACCTGATTCGTAAG |
| serS2mut-BamH-R | AACTGGATCCCTAGTGATGATGATGATGATGACGCTTGGGGCGGACCAG |
| EcserS-F | CTACCGTCTAGATTAATGTTATCGTTGCGG |
| EcserS-R | GAGGAGGAATTCTAATTATTGTTCGGTATC |
Complementation of temperature-sensitive E. coli strain K28.
E. coli strain K28 (5, 33) was transformed with the three pUC plasmids carrying the Streptomyces serS1 and serS2 and E. coli serS genes. To test complementation, a single colony of each transformant was inoculated in low-salt LB (0.05% NaCl) liquid medium with 100 μg/ml ampicillin and grown overnight. The culture was then diluted to an A600 of ∼0.01, and 5 μl of the dilution was dispensed onto low-salt LB agar plates with ampicillin. The plates were grown at the permissive (30°C) or nonpermissive (40°C) temperature for 15 h.
Agar diffusion assay for measuring albomycin susceptibility.
An overnight culture (with an antibiotic added if required for maintaining a plasmid) of a tester bacterial strain was diluted with saline to an optical density at 600 nm of ∼0.08. A cotton swab tip was saturated with this bacterial dilution and swiped across a Mueller-Hinton agar (BD Biosciences) plate that would grow a lawn of bacteria. Holes of ∼0.4 cm in diameter within an appropriate distance were punched in the agar. An albomycin dilution (20 μl) was pipetted into the holes. The plate was incubated at 37°C for 16 to 24 h. The size of the growth inhibition zone surrounding the holes indicates the sensitivity of the tester organism to the antibiotic at the concentration tested.
Overexpression and purification of His6-tagged SerRS1, SerRS2, and SerRS2 H270G proteins.
Fresh transformants of E. coli BL21 harboring the desired plasmids were grown overnight in LB medium supplemented with antibiotics at 37°C and then diluted 100-fold into fresh LB medium with the antibiotic. The culture was grown at 37°C until the A600 reached ∼0.6, whereupon isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 0.5 mM. The culture was then shifted to 18°C and grown overnight to achieve a high yield of soluble proteins. Purification and analysis of the His6-tagged SerRS2, SerRS2 H270G mutant, and SerRS1 were carried out with Ni-nitriloacetic acid spin columns (Qiagen) according to the manufacturer's protocols (native conditions). Then proteins were dialyzed and concentrated using an Amicon Microcon YM-10 ultrafiltration membrane (nominal molecular weight limit, 10,000). Protein concentrations were determined by the Bio-Rad protein assay (Bio-Rad). The purified proteins were stored at −20°C in 10 mM Tris buffer (pH 7.5) containing 15% glycerol and 10 mM dithiothreitol. SerRS2 and SerRS1 were produced at approximately 5.6 mg and 1.8 mg per liter of culture, respectively. By denaturing protein gel analysis, the purified proteins exhibited the molecular weights expected for the monomers and appeared to be >95% pure. In the following enzymatic kinetic analyses, we used the active-site titration assay to quantitatively assess the active concentration of the purified proteins.
Active-site titration and filter-binding assays.
The active-site concentration was determined by formation of the seryl-AMP-enzyme complex, using a procedure modified from a published protocol (36). aaRS-catalyzed reactions go through a two-step reaction sequence. First, the amino acid and ATP react to form an aminoacyl adenylate intermediate; then, in the second step, the amino acid is transferred to the 3′-OH group (in the case of SerRS) of the 3′-terminal ribose of the tRNA. In the absence of tRNA but the presence of pyrophosphatase (PPiase), the serine-AMP will be trapped in the enzyme and no turnover will occur. When the reaction mixture is passed through a nylon filter (Whatman Protran BA85), the seryl-AMP-enzyme complex will be retained on the filter. By determining the amount of the radioactive serine left on the filter, the active sites in an enzyme preparation can be quantified. Since SB-217452 is a mimic of Ser-AMP and it competes for the binding sites on SerRS1, this assay can be used in the presence of the inhibitor to estimate available active sites in the enzyme. The reaction was performed in a 50-μl volume at 37°C in 100 mM HEPES (pH 7.2), 10 mM KCl, 10 mM MgCl2, 4 mM ATP, 20 μM [14C]serine (∼170 cpm/pmol), 0.25 to 2.3 μM SerRS enzyme, and 5 μl PPiase (Roche). The inhibitor (SB-217452) was added when necessary, the mixture was incubated for 10 min, and then an aliquot was spotted on the filter under vacuum. The filter was immediately washed with 5 ml of a cold buffer containing 100 mM HEPES (pH 7.2), 10 mM KCl, and 10 mM MgCl2. The filter was then dried at 80°C for 25 min, followed by liquid scintillation counting.
ATP-32PPi exchange assay.
The reaction was carried out at 30°C in an 80-μl volume containing 100 mM Na-HEPES (pH 7.2), 30 mM KCl, 10 mM MgCl2, 2 mM NaF, 2 mM ATP, 2.5 mM dithiothreitol, 2 mM 32PPi (∼2 cpm/pmol; PerkinElmer), and 100 to 200 nM enzyme. The concentrations of l-serine ranged from 25 to 1,600 μM. After various incubation times, 18 μl reaction mixture was withdrawn and quenched by adding the mixture to 980 μl of a solution containing 1% charcoal, 5.6% perchloric acid, and 75 mM PPi. The radiolabeled ATP bound to the charcoal was filtered through a Whatman 3MM filter paper under vacuum and washed three times with 5 ml water and once with 5 ml ethanol. The filters were dried at 80°C for 15 min, and the radioactivity was counted by Ecolite (MP Biomedicals) liquid scintillation counting.
Aminoacylation of tRNA and IC50 determination.
Seryl-tRNA formation was measured as trichloroacetic acid (TCA)-precipitable radioactivity in 40-μl reaction mixtures containing 0.1 M HEPES (pH 7.2), 10 mM MgCl2, 30 mM KCl, 4 mM ATP, 1 to 1,200 μM [14C]serine, 4 mg/ml E. coli total tRNA, and 20 to 50 nM enzyme that gave linear initial velocities (Vi). Reactions were carried out at 37°C or 30°C for 1 to 8 min and stopped by adding the mixture to a 5% TCA-soaked 3MM filter paper. The filter paper was washed three times with 5% TCA (5 min each) and once with ethanol (15 min). After the filter paper was dried at 80°C for 15 min, the radioactive Ser-tRNA was counted by liquid scintillation counting and used to calculate the Vi. The steady-state kinetic constants Km and kcat were calculated in KaleidaGraph 3.0 using a Michaelis-Menten equation fit. For IC50 measurements of inhibitors, the procedures and conditions were similar to those described above. [14C]serine was at 8.5 μM (close to its Km) in the SerRS1 reactions and 250 μM (close to its Km) in the SerRS2 reactions. SerRS1 (50 nM) or SerRS2 (20 nM) was added to each reaction mixture. SB-217452 (0.01 μM to 10 μM) was incubated with other components before the enzyme was added. Reactions were carried out at 30°C. The Vi within 4 or 8 min was calculated and normalized to the rate of the control reaction. The percent reduction of Vi was plotted against the concentrations of the inhibitor.
Enzymatic generation and purification of SB-217452.
For the purification of albomycin, we followed the published procedure (7) and used an Agilent 1100 series high-pressure liquid chromatography (HPLC) system. The purified albomycin was eluted from a Phenomenex Spherclone C18-HPLC column at 15 to 20% acetonitrile in water. Starting with 500 μg pure albomycin, a desferric compound was obtained after treatment with 8-hydroxyquinoline (35). The linear peptide was digested by pronase E (final concentration, 1 mg/ml; Sigma) for 2 days at 37°C, and FeCl3 was later added to 1 mM. The reaction mixture was first fractionated using a Biogel P-2 size exclusion column (diameter, 1.2 mm; height, 95 cm) with 20 mM NaCl buffer. Fractions that inhibited the aminoacylation of tRNASer were retained on the column longer than the albomycin fractions. The inhibitory fractions were collected, concentrated with a freeze-dryer, and further separated by HPLC with a polar C18 Ascentis RP-Amide column (4.6 mm by 15 mm; 5-μm particle size; Sigma). The fractions were eluted with two solvents: H2O plus 0.1% trifluoroacetic acid (TFA) (solvent A) and acetonitrile plus 0.1% TFA (solvent B). Proportions of solvent B to solvent A in the mixture at various times were as follows: 0 to 7 min, 0.1% B; 8 to 15 min, 0.1% to 70% B; and 15 to 17 min, 70% B. The flow rate was 1.5 ml/min. Elution was monitored with 306-nm- or 254-nm-wavelength UV light. SB-217452 was isocratically eluted at ∼6 min as a single peak in water with 0.1% TFA. Electrospray ionization-mass spectrometry (ESI-MS) confirmed its identity and showed that the preparation was free of serine and other peptides. The SB-217452 concentration was estimated based on an extinction coefficient (ɛ) at 306 nm of 14,791 mM−1 cm−1.
Nucleotide sequence accession numbers.
The DNA sequences of the Streptomyces serS1 and serS2 genes were deposited in GenBank, National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), with accession numbers GQ465789 and GQ465790, respectively.
RESULTS
Sequence comparison of two SerRS paralogs and expression and purification of the enzymes.
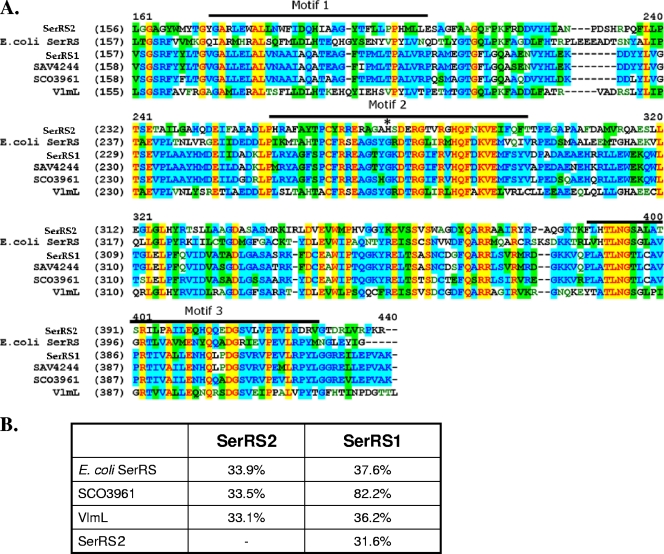
In the recently identified albomycin biosynthetic gene cluster (alb) of Streptomyces arenicola strain ATCC 700974 (38), the alb10 gene encodes a protein showing homology to several bacterial SerRSs, with the highest identity (68%) to an actinomycete SerRS, Salinispora sp. SerRS. Only one serS gene (SCO3961) has been found in the Streptomyces coelicolor genome. This serS gene is highly conserved (>80% homology) in several other Streptomyces species, such as SAV4244 in Streptomyces avermitilis. SCO3961 should encode proteins that are involved in primary metabolism and have housekeeping functions, while alb10 is located in the alb gene cluster and may have been acquired through horizontal gene transfer. The ∼35-kbp DNA region containing alb10 is completely absent in the four Streptomyces genomes in GenBank. This suggests that another, more conserved, serS gene exists in the genome of the albomycin producer in addition to alb10. A series of primers based on the regions surrounding the serS gene in S. coelicolor were used to amplify a PCR product that had about the same length as SCO3961. An open reading frame designated SerRS1 was identified in the DNA sequence and translated into protein for comparison with Alb10 and other bacterial SerRSs by BLAST and sequence alignment. SerRS1 displays a stronger degree of similarity to other housekeeping Streptomyces SerRSs (Fig. 2A). Like the pathway-specific VmlL, Alb10 (named SerRS2 herein) exhibits only minimal identity (∼33%) to other bacterial SerRSs (Fig. 2B). Both SerRS1 and SerRS2 contain three motifs that are characteristic of class II aaRSs, including all SerRSs. Motif 3 seems most conserved, while motif 1 is more divergent between a Streptomyces housekeeping SerRS and E. coli SerRSs (Fig. 2A). In motif 2, where the protein interacts with its cognate tRNA, all the compared SerRSs except SerRS2 are similar. A SerRS2 H270G mutant was generated to better characterize the enzyme's activity (see below). The corresponding genes were cloned into pET vectors for overexpression as C-terminal His6-tagged proteins in E. coli BL21. In conclusion, two SerRS paralogs were identified in the Streptomyces sp. strain ATCC 700974 genome. We then investigated whether they are both biochemically functional and if their low sequence homology correlates with functional divergence.
FIG. 2.
Sequence alignment of bacterial SerRSs (A) and comparison of amino acid identities between select enzymes (B). (A) The SerRS amino acid sequences are partially shown. Numbers in parentheses indicate the position of the beginning amino acid residue in each protein sequence. Numbers above the alignment are the positions of the consensus sequences. Conserved class II motifs 1, 2, and 3 are labeled. The histidine residue marked with an asterisk (*) in SerRS2 was mutated to a glycine to make the H270G mutant. SerRS1 and SerRS2 are discussed in the text. SAV4244 is from Streptomyces avermitilis (GenBank accession no. NP_825421). SCO3961 is from Streptomcyes coelicolor (NP_628145). VlmL is from Streptomyces viridifaciens (AAN10249). Escherichia coli SerRS is under accession no. AAC73979.
The two SerRS paralogs display divergence in the binding of serine.
The ATP-pyrophosphate (PPi) exchange assay was first used to confirm that SerRS1 and SerRS2 can exchange the radiolabel from 32PPi to ATP in the presence of serine and the absence of tRNA substrate. PPi exchange measures the reversible formation of the seryl-adenylate with serine and ATP as substrates. With an excess of ATP, the kinetic parameters of SerRS1 for serine obtained in this assay are a kcat of 420 ± 30 min−1 and a Km of 400 ± 37 μM; for SerRS2, a kcat of 106 ± 30 min−1 and a Km of 260 ± 9 μM. In the aminoacylation assay, since all tRNASer species possess a long variable arm and it has been shown that bacterial SerRSs can charge heterologous tRNASer, E. coli tRNASer from total tRNA was used in the reaction as a substrate. The kinetic parameters are displayed in Table 2 and are compared with the data for yeast SerRS with serine in charging its cognate tRNASer. In summary, while the catalytic efficiencies (kcat/Km) of SerRS1 and SerRS2 are generally comparable in these two assays, there are a number of differences between the two enzymes. Increasing the temperature of the aminoacylation reaction mixtures from 30°C to 37°C increased the tRNA charging efficiency of SerRS1 by approximately threefold but had a negligible effect on SerRS2. When the Kms of the assays were compared, the affinity of serine to SerRS1 was lowered dramatically (∼40-fold) in the presence of tRNA but that of serine to SerRS2 remained almost unchanged. The most striking difference is that SerRS2 required at least 20 times more serine to reach its maximum velocity in the aminoacylation assay, which was offset by the much higher turnover rate (kcat) of SerRS2 compared to SerRS1. The aminoacylation Km of SerRS1 for serine is close to those of other Streptomyces housekeeping aaRS (e.g., the Km of TrpRS1 for tryptophan in Streptomyces coelicolor is ∼3.7 μM) but significantly lower than that of yeast SerRS (∼63 μM) and E. coli SerRS (∼110 μM). The SerRS2 H270G mutant was ninefold less efficient than the wild type, which suggests that the integrity of motif 2 is important to the function of this SerRS.
TABLE 2.
Kinetic parameters of SerRS1, SerRS2 from Streptomyces sp. strain ATCC 700974, the SerRS2 H270G mutant, and yeast SerRSa obtained in aminoacylation assay
| SerRS and assay temp | Km (μM) | kcat (min−1) | kcat/Km(min−1 μM−1) |
|---|---|---|---|
| SerRS1 | |||
| 37°C | 8.6 ± 0.5 | 7.5 ± 1.1 | 0.9 ± 0.07 |
| 30°C | 12 ± 0.4 | 3.4 ± 0.7 | 0.3 ± 0.05 |
| SerRS2 (30°C) | 240 ± 65 | 82 ± 55 | 0.3 ± 0.08b |
| SerRS2 H270G mutant (30°C) | 410 | 11 | 0.03c |
| Yeast SerRS (30°C) | 63.00 | 58.20 | 0.92 |
Yeast data taken from a report by Lenhard et al. (16).
At 37°C, kcat/Km = 0.4 ± 0.07 min−1 μM−1.
At 37°C, kcat/Km = 0.10 min−1 μM−1.
Examination of the activity of SerRS1 and SerRS2 in vivo.
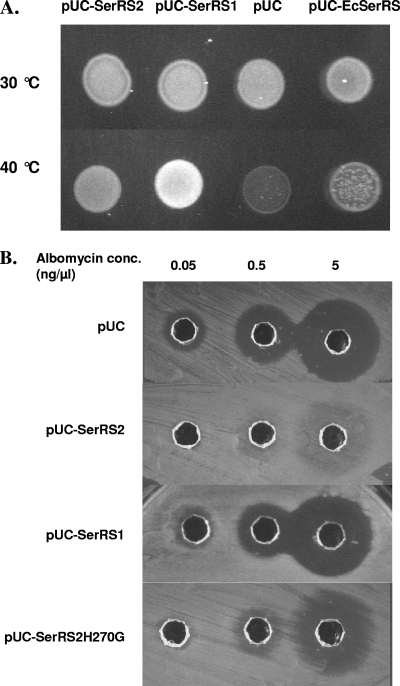
Genetic complementation was used to test the in vivo activities of SerRS1 and SerRS2. The host strain was an E. coli mutant (K28) that is ineffective at making its own serS gene product at high temperature (5, 33). serS1 and serS2 were separately cloned into a high-copy vector, pUC19, under the control of a constitutive glnS′ promoter, which is a glnS mutant promoter used for aaRS expression in E. coli (27), and the resulting plasmids were transformed into E. coli K28. At the permissive temperature (30°C), strains carrying different plasmids grew at similar rates (Fig. 3A). At the nonpermissive temperature (40°C), cells producing SerRS1 were able to grow to a high density while cells containing SerRS2 or empty vector had weaker or no growth (Fig. 3A). This suggests that SerRS1 could charge E. coli tRNASer in vivo and genetically complement the loss of E. coli SerRS while SerRS2 was less competent at the high temperature. A plasmid containing the wild-type E. coli serS with its upstream and downstream ∼150-bp regions did not seem to support healthy growth of the cells at 40°C, suggesting that overexpression of serS in its native host generated some toxicity under these conditions.
FIG. 3.
In vivo activities of Streptomyces SerRS1 and SerRS2. serS1, serS2, E. coli serS, and the serS2(H270G) mutant were cloned into pUC19 (pUC) vector under the control of the glnS′ promoter (25). (A) The plasmids were transformed into an E. coli temperature-sensitive (ts) mutant strain K28 that cannot produce chromosomal SerRS at 40°C. At 30°C, the chromosomal SerRS functions normally (pUC control). The wild-type E. coli serS gene in the pUC vector was able to complement the ts mutation but shows some toxicity when highly expressed at 40°C. serRS1 and serRS2 could also genetically complement the mutant and enable growth at the higher temperature. (B) The plasmids indicated were transformed into wild-type E. coli JM109. The transformants were used as test organisms in an agar diffusion assay with albomycin dilutions (see Materials and Methods). The size of the inhibition zone indicates the susceptibility of the transformants.
We used a wild-type E. coli strain (JM109) for testing albomycin susceptibility when the SerRS-encoding plasmids were present. The transformants were used as tester organisms in an agar diffusion assay for measuring susceptibility to the purified albomycin (Fig. 3B). When SerRS2 was overproduced, it increased the albomycin resistance level at least 100- to 1,000-fold. To confirm the role of SerRS2 and identify the resistance determinant at the amino acid level, we introduced mutations into the serS2 gene. In the cocrystal structure of Thermus thermophilus SerRS, a seryl-adenylate analog (Ser-AMS) apparently has substantial interaction with the amino acid residues of motif 2, a structural motif characterizing class II aaRSs (8, 16). This motif is relatively well conserved among the Streptomyces SerRSs, including VmlL, except for SerRS2 (Fig. 2). We focused on the G268-to-S271 region of SerRS2 that is a loop linked by two conserved β sheets in the catalytic core. A single amino acid change, H270G, reduced the albomycin resistance of the cells carrying the mutant to 0.5 ng/μl (∼0.5 μM) (Fig. 3B). Although the SerRS2 H270G mutant is still active, it maintains only one-ninth the aminoacylation activity of the wild type (see above). From the crystal structure, this residue does not seem to interact with Ser-AMS. It is unclear how SerRS2 folds differently from the T. thermophilus SerRSs model in the local structure. At the very least, the resistance of SerRS2 does not result from mutations of SerRS1 in this region. In summary, serS2 encodes a protein with the biochemical properties necessary to convey antibiotic self-resistance in the albomycin-producing Streptomyces strain. SerRS2 His270 in motif 2 participates in binding the substrates and possibly discriminates against the SB-217452 inhibitor. More-detailed analyses of the resistance determinants in SerRS2 and delineation of the binding of the tRNA, substrates, or inhibitor to SerRS2 at motif 2 now require structural investigation.
Inhibition analysis of SerRS1 and SerRS2 with enzymatically generated SB-217452 and serine hydroxamate (SHX).
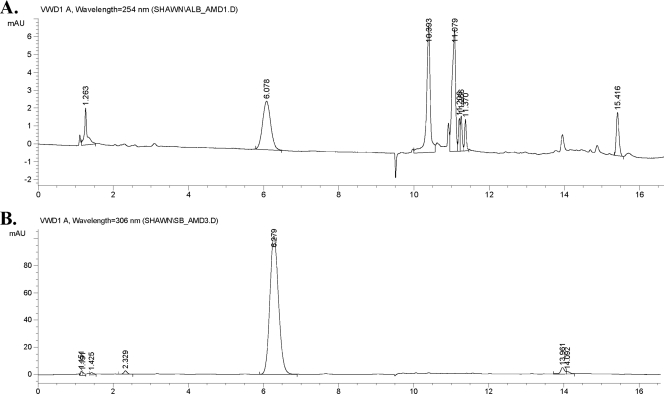
SB-217452 could be isolated from large-scale fermentations of Streptomyces as a by-product of albomycin biosynthesis (31). It could also be enzymatically generated by digestion of albomycin with many peptidases in vitro (2). Pronase E, a mixture of at least three proteolytic activities, including an extracellular serine protease, was shown to degrade albomycin to SB-217452 as the only UV-detectable product. No other structure related to albomycin could be isolated. We utilized this approach starting with the desferric compound from 500 μg pure albomycin. After incubation with pronase E for 3 days, Fe3+ was added back into the reaction mixture to reconstitute albomycin from the undigested peptidyl nucleoside. The mixture was first fractionated by size exclusion chromatography (Biogel P-2). Albomycin was tracked by its bioactivity, and compounds inhibiting the charging of tRNASer were monitored by the aminoacylation assay. Assays of fractions during purification showed that the inhibition activity lagged behind albomycin by 1/10 of the bed volume. The concentrated inhibiting fractions were further separated on a reverse-phase HPLC column monitored by UV absorbance (Fig. 4). SB-217452 was eluted as a symmetric peak at ∼6.1 min, and ESI-MS confirmed that it was the major product and that the sample was devoid of serine. The final yield of SB-217452 was estimated to be ∼10 μg (2% recovery). Desferri-albomycin and SB-217452 did not show any bioactivity against E. coli whole cells, confirming that the hydroxamate-ferric complex structure is essential for the activity of albomycin.
FIG. 4.
Reverse-phase HPLC profile of SB-217452. The column is an Ascentis RP-Amide HPLC column (4.6 mm by 15 mm, 5-μm particle size). The separation conditions are described in Materials and Methods. SB-214752 was eluted at ∼6.1 min, and albomycin was eluted at ∼10.4 min. (A) Digestion reaction mixture of pure albomycin with pronase E. (B) After size exclusion chromatography, the concentrated active fractions (as described in the text) were further separated by HPLC. The peak at ∼6.1 min was collected as SB-217452 and confirmed by ESI-MS; m/z = 477.14 [M + H]+ or 475.15 [M − H]−.
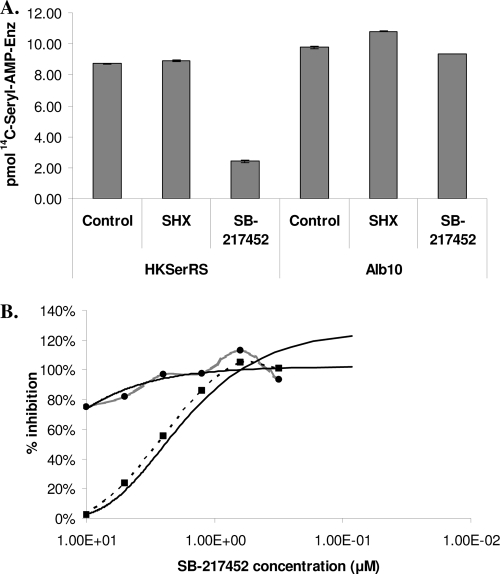
We then analyzed the inhibition of SerRS1 and SerRS2 with the purified SB-217452 and SHX. The active-site titration assay is essentially a filter-binding assay that measures the amount of radioactive seryl-AMP-enzyme complex retained on a nylon membrane filter after extensive washing. In the presence of a competitive inhibitor, the reaction will produce less of the complex than the control reaction with the same amount of enzyme. SHX is a known bacterial SerRS inhibitor with an inhibition constant (Ki) of 30 μM for E. coli SerRS (32) and 2.7 mM for Saccharomyces cerevisiae SerRS (33). In this experiment, SHX did not have any inhibitory effect on the binding of serine to the Streptomyces SerRSs (Fig. 5A) at a 1 mM concentration. We further tested SHX in the aminoacylation assay with SerRSs at lower concentrations (∼10 to 50 nM). SHX at concentrations up to 500 μM did not change the initial velocity of the reaction (data not shown). In contrast, 2 μM SB-217452 significantly inhibited the formation of the seryl-AMP-SerRS1 complex but had no effect on the seryl-AMP-SerRS2 complex (Fig. 5A). The inhibitory activity of this compound was also analyzed by the aminoacylation assay with SerRS1 at 50 nM and SerRS2 at 20 nM. The IC50 of SB-217452 against SerRS1 was estimated to be ∼1 to 2 μM (Fig. 5B). SerRS2 was not significantly inhibited at the concentrations tested, which agrees with our previous finding that serS2 (alb10) is the albomycin resistance gene. In summary, SB-217452 is an effective competitive inhibitor of the Streptomyces housekeeping SerRS.
FIG. 5.
In vitro inhibition of the two SerRSs. (A) Filter-binding assay of the inhibition of SerRS1 and SerRS2 with SHX and SB-217452. In a 50-μl reaction mixture, SerRS1 was at 230 nM and SerRS2 at 200 nM. SHX was added to 1 mM and SB-217452 was added to an ∼2 μM final concentration. (B) IC50 determination. The SB-217452 inhibitor concentration is shown in logarithmic scale. About 2 μM inhibitor was required in the assay (see Materials and Methods) with SerRS1 to achieve its 50% inhibitory activity. The assay was repeated three times, and a representative set of data is shown. Dashed line with squares, assay with SerRS1; gray line with circles, assay with SerRS2; solid lines, predicted trend lines.
DISCUSSION
Genomes of Streptomyces species, high-GC gram-positive bacteria, harbor many duplicated aaRS genes that would otherwise be essential when presented as single-copy genes in pathogenic gram-negative and most low-GC gram-positive bacteria. These genes may have physiological roles in dealing with harsh environments or be directly involved in biosynthetic pathways given the huge metabolic potential of this group of bacteria. For example, several structurally complex natural products from actinomycetes are potent aaRS inhibitors, and functional genomic analyses of the corresponding strains can offer a means to explore the relationship between duplicated aaRSs and identify antibiotic biosynthetic gene clusters (12, 29, 30). The identification of two serS genes in the antibiotic albomycin producer Streptomyces sp. strain ATCC 700974 provided a means to explore the relationship between aaRS duplication and antibiotic biosynthesis metabolism. Comparison of the two serS genes we identified revealed sequence and structural differences consistent with significant functional divergence between the two corresponding enzymes, SerRS1 and SerRS2. SerRS1 has high sequence homology to other Streptomyces housekeeping SerRSs, and its kinetic parameters are similar to those of other Streptomyces aaRSs involved in primary metabolism. Upon the binding of tRNASer, the affinity of serine to SerRS1 is dramatically increased. serS2 is the alb pathway-specific gene that was likely acquired through horizontal gene transfer. Seven genes in the alb cluster have been deleted in the wild-type strain, and the resulting mutants do not produce albomycin or albomycin analogs (S. Chen, unpublished data). This is consistent with the finding here that heterologous expression of serS2 confers albomycin resistance to the host cells, supporting the proposal that the ∼35-kb DNA sequences we cloned are indeed the albomycin biosynthetic genes, as antibiotic resistance genes are often clustered with the corresponding biosynthetic genes. This does not rule out an additional role for SerRS2, such as generating Ser-tRNASer for albomycin biosynthesis, since the source of the carrier molecule that provides serine during antibiotic peptide bond formation has not yet been identified.
Although SerRS1 and SerRS2 are comparable in their overall catalytic efficiencies, the aminoacylation Km of serine for SerRS2 is ∼20 times higher than that for SerRS1. Most genes involved in antibiotic biosynthesis in Streptomyces were likely acquired through late evolutionary events, and biosynthetic processes are often initiated in the nutrient-depleted growth phase, when the genes in biosynthetic operons are turned on by pathway-specific regulators. Albomycin activity was detectable only at 40 hours after inoculation in a fermentation study (7). Thus, the expression of the biosynthetic enzymes is apparently regulated. It would be interesting to investigate if serS2 is expressed when serS1 is also expressed, as well as to determine the ratio of the two proteins when albomycin is produced. How SerRS2 interacts with other Alb biosynthetic proteins, perhaps in an enzymatic complex, remains an intriguing question. As the affinity of serine to SerRS2 is much lower than that to SerRS1, the cells could use these as regulatory mechanisms to control the amount of albomycin produced under different growth conditions. An improved understanding of the physiological constraints on antibiotic production may have important practical implications. The lack of high-titer strains for industrial-scale production was a deciding factor in the decision to abandon the development of albomycin for clinical use (24). An improved understanding of the Streptomyces self-resistance mechanism and the related Alb biosynthetic enzymes now provides a basis for possible future improvements in albomycin production.
From an evolutionary perspective, high-GC gram-positive bacteria form a phylogenetic clade distinct from most human pathogens that are low-GC gram-positive or gram-negative bacteria. However, Streptomyces species and the pathogen Mycobacterium tuberculosis are associated in the order Actinomycetales. There are several examples of enzymes involved in primary metabolism in actinomycetes that have a molecular structure different from those of their counterparts in other bacteria such as E. coli (28). Such enzymes are potentially attractive as antimycobacterial drug targets (1). In the case of SerRS, Mycobacterium tuberculosis has only one serS gene in its genome, and the similarity of Mycobacterium SerRS to SerRS1 is 72%, compared to 50% between Mycobacterium and E. coli SerRSs. Given that Streptomyces housekeeping SerRS1 and E. coli SerRS display differences in SHX discrimination, it is tempting to speculate that Mycobacterium SerRS might also show exploitable specifics in the recognition of serine substrate and reaction intermediate mimics such as albomycin. The efficacy of albomycin in treating mycobacterial infection has never been studied, and Mycobacterium SerRS could be another attractive antimycobacterial drug target for designing genus-specific enzyme inhibitors. Future studies will now focus on attempting to test whether SB-217452 can inhibit Mycobacterium SerRS in vitro and in cell-based assays.
Acknowledgments
We thank Zhixin Miao, Shannon Cook, Hao Chen, and Glen Jackson in the Department of Chemistry at Ohio University for providing ESI-MS analysis.
The research in the laboratory of S.C. is currently supported by a start-up fund from Ohio University. Work on antibiotic resistance in the laboratory of M.I. is supported by NIGMS (grant no. GM65183).
Footnotes
Published ahead of print on 31 August 2009.
REFERENCES
- 1.Alhamadsheh, M. M., F. N. Musayev, A. A. Komissarov, S. Sachdeva, H. T. Wright, J. N. Scarsdale, G. Florova, and K. A. Reynolds. 2007. Alkyl-CoA disulfides as inhibitors and mechanistic probes for FabH enzymes. Chem. Biol. 14:513-524. [DOI] [PubMed] [Google Scholar]
- 2.Benz, G. 1984. Enzymatische Spaltung der Desferriform der Albomycine δ1, δ2. Liebigs Ann. Chem. 8:1399-1407. [Google Scholar]
- 3.Benz, G., T. Schroder, J. Kurz, C. Wunsche, W. Karl, G. Steffens, J. Pfitzner, and D. Schmidt. 1982. Constitution of the deferriform of the albomycin δ1, δ2, and ɛ. Angew. Chem. Int. Ed. Engl. 21:527-528. [Google Scholar]
- 4.Braun, V., K. Günthner, K. Hantke, and L. Zimmermann. 1983. Intracellular activation of albomycin in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 156:308-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, S. J., B. Low, and W. Konigsberg. 1973. Isolation and characterization of a regulatory mutant of an aminoacyl-transfer ribonucleic acid synthetase in Escherichia coli K-12. J. Bacteriol. 113:1096-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Sayed, A. K., J. Hothersall, S. M. Cooper, E. Stephens, T. J. Simpson, and C. M. Thomas. 2003. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 10:419-430. [DOI] [PubMed] [Google Scholar]
- 7.Fiedler, H. P., F. Walz, A. Dohle, and H. Zahner. 1985. Albomycin: studies on fermentation, isolation and quantitative determination. Appl. Microbiol. Biotechnol. 21:341-347. [Google Scholar]
- 8.Fujinaga, M., C. Berthet-Colominas, A. D. Yaremchuk, M. A. Tukalo, and S. Cusack. 1993. Refined crystal structure of the seryl-tRNA synthetase from Thermus thermophilus at 2.5 Å resolution. J. Mol. Biol. 234:222-233. [DOI] [PubMed] [Google Scholar]
- 9.Garg, R. P., J. M. Gonzalez, and R. J. Parry. 2006. Biochemical characterization of VlmL, a seryl-tRNA synthetase encoded by the valanimycin biosynthetic gene cluster. J. Biol. Chem. 281:26785-26791. [DOI] [PubMed] [Google Scholar]
- 10.Garg, R. P., X. L. Qian, L. B. Alemany, S. Moran, and R. J. Parry. 2008. Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA. Proc. Natl. Acad. Sci. USA 105:6543-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmann, A., H.-P. Fiedler, and V. Braun. 1979. Uptake and conversion of the antibiotic albomycin by Escherichia coli K-12. Eur. J. Biochem. 99:517-524. [DOI] [PubMed] [Google Scholar]
- 12.Hornemann, U., L. H. Hurley, M. K. Speedie, and H. G. Floss. 1971. The biosynthesis of indolmycin. J. Am. Chem. Soc. 93:3028-3035. [DOI] [PubMed] [Google Scholar]
- 13.Ibba, M., and D. Söll. 2000. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69:617-650. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J.-G., B. K. Park, S.-U. Kim, D. Choi, B. H. Nahm, J. S. Moon, J. S. Reader, S. K. Farrand, and I. Hwang. 2006. Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc. Natl. Acad. Sci. USA 103:8846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitabatake, M., K. Ali, A. Demain, K. Sakamoto, S. Yokoyama, and D. Söll. 2002. Indolmycin resistance of Streptomyces coelicolor A3(2) by induced expression of one of its two tryptophanyl-tRNA synthetases. J. Biol. Chem. 277:23882-23887. [DOI] [PubMed] [Google Scholar]
- 16.Lenhard, B., S. Filipic, I. Landeka, I. Skrtic, D. Soll, and I. Weygand-Durasevic. 1997. Defining the active site of yeast seryl-tRNA synthetase. Mutations in motif 2 loop residues affect tRNA-dependent amino acid recognition. J. Biol. Chem. 272:1136-1141. [DOI] [PubMed] [Google Scholar]
- 17.Metlitskaya, A., T. Kazakov, A. Kommer, O. Pavlova, M. Praetorius-Ibba, M. Ibba, I. Krasheninnikov, V. Kolb, I. Khmel, and K. Severinov. 2006. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic Microcin C. J. Biol. Chem. 281:18033-18042. [DOI] [PubMed] [Google Scholar]
- 18.Ochsner, U. A., X. Sun, T. Jarvis, I. Critchley, and N. Janjic. 2007. Aminoacyl-tRNA synthetases: essential and still promising targets for new anti-infective agents. Exp. Opin. Investig. Drugs 16:573-593. [DOI] [PubMed] [Google Scholar]
- 19.O'Donoghue, P., and Z. Luthey-Schulten. 2003. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol. Mol. Biol. Rev. 67:550-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olano, C., B. Wilkinson, C. Sánchez, S. J. Moss, R. Sheridan, V. Math, A. J. Weston, A. F. Braña, C. J. Martin, M. Oliynyk, C. Méndez, P. F. Leadlay, and J. A. Salas. 2004. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: cluster analysis and assignment of functions. Chem. Biol. 11:87-97. [DOI] [PubMed] [Google Scholar]
- 21.Oliynyk, M., M. Samborskyy, J. B. Lester, T. Mironenko, N. Scott, S. Dickens, S. F. Haydock, and P. F. Leadlay. 2007. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 25:447-453. [DOI] [PubMed] [Google Scholar]
- 22.Payne, D. J., M. N. Gwynn, D. J. Holmes, and D. L. Pompliano. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 23.Pramanik, A., and V. Braun. 2006. Albomycin uptake via a ferric hydroxamate transport system of Streptococcus pneumoniae R6. J. Bacteriol. 188:3878-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pramanik, A., U. H. Stroeher, J. Krejci, A. J. Standish, E. Bohn, J. C. Paton, I. B. Autenrieth, and V. Braun. 2007. Albomycin is an effective antibiotic, as exemplified with Yersinia enterocolitica and Streptococcus pneumoniae. Int. J. Med. Microbiol. 297:459-469. [DOI] [PubMed] [Google Scholar]
- 25.RajBhandary, U. L., and D. Söll. 2008. Aminoacyl-tRNAs, the bacterial cell envelope, and antibiotics. Proc. Natl. Acad. Sci. USA 105:5285-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rock, F. L., W. Mao, A. Yaremchuk, M. Tukalo, T. Crepin, H. Zhou, Y.-K. Zhang, V. Hernandez, T. Akama, S. J. Baker, J. J. Plattner, L. Shapiro, S. A. Martinis, S. J. Benkovic, S. Cusack, and M. R. K. Alley. 2007. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316:1759-1761. [DOI] [PubMed] [Google Scholar]
- 27.Ryu, Y., and P. G. Schultz. 2006. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods 3:263-265. [DOI] [PubMed] [Google Scholar]
- 28.Scarsdale, J. N., G. Kazanina, X. He, K. A. Reynolds, and H. T. Wright. 2001. Crystal structure of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III. J. Biol. Chem. 276:20516-20522. [DOI] [PubMed] [Google Scholar]
- 29.Stefanska, A. L., R. Cassels, S. J. Ready, and S. R. Warr. 2000. SB-203207 and SB-203208, two novel isoleucyl tRNA synthetase inhibitors from a Streptomyces sp. I. Fermentation, isolation and properties. J. Antibiot. (Tokyo) 53:357-363. [DOI] [PubMed] [Google Scholar]
- 30.Stefanska, A. L., N. J. Coates, L. M. Mensah, A. J. Pope, S. J. Ready, and S. R. Warr. 2000. SB-219383, a novel tyrosyl tRNA synthetase inhibitor from a Micromonospora sp. I. Fermentation, isolation and properties. J. Antibiot. (Tokyo) 53:345-350. [DOI] [PubMed] [Google Scholar]
- 31.Stefanska, A. L., M. Fulston, C. S. V. Houge-Frydrych, J. J. Jones, and S. R. Warr. 2000. A potent seryl tRNA synthetase inhibitor SB-217452 isolated from a Streptomyces species. J. Antibiot. 53:1346-1353. [DOI] [PubMed] [Google Scholar]
- 32.Tosa, T., and L. I. Pizer. 1971. Biochemical bases for the antimetabolite action of l-serine hydroxamate. J. Bacteriol. 106:972-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weygand-Durasevic, I., N. Ban, D. Jahn, and D. Söll. 1993. Yeast seryl-tRNA synthetase expressed in Escherichia coli recognizes bacterial serine-specific tRNAs in vivo. Eur. J. Biochem. 214:869-877. [DOI] [PubMed] [Google Scholar]
- 34.Weygand-Durasevic, I., and S. Cusack. 2005. Seryl-tRNA synthetases, p. 177-192. In M. Ibba, C. Francklyn, and S. Cusack (ed.), The aminoacyl-tRNA synthetases. Landes Bioscience, Austin, TX.
- 35.Wiebe, C., and G. Winkelmann. 1975. Kinetic studies on the specificity of chelate-iron uptake in Aspergillus. J. Bacteriol. 123:837-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson, A. J., A. R. Fersht, D. M. Blow, and G. Winter. 1983. Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthetase cysteine-35 to glycine-35 mutation. Biochemistry 22:3581-3586. [DOI] [PubMed] [Google Scholar]
- 37.Yanagisawa, T., and M. Kawakami. 2003. How does Pseudomonas fluorescens avoid suicide from its antibiotic pseudomonic acid? J. Biol. Chem. 278:25887-25894. [DOI] [PubMed] [Google Scholar]
- 38.Zeng, Y., and S. Chen. 2008. Biosynthesis of albomycin—a Trojan horse antibiotic, abstr. p. 10. 7th U.S.-Japan Seminar on the Biosynthesis of Natural Products: Enzymology, Structural Biology, and Drug Discovery, La Jolla, CA.