Abstract
Etravirine (ETR) is a second-generation nonnucleoside reverse transcriptase (RT) inhibitor (NNRTI) active against common human immunodeficiency virus type 1 (HIV-1) drug-resistant strains. This study was designed to determine the extent to which each of the Y181C or G190A mutations in RT might confer resistance to ETR and other members of the NNRTI family of drugs. Recombinant HIV-1 RT enzymes containing either the Y181C or the G190A mutation, or both mutations in tandem, were purified. Both RNA- and DNA-dependent DNA polymerase assays were performed in order to determine the extent to which each of these mutations might confer resistance in cell-free biochemical assays against each of ETR, efavirenz, and nevirapine. Both the biochemical and the cell-based phenotypic assays confirmed the susceptibility of G190A-containing enzymes and viruses to ETR. The results of this study indicate that the G190A mutation is not associated with resistance to ETR.
Etravirine (ETR; formerly known as TMC-125) is a second-generation nonnucleoside reverse transcriptase (RT) inhibitor (NNRTI) that retains activity against human immunodeficiency virus type 1 (HIV-1) variants containing common resistance mutations conferring resistance to nevirapine (NVP). In particular, ETR is a highly flexible diarylpyrimidine compound able to adapt its orientation and overcome common NNRTI-associated mutations, including K103N, which is present in 40 to 60% of patients failing NNRTI-containing treatment regimens (10, 17). Furthermore, ETR displays a high genetic barrier to resistance, requiring the accumulation of several NNRTI-associated mutations for high-level resistance to become manifest (10, 14, 17, 18).
The DUET-1 and DUET-2 clinical trials have identified an array of 17 resistance-associated mutations (RAMs) that confer diminished sensitivity to ETR, including V90I, A98G, L100I, K101E/H/P, V106I, E138A, V179D/F/T, Y181C/I/V, G190A/S, and M230L (3, 4). Many of these mutations may represent preexisting resistance in NNRTI-experienced patients, with Y181C-G190A (27%) and K101E-G190A (12.5%) being the most prevalent RAM combinations (14, 17, 18).
Further studies are needed to determine the individual and interactive roles of RAMs in conferring diminished sensitivity to ETR, as is evident from poor concordance among different genotypic interpretative algorithms in regard to the role that individual mutations play (1, 5, 6, 15). It is also important to accurately describe the mutations tolerated by ETR, i.e., mutations that result in no effect, reduced efficacy, or a lack of a virological response.
To determine the role of G190A in ETR resistance, we expressed purified recombinant RT enzymes containing this mutation alone or in tandem with Y181C. The latter mutation was chosen because of previous reports that it confers resistance to each of NVP, efavirenz (EFV), and, to a much lesser extent, ETR (15). As an additional control, we also studied dapivirine (DAP; formerly known as TMC-120), a compound that Tibotec Pharmaceuticals has licensed to the International Partnership for Microbicides for possible development as a vaginal microbicide.
MATERIALS AND METHODS
Site-directed mutagenesis.
The G190A, Y181C, and G190A plus Y181C mutations were introduced into the subtype B HIV-1 RT heterodimer expression plasmid pRT6H_PROT, kindly provided by S. F. LeGrice (12), by using a QuikChange II XL site-directed mutagenesis kit (Stratagene). DNA sequencing was performed in both directions across the entire RT-encoding region to verify the absence of spurious mutations and the presence of the desired mutation.
Purification of recombinant HIV-1 RTs and activity determination.
Recombinant wild-type (WT) and mutated RTs were expressed and purified as described previously (11). The protein concentration was measured by use of a Bradford protein assay kit (Bio-Rad Laboratories), and the purities of the recombinant RT preparations were verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Quantification of RT DNA polymerase activity was performed as described previously (16). An active unit of RT was defined as the amount of enzyme that incorporates 1 pmol of dTTP in 10 min at 37°C.
NNRTI inhibition of RDDP activity.
Reactions to determine NNRTI inhibition of RNA-dependent DNA polymerase (RDDP) activity were performed as reported previously (13, 16, 20). Briefly, RT reaction buffer containing 50 mM Tris (pH 7.8), 5 mM MgC12, 60 mM KCl, 10 mM dithiothreitol, 10 μM of dTTP with 2.5 μCi of [3H]dTTP (70 to 90 mCi/mM), 5 U of template/primer poly(rA)/oligo(dT)12-18 (Amersham), 5 U of recombinant RTs, and various amounts of the RT inhibitors (NVP, EFV, ETR, and DAP) were included in 50-μl reaction volumes, which were incubated at 37°C for 30 min. The reactions were terminated with 0.2 ml of 10% cold trichloroacetic acid (TCA) and 20 mM sodium pyrophosphate. After 30 min on ice, the precipitated products were filtered onto a 96-well plate with glass fiber filters (Millipore) and sequentially washed with 10% TCA and 95% ethanol. The radioactivity of the incorporated products was analyzed by liquid scintillation spectrometry. The 50% inhibitory concentration (IC50) of each NNRTI was determined by curve fit analysis with GraphPad Prism 4.0 software, version 7.
NNRTI inhibition of DNA-dependent DNA polymerase (DDDP) activity.
The primer-template (ppt18-ppt57) substrates used to study the inhibition of DNA synthesis by NNRTIs were derived from the polypurine tract of the HIV-1 genome. The ppt18 primer was radiolabeled at its 5′ end with [γ-32P]ATP and annealed to the ppt57 template as described previously (8). Catalysis by the WT and mutant RT enzymes was determined by measuring the extension of the labeled ppt18 primer on the ppt57 template. Each of the NNRTI compounds to be evaluated, including NVP, EFV, ETR, and DAP, was serially diluted in 50% dimethyl sulfoxide (DMSO). The reaction mixtures contained 150 nM labeled primer-template (calculated as the primer concentration), 5 U of recombinant RTs, 50 mM Tris (pH 7.8), 5 mM MgC12, 60 mM KCl, 10 mM dithiothreitol, and 5% DMSO in a total volume of 20 μl. Initiation of the reaction was performed by adding 100 μM of each of the deoxynucleoside triphosphates, excluding dATP, so that primer extension would be restricted to 4 nucleotides for better resolution and quantification. After 15 min at 37°C, an equal volume of formamide sample buffer was added and the heat-denatured samples were resolved in a 6% polyacrylamide-7 M urea gel, followed by phosphorimaging. Band intensities were analyzed with ImageQuant software (GE Healthcare). The IC50 of each NNRTI was determined with GraphPad Prism 4.0 software from plots of the percentages of the level of primer extension relative to the inhibitor concentration.
Phenotypic analysis of NNRTI drug susceptibilities of G190A-containing viruses in vitro.
Three WT viruses and three viruses harboring G190A and A98S, obtained from our primary HIV infection cohort, were amplified as described previously (3, 5). Drug susceptibility was measured in cell culture-based phenotypic assays to determine the extent to which the NNRTI drugs inhibited HIV replication in vitro. Briefly, cord blood mononuclear cells were infected with different viral isolates; and the 50% drug effective concentrations (EC50s) of each of NVP, EFV, ETR, and DAP were ascertained by monitoring the cells for the production of the p24 antigen, as described previously (3).
RESULTS
Purification of recombinant HIV-1 RT.
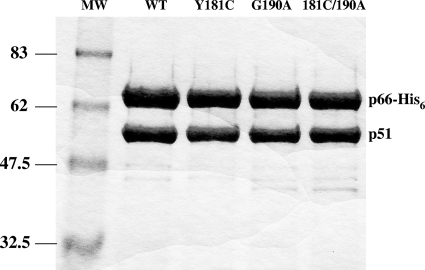
Recombinant WT heterodimeric (p66/p51) RT and RT enzymes containing the G190A, the Y181C, and both the G190A plus Y181C substitutions were purified to >95% homogeneity; the RT subunits p66 and p51 were processed to similar molar ratios, as determined by SDS-PAGE analysis (Fig. 1). The mutations introduced into the recombinant HIV-1 RT did not interfere with either heterodimer formation or enzyme purification.
FIG. 1.
Coomassie brilliant blue staining of purified heterodimer RTs after purification on 8% SDS-polyacrylamide gels. Lanes MW, molecular mass standards (in kilodaltons). The lanes with the purified recombinant RT heterodimers (WT, G190A, Y181C, or both G190A and Y181C) are indicated.
Inhibitory effects of NNRTIs determined by RDDP and DDDP assays.
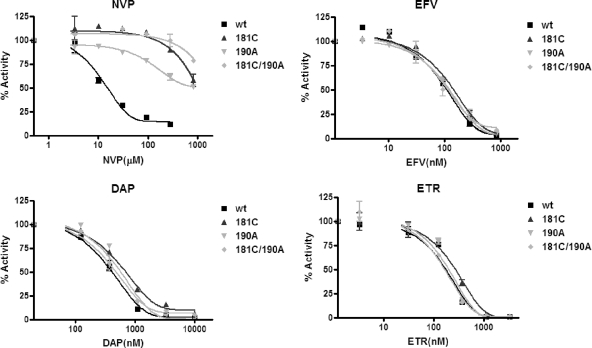
The inhibitory effects of various NNRTIs on the RDDP activity of the HIV-1 WT and mutant RTs were measured by a filtration RT assay. The sensitivities of the WT and the mutant RTs to each of NVP, EFV, ETR, and DAP were determined. The results presented in Fig. 2 show that each of the mutated RTs displayed high-level resistance to NVP but that all of the enzymes remained fully susceptible to each of EFV, DAP, and ETR. The IC50s of each drug tested with each of the RTs are shown in Table 1.
FIG. 2.
Inhibition of RNA-dependent DNA polymerase RT activity by NNRTIs determined by filtration RT assay. The inhibition of WT and mutant RT activity was determined in the absence and the presence of various amounts of NVP, EFV, DAP, or ETR.
TABLE 1.
IC50s of recombinant RTs for NNRTIs by RDDP filter-based assay and DDDP gel-based assay
| Assay and RT mutation | Drug susceptibility (IC50)a |
|||
|---|---|---|---|---|
| ETR (nM) | DAP (nM) | EFV (nM) | NVP (μM) | |
| RDDP | ||||
| WT | 175 ± 32 | 370 ± 32 | 93 ± 14 | 11 ± 2 |
| G190A | 191 ± 25 | 517 ± 49 | 112 ± 12 | >1,000 |
| Y181C | 257 ± 35 | 570 ± 69 | 126 ± 23 | >1,000 |
| G190A-181C | 162 ± 10 | 404 ± 31 | 100 ± 18 | >1,000 |
| DDDP | ||||
| WT | 97 ± 13 | 322 ± 45 | 18 ± 3 | 0.7 ± 0.1 |
| G190A | 129 ± 22 | 316 ± 41 | 216 ± 31 | >67 |
| Y181C | 107 ± 16 | 430 ± 52 | 36 ± 5 | >67 |
| 190A-81C | 279 ± 25 | 483 ± 33 | 380 ± 43 | >67 |
Data represent the means ± standard deviations of three separate determinations.
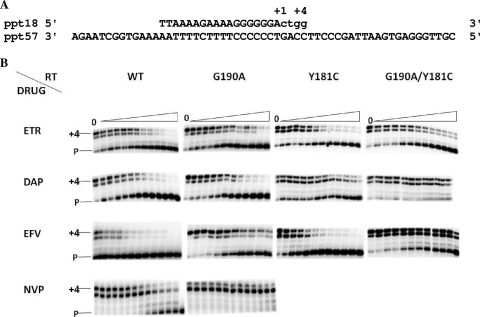
The inhibitory effects of the NNRTIs on DDDP activity were measured by a gel-based primer extension assay (Fig. 3A). Figure 3B presents representative gels showing the dose-dependent inhibition of DNA polymerase activity by NNRTIs. In the case of NVP, only the results of the analyses performed with the RT with the G190A mutation and the WT RT are shown, since the other two mutant RTs were highly resistant to NVP. The IC50s were determined and are summarized in Table 1. For ETR and DAP, the differences in the IC50s between the WT and the mutant RTs were less than threefold, indicating that these two drugs remained potent inhibitors of the mutated RTs. The IC50 of NVP for all mutant RTs was increased by more than 100-fold relative to that for the WT RT. Each of the RTs with the Y181C mutation and the Y181C and G190A double mutations were highly resistant to NVP. Y181C conferred only slight resistance (≈2-fold) to EFV, while the RT with the G190A and the RT with the G190A and Y181C mutations were ≈12- and 21-fold resistant to EFV, respectively.
FIG. 3.
Inhibition of DNA-dependent DNA polymerase RT activity by NNRTIs determined by gel-based RT assay. (A) Graphic representation of the primer-template system (ppt18-ppt57) used to monitor the inhibition of HIV-1 RT DNA polymerase activity by NNRTIs. +1 and +4, the positions of the first and the last nucleotide incorporated, respectively. (B) Dose-dependent inhibition of DNA polymerase activity by NNRTIs. Reactions were performed with increasing concentrations of the NNRTIs. The positions of the labeled primer (P) and the full-length extension product (+4) are indicated on the left. The concentrations of the NNRTIs used are as follows: for ETR, 0, 17, 26, 39, 58.5, 87.8, 131, 197.5, 296, 444, and 666 nM and 1 and 10 μM; for DAP 0, 23, 34, 52, 78, 117, 175, 260, 395, 590, 888, 1,330, and 2,000 nM; for EFV, 0, 4, 8, 16, 32, 64, 128, 180, 250, 352, and 493 nM and 10 and 100 μM; and for NVP, 0, 10, 31, 95, 285, and 857 nM; 2.5, 7.7, 23, 69, 208, and 625 μM; and 1.87 mM.
Drug susceptibilities of viruses with the G190A and Y181C mutations determined by cell-based phenotypic assays.
Previous studies have shown that G190A is present in approximately 3% of treatment-naïve newly infected persons (13). Three treatment-naïve viruses containing G190A were isolated and compared to WT viruses for their susceptibilities to NVP, EFV, ETR, and DAP. Table 2 shows that viruses harboring G190A showed high-level resistance to NVP and low-level resistance to EFV, while they remained susceptible to ETR and DAP. Moreover, both moderate and extensive hypersensitivity of the mutants with the A98S and G190A double mutation to EFV and DAP, respectively, were observed in the cell-based phenotypic assay (Table 2).
TABLE 2.
Susceptibilities of viruses harboring G190A to NNRTIs in cell-based phenotypic assays
| Isolate (mutation) | Drug susceptibility (EC50)a |
|||
|---|---|---|---|---|
| NVP | EFV | ETR | DAP | |
| 5269 (WT) | 0.007 | 0.00025 | 0.00090 | 0.0003 |
| 5331 (WT) | 0.010 | 0.00044 | 0.00251 | 0.0040 |
| 5512 (WT) | 0.020 | 0.00019 | 0.00226 | 0.0105 |
| Mean | 0.012 ± 0.003 | 0.00029 ± 0.00008 | 0.00189 ± 0.00050 | 0.0049 ± 0.0029 |
| 8116 (A98S, G190A) | 0.852 (71) | 0.00103 (3.6) | 0.00153 (0.8) | 0.0001 (0.02) |
| 8117 (A98S, G190A) | 0.703 (58) | 0.00030 (1.0) | 0.00065 (1.3) | 0.0002 (0.04) |
| 9225 (A98S, G190A) | 0.997 (83) | 0.00085 (2.9) | 0.00017 (0.9) | 0.00035 (0.07) |
| Mean | 0.850 ± 0.084 (70) | 0.00072 ± 0.00022 (2.5) | 0.00078 ± 0.00039 (0.4) | 0.0002 ± 0.0001 (0.04) |
The drug susceptibilities of WT viruses and resistant viruses harboring the A98S and G190A mutations were determined in two to three separate experiments. The overall susceptibilities of the WT RT and the RT with the G190A mutation are expressed as the means ± standard errors of the means. The values in parentheses are the fold resistance.
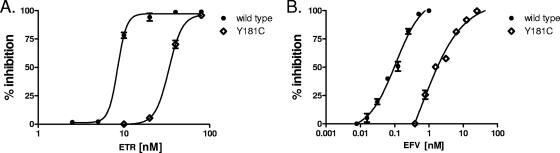
The relative susceptibility of the RT with the Y181C mutation was also determined by using recombinant single-cycle assays with TZM-bl cells. As demonstrated in Fig. 4, viruses with the Y181C RT mutation showed low-level (approximately threefold) phenotypic resistance to ETR and approximately fivefold resistance to EFV.
FIG. 4.
The RT mutation Y181C confers approximately three- to fourfold resistance to ETR in short-term replication assays. NNRTIs were diluted twofold on TZM-bl cells 1 h prior to infection with WT HIV-1 (•) or HIV-1 with the Y181C mutation (224) derived from molecular clones. After 48 h, the cells were rinsed and lysed and the luciferase activity was quantified. The data points depict the means and standard deviations of two independent experiments, in which each drug dilution was performed and analyzed in duplicate. The percent inhibition of replication of WT virus and virus with the Y181C mutation is shown on the y axis, while the x axis denotes the dose of ETR (A) or EFV (B) tested. The ETR EC50s for inhibition of replication of WT HIV-1 and HIV-1 with the Y181C mutation were determined to be 8.53 ± 0.0134 nM and 33.5 ± 1.68 nM, respectively. The EFV IC50s were 106 ± 1.27 μM and 910 ± 2.62 μM for WT HIV-1 and HIV-1 with the Y181C mutation, respectively. These values translate to approximately fourfold resistance to ETR and ninefold resistance to EFV.
DISCUSSION
ETR is a recently approved NNRTI that can overcome single point mutations such as K103N that confer cross-resistance to both NVP and EFV. In general, the high genetic barrier of ETR requires the accumulation of more than three resistance-associated mutations in order for diminished drug efficacy to result, and the relative roles of each of these mutations in the development of such resistance are unclear. The present study was initiated to determine the extent to which the G190A and Y181C mutations might affect viral susceptibility to ETR and other NNRTIs. Our findings indicate that neither G190A nor Y181C can, on its own, confer high-level resistance to ETR. We believe that these two mutations might currently be weighted inappropriately in drug resistance interpretation algorithms and that this might lead to ETR being potentially excluded from use in certain therapeutic situations.
HIV resistance to EFV and NVP is responsible for a high proportion of treatment failures, since single point mutations such as K103N can confer very high level resistance to these drugs. ETR has the distinct advantage of showing virological activity in patients who have failed NVP- and EFV-based regimens, despite the fact that their viruses commonly harbor such mutations as K103N. Although Y181C and G190A have been included as ETR mutations in genotypic resistance algorithms, our findings and those of others are of importance in showing that these two mutations confer only low-level resistance that borders on clinical cutoff values (14). The potential of mutated viruses to remain susceptible to ETR is of clinical relevance, given the high frequency of transmitted G190A resistance in drug-naïve subjects as well as the high frequency of Y181C in NVP-experienced individuals.
Previous data have shown that the Y181C mutation in RT is associated with partial resistance to ETR but is insufficient, on its own, to eliminate the antiviral activity of this compound (15). This result was confirmed in the present study on the basis of the results of recombinant RT assays performed with RNA and DNA templates and through the use of both filter- and gel-based assays. Our results also demonstrate that the G190A substitution in RT, while it is associated with resistance to each of NVP and EFV, does not confer resistance to either ETR or a related NNRTI, DAP, which is currently undergoing consideration for possible development as an anti-HIV microbicide. Our results also demonstrate that the presence of both the Y181C and the G190A substitutions in tandem resulted in higher-level resistance to both NVP and EFV than the level of resistance achieved with either mutation acting alone. However, the simultaneous presence of both mutations did not significantly add to the levels of resistance to ETR and DAP obtained with either mutation as a single substitution.
It is also instructive that high-level Y181C-mediated resistance to NVP was documented by each of the filter-based and gel-based RT assays, which were performed with RNA and DNA templates, respectively. The use of the DNA template also revealed resistance to EFV, but this result was not obtained by the RNA-dependent DNA polymerase assay. The changes in the levels of Y181C-mediated resistance to ETR in the DNA- and RNA-dependent polymerase assays were 2-fold and 1.35-fold, respectively. This is consistent with our findings of an approximately threefold phenotypic resistance on the part of Y181C to ETR and an approximately fivefold resistance to EFV, similar to data reported by others (14). It is also instructive that high-level Y181C-mediated resistance to NVP was documented by each of the filter-based and the gel-based RT assays that were performed with RNA and DNA templates, respectively. The use of the DNA template also revealed resistance to EFV, but this result was not obtained by the RDDP assay. This finding illustrates that RNA-based and DNA-based RT assays can sometimes yield qualitatively different results (2). Further studies should be performed with G190A together with the ETR-associated major mutation G190S, which confers ≈8- to 23-fold resistance to ETR when G190S occurs in association with Y181C (14).
Many of the clinical data sets regarding ETR resistance are based on the DUET studies, which involved highly treatment-experienced patients, including 14% who possessed approximately three preexisting NNRTI resistance-associated mutations (5). The frequency of NNRTI resistance-associated polymorphisms at the baseline may be different in treatment-naïve versus treatment-experienced individuals (1). ETR may demonstrate antiviral activity in treatment-naïve individuals who harbor transmitted NNRTI resistance-associated mutations, including K103N and G190A. As shown here in enzymatic and cell-based phenotypic assays, viruses harboring the G190A substitution retained susceptibility to both ETR and DAP.
Others have reported 3- and 3.3-fold resistance to ETR when the Y181C mutation and the Y181C and G190A mutations, respectively, are present (14). Our cell culture phenotypic data show that viruses with the Y181C mutation and viruses with the G190A mutation show approximately three- and fivefold resistance to ETR and EFV, respectively, whereas they show a >100-fold resistance to NVP. Data from the Stanford HIV-1 resistance database (http.//hivdb.stanford.edu) show 21.5% and 3.6% comparative frequencies of Y181C in NVP- and EFV-experienced persons, respectively. This suggests that Y181C is a major mutation associated with resistance to NVP but not to EFV.
On the basis of these data, we conclude that G190A and Y181C are not directly associated with resistance to ETR. Our data are also supported by separate analyses of ETR resistance involving Y181C and G190A in the DUET clinical study (19). This is not unexpected, given the conformational flexibility of ETR and its ability to reorient when single mutations are present in the RT binding pocket (7). ETR, with its high genetic barrier and low-level (approximately threefold) resistance, may remain an NNRTI option for patients harboring viruses with Y181C and G190A mutations, in the absence of other ETR-associated NNRTI mutations.
The relative weighting of resistance mutations has led to discordance among a variety of interpretative algorithms, including those of the Agence Nationale de Recherche sur le Sida, the International AIDS Society—USA, Monogram Diagnostics, and Virco Diagnostics, and in some cases has led to the exclusion of ETR as a potentially useful drug (1, 5, 9). Additional clinical data will be necessary to reevaluate the role of certain mutations in regard to the durability of ETR-based therapy in NNRTI-experienced patients. In all likelihood, additional studies based on cell culture selection, enzymatic analysis, and site-directed mutagenesis performed with viruses with additional mutations may further define the profile of resistance to ETR.
Acknowledgments
We thank Stuart Le Grice for providing the pRT6H-PROT DNA construct.
This research was supported by grants from the Canadian Institutes of Health Research, the Fonds de la Recherche en Santé du Québec-Réseau SIDA, and the International Partnership for Microbicides.
Footnotes
Published ahead of print on 24 August 2009.
REFERENCES
- 1.Benhamida, J., C. Chappey, E. Coakley, and N. T. Parkin. 2008. HIV-1 genotype algorithms for prediction of etravirine susceptibility. Novel mutations and weighting factors identified through correlations to phenotype. Antivir. Ther. 13(Suppl. 3):A130. [Google Scholar]
- 2.Borkow, G., R. S. Fletcher, J. Barnard, D. Arion, D. Motakis, G. I. Dmitrienko, and M. A. Parniak. 1997. Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry 36:3179-3185. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, B. G., M. Oliveira, F. Doualla-Bell, D. D. Moisi, M. Ntemgwa, F. Frankel, M. Essex, and M. A. Wainberg. 2006. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS 50:2210-2213. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, B. G., M. Roger, D. D. Moisi, M. Oliveira, I. Hardy, R. Turgel, H. Charest, J. P. Routy, and M. A. Wainberg. 2008. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS 22:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotte, L., M. A. Trabaud, J. C. Tardy, C. Brochier, R. P. Gilibert, P. Miailhes, C. Trépo, and P. André. 2000. Prediction of the virological response to etravirine in clinical practice: comparison of three genotype algorithms. J. Med. Virol. 81:672-677. [DOI] [PubMed] [Google Scholar]
- 6.Daar, E. S. 2008. Emerging resistance profiles of newly approved antiretroviral drugs. Top. HIV Med. 16:110-116. [PubMed] [Google Scholar]
- 7.Das, K., A. D. Clark, Jr., P. J. Lewi, J. Heeres, M. R. De Jonge, L. M. Koymans, H. M. Vinkers, F. Daeyaert, D. W. Ludovici, M. J. Kukla, B. De Corte, R. W. Kavash, C. Y. Ho, H. Ye, M. A. Lichtenstein, K. Andries, R. Pauwels, M. P. De Béthune, P. L. Boyer, P. Clark, S. H. Hughes, P. A. Janssen, and E. Arnold. 2004. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 47:2550-2560. [DOI] [PubMed] [Google Scholar]
- 8.Diallo, K., B. Marchand, X. Wei, L. Cellai, M. Gotte, and M. A. Wainberg. 2003. Diminished RNA primer usage associated with the L74V and M184V mutations in the reverse transcriptase of human immunodeficiency virus type 1 provides a possible mechanism for diminished viral replication capacity. J. Virol. 77:8621-8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geretti, A. M. 2008. Shifting paradigms: the resistance profile of etravirine. J. Antimicrob. Chemother. 62:643-647. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, L. B., and L. D. Saravolatz. 2009. Etravirine, a next generation nonnucleoside reverse transcriptase inhibitor. Clin. Infect. Dis. 48:1123-1128. [DOI] [PubMed] [Google Scholar]
- 11.Le Grice, S. F., C. E. Cameron, and S. J. Benkovic. 1995. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 262:130-144. [DOI] [PubMed] [Google Scholar]
- 12.Le Grice, S. F., and F. Grüninger-Leitch. 1990. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem. 187:307-314. [DOI] [PubMed] [Google Scholar]
- 13.Munshi, V., M. Lu, P. Felock, R. J. Barnard, D. J. Hazuda, M. D. Miller, and M. T. Lai. 2008. Monitoring the development of non-nucleoside reverse transcriptase inhibitor-associated resistant HIV-1 using an electro chemiluminescence-based reverse transcriptase polymerase assay. Anal. Biochem. 374:121-132. [DOI] [PubMed] [Google Scholar]
- 14.Poveda, E., C. de Mendoza, T. Pattery, M. González Mdel, J. Villacian, and V. Soriano. 2008. Phenotypic impact of resistance mutations on etravirine susceptibility in HIV patients with prior failure to nonnucleoside analogues. AIDS 22:2395-2398. [DOI] [PubMed] [Google Scholar]
- 15.Quan, Y., B. G. Brenner, R. G. Marlink, M. Essex, T. Kurimura, and M. A. Wainberg. 2003. Drug resistance profiles of recombinant reverse transcriptases from human immunodeficiency virus type 1 subtypes A/E, B, and C. AIDS Res. Hum. Retrovir. 19:743-753. [DOI] [PubMed] [Google Scholar]
- 16.Schiller, D. S., and M. Youssef-Bessler. 2009. Etravirine: a second-generation nonnucleoside reverse transcriptase inhibitor (NNRTI) active against NNRTI-resistant strains of HIV. Clin. Ther. 31:692-704. [DOI] [PubMed] [Google Scholar]
- 17.Vingerhoets, J., M. Peeters, H. Azijn, L. Tambuyzer, A. Hoogstoel, N. Nijs, M. P. de Bethune, and G. Picchio. 2008. An update of the list of NNRTI mutations associated with decreased virological response to etravirine (ETR): multivariate analysis on the pooled DUET-1 and DUET-2 clinical trial data. Antivir. Ther. 13(Suppl. 3):A26. [Google Scholar]
- 18.Vingerhoets, J., H. Azijn, E. Fransen, I. De Baere, L. Smeulders, D. Jochmans, K. Andries, R. Pauwels, and M.-P. de Bethune. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vingerhoets, J., B. Clotet, M. Peeters, G. Picchio, L. Tambuyzer, K. Cao-Van, G. De Smedt, B. Woodfall, and M. P. de Béthune. 2007. Impact of baseline NNRTI mutations on the virological response to TMC125 (etravirine; ETR) in the DUET-1 and DUET-2 phase III clinical trials, abstr. P7.3/05. Abstr. 11th Eur. AIDS Conf.
- 20.Zhang, Z., M. Walker, W. Xu, J. H. Shim, J. L. Girardet, R. K. Hamatake, and Z. Hong. 2006. Novel nonnucleoside inhibitors that select nucleoside inhibitor resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 50:2772-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]