Abstract
Summary: Treatment of infectious diseases becomes more challenging with each passing year. This is especially true for infections caused by the opportunistic pathogen Pseudomonas aeruginosa, with its ability to rapidly develop resistance to multiple classes of antibiotics. Although the import of resistance mechanisms on mobile genetic elements is always a concern, the most difficult challenge we face with P. aeruginosa is its ability to rapidly develop resistance during the course of treating an infection. The chromosomally encoded AmpC cephalosporinase, the outer membrane porin OprD, and the multidrug efflux pumps are particularly relevant to this therapeutic challenge. The discussion presented in this review highlights the clinical significance of these chromosomally encoded resistance mechanisms, as well as the complex mechanisms/pathways by which P. aeruginosa regulates their expression. Although a great deal of knowledge has been gained toward understanding the regulation of AmpC, OprD, and efflux pumps in P. aeruginosa, it is clear that we have much to learn about how this resourceful pathogen coregulates different resistance mechanisms to overcome the antibacterial challenges it faces.
INTRODUCTION
Infectious diseases have been an important cause of morbidity and mortality throughout our history. With the expansion of the antibiotic era during the 20th century, there was a growing confidence that the need for infectious disease specialists would all but disappear. However, no one could have predicted the impact that an increasing immunocompromised population would have on the resurgence of infectious diseases during the last 3 decades. Furthermore, the ability of bacterial pathogens to adapt and to overcome the challenges of antibiotics in their environment has been nothing short of impressive. We are now faced with a growing population of pan-resistant bacteria that threaten to move us into what some consider the “postantibiotic era” of infectious diseases.
Some of the more problematic drug-resistant pathogens encountered today include methicillin-resistant Staphylococcus aureus, multidrug-resistant Streptococcus pneumoniae, and vancomycin-resistant Enterococcus spp. among the gram-positive bacteria and multidrug-resistant Acinetobacter baumannii, Klebsiella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa among the gram-negative bacteria. This review focuses specifically on the resistance problems associated with P. aeruginosa, with a special emphasis on the complexity by which key chromosomally encoded resistance mechanisms are regulated and coregulated to make P. aeruginosa one of our greatest therapeutic challenges.
HISTORICAL AND CLINICAL SIGNIFICANCE OF P. AERUGINOSA
The opportunistic bacterial pathogen currently known as P. aeruginosa has received several names throughout its history based on the characteristic blue-green coloration produced during culture. Sédillot in 1850 was first to observe that the discoloration of surgical wound dressings was associated with a transferable agent (196). The pigment responsible for the blue coloration was extracted by Fordos in 1860, and in 1862 Lucke was the first to associate this pigment with rod-shaped organisms (196). P. aeruginosa was not successfully isolated in pure culture until 1882, when Carle Gessard reported in a publication entitled “On the Blue and Green Coloration of Bandages” the growth of the organism from cutaneous wounds of two patients with bluish-green pus (65). In several additional reports between 1889 and 1894, P. aeruginosa (Bacillus pyocyaneus) was described as the causative agent of blue-green purulence in the wounds of patients (261). A more thorough presentation on the routes of invasion and dissemination of P. aeruginosa leading to acute or chronic infection was provided by Freeman in a 1916 article (56).
P. aeruginosa is a ubiquitous organism present in many diverse environmental settings, and it can be isolated from various living sources, including plants, animals, and humans. The ability of P. aeruginosa to survive on minimal nutritional requirements and to tolerate a variety of physical conditions has allowed this organism to persist in both community and hospital settings. In the hospital, P. aeruginosa can be isolated from a variety of sources, including respiratory therapy equipment, antiseptics, soap, sinks, mops, medicines, and physiotherapy and hydrotherapy pools (199). Community reservoirs of this organism include swimming pools, whirlpools, hot tubs, contact lens solution, home humidifiers, soil and rhizosphere, and vegetables (77, 196, 199).
P. aeruginosa is seldom a member of the normal microbial flora in humans. Representative colonization rates for specific sites in humans are 0 to 2% for skin, 0 to 3.3% for the nasal mucosa, 0 to 6.6% for the throat, and 2.6 to 24% for fecal samples (164). However, colonization rates may exceed 50% during hospitalization (199), especially among patients who have experienced trauma to or a breach in cutaneous or mucosal barriers by mechanical ventilation, tracheostomy, catheters, surgery, or severe burns (17, 49, 182, 252, 257). Patients with impaired immunity have higher risks for colonization by this organism (164, 199), and disruption in the normal microbial flora as a result of antimicrobial therapy has also been shown to increase colonization by P. aeruginosa (17, 18, 250).
Despite the wide distribution of P. aeruginosa in nature and the potential for community-acquired infections, serious infections with P. aeruginosa are predominantly hospital acquired. A review of surveillance data collected by the CDC National Nosocomial Infections Surveillance System from 1986 to 1998 shows that P. aeruginosa was identified as the fifth most frequently isolated nosocomial pathogen, accounting for 9% of all hospital-acquired infections in the United States (48, 171). P. aeruginosa was also the second leading cause of nosocomial pneumonia (14 to 16%), third most common cause of urinary tract infections (7 to 11%), fourth most frequently isolated pathogen in surgical site infections (8%), and seventh leading contributor to bloodstream infections (2 to 6%). Data from more recent studies continue to show P. aeruginosa as the second most common cause of nosocomial pneumonia, health care-associated pneumonia, and ventilator-associated pneumonia (64, 106) and the leading cause of pneumonia among pediatric patients in the intensive care unit (ICU) (214).
P. aeruginosa is especially problematic for seriously ill patients in ICUs. From 1992 to 1997, data from the National Nosocomial Infections Surveillance System showed that P. aeruginosa was responsible for 21% of pneumonias, 10% of urinary tract infections, 3% of bloodstream infections, and 13% of eye, ear, nose, and throat infections within ICUs in the United States (213). A similar study conducted in Europe identified P. aeruginosa as the second most frequently isolated organism in reported cases of ICU-acquired infections (242). In this surveillance study, P. aeruginosa was accountable for 30% of pneumonias, 19% of urinary tract infections, and 10% of bloodstream infections.
RESISTANCE CHALLENGES FOR TREATMENT OF P. AERUGINOSA
P. aeruginosa presents a serious therapeutic challenge for treatment of both community-acquired and nosocomial infections, and selection of the appropriate antibiotic to initiate therapy is essential to optimizing the clinical outcome (15, 156). Unfortunately, selection of the most appropriate antibiotic is complicated by the ability of P. aeruginosa to develop resistance to multiple classes of antibacterial agents, even during the course of treating an infection. Epidemiological outcome studies have shown that infections caused by drug-resistant P. aeruginosa are associated with significant increases in morbidity, mortality, need for surgical intervention, length of hospital stay and chronic care, and overall cost of treating the infection (7, 25, 62). Even more problematic is the development of resistance during the course of therapy, a complication which has been shown to double the length of hospitalization and overall cost of patient care (41). P. aeruginosa can develop resistance to antibacterials either through the acquisition of resistance genes on mobile genetic elements (i.e., plasmids) or through mutational processes that alter the expression and/or function of chromosomally encoded mechanisms. Both strategies for developing drug resistance can severely limit the therapeutic options for treatment of serious infections.
Antibacterial Resistance Trends
Presented in Table 1 are rates of P. aeruginosa resistance to several antipseudomonal drugs (54, 95, 99, 100, 178, 211, 212). This summary is not meant to be inclusive of all of the published literature, but rather highlights data reported for isolates from several U.S. surveillance studies since January 2000. If multiple years were included in a study, the resistance rates for the most recent year are presented in Table 1.
TABLE 1.
Rates of antibacterial resistance among P. aeruginosa isolates from hospitals and ICUs
| Antibiotic | % of strains exhibiting resistancea |
|||||||
|---|---|---|---|---|---|---|---|---|
| Hospital study, 2006 (n = 606) (211) | Hospital study, 2005 (n = 589) (212) | Hospital study, 2002 (n = 9,896) (54) | ICU study, 2002 (n = 951) (178) | ICU study, 2000-2002 (n ≥ 7,500) (95) | Hospital study, 2001 (n ≥ 2,157) (99) | ICU study, 2001 (n ≥ 543) (99) | Hospital study, 2000 (n = 882) (100) | |
| β-Lactams | ||||||||
| Cefepime | 6 | 5 | 9 | 25 | 12 | 8 | 10 | 9 |
| Ceftazidime | 13 | 10 | 13 | 19 | 17 | 9 | 9 | 13 |
| Piperacillin-tazobactam | 11 | 9 | 11 | 10 | 14 | 8 | 8 | 13 |
| Aztreonam | 12 | 32 | ||||||
| Imipenem | 11 | 7 | 16 | 23 | 22 | 12 | 16 | 16 |
| Meropenem | 6 | 7 | 18 | 11 | 16 | 10 | ||
| Fluoroquinolones | ||||||||
| Ciprofloxacin | 21 | 22 | 35 | 32 | 33 | 26 | 25 | 25 |
| Levofloxacin | 22 | 22 | 34 | 32 | 27 | 25 | 27 | |
| Aminoglycosides | ||||||||
| Amikacin | 5 | 10 | 4 | 3 | ||||
| Tobramycin | 8 | 10 | 12 | 16 | ||||
| Gentamicin | 12 | 12 | 16 | 22 | 15 | 15 | 14 | |
Based upon CLSI interpretive breakpoints.
P. aeruginosa exhibits the highest rates of resistance for the fluoroquinolones, with resistance to ciprofloxacin and levofloxacin ranging from 20 to 35%. Although not surprising, the highest rates were reported for isolates obtained from patients in ICUs (Table 1). P. aeruginosa isolates from ICU patients also trend toward higher rates of β-lactam resistance than general trends for hospitalized patients. Based on the data in Table 1, it is difficult to draw any strong conclusions about trends of resistance to various β-lactams. Among the aminoglycosides, most studies have focused on gentamicin, with resistance rates ranging from 12 to 22%. Gentamicin was the least active of the aminoglycosides, with lower rates of resistance being reported for tobramycin and amikacin in most studies (Table 1).
Although the resistance trends from large national surveillance studies provide important data for consideration, these studies do not address the potential for much higher rates of resistance within individual communities and hospitals. For example, during the years 2001 and 2006, rates of nonsusceptibility among P. aeruginosa isolates in Brooklyn, NY, ranged from 27 to 29% for cefepime, 30 to 31% for imipenem, 23% for meropenem, and 41 to 44% for ciprofloxacin (113). These rates are substantially higher than national trends focusing on all hospital isolates of P. aeruginosa (Table 1).
Not only are rates of resistance to individual drugs or drug classes a concern, but the prevalence of multidrug-resistant strains (resistant to three or more drug classes) is an even more serious therapeutic challenge. A national surveillance of 13,999 nonduplicate P. aeruginosa isolates from ICU patients showed that multidrug resistance increased significantly, from 4% in 1993 to 14% in 2002 (Fig. 1A) (178). For comparison, another ICU surveillance study evaluated over 37,000 P. aeruginosa isolates from 1997 to 2002 and reported an increase in prevalence of multidrug-resistant strains from 13% to 21% (Fig. 1B) (132). Finally, Flamm et al. reported rates of multidrug-resistant P. aeruginosa ranging from 23 to 26% among 52,000 P. aeruginosa isolates collected in the United States from 1999 to 2002 (54). The highest prevalence of multidrug-resistant strains was observed among isolates from lower respiratory tract infections, whereas the lowest prevalence was observed among isolates from upper respiratory tract infections. Not surprisingly, multidrug-resistant strains were isolated more frequently from ICU and nursing home patients.
FIG. 1.
Increasing prevalence of multidrug resistance among P. aeruginosa isolates from ICU patients in the United States. (A) Data for 13,999 nonduplicate isolates collected from 1993 to 2002 (178); (B) data for 37,390 isolates collected from 1997 to 2000 (132). Data represent the percentage of P. aeruginosa isolates that expressed a phenotype of multidrug resistance (resistance to three or more drug classes) during each year of the studies. (Panel A is adapted from reference 178 with permission; panel B is based on data from reference 132.)
A multidrug-resistant phenotype can arise in P. aeruginosa through the acquisition of multiple imported resistance mechanisms on mobile genetic elements, a combination of imported and chromosomally encoded resistance mechanisms, accumulation of multiple chromosomal changes over time, and/or a single mutational event leading to the overexpression of a multidrug resistance mechanism, i.e., an efflux pump. This review focuses primarily on the key chromosomally encoded resistance mechanisms, their clinical significance, and their complex mechanisms of regulation. However, a brief overview of important imported resistance mechanisms is presented first.
Imported Resistance Mechanisms
In relation to the antipseudomonal drug classes presented in Table 1, imported resistance among P. aeruginosa isolates impacts the β-lactams and aminoglycosides but not the fluoroquinolones. As discussed below, fluoroquinolone resistance among P. aeruginosa isolates has been linked only to chromosomal genes, with mutational changes in the fluoroquinolone targets DNA gyrase (gyrA and gyrB) and/or topoisomerase IV (parC and parE) and/or overexpression of multidrug efflux pumps (Fig. 2). Although the plasmid-encoded DNA gyrase protection protein Qnr and the fluoroquinolone-modifying enzyme AAC(6′)Ib-cr can contribute to fluoroquinolone resistance among strains of Enterobacteriaceae (89, 186, 215), these two plasmid-encoded mechanisms have not been found in clinical isolates of P. aeruginosa (89, 198).
FIG. 2.
Mutational resistance to fluoroquinolones and carbapenems involving chromosomally encoded mechanisms expressed by P. aeruginosa. (A) Interactions of fluoroquinolones and carbapenems with “wild-type” susceptible P. aeruginosa expressing basal levels of AmpC, OprD, and nonmutated fluoroquinolone target genes (gyrA, gyrB, parC, and parE). Fluoroquinolone molecules pass through the outer membrane, peptidoglycan, periplasmic space, and cytoplasmic membrane and interact with DNA gyrase and topoisomerase IV (Topo IV) targets in the cytoplasm when these enzymes are complexed with DNA. Carbapenem molecules pass through the outer membrane-specific porin OprD and interact with their target PBPs, located on the outside of the cytoplasmic membrane. (B) Chromosomally encoded mechanisms of resistance to fluoroquinolones and carbapenems. Fluoroquinolone resistance is mediated by (i) overexpression of RND efflux pumps extruding the drug molecules from the periplasmic and cytoplasmic spaces and/or (ii) mutational changes within the target genes. Locations of the QRDRs within target genes are highlighted in yellow. Carbapenem resistance is mediated primarily by (i) decreased production or loss of functional OprD in the outer membrane and/or (ii) overproduction of RND efflux pumps (with the exception of imipenem). Minor changes in susceptibility can be observed due to overexpression of AmpC, adding to the resistance potential.
Imported resistance to the β-lactams involves the production of inactivating β-lactamases, for which several families have been identified among clinical isolates of P. aeruginosa. The variety, prevalence, and clinical significance of imported β-lactamases in P. aeruginosa have been addressed in several reviews over the last decade (74, 132-134). The most common imported β-lactamases found among P. aeruginosa isolates are penicillinases belonging to the molecular class A serine β-lactamases (PSE, CARB, and TEM families). Within this group, enzymes belonging to the PSE family appear to be the most prevalent (14). The therapeutic impact of these penicillinases is relatively limited since they do not impact the clinical efficacy of extended-spectrum cephalosporins, monobactams, or carbapenems. Although less frequent, class A extended-spectrum β-lactamases have also been detected in strains of P. aeruginosa and have included enzymes from the TEM, SHV, CTX-M, PER, VEB, GES, and IBC families (27, 36, 155, 175, 192, 197, 268, 284). Extended-spectrum β-lactamases from the class D, OXA-type enzymes have also been encountered within P. aeruginosa (170, 191).
Similar to the case for the Enterobacteriaceae, extended-spectrum β-lactamases alone do not provide P. aeruginosa resistance to the carbapenems. However, the prevalence of different classes of carbapenem-hydrolyzing enzymes has been increasing globally. The first class B metallo-β-lactamases in P. aeruginosa were identified in 1991 in Japan (266). Since that initial report, metallo-β-lactamases have been reported for P. aeruginosa isolates from nearly all regions of the globe (57, 96, 117, 176, 187, 281), and four major families have been identified (IMP, VIM, SPM, and GIM families) (28, 59, 135, 176, 253). Recently, class A carbapenemases of the KPC family have been identified. The first characterized KPC-producing P. aeruginosa isolate was collected in Colombia and reported in 2007 (262). The most recent report identifies the spread of KPC genes into clonally related and unrelated strains of P. aeruginosa from Puerto Rico (281). The first identification of an imported OXA-type carbapenemase in P. aeruginosa was reported in 2008, and it was shown to be the same OXA-40 carbapenemase previously described for A. baumannii (237).
Imported resistance to aminoglycosides most commonly involves enzymatic inactivation of the drug molecule through chemical modification. The history, molecular characterization, prevalence, and clinical significance of aminoglycoside-inactivating enzymes in P. aeruginosa were recently covered in an excellent review (200). These enzymes are categorized into the following three families, based upon the chemical modification they mediate: (i) aminoglycoside phosphoryltransferase enzymes phosphorylate the drug molecule, (ii) aminoglycoside acetyltransferase enzymes acetylate the drug molecule, and (iii) aminoglycoside nucleotidyltransferase enzymes adenylate the drug molecule. Although the range of aminoglycosides inactivated by specific enzymes within this family can differ, the ability of P. aeruginosa to carry the genes for multiple aminoglycoside-inactivating enzymes provides individual strains with the potential to develop resistance to all aminoglycosides.
In addition to the variety of aminoglycoside-modifying enzymes, high-level resistance to multiple aminoglycosides can be associated with methylation of the 16S rRNA. This mechanism was first reported for P. aeruginosa in 1993, and the methylase-encoding gene was designated rmtA (287). There are currently five characterized ribosomal methyltransferase enzymes (RmtA, RmtB, RmtC, RmtD, and ArmA) that have been found worldwide among clinical isolates of P. aeruginosa and Enterobacteriaceae (42, 60, 61, 265, 286, 287). Although methyltransferases do not appear to be as common as the aminoglycoside-modifying enzymes, all of the genes encoding these enzymes have been associated with mobile genetic elements (43, 61, 68, 264, 265, 285), raising concern about their widespread dissemination among P. aeruginosa isolates and other gram-negative bacilli.
Chromosomally Encoded Resistance Mechanisms
Similar to imported resistance mechanisms, there are a variety of resistance mechanisms encoded on the P. aeruginosa chromosome. These mechanisms include several aminoglycoside-inactivating enzymes (200) and a class D oxacillinase, OXA-50 (67). As mentioned above, characterized mechanisms of fluoroquinolone resistance among P. aeruginosa isolates have been restricted to chromosomal genes, including target mutations and active efflux (Fig. 2). Similar to the case for other gram-negative bacteria, DNA gyrase is the primary target for the fluoroquinolones in P. aeruginosa (84). Therefore, the first target-specific mutations are typically observed within the quinolone resistance determining region (QRDR) of gyrA (6, 116, 165, 249, 283). The highest levels of resistance are observed in strains that have mutations in the QRDR of both gyrA and the topoisomerase IV gene parC (6, 90, 91, 116, 165). Although mutational changes within the other two genes, gyrB and parE, have been described, the prevalence of these mutations appears to be much lower (6). Recent studies involving screening of the Harvard PA14 library of P. aeruginosa mutants have identified a number of other chromosomal genes that may be involved in antibacterial resistance or in increasing the frequency of mutation to resistance (20, 44, 233, 271).
The three most intensely studied chromosomally encoded resistance mechanisms in P. aeruginosa are the AmpC cephalosporinase, the OprD outer membrane porin, and the multidrug efflux pumps (Fig. 2). The remainder of this review focuses specifically on the clinical relevance of these three resistance mechanisms and the complex pathways P. aeruginosa utilizes to regulate their expression. The ability of P. aeruginosa to coregulate different resistance mechanisms makes this pathogen a constantly moving target that continues to challenge therapeutic strategies.
AmpC-MEDIATED RESISTANCE
AmpC and Resistance to β-Lactams
P. aeruginosa carries an inducible AmpC cephalosporinase which is similar to the chromosomally encoded AmpC found in several members of the Enterobacteriaceae (23, 34, 76, 222, 227). Wild-type strains of P. aeruginosa produce only low basal levels of AmpC and are susceptible to the antipseudomonal penicillins, penicillin-inhibitor combinations, cephalosporins, and carbapenems (227). However, when AmpC production is significantly increased, P. aeruginosa develops resistance to all β-lactams, with the exception of the carbapenems, to be discussed later (34, 227). This is in contrast to some AmpC-overproducing members of the Enterobacteriaceae, which remain susceptible to cefepime and require additional mechanisms to develop cefepime resistance (i.e., downregulation of porin production) (13, 45, 78, 194, 223). Although it is possible that variability in the hydrolytic activity of AmpCs from P. aeruginosa and the Enterobacteriaceae could play a role in these differences, cefepime hydrolysis data obtained with purified AmpC enzymes do not support this hypothesis (208). Rather, the greater impermeability of the P. aeruginosa outer membrane may play an important role in allowing AmpC overproduction to push cefepime MICs above the resistance breakpoint (75).
Whereas resistance to most of the β-lactams emerges as a result of AmpC overproduction, a definitive relationship between P. aeruginosa AmpC and carbapenem susceptibility remains convoluted. AmpC-deficient strains of P. aeruginosa created through allelic exchange exhibit significant, ≥4-fold increases in susceptibility to imipenem and panipenem but not to meropenem (148). Additional P. aeruginosa isolates, defined as AmpC deficient yet still producing AmpC, also exhibit increased susceptibility to imipenem and doripenem but not to meropenem (131, 136, 169). These data suggest that AmpC may play a role in the level of intrinsic susceptibility of P. aeruginosa to carbapenems. In contrast, published data have suggested that overproduction of AmpC does not play a discernible role in the development of carbapenem resistance among P. aeruginosa isolates. AmpC overproduction among isogenic mutants selected with β-lactams does not significantly decrease P. aeruginosa susceptibility to carbapenems (63, 131, 169, 274). In addition, AmpC overproduction in carbapenem-susceptible clinical isolates of P. aeruginosa has been reported. Data from one study of 47 characterized AmpC-overproducing clinical isolates showed that only 7 (15%) were resistant to imipenem (calculated from the data in Table 2 of reference 225). Gutierrez et al. recently reported that 51% of carbapenem-resistant clinical isolates of P. aeruginosa in their study overproduced AmpC (73). Although statistical analysis suggested that meropenem-resistant strains were more likely to overproduce AmpC than meropenem-susceptible strains, Gutierrez et al. concluded that “AmpC hyperproduction is neither sufficient nor necessary for meropenem clinical resistance”.
TABLE 2.
Genetic knockout of ampD homologue genes and ampC expression in P. aeruginosa PAO1a
| Strain | Avg level of ampC expression ± SDb |
|
|---|---|---|
| Basal | Induced | |
| PAO1 | 1c | 43 ± 9 |
| PAΔD | 60 ± 19 | 152 ± 38 |
| PAΔDh2 | 1c | 48 ± 15 |
| PAΔDh3 | 1c | 55 ± 5 |
| PAΔDh2Dh3 | 2 ± 0.14 | 81 ± 26 |
| PAΔDDh2 | 62 ± 9 | 234 ± 58 |
| PAΔDDh3 | 191 ± 52 | 1,014 ± 297 |
| PAΔDDh2Dh3 | 1,020 ± 87 | 1,105 ± 88 |
Modified from reference 91 with permission.
Real-time reverse transcription-PCR was used to measure ampC expression in P. aeruginosa grown in the presence of 50 μg/ml of cefoxitin (induced) or the absence of cefoxitin (basal).
Actual ampC expression data were not presented in the original report (91). Based on the methodology described and interpretation of the data presented, basal ampC expression was set to a value of 1 in this table.
The challenge of specifically linking AmpC overproduction to carbapenem resistance is our own limited knowledge of the complex interplay between resistance mechanisms in P. aeruginosa and the multitude of pathways by which P. aeruginosa coregulates resistance mechanisms. How do we specifically link AmpC overproduction to carbapenem resistance among uncontrolled clinical isolates with diverse genetic and environmental backgrounds? Even with isogenic laboratory strains, how do we specifically link AmpC production to a particular phenotype, knowing that altered AmpC production could be accompanied by changes in the expression of other resistance mechanisms through coregulatory pathways? Despite how much we have yet to learn about the resistance potential of P. aeruginosa, the data generated thus far suggest that AmpC overproduction alone does not significantly alter P. aeruginosa's susceptibility to the carbapenems but could certainly contribute to resistance if accompanied by additional resistance mechanisms (e.g., efflux pump overproduction, decreased OprD, and/or production of a class A/class B carbapenemase). Adding even more complexity is the potential for mutational variants of the chromosomally encoded AmpC enzyme (extended-spectrum AmpC) to provide P. aeruginosa with carbapenem resistance (216). Extended-spectrum AmpCs were first identified in Serratia marcescens and Enterobacter spp. (10, 143, 153) and, most recently, in E. coli (141, 142). Amino acid modifications near the active sites of these enzymes can lead to increased hydrolytic activity against cefepime, ceftazidime, and imipenem. However, increased catalytic activity of these enzymes only reduces susceptibility to cefepime and imipenem. Overproduction of these extended-spectrum AmpCs seems to be a requirement for cefepime (10) and/or carbapenem (216) resistance.
Clinical Significance of AmpC Overproduction
Similar to the discussion above, the clinical impact of AmpC overproduction by P. aeruginosa is difficult to assess due to the complex interplay of multiple resistance mechanisms. Nevertheless, Tam et al. reported that patients were 67.5 times more likely to be given inappropriate antibiotics when the infections were caused by AmpC-overproducing P. aeruginosa than when they were caused by P. aeruginosa that did not overproduce AmpC (251). In addition, these patients were more likely to have persistent bacteremia (45% versus 6%), underscoring the need to rapidly identify AmpC-overproducing P. aeruginosa isolates in the clinical laboratory and to understand the mechanisms of AmpC overproduction in this pathogen.
The ability of resistant P. aeruginosa to emerge during the course of therapy presents an even greater challenge. In this scenario, patients are treated with an appropriate β-lactam based on initial susceptibility data, only to fail therapy due to the emergence of AmpC-mediated resistance. This phenomenon has been observed in 14 to 56% of patients treated with antipseudomonal penicillins, penicillin-inhibitor combinations, aztreonam, and extended-spectrum cephalosporins (50, 71, 94, 97, 118, 147, 226, 228, 235), and emergence of resistance/clinical failure is observed most frequently with infections outside the urinary tract and in patients with underlying cystic fibrosis and neutropenia. Although combining an aminoglycoside with an antipseudomonal β-lactam is one strategy for preventing the emergence of AmpC-mediated resistance, this combination is not always effective in achieving that goal (97, 118, 147, 226, 228, 235).
As an initial step toward the future identification of novel strategies for combating AmpC-mediated resistance, it is essential that we elucidate the mechanisms by which the expression of this resistance mechanism is regulated. The following sections review the current understanding of mechanisms involved in regulation of AmpC-mediated resistance among P. aeruginosa isolates and, where appropriate, compare and contrast them with mechanisms of AmpC regulation among Enterobacteriaceae.
Pathways for AmpC Overproduction
Overproduction of the chromosomally encoded AmpC enzyme in P. aeruginosa and some members of the Enterobacteriaceae can occur either by induction of the ampC gene or through a process of derepression leading to constitutive high-level expression. Overexpression through the induction pathway occurs during exposure to specific β-lactams and β-lactamase inhibitors (e.g., cefoxitin, imipenem, and clavulanate) (112, 123, 128, 247, 267), but the process is reversible after removal of the inducing agent. In contrast, AmpC derepression occurs when proteins involved in the induction pathway are compromised through chromosomal mutations (83, 97, 108, 111, 114, 124) and the cephalosporinase is constitutively produced at an elevated level, even in the absence of an inducing β-lactam (9, 83, 114, 115). As discussed later in this section, phenotypes of AmpC derepression are more diverse among P. aeruginosa isolates than among the Enterobacteriaceae. Strains of P. aeruginosa can transition through a phenotype of partial derepession before achieving full derepression of AmpC.
Factors Involved in Regulation of ampC Expression
Figure 3 illustrates the key components involved in the regulation of ampC expression. The ampC induction pathway involves the following three major gene products: (i) an inner membrane permease known as AmpG; (ii) a cytosolic amidase, AmpD; and (iii) a transcription factor, AmpR, belonging to the LysR family of regulatory proteins (11, 138, 139). These three proteins are required for induction of the ampC gene in both Enterobacteriaceae and P. aeruginosa, although there is no direct evidence linking AmpG to the ampC induction pathway of P. aeruginosa (11, 81, 88, 109, 110, 127, 138, 139).
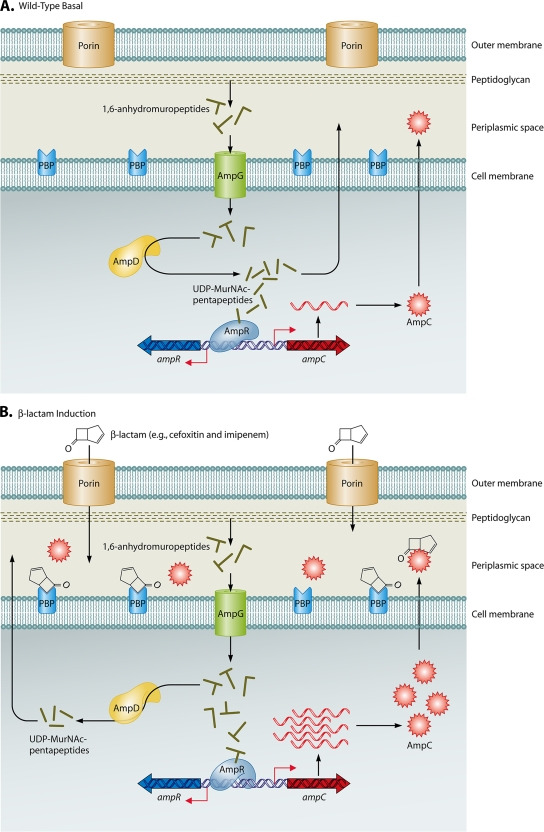
FIG. 3.
Mechanisms involved in regulation of ampC expression. These figures represent the current knowledge obtained from studies with members of the Enterobacteriaceae and appear to parallel events in P. aeruginosa. (A) Wild-type basal expression of ampC. During normal cell wall recycling, 1,6-anhydromuropeptides are removed from the cell wall and transported into the cytoplasm via the AmpG permease. The 1,6-anhydromuropeptides are cleaved by AmpD to generate free tripeptides, which are later converted into UDP-MurNAc-pentapeptides. UDP-MurNAc-pentapeptide interacts with AmpR bound to the ampR-ampC intergenic region, creating a conformation that represses transcription of ampC. Low basal levels of AmpC are produced, and the enzyme is localized to the periplasmic space. (B) β-Lactam induction of ampC expression. Inducing β-lactams, such as cefoxitin and imipenem, cross the outer membrane through porins, enter the periplasmic space, and interact with target PBPs. An increase in pools of 1,6-anhydromuropeptides is observed, and AmpD is unable to efficiently process the higher levels of cell wall fragments. The anhydro-MurNAc-peptides (inducing peptides) replace UDP-MurNAc-pentapeptides (suppressing peptides) bound to AmpR, causing a conformational change in the protein. AmpR is converted into a transcriptional activator, ampC is expressed at higher levels, and levels of AmpC increase in the periplasmic space. When the amount of β-lactam decreases below “inducing levels,” the cytoplasmic pool of anhydro-MurNAc-peptides also decreases, and AmpD is able to efficiently cleave these peptides, restoring wild-type ampC expression, as shown in panel A. (C) AmpD-associated derepression of ampC expression. Mutations leading to the inactivation of AmpD or decreased expression of ampD impair the processing of cell wall recycled products and lead to increased levels of anhydro-MurNAc-peptides (inducing peptides) in the cytoplasm. As a result, the binding of inducing peptides to AmpR is favored, AmpR is “locked” in a conformation for transcriptional activation of ampC expression, and high-level constitutive expression of ampC is observed.
The P. aeruginosa ampC and ampR genes and their corresponding intergenic region were first described in 1990 (139), and the gene organization is identical to that for members of the Enterobacteriaceae with inducible ampC genes. In both P. aeruginosa and Enterobacteriaceae, the ampR and ampC genes are divergently transcribed, and the binding of AmpR to the intergenic region between ampC and ampR is required for ampC induction (Fig. 3) (12, 16, 86, 126, 138, 139, 217). In the Enterobacteriaceae, AmpR negatively regulates ampC expression as well as the expression of its own gene, ampR (125, 126). In studies with P. aeruginosa, the data have been more conflicting and difficult to compare. Using an ampR knockout of P. aeruginosa PAO1, Kong et al. reported a 12-fold increase in nitrocefin hydrolysis in the absence of AmpR (107). However, interpretation of these data was complicated by the presence of another chromosomal β-lactamase, PoxB, which also hydrolyzes nitrocefin and appears to be regulated by AmpR (107). Further experiments with a PampC-lacZ promoter gene construct inserted into the chromosome of P. aeruginosa suggested that AmpR does not negatively regulate ampC (107). More recently, however, Moya et al. concluded that AmpR is a negative regulator of ampC, based upon their more direct analysis of ampC expression in an ampR knockout of P. aeruginosa PAO1 (166).
Recent reporter gene studies have also suggested that P. aeruginosa AmpR is a global regulator affecting the expression of multiple genes (lasB, rhlR, poxB, lasA, lasI, and lasR) in addition to ampC (107). Although direct RNA expression experiments are needed to confirm a global regulatory function, these data raise the possibility that regulation by AmpR may be more complex in P. aeruginosa than what has been shown for members of the Enterobacteriaceae (107).
Mechanism of ampC Induction
Whether ampC is expressed at a low constitutive level or elevated through induction is dependent upon the type of cofactor (i.e., cell wall precursor peptide) that binds to AmpR (Fig. 3). In this respect, the AmpC regulatory pathway is intimately linked to the cell wall recycling pathway via AmpD and AmpR. The general mechanism for regulation of low basal ampC expression in a wild-type P. aeruginosa strain is extrapolated from data obtained from the Enterobacteriaceae and shown in Fig. 3A. During normal growth, cell wall synthesis requires the addition and subtraction of cell wall components, resulting in the release of muropeptides (39, 40, 69, 80, 87). These muropeptides are transported into the cell via the AmpG permease (30, 87). Once inside the cell, these muropeptides are modified by AmpD into free peptide and anhydromuramic acid, with the resulting peptide recycled back into the cell wall synthesis pathway (80, 81, 87, 88). During peptide processing for use in cell wall synthesis, UDP is added to the pentapeptide (80). Excess “repressing” UDP-pentapeptide binds to AmpR and keeps AmpR in a conformational state that does not allow efficient transcription from the ampC promoter (86). It has been hypothesized that this conformation of AmpR prevents RNA polymerase from interacting efficiently with the ampC promoter, resulting in a low basal level of ampC expression.
The process of AmpC induction requires binding of an inducing β-lactam or β-lactamase inhibitor (e.g., cefoxitin, imipenem, or clavulanate) to penicillin binding proteins (PBPs) (Fig. 3B) (183, 190, 224). Since induction of AmpC does not result from the interactions of all β-lactams with PBPs, there clearly is something specific about the interactions of inducing compounds such as cefoxitin, imipenem, and clavulanate. Studies with the Enterobacteriaceae have shown that inducing β-lactams have a higher affinity for the low-molecular-weight PBPs (72, 82, 177), and genetic knockout of the genes for these PBPs provides additional support for their role (183, 190, 224). However, these studies were performed in E. coli, which does not possess an intrinsic inducible ampC system, making the data difficult to interpret (1). Data from a recently published study with P. aeruginosa have demonstrated that loss of the nonessential low-molecular-weight protein PBP4 is associated with increased expression of ampC and partial derepression of ampC (166). Although these experiments clearly demonstrated an association between PBP4 and derepression of ampC (described in more detail later in the review), the authors concluded from their study that PBP4 plays a role in ampC induction as well. However, strains lacking functional PBP4 still demonstrated induction of ampC, suggesting that PBP4 is not essential for the induction pathway (166). The role of other low-molecular-weight PBPs was not addressed in this study and should still be evaluated to determine their selective role in induction of ampC by inducing β-lactams.
Regardless of the precise mechanism responsible for selective AmpC induction with specific β-lactams, the result is an increase in the concentration of “inducing” muropeptides compared to the amount of “repressing” UDP-pentapeptide found in the cytoplasm (39, 40, 87). The “repressing” UDP-pentapeptide bound to AmpR is believed to be replaced by the “inducing” muropeptide form, changing the conformation of AmpR as it binds to the ampC promoter (76, 86). This conformational change has been suggested to provide a more efficient interaction with RNA polymerase, resulting in a significant increase in ampC expression (86). When the inducing β-lactam or β-lactamase inhibitor is removed, normal cell wall synthesis and cell wall recycling are restored. The result is a restoration of basal ampC expression through the replacement of the AmpR-bound “inducing” muropeptide with the “repressing” UDP-pentapeptide that is now present at a higher concentration.
Mechanisms of ampC Derepression
Modification of any protein involved in the induction pathway or modification of the ampC promoter could conceivably lead to derepression of ampC expression. Although studies with the Enterobacteriaceae have identified mutations within ampR (111), the majority of changes observed in clinical isolates have been associated with ampD (46, 83, 108, 231, 246). AmpD-associated derepression has been linked to mutations within the ampD structural gene and mutations leading to decreased ampD expression (231). In these strains, the AmpD amidase that cleaves the muropeptides entering the cell during cell wall synthesis has been modified or decreased, and normal processing of the muropeptides would be compromised (Fig. 3C). As a result, the concentration of “inducing” muropeptides in the cytoplasm would permanently increase, which favors their binding to AmpR due to the stoichiometric effect described above for the pathway of induction (39, 40, 86, 87). This culminates in a constitutive elevation of ampC expression.
Paralleling what is observed for the Enterobacteriaceae, P. aeruginosa AmpD has been characterized as a negative regulator of AmpC (114), and ampD mutations are an important mechanism of ampC derepression (9, 97, 98, 115, 232). Although not as common as ampD mutations, the potential role of mutated AmpR in P. aeruginosa derepression of AmpC has been described and correlated with a similar mutation in Enterobacter cloacae (9, 111, 232). In contrast to the case for the Enterobacteriaceae, full derepression of AmpC in P. aeruginosa is not always a single-step process. Campbell et al. have described the following three phenotypes of ampC expression: (i) low-basal-level expression that is inducible (wild type), (ii) moderate-basal-level expression that is inducible (partial derepression), and (iii) high-basal-level expression that is constitutive (full derepression) (23). A further complexity is that some AmpC-overproducing P. aeruginosa strains do not exhibit mutations in either ampR, ampD, or the ampR-ampC intergenic region and do not exhibit changes in the level of ampD expression (9, 23, 97, 232, 282). In one of these studies, mutants that expressed significant increases in the basal level of ampC were selected from a partially derepressed P. aeruginosa strain that lacked a functional AmpD protein (282). The mechanism of increased ampC expression did not involve additional mutations in ampR, ampD, or the ampR-ampC intergenic region or changes in ampD expression. However, a functional ampD gene was able to completely transcomplement and restore the wild-type phenotype for ampC expression and ceftazidime susceptibility (282). These observations suggest that undiscovered factors or pathways likely contribute to the regulation of ampC and the derepressed phenotype.
Additional candidates for regulation of ampC expression include AmpE, homologues of AmpD, and PBP4. In P. aeruginosa and the Enterobacteriaceae, expression of ampD is linked to the ampE gene (83, 114). Although some publications have indicated a potential role for AmpE in the AmpC regulatory pathway (83, 98), a specific role has yet to be established. Other studies suggest that AmpE plays no role in the regulation of AmpC (114, 166).
AmpD Homologues and Regulation of ampC Expression
The entire genome of P. aeruginosa PAO1 has been sequenced, opening the door for further investigations into the mechanisms involved in the regulation of ampC expression. In 2006, Juan et al. reported the identification of two additional P. aeruginosa AmpD homologues (AmpDh2 and AmpDh3) (98). E. coli possesses an outer membrane lipoprotein designated AmiD that was suggested to be a homologue of AmpDh2 by the same investigators (167), and this was later confirmed by Schmidtke and Hanson (232). There are only two N-acetyl-anhydro-muramyl-l-alanine amidases in the E. coli genome, so no homologue for AmpDh3 has been determined. Although the level of identity between AmpDh2 or AmpDh3 and the original AmpD protein is only 25 to 27% at the amino acid level, this may be attributed to the fact that the two new homologues possess membrane-spanning tails not observed with AmpD (232). However, AmpDh2 and AmpDh3 retain necessary conserved sequences within the active site compared with AmpD and are able to transcomplement an AmpD-deficient strain of P. aeruginosa PAO1 (98).
Using a series of ampD homologue knockout mutants, Juan et al. investigated the relative impact of each homologue on expression of ampC (98). Their observations are summarized in Table 2. The wild-type laboratory strain P. aeruginosa PAO1 served as the parent strain in this study and exhibited the expected low-basal-level inducible phenotype. The loss of ampD from P. aeruginosa PAO1 changed the phenotype to moderate-basal-level inducible expression of ampC (partial derepression). Surprisingly, deletion of ampDh2 or ampDh3 alone or in combination with each other did not alter the low-basal-level inducible phenotype. Although deletion of ampD in combination with deletion of ampDh2 or ampDh3 did not alter the partially derepressed phenotype observed with ampD deletion alone, the double knockout of ampD and ampDh3 did exhibit significant increases in both basal and cefoxitin-induced ampC expression. Finally, full derepression to high-basal-level, constitutive expression of ampC required the combined deletion of all three homologues. From these data, Juan et al. concluded that AmpD exhibits the greatest influence on ampC expression, followed by AmpDh3 and then AmpDh2.
The discovery and characterization of the ampD homologues, as well as their coordinated regulation of ampC expression, were important contributions to understanding the complex regulation of ampC expression in this pathogen. Not surprisingly, data from the analysis of clinical isolates suggest the use of additional pathways for regulation of ampC. For example, the same researchers analyzed 10 clinical isolates of P. aeruginosa that overproduced AmpC and found that 4 of the isolates did not exhibit any mutations in ampD (97), suggesting that loss of ampD is not an absolute requirement for ampC derepression in P. aeruginosa. Although potential decreases in ampD homologue expression were not assessed, ampC derepression in these four strains was later shown to be associated with mutations in the nonessential low-molecular-weight PBP4 gene, dacB (see below) (166). Schmidtke and Hanson reported that three clinical isolates of P. aeruginosa exhibiting full derepression of ampC (constitutive high-basal-level expression) exhibited inactivating mutations within ampD only, with no mutations or changes in the expression of ampDh2 or ampDh3 (232). Although the potential role of PBP4 was not evaluated by Schmidtke and Hanson, full derepression of ampC has been observed in a spontaneous dacB (PBP4) mutant of P. aeruginosa PAO1 with subsequent knockout of ampD (166).
PBP4 and Regulation of ampC Expression
PBP4 was recently identified as an important component of ampC regulation (166). Genetic knockout of dacB, which encodes PBP4, results in a partially derepressed phenotype in P. aeruginosa PAO1, similar to that of a knockout mutant of ampD (166). Furthermore, PBP4 mutations have also been associated with derepression of ampC in clinical isolates, and the association appears to be at least as common as mutations in ampD (166).
PBP4-associated derepression of ampC is also associated with the two-component global regulator CreBC, which is a homologue of the Aeromonas sp. regulator BrlAB (8, 174). Laboratory mutants and clinical isolates that exhibit PBP4-associated derepression of ampC also exhibit a significant increase in the expression of creD (166), a gene regulated by CreBC. However, the protein encoded by creD has not been characterized, and its role in ampC regulation is questionable. Elimination of creD in PBP4-associated derepressed mutants did not significantly alter β-lactam susceptibility (166). In contrast, the knockout of creBC significantly increased susceptibility to penicillins, cephalosporins, and aztreonam in ampC-derepressed dacB mutants. This link between CreBC and ampC regulation appears to be specific for PBP4, as no correlation was noted for ampD-associated derepressed mutants. Perhaps the most interesting observation is that increased β-lactam susceptibility was not associated with any changes in the level of ampC expression, suggesting that CreBC enhances resistance through a pathway other than AmpC.
Each of the studies discussed above provides important information on how P. aeruginosa regulates expression of ampC. However, it is clear that we have much to learn about the complex mechanisms by which AmpR, AmpD homologues, and PBP4 interact to control this important resistance mechanism. Furthermore, the pathway that P. aeruginosa uses to selectively induce ampC in the presence of β-lactams such as cefoxitin and imipenem remains uncharacterized, despite our advances in understanding the steps to derepression. Of particular interest would be further investigation into other low-molecular-weight PBPs and their role as initial triggers for ampC induction. A more complete understanding of the factors and complex pathways regulating ampC expression could lead to the identification of potential targets for controlling AmpC-mediated resistance and preserving the antibacterial activity of the β-lactam class.
OprD-MEDIATED RESISTANCE
The outer membrane of gram-negative bacteria constitutes a semipermeable barrier that slows the penetration of antibiotics, and specific to this review, the outer membrane of P. aeruginosa is only 8% as permeable as the outer membrane of Escherichia coli (75). However, in order to survive, P. aeruginosa must allow the passage of nutrients into the cell, and this is accomplished through a collection of water-filled protein channels called porins. Sequence analysis of the P. aeruginosa genome has identified 163 known or predicted outer membrane proteins, with 64 of these outer membrane proteins grouped into three families of porins (75). These porins play an important physiological role in the transport of sugars, amino acids, phosphates, divalent cations, and siderophores (75). Certain hydrophilic antibiotics, such as β-lactams, aminoglycosides, tetracyclines, and some fluoroquinolones, have also been shown to transverse the outer membrane through porin channels (173, 289). Not surprisingly, the loss of specific porin channels can decrease the susceptibility of P. aeruginosa to certain antibacterial agents. This section of the review focuses specifically on the association of the outer membrane porin OprD and the susceptibility of P. aeruginosa to carbapenem antibiotics.
OprD and P. aeruginosa Susceptibility to Carbapenems
The P. aeruginosa porin OprD is a substrate-specific porin that has been shown to facilitate the diffusion of basic amino acids, small peptides that contain these amino acids, and carbapenems into the cell (254, 255). This aqueous porin shares close homology to the nonspecific porin OmpF in E. coli. Pirnay et al. evaluated oprD genes from 55 strains of P. aeruginosa (environmental and clinical) and found evidence that the genetic sequence of oprD and the amino acid sequence of OprD are diverse across individual strains (195). DNA sequence identities ranged from 91 to 93%, whereas amino acid sequence identities varied from 88 to 93%. There was evidence of intraspecies recombinational events.
OprD appears to serve as the preferred portal of entry for the carbapenems, and loss of OprD from the outer membrane significantly decreases the susceptibility of P. aeruginosa to available carbapenems (103, 132, 140, 144, 169, 209, 210, 221, 229, 254). However, data presented later in this section highlight how much we have yet to learn about the dynamic interactions of carbapenems with P. aeruginosa.
The impact of OprD-mediated resistance on the carbapenems can be analyzed in two ways. The first consideration is the relative impact on the antibacterial potency of the carbapenems, as measured by increases in MICs. Data from a recent study of isogenic “wild-type” and OprD-deficient mutant pairs demonstrated that the loss of OprD decreases the susceptibility of P. aeruginosa to meropenem 4- to 32-fold, compared to 4- to 16-fold for imipenem and 8- to 32-fold for doripenem (221). For several OprD-deficient mutants in this study, the impact on meropenem potency was greater than that on the potency of other carbapenems. These data appear to conflict with the conclusions of Perez et al., who suggested that meropenem utilizes alternative pathways for entry across the outer membrane of P. aeruginosa (188). However, Perez et al. did not evaluate isogenic mutant pairs in their analysis of OprD deficiency and meropenem susceptibility. Rather, these investigators focused their study on unrelated clinical isolates that exhibited a phenotype of imipenem resistance and meropenem susceptibility. Although the P. aeruginosa isolates were susceptible to meropenem, MICs ranged from 2 to 4 μg/ml, well above expected meropenem MICs against “wild-type” strains (21, 256). Therefore, it is likely that decreased OprD in the clinical isolates was in fact responsible for elevating meropenem MICs to the susceptible breakpoint.
The second aspect of OprD-mediated resistance to consider is the clinical impact on the carbapenems. Although loss of OprD may impact susceptibility to imipenem less than that to meropenem (based on changes in MICs), this resistance mechanism frequently pushes MICs of imipenem above the resistance breakpoint. For example, in a study by Sakyo et al., all 10 OprD-deficient mutants lost susceptibility to imipenem, with MICs of >4 μg/ml (221). This is not surprising, since the MIC50 for imipenem against P. aeruginosa is already 1 μg/ml (21, 256), requiring only an eightfold decrease in potency to push the MIC into the intermediate range. In contrast to the case for imipenem, MICs for meropenem remained below 4 μg/ml for 4 of 10 mutants, and MICs for doripenem remained below 4 μg/ml for 8 of 10 mutants (221). Meropenem and doripenem exhibit an intrinsic potency that is fourfold greater than that of imipenem (21, 256). Therefore, the impact of OprD deficiency on the potency of these carbapenems does not always push the MICs above the susceptible breakpoint, and additional resistance mechanisms (efflux pump and/or carbapenemase) may be required to provide resistance to these two carbapenems.
The following sections review the current understanding of how P. aeruginosa regulates the expression of oprD. Published data to this point have identified mechanisms influencing the transcriptional expression of oprD and the translation of a functional porin protein.
Characterization of oprD Promoter Elements
Although the relationship between OprD and resistance to the carbapenems has been recognized for 2 decades, characterization of the oprD promoter and understanding of the regulation of OprD remain limited compared to our understanding of AmpC- and efflux-mediated resistance mechanisms. Figure 4 depicts the key promoter elements for oprD, based upon 5′ rapid amplification of cDNA ends of the oprD promoter of P. aeruginosa PAO1 grown in Mueller-Hinton broth (278). In these studies, two start sites for oprD transcription were identified, with transcription initiating at similar frequencies from an adenine 71 bases and a thymine 23 bases upstream of the translational start codon for oprD. Putative −10 and −35 elements are also presented in Fig. 4.
FIG. 4.
Characterization of oprD promoter elements. Transcription of oprD in P. aeruginosa PAO1 initiates with equal frequencies from two start sites, located 23 bases (SS1) and 71 bases (SS2) upstream of the structural gene (ATG translation start codon is highlighted in orange). The putative −10 and −35 promoter elements for SS1 are highlighted in red, and the putative −10 and −35 promoter elements for SS2 are highlighted in blue.
The transcriptional start sites and putative promoter elements observed for P. aeruginosa grown in Mueller-Hinton broth are different from those described by Ochs et al. (179). When P. aeruginosa was cultured with arginine, histidine, glutamate, or alanine as the sole source of carbon and with ammonium sulfate as the nitrogen source, the primary start site of transcription was reported to be a thymine located 89 bases upstream of the oprD translational start codon. Furthermore, induction of oprD expression by arginine was shown to be dependent on the arginine-responsive regulatory protein ArgR, which binds to an operator in the promoter region of oprD (179). In contrast, induction with the amino acid glutamate was independent of ArgR activation. Therefore, it appears that P. aeruginosa utilizes multiple transcriptional start sites and mechanisms to regulate expression of oprD, depending upon the growth conditions encountered.
Molecular Mechanisms of OprD-Mediated Resistance
The pathway to OprD-mediated resistance can involve mechanisms that decrease the transcriptional expression of oprD and/or mutations that disrupt the translational production of a functional porin for the outer membrane. At the level of oprD transcription, characterized mechanisms include (i) disruptions of the oprD promoter, (ii) premature termination of oprD transcription, (iii) coregulation with mechanisms of trace metal resistance, (iv) salicylate-mediated reduction, and (v) decreased transcriptional expression through mechanisms of coregulation with the multidrug efflux pump encoded by mexEF-oprN. oprD promoter disruptions have occurred as a result of deletions or insertions within the upstream region of oprD. Yoneyama and Nakae reported the association of a large deletion encompassing the putative promoter and initiation codon that prevented transcription of oprD (288). IS1394 and an ISPa16-like insertion (IS) element have been described upstream of the oprD coding region for imipenem-resistant isolates of P. aeruginosa exhibiting decreased oprD expression (273, 281).
El Amin et al. evaluated the transcription of oprD among clinical isolates of P. aeruginosa by using two sets of primers that amplified either the upstream or downstream regions of the structural gene (47). Two strains of P. aeruginosa exhibited normal levels of oprD transcription in experiments with the upstream primers, but oprD transcripts were undetectable with the downstream primers. Four additional strains demonstrated significant differences in the amounts of oprD transcript measured with the two primer sets. El Amin et al. concluded that premature termination of transcription was occurring in these strains, potentially due to mutations within the structural gene sequence.
The trace metals zinc and copper have been shown to decrease the expression of oprD in P. aeruginosa, leading to imipenem resistance (22, 189). This negative regulation is mediated through the regulatory proteins CzcR and CopR, which respond to the presence of zinc and copper, respectively, to activate mechanisms of metal resistance. CzcR and CopR both downregulate oprD transcription directly or indirectly, through unidentified factors. The weak aromatic acid salicylate was also shown to repress the transcription of oprD through an uncharacterized mechanism, leading to imipenem resistance among salicylate-exposed P. aeruginosa isolates (180).
Perhaps the most complex and intriguing mechanisms impacting the transcription of oprD are those that are linked to the regulation of expression of the mexEF-oprN efflux pump (101, 180). These mechanisms of coregulation are discussed in detail later in the review, but they highlight the complexity by which P. aeruginosa is able to regulate expression of resistance mechanisms and why it is sometimes so difficult to definitively link phenotypes to changes in one specific mechanism. Finally, mechanisms of OprD deficiency related to translation of an active porin include (i) mutations, insertions, and/or deletions creating frameshifts and premature stop codons (195) and (ii) disruption of the oprD structural gene by insertion of large IS elements (51, 277).
Discordance between oprD Expression and Susceptibility to Imipenem
Although the relationship between OprD deficiency and imipenem resistance has been well established in the literature, it should not come as a surprise that P. aeruginosa does not always follow expected rules. The genetic versatility of this pathogen and its ability to coregulate multiple resistance mechanisms make P. aeruginosa a constantly moving target and one of our greatest therapeutic challenges. Studies from our laboratory have identified intriguing strains that exhibit discordance between oprD expression and susceptibility to carbapenems.
The first example is an isogenic mutant, P. aeruginosa 410L, that was selected with levofloxacin from P. aeruginosa Tokai#1 (275). As background, strain Tokai#1 is an isogenic mutant of P. aeruginosa PAO1 that lacks susceptibility to both levofloxacin and imipenem (145). The MIC for imipenem against Tokai#1 is 8 μg/ml, compared to 1 μg/ml against the parental PAO1 strain, and this decreased susceptibility correlates with a fivefold decrease in the level of oprD expression (275). Expression of oprD in mutant strain 410L decreased further, to a level 3-fold below that of Tokai#1 and 17-fold below that of the wild-type parent, PAO1. However, despite the further decrease in oprD expression and the inability to detect OprD in outer membrane preparations, mutant 410L lost its resistance to imipenem and reverted back to a level of susceptibility similar to that of the original PAO1 parent.
The second example is an isogenic imipenem-hypersusceptible mutant, P. aeruginosa 244-921C, that was selected from an imipenem-resistant clinical isolate, P. aeruginosa strain 244, using ciprofloxacin (274). The mechanism of imipenem resistance for P. aeruginosa 244 (MIC = 16 μg/ml) was shown to be a base transition creating a premature stop codon and preventing translation of full-length OprD. This mutation was retained in the isogenic 244-921C mutant. Therefore, the eightfold increase in susceptibility of P. aeruginosa 244-921C to imipenem (MIC = 2 μg/ml) occurred despite the absence of an active OprD porin in the outer membrane.
The discordance between OprD and susceptibility to imipenem described above highlights how much we have yet to learn about the dynamic interactions of carbapenems with P. aeruginosa. The mechanism(s) responsible for increased susceptibility to imipenem in these strains has yet to be elucidated, but characterization of these mechanisms could provide pathways for development of therapeutic strategies to enhance the efficacy of carbapenems.
EFFLUX-MEDIATED RESISTANCE
While the loss of porins such as OprD represents an effective barrier for drug entry into the cell, a reduction in drug accumulation can also be achieved through active export by membrane-associated pumps. Efflux pumps have been categorized into five superfamilies, based primarily on amino acid sequence identity, the energy source required to drive export, and substrate specificities of the different pumps (218, 259). The superfamilies include (i) the ATP-binding cassette (ABC) family, (ii) the small multidrug resistance family, (iii) the major facilitator superfamily, (iv) the resistance-nodulation-division (RND) family, and (v) the multidrug and toxic compound extrusion family. Although sequence analysis of the P. aeruginosa genome has revealed the presence of efflux systems from all five superfamilies, the largest number of predicted pumps belong to the RND family, with a total of 12 RND systems (including two divalent metal cation transporters) (248). Unlike the primary active transporters of the ABC superfamily, which utilize ATP hydrolysis for energy, the RND family (as well as the remaining superfamilies) are secondary active transporters (symporters, antiporters, and uniporters) that derive the energy required for compound extrusion by proton motive force. Disruption of the proton gradient through the addition of a proton uncoupler, carbonyl cyanide m-chlorophenylhydrazone, increases the accumulation in these bacteria of substrates that are normally exported (172, 201).
RND pumps typically exist as a tripartite system consisting of a periplasmic membrane fusion protein (MFP), an outer membrane factor (OMF), and a cytoplasmic membrane (RND) transporter (Fig. 5). This complex forms a channel spanning the entire membrane, allowing for the transportation of lipophilic and amphiphilic drugs from the periplasmic space and cytoplasm to the extracellular environment. The genes encoding these pumps are organized into operons on the P. aeruginosa chromosome (Fig. 6). Each operon is composed of at least two genes, coding for the MFP and the RND transporter. Six of the 12 operons possess an OMF gene, completing the tripartite system, while the remaining operons are devoid of an OMF gene. Several operons have an adjacent regulatory gene transcribed in the same orientation or divergently from the operon, whose product functions as a repressor or activator of pump expression. Operons may contain additional genes besides those coding for the efflux pump. For example, mexG in the mexGHI-opmD operon encodes an integral membrane protein, and PA2528-PA2527-PA2526-opmB possesses a second RND transporter gene, PA2526.
FIG. 5.
Structure and function of RND efflux pumps in P. aeruginosa. RND pumps typically exist in a tripartite system consisting of an RND cytoplasmic membrane transporter (RND), an MFP, and an OMF. This complex forms a channel spanning the entire membrane, allowing for the proton-driven transport of lipophilic and amphiphilic drugs from the cytoplasm of the cell across the cytoplasmic membrane, through the periplasmic space, across the peptidoglycan, and across the outer membrane. The RND efflux pumps can also extrude drugs from the periplasmic space before they cross the cytoplasmic membrane.
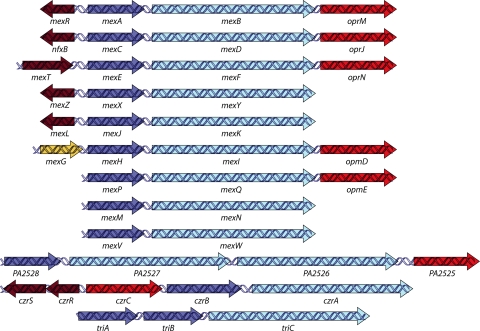
FIG. 6.
RND efflux operons in P. aeruginosa. Genes which encode protein components or characterized pumps are denoted by their gene names, and genes encoding protein components of uncharacterized pumps are designated with the P. aeruginosa (PA) numbers assigned in the annotated P. aeruginosa genome sequence (GenBank). Genes are depicted with the following color scheme: dark red arrow, transcriptional regulator-encoding gene; dark blue arrow, membrane fusion protein-encoding gene; light blue arrow, RND transporter-encoding gene; red arrow, outer membrane protein-encoding gene; and gold arrow, gene encoding a protein with unknown function. (Adapted from reference 234 with permission of the publisher.)
The 10 RND pumps in P. aeruginosa (excluding the metal cation transporters) are named MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexXY, MexJK, MexGHI-OpmD, MexVW, MexPQ-OpmE, MexMN, and TriABC (Table 3). Mex is an acronym for multiple efflux, and “Tri” refers to triclosan efflux. While several of these pumps share common substrates, they are also responsible for unique phenotypes inherent to their expression. Substrates of these pumps include antibiotics, biocides, dyes, detergents, organic solvents, aromatic hydrocarbons, and homoserine lactones (Table 3) (205, 234). Although not discussed in this review, these pumps may also have a physiological role in P. aeruginosa (e.g., cell-to-cell communication and pathogenicity) (193), besides their protective effects against antimicrobials. This section of the review focuses on the complex regulation of these pumps and the antibiotic phenotypes associated with their expression in planktonic cells. While P. aeruginosa can also live as a community encompassed in an exopolysaccharide matrix (i.e., biofilm), recent evidence has suggested that the RND efflux pumps do not participate in biofilm resistance to typical antipseudomonal agents (38) but may be involved with azithromycin resistance in biofilms (66).
TABLE 3.
Characteristics of RND efflux pumps in P. aeruginosa
| Operon | Component | Functiona | Regulatore |
Substrate(s) |
||
|---|---|---|---|---|---|---|
| Primaryb | Secondaryc | Antibiotics | Additional compounds | |||
| mexAB-oprM | MexA | MFP | MexR | NalC | Fluoroquinolones, β-lactams, β-lactamase inhibitors, tetracyclines, chloramphenicol, macrolides, novobiocin, trimethoprim, sulfonamides | Biocides (e.g., triclosan), detergents, dyes, HSL,f aromatic hydrocarbons |
| MexB | RND | NalD | ||||
| OprM | OMF | |||||
| mexCD-oprJ | MexC | MFP | NfxB | Fluoroquinolones, β-lactams, tetracycline, chloramphenicol, macrolides, trimethoprim, novobiocin | Biocides (e.g., triclosan), detergents, dyes, aromatic hydrocarbons | |
| MexD | RND | |||||
| OprJ | OMF | |||||
| mexEF-oprN | MexE | MFP | MexT | MexS | Fluoroquinolones, chloramphenicol, trimethoprim | Biocides (e.g., triclosan), aromatic hydrocarbons |
| MexF | RND | MvaT | ||||
| OprN | OMF | |||||
| mexXY | MexX | MFP | MexZ | Fluoroquinolones, β-lactams, tetracycline, aminoglycosides, macrolides, chloramphenicol | ||
| MexY | RND | |||||
| OprM/Opm-d | OMF | |||||
| mexJK | MexJ | MFP | MexL | Tetracycline, erythromycin | Biocides (e.g., triclosan) | |
| MexK | RND | |||||
| OprM/OpmH | OMF | |||||
| mexGHI-opmD | MexG | ? | SoxR | Fluoroquinolones | Vanadium | |
| MexH | MFP | |||||
| MexI | RND | |||||
| OpmD | OMF | |||||
| mexVW | MexV | MFP | — | Fluoroquinolones, tetracycline, chloramphenicol, erythromycin | ||
| MexW | RND | |||||
| OprM | OMF | |||||
| mexPQ-opmE | MexP | MFP | — | Fluoroquinolones, tetracycline, chloramphenicol, macrolides | ||
| MexQ | RND | |||||
| OpmE | OMF | |||||
| mexMN | MexM | MFP | — | Chloramphenicol, thiamphenicol | ||
| MexN | RND | |||||
| OprM | OMF | |||||
| triABC | TriA | MFP | — | Triclosan | ||
| TriB | MFP | |||||
| TriC | RND | |||||
| OpmH | OMF | |||||
MFP, membrane fusion protein; RND, resistance-nodulation-division transporter; OMF, outer membrane factor; ?, encodes a protein of unknown function.
Regulatory proteins that directly control expression of the efflux operons.
Proteins that indirectly activate operon expression when mutated.
MexXY may utilize OpmB, OpmG, OpmH, and/or OmpI as OMFs.
—, not identified.
HSL, homoserine lactones.
MexAB-OprM Efflux Pump
During a study examining siderophore-mediated iron transport, the first multidrug efflux pump in P. aeruginosa, MexAB-OprM, was discovered by Poole et al. (204). MexAB-OprM is able to export drugs from several different classes, including fluoroquinolones, tetracyclines, chloramphenicol, β-lactams and β-lactamase inhibitors, macrolides, novobiocin, trimethoprim, and sulfonamides (102, 120, 121, 243, 244). Of the RND efflux pumps, MexAB-OprM has the broadest substrate profile for the β-lactam class, with an ability to export β-lactams such as the carboxypenicillins, aztreonam, extended-spectrum cephalosporins (e.g., ceftazidime and cefotaxime), penems (e.g., faropenem), and the carbapenems meropenem and panipenem (but not imipenem and biapenem). MexAB-OprM participates in the intrinsic resistance of P. aeruginosa to the agents listed above through its constitutive production in wild-type cells (205). Knockout studies of mexAB-oprM alone or in combination with other resistance mechanisms have confirmed the pump's role in intrinsic resistance, as these mutants become hypersensitive (120, 148, 161).
Two additional characteristics are associated with mexAB-oprM expression. First, growth-phase-dependent expression of mexAB-oprM has been demonstrated (53). As the growth cycle progressed and cell density increased, mexAB-oprM transcription also increased until maximum expression occurred in late log phase/early stationary phase. The growth-phase-dependent upregulation was suggested to involve a quorum sensing signal. Quorum sensing is a mechanism by which bacteria monitor cell density through cell-to-cell communication, allowing for the coordinated expression of certain genes (e.g., virulence factors) in a cell density-dependent manner (260). Cell-to-cell signaling is mediated by diffusible autoinducers, known as homoserine lactone molecules, which interact with their cognate receptors to activate gene expression. N-Butyryl-l-homoserine lactone (C4-HSL) is synthesized as part of the rhl quorum sensing system and was shown to enhance expression of mexAB-oprM (146, 230). Second, the OMF gene, oprM, was shown to be expressed independently of mexAB (290). A weak promoter was discovered upstream of oprM in the coding region of mexB. This promoter was suggested to be less active than the promoter upstream of mexA, contributing to only a fraction of the total amount of transcript. OprM may serve as an OMF for the MexXY (5, 152, 159), MexJK (33), MexVW (122), and MexMN (158) systems, and possibly other RND pumps. Independent expression of oprM may allow for sufficient levels of OMF to accommodate multiple pumps or to ensure its presence in case expression from the mexAB-oprM promoter is compromised.
Several regulatory loci influence the expression of the mexAB-oprM operon. The mexR gene is located directly upstream of but transcribed divergently from mexAB-oprM and encodes a repressor belonging to the MarR family of regulatory proteins (Fig. 6) (206). MexR binds as a stable homodimer to two sites, each consisting of inverted repeat sequences, within the mexR-mexA intergenic region (52). The binding region encompasses overlapping mexR and mexA promoters, and association of MexR dimers with these sites represses transcription of the mexAB-oprM operon and negatively autoregulates its own expression (52, 220). A second regulatory factor of the mexAB-oprM operon, NalD, was discovered following insertional mutagenesis of a gene, nalD, located adjacent to a putative pump gene of the major facilitator superfamily (239). NalD is a repressor of the TetR family that binds to a sequence upstream of mexAB-oprM but downstream of the mexR binding sites (160). The NalD operator overlaps a second promoter for mexAB-oprM, and binding of NalD restricts expression from this proximal mexA promoter. The C4-HSL-mediated growth phase regulation of mexAB-oprM described above occurs independently of MexR (230). However, the possibility that C4-HSL directly or indirectly influences NalD-mediated regulation of mexAB-oprM to promote growth phase expression has yet to be determined experimentally.
Overexpression of mexAB-oprM has been detected in nalB-, nalC-, and nalD-type multidrug-resistant mutants and selected both in vivo (2, 137, 181, 184, 239, 273) and in vitro (2, 24, 239, 245). In nalB-type strains, mutations within mexR disrupt the translation of full-length protein or compromise the repressor activity of MexR by causing a loss of dimerization, defects in DNA binding, or, possibly, instability of the protein (2, 219). Several mutations in mexR have been described, including nucleotide changes (e.g., base substitutions, deletions, and insertions) and insertion of an IS element (2, 19, 219, 245). The absence of MexR from its operators leads to pump overexpression from the distal mexA promoter (52). nalC-type strains hyperexpress mexAB-oprM, but at lower levels than those in nalB-type mutants (245). Transposon insertional mutagenesis identified the site for nalC-type mutations in the PA3721 gene (renamed nalC) (24). nalC encodes a putative repressor of the TetR/AcrR family, whose genes are located upstream of an operon, PA3720-PA3719, that is negatively regulated by NalC (24). Loss of NalC resulted in overexpression of PA3720-PA3719, and subsequent experiments demonstrated that PA3719 (renamed ArmR) upregulates mexAB-oprM by interacting with MexR (24, 35). ArmR, a 53-amino-acid antirepressor, allosterically inhibits MexR dimer-DNA binding by occupying a hydrophobic binding cavity within the center of the MexR dimer (272). As implied by the name, overexpression of mexAB-oprM in nalD-type mutants occurs in response to disruption of NalD (239). Complementation of nalD-type mutants with a functional nalD gene reduced mexAB-oprM hyperexpression and drug resistance (239). Mutations within NalD were believed to alleviate repression of the proximal mexA promoter (160), presumably by an inability to bind to its operator. Interestingly, maximum expression from the proximal mexA promoter in NalD-negative strains requires the presence of MexR (160).
A mutational event within one of the known regulatory genes may not be the sole mechanism to increase mexAB-oprM transcription. Chen et al. recently described the effect of oxidative stress on operon expression (29). Oxidation of two cysteine residues within MexR causes the formation of an intermonomer disulfide bond which alters the conformation of MexR. Oxidized MexR dissociates from the mexR-mexA intergenic region, allowing access of RNA polymerase to the distal mexA promoter. In this situation, the MexR protein serves as a sensor of oxidative stress, and the response culminates in activation of the efflux defense mechanism, possibly to remove the agent responsible for inducing the oxidative stress.
MexCD-OprJ Efflux Pump
The operon coding for MexCD-OprJ was cloned and sequenced by Poole et al. in 1996 and showed a high degree of homology to MexAB-OprM (203). MexCD-OprJ can extrude a variety of antimicrobial agents, including fluoroquinolones, β-lactams, chloramphenicol, tetracycline, novobiocin, trimethoprim, and macrolides (70, 102, 149, 243). Unlike MexAB-OprM, MexCD-OprJ does not have an extensive substrate profile for the β-lactams, but rather, it preferentially exports the fourth-generation cephalosporins (e.g., cefepime, cefpirome, and cefozopran) (149, 203). Transcription of mexCD-oprJ can be observed in wild-type cells (276), but the levels are most likely not sufficient to produce detectable levels of protein (70, 203). In addition, deletion of mexCD-oprJ has no impact on wild-type susceptibility, indicating that this pump does not contribute to intrinsic resistance (162, 244). Expression of mexCD-oprJ was shown to be inducible in response to benzalkonium chloride, chlorhexidine gluconate, tetraphenylphosphonium chloride, ethidium bromide, rhodamine 6G, and acriflavine but not in response to clinically relevant antibiotics (162). Induction of mexCD-oprJ by membrane damaging agents (i.e., chlorhexidine) was dependent upon the stress response sigma factor AlgU (55).
Expression of mexCD-oprJ is governed by the product of a gene, nfxB, located upstream of mexCD-oprJ but transcribed divergently from the operon (Fig. 6) (203). NfxB displays similarity to proteins of the LacI-GalR family who possess a helix-loop-helix motif characteristic of DNA binding (203). NfxB negatively regulates expression of mexCD-oprJ, as well as its own expression (203, 238), by binding to a site composed of two 39-bp repeats within the nfxB-mexC intergenic region (238). Mutations within nfxB are suggested to alter the repressor activity of NfxB, leading to hyperexpression of mexCD-oprJ in so-called nfxB-type mutants (203, 238). Several mutations have been described for nfxB from laboratory strains and clinical isolates, such as base substitutions, deletions, and interruption with an IS element (37, 90, 203). Two types of hyperexpressing mutants, types A and B, have been identified in P. aeruginosa, with differing levels of MexCD-OprJ production and susceptibility (149). Type B mutants produce larger amounts of MexCD-OprJ and have more substantial changes in susceptibility than do type A mutants. Complementation of both types of mutants with wild-type nfxB restored susceptibilities to wild-type levels, suggesting that mutations in NfxB were responsible (149). Regulation of mexCD-oprJ expression may not be limited to NfxB. In mexAB-oprM and oprM deletion derivatives of wild-type strain PAO1, expression of mexCD-oprJ markedly increased, indicating that MexAB-OprM may influence the expression of mexCD-oprJ (119). The mechanism responsible for mexCD-oprJ upregulation in MexAB-OprM-deficient strains has yet to be determined.
Whereas the overproduction of MexCD-OprJ in P. aeruginosa provides resistance to several antibacterial drugs, an interesting phenotype of hypersusceptibility (≥4-fold increase in susceptibility) to some β-lactams and aminoglycosides is observed in the same strains. The correlation between MexCD-OprJ overproduction and hypersusceptibility and the mechanisms responsible for hypersusceptibility are discussed later in this section.
MexEF-OprN Efflux Pump
Three open reading frames in an operon named mexEF-oprN were characterized by Kohler et al., and the products of mexEF-oprN exhibit amino acid identity to the components of MexAB-OprM and MexCD-OprJ (104). Substrates recognized by MexEF-OprN include fluoroquinolones, chloramphenicol, and trimethoprim (104), but the pump has no apparent affinity for currently available β-lactams. Expression of mexEF-oprN was originally reported as quiescent (104), but later studies detected low-level expression in strains with wild-type susceptible phenotypes (119, 276). Disruption of mexEF-oprN in wild-type strains did not affect susceptibility, excluding its participation in intrinsic resistance (104).
Regulation of mexEF-oprN differs from that of mexAB-oprM and mexCD-oprJ in that operon expression is not suppressed by a negative regulator. Multiple factors are capable of controlling mexEF-oprN transcription. mexT, located upstream of and transcribed in the same orientation as mexEF-oprN (Fig. 6 and 7A), encodes a protein belonging to the LysR family of transcriptional activators that is capable of positively regulating mexEF-oprN expression (Fig. 7A) (101, 180). Because of sequence variations within mexT genes of different wild-type P. aeruginosa strains, activation of MexT can occur through different pathways (145). In some P. aeruginosa strains, MexT exists in a dormant form because of suppressing mutations within the coding region, but additional cis-acting mutations or deletions within mexT convert inactive MexT into an active form (145).
FIG. 7.
Coregulation of mexEF-oprN and oprD in P. aeruginosa. The models represent the proposed mechanisms of coregulation of mexEF-oprN and oprD in P. aeruginosa. Each panel represents the chromosome of P. aeruginosa, highlighting the mexE, mexF, oprN, and oprD structural genes and the proposed genes involved in coregulation, mexT, mexS, and mvaT. (A) Basal expression of mexEF-oprN and oprD in wild-type P. aeruginosa. In wild-type P. aeruginosa, MexT is functionally inactive due to either the presence of suppressing mutations or the lack of a secondary effector molecule. As a result, expression of mexEF-oprN occurs at a low basal level, and expression of oprD occurs at a level sufficient to provide quantities of OprD in the outer membrane sufficient for normal cellular function. (B) MexT-associated coregulation of mexEF-oprN and oprD. In nfxC-type mutants, MexT becomes active through a mutation within the structural gene. The activated MexT protein positively regulates (green arrow) transcription of mexEF-oprN, leading to overexpression of the efflux operon and overproduction of the MexEF-OprN efflux pump. Simultaneously, MexT negatively regulates (red arrow) oprD at the transcriptional and posttranscriptional levels, leading to decreased production of OprD. (C) MexS-associated coregulation of mexEF-oprN and oprD. Loss of MexS, a putative oxidoreductase/dehydrogenase, has been suggested to cause a buildup of secondary metabolites which may serve as effector molecules for MexT. These effector molecules could bind to MexT, alter the conformational state of the regulatory protein, and transform MexT into an activating transcriptional regulator. As a result, MexT can positively regulate (green arrow) the expression of mexEF-oprN and negatively regulate (red arrow) the expression of oprD, similar to what is described for panel B. (D) MvaT-associated coregulation of mexEF-oprN and oprD. Loss of the global regulatory protein MvaT is also associated with the upregulation of the mexEF-oprN operon. The mechanism of MvaT-associated regulation has not been elucidated, but it functions independent of MexT and MexS. In contrast to the case for the MexT- and MexS-associated regulatory pathways, loss of MvaT causes an upregulation of both mexEF-oprN and oprD expression.
In other strains, suppressing mutations are not present within mexT (145), but stimulation of MexT requires potential changes in the levels of cognate effector molecules (101, 145). Mutations in these strains occur within a gene, mexS, located upstream of mexT that encodes a putative oxidoreductase/dehydrogenase homologue (Fig. 7C) (241). Inactivation of MexS is believed to cause a buildup of metabolites that serve as effector molecules for MexT, which, in turn, upregulates mexEF-oprN expression to remove the toxic metabolites (241). Therefore, an active MexT protein is required for upregulation of mexEF-oprN by this pathway. While mexEF-oprN-overexpressing strains have been encountered among nosocomial P. aeruginosa strains (47, 181, 207, 273), a correlation between pump overexpression and genetic changes within mexT and/or mexS in clinical isolates has, to the best of our knowledge, been lacking.
Expression of mexEF-oprN is also controlled by a member of the histone-like nucleoid structuring protein family, MvaT, which serves as a global regulator of genes involved in virulence, housekeeping, and biofilm formation (Fig. 7D) (26, 35, 258, 270). MvaT binds to and oligomerizes across AT-rich regions of DNA with a high affinity and, as a result, silences the expression of certain genes (26). Deletion of mvaT caused an increase in mexEF-oprN expression that was not dependent on mexT or mexS (270). MvaT does not bind upstream of mexEF-oprN (270). Thus, the loss of this regulatory protein most likely has an indirect effect on regulating mexEF-oprN expression.
Data from a more recent study suggest that regulation of mexEF-oprN is even more complex and that more uncharacterized pathways are likely operative in P. aeruginosa (279). In this study, an isogenic mutant overexpressing mexEF-oprN was selected from a clinical isolate of P. aeruginosa by use of levofloxacin. Analysis of gene sequences and transcriptional expression demonstrated that the overexpression of mexEF-oprN did not involve changes in mexT, mexS, or mvaT. The mechanism of mexEF-oprN regulation in this isogenic mutant remains to be characterized.
Similar to nfxB-type mutants, nfxC-type mutants become hypersusceptible to certain β-lactams and aminoglycosides (58, 202). β-Lactam hypersusceptibility was associated with repression of the quorum sensing-mediated enhancement of mexAB-oprM expression by MexT (146). Transcription of the C4-HSL autoinducer synthase gene, rhlI, was previously shown to be reduced in nfxC-type mutants (105). As mentioned above, growth phase expression of mexAB-oprM is dependent on this quorum sensing molecule. Therefore, MexT appears to act as a global regulator controlling expression of mexEF-oprN, oprD, and genes regulated by the autoinducer C4-HSL, such as mexAB-oprM.
MexXY Efflux Pump
mexXY was cloned from the P. aeruginosa chromosome by Mine et al. in 1999, and the pump displays properties similar to those of the efflux systems described above (159). Unlike the operons described above, mexXY lacks a gene coding for an outer membrane protein (Fig. 6). Instead, MexXY is able to associate with OprM (5, 32, 159), and possibly other outer membrane proteins, such as OpmB, OpmG, OpmH, and OpmI, to form a functional tripartite system (32, 168). Fluoroquinolones, specific β-lactams (i.e., cefepime), aminoglycosides, tetracycline, chloramphenicol, and erythromycin are all substrates for MexXY (202, 234). mexXY expression is induced when cells are grown in the presence of tetracycline, erythromycin, and gentamicin (152). Deletion of mexXY in wild-type strains increases their susceptibility to these antibiotics, suggesting that this pump contributes to intrinsic resistance (152, 161). MexXY does not contribute to the intrinsic resistance to fluoroquinolones, a pump substrate, because these agents fail to induce mexXY expression (152).
The product of a gene, mexZ, located upstream of but transcribed divergently from mexXY, negatively regulates the expression of the operon (Fig. 6) (5, 154). MexZ belongs to the TetR family of transcriptional regulators and contains a characteristic N-terminal helix-turn-helix DNA binding domain. MexZ binds as a homodimer to an inverted repeat region, located in the mexZ-mexX intergenic region, which encompasses the putative mexXY promoter (154). Mutations in mexZ or the mexZ-mexX intergenic region have been associated with hyperexpression of mexXY (85, 263). However, overexpression of mexXY has also been detected in mutants not harboring mutations within mexZ or the mexZ-mexX intergenic region (137, 240, 269). Llanes et al. proposed the names agrZ-type and agrW-type mutants to describe mutants hyperexpressing mexXY with alterations within mexZ and outside mexZ, respectively (137). Thus, mexXY may be regulated by multiple factors, similar to mexAB-oprM, and mutations within as yet unidentified genes are necessary for mexXY overexpression. MexXY hyperexpression has been well documented for agrZ-type and agrW-type clinical isolates resistant to fluoroquinolones, cefepime, and aminoglycosides (79, 137, 240, 263, 283).
Multiple pathways also participate in the regulation of mexXY induction. Interaction of bacterial ribosomes with protein synthesis inhibitors (e.g., tetracycline, chloramphenicol, and aminoglycosides) is necessary to upregulate mexXY expression, as plasmid-encoded ribosomal protection mechanisms reduce mexXY drug inducibility (93). Induction may not always require the presence of MexZ (93, 163). Inducing antibiotics had no effect on the binding of MexZ to its operator (154) and could further enhance mexXY expression in a mexZ-deficient mutant (93), suggesting that mexXY induction was independent of MexZ. A more recent study by Morita et al. implicated the PA5471 gene in mediating mexXY induction (163). Disruption of PA5471 compromised mexXY drug inducibility, while increased expression of PA5471 alone stimulated mexXY expression. PA5471 transcription is also induced by ribosome inhibitors (the same drugs which induce mexXY), and PA5471 induction indirectly or directly alters MexZ activity, but not mexZ expression, resulting in mexXY upregulation (163).
MexJK Efflux Pump
The MexJK efflux pump was discovered in a ΔmexAB-oprM ΔmexCD-oprJ background strain following selection with the broad-spectrum biocide triclosan (33). An OMF-encoding gene is also absent from the mexJK operon (Fig. 6), but a three-component system is formed with the addition of either OprM (32, 33) or OpmH (32). MexJK-OprM exports erythromycin and tetracycline from the cell, while MexJK-OpmH removes triclosan (32). mexJK was reported not to be expressed in wild-type cells (33).
A member of the TetR regulatory family, mexL, is present upstream of mexJK but transcribed in the opposite orientation (Fig. 6) (33). MexL binds as a multimer to inverted repeats within the mexL-mexJ intergenic region encompassing overlapping mexJK and mexL promoters (31). MexL binding to this site represses the transcription of both mexJK and mexL, thereby negatively autoregulating its own expression. A mutation within the helix-turn-helix motif of MexL prevented binding to its operator site and led to mexJK overexpression in a laboratory mutant (31). Overexpression of mexJK has also been detected in a few clinical isolates, but the pump's contribution to resistance and potential mutations in mexL were not evaluated (79).
Additional RND Efflux Pumps
The contributions of the remaining RND efflux pumps to resistance and the factor(s) governing their expression have just begun to be elucidated. Their up- or downregulation in clinical isolates with various phenotypes has yet to be reported. mexGHI-opmD expression is detectable in wild-type cells and is under the control of a redox-active protein, SoxR, belonging to the MerR family of transcriptional regulators (4, 185). Norfloxacin was the only antibiotic reported to be a substrate of MexGHI-OpmD (236), but this pump was also implicated in the export of other nonantibiotic compounds, including vanadium (4). Interestingly, a mutant incapable of expressing mexGHI-opmD showed decreased susceptibility to tetracycline and ticarcillin-clavulanate (4). mexI and opmD knockout mutants in a subsequent study also exhibited decreased susceptibility to kanamycin, spectinomycin, carbenicillin, nalidixic acid, tetracycline, and chloramphenicol (3). Perhaps the loss of mexGHI-opmD triggered the overexpression of another RND efflux pump, similar to the overexpression of mexCD-oprJ and mexEF-oprN observed in P. aeruginosa knockout mutants of mexAB-oprM (119).
The substrate profiles of MexVW, MexPQ-OpmE, and MexMN were all examined in an efflux-deficient strain (ΔmexAB-oprM ΔmexCD-oprJ ΔmexEF-oprN ΔmexXY) harboring a plasmid coding for each system individually (122, 158). MexVW used OprM and other, as yet unidentified OMF proteins for the export of fluoroquinolones, tetracycline, chloramphenicol, and erythromycin (122). MexPQ-OpmE elevated the MICs of fluoroquinolones, tetracycline, chloramphenicol, and several macrolides in the recipient strain (158). Susceptibility to chloramphenicol and thiamphenicol was decreased in the recipient containing MexMN in combination with OprM (158).
TriABC was the last RND efflux pump to be characterized for P. aeruginosa. triABC differs from the other RND operons in that triA and triB each code for MFPs, both of which are required for substrate export (157). The OMF OpmH associates with TriABC to assemble a functional pump capable of exporting triclosan (157). No antibiotics were reported as substrates for this pump.
COREGULATION OF RESISTANCE MECHANISMS
MexCD-OprJ Overproduction and Hypersusceptibility to Antibacterials
While the overexpression of mexCD-oprJ provides resistance to several classes of antibacterial drugs, hypersusceptibility to aminoglycosides and other β-lactams is also observed (149, 203). Hypersusceptibility is defined as a fourfold or greater increase in susceptibility, and the level of hypersusceptibility is related to the level of MexCD-OprJ production. Masuda et al. demonstrated that type B mutants (most production of MexCD-OprJ) exhibit larger increases in susceptibility than do type A mutants (moderate production of MexCD-OprJ) (149). However, the mechanism(s) responsible for hypersusceptibility is much more complex than simply being associated with production of MexCD-OprJ. Phenotypic reversion studies with mexCD-oprJ-overexpressing mutants have demonstrated that hypersusceptibility to some antibiotics is not always associated with the level of mexCD-oprJ overexpression (280). Furthermore, as discussed below, a single mechanism does not account for hypersusceptibility to all affected antibiotics, and the mechanism(s) responsible for hypersusceptibility to imipenem remains uncharacterized.
Hypersusceptibility of laboratory-generated nfxB-type mutants to carbenicillin and aztreonam (MexAB-OprM substrates) has previously been linked to a concurrent downregulation of MexAB-OprM production (70). However, during the analysis of nfxB-type clinical isolates, mexB transcript and MexB protein levels were not downregulated, despite hypersusceptibility to these drugs (92). Instead, a decrease in MexAB-OprM pump activity in these strains was proposed as the mechanism (92). These results do not necessarily exclude a possible coregulation of mexCD-oprJ and oprM expression, since transcription of oprM can occur independently of that of mexAB (290).
MexCD-OprJ-overproducing mutants become hypersusceptible to additional β-lactams, including sulbenicillin, cefpodoxime, ceftriaxone, imipenem, and biapenem (151), but none of the aforementioned drugs are exported by MexAB-OprM, implying an additional mechanism(s). A disruption in the induction pathway of AmpC was suggested to cause hypersusceptibility to these β-lactams (151). Although data from our own studies also showed decreased induction of ampC expression associated with overexpression of mexCD-oprJ (274), this association was observed only in isogenic mutants from a clinical isolate with wild-type ampC expression. When the parent strains were derepressed for ampC expression, overexpression of mexC was not associated with any change in ampC expression among the isogenic mutants selected (274).
The mechanism(s) involved in hypersusceptibility to imipenem remains uncharacterized. Although other investigators have suggested that imipenem hypersusceptibility is a result of disrupted ampC induction (151), the observation that AmpC overproduction does not significantly increase imipenem MICs (63, 131, 169, 274) argues against this hypothesis. Furthermore, our own studies have shown that imipenem-hypersusceptible mutants can be selected from P. aeruginosa parent strains that are partially and fully derepressed for ampC expression, without any changes in either the expression of ampC or AmpC hydrolysis activity (274). These studies also demonstrated that imipenem hypersusceptibility is unrelated to the level of OprD, as hypersusceptible mutants failed to exhibit any changes in oprD expression or OprD protein in the outer membrane (274). Furthermore, imipenem-hypersusceptible mutants were selected from a P. aeruginosa strain that lacked a functional OprD protein (274). Characterization of the mechanism(s) responsible for imipenem hypersusceptibility among mexCD-oprJ-overexpressing P. aeruginosa strains may uncover potential drug targets to enhance the activity of carbapenems.
The mechanism responsible for hypersusceptibility to aminoglycosides remains unknown. As previously discussed, aminoglycosides are substrates for the MexXY efflux pump, and suppression of MexXY in strains overproducing MexCD-OprJ has been suggested (205), but this inverse relationship has not been observed in clinical isolates (92).
Coregulation of MexEF-OprN and OprD
One of the most intriguing and complex coregulatory pathways involves the concurrent overproduction of the MexEF-OprN efflux pump and downregulation/upregulation of the OprD porin. Clinically, this coregulatory process allows a single mutational process to impact the potencies of both the fluoroquinolones and carbapenems. To date, three proteins, MexT, MexS, and MvaT, have been linked directly or indirectly to the coregulation of MexEF-OprN and OprD in P. aeruginosa (Fig. 7).
P. aeruginosa strains that overexpress mexEF-oprN through MexT become resistant to antibiotics which are substrates for the pump (listed above) but also lose susceptibility to imipenem, which is not extruded by MexEF-OprN. Rather, the loss of susceptibility to imipenem is associated with a concurrent decrease of oprD expression and OprD (104, 150, 180). The link between pump hyperexpression and porin loss involves coregulation by MexT (Fig. 7B). Studies have shown that MexT alone is capable of downregulating oprD at the transcriptional and posttranscriptional levels, causing a significant reduction in the amount of OprD (101, 180). Further study into this mechanism of coregulation has shown that neither premature termination of transcription nor changes in transcript stability are responsible for the reduced levels of oprD transcript (278). Instead, the MexT-associated decrease in oprD expression involves decreased transcription initiation from the start-site-proximal oprD promoter (Fig. 8) (278). It is possible that MexT or another regulatory factor(s) and/or cofactors bind to the region between SS1 and SS2 and inhibit transcription initiation from SS1.
FIG. 8.
MexT-associated downregulation of oprD expression. Transcription of oprD in wild-type P. aeruginosa PAO1 initiates from two start sites, SS1 and SS2. MexT-associated downregulation of oprD expression is associated with a selective inhibition of transcription from SS1 (278). The model proposed in this figure involves the binding of a regulatory protein, potentially MexT, with or without cofactors, within the promoter region between SS1 and SS2, blocking efficient initiation of transcription from SS1.
Mutations within mexS, encoding a putative oxidoreductase/dehydrogenase, are also associated with concurrent overexpression of mexEF-oprN and downregulation of oprD expression (Fig. 7C) (241). As discussed above for regulation of MexEF-OprN, inactivation of MexS is believed to cause a buildup of metabolites that serve as effector molecules for MexT (241), which mediates the downregulation of oprD expression following activation. In contrast to MexT- and MexS-associated downregulation of oprD expression, inactivation of the global regulator MvaT causes an upregulation in both mexEF-oprN and oprD transcription (Fig. 7D) (270). However, the mechanistic pathway responsible for oprD upregulation has not been characterized.
Regulation of mexEF-oprN expression by MexT (with or without mexS inactivation) and MvaT can impact susceptibility to imipenem through oprD coregulation. However, coregulation of mexEF-oprN and oprD is not always observed, further highlighting the complexity of resistance mechanism regulation in P. aeruginosa. For example, in a recent study, an isogenic mutant overexpressing mexEF-oprN showed no phenotypic change in imipenem susceptibility, oprD transcript levels, or levels of OprD in the outer membrane (279). Sequence and expression analysis of the regulatory genes mexT, mexS, and mvaT failed to show any genetic changes in the mutant, suggesting an alternative pathway of mexEF-oprN regulation without concurrent oprD regulation (279). Reminiscent of increased mexCD-oprJ expression in mexAB-oprM and oprM knockout mutants, mexEF-oprN was also hyperexpressed in mexAB-oprM-inactivated mutants, but a change in imipenem susceptibility was not observed in the deletion mutants (119).
PREVENTING EMERGENCE OF CHROMOSOMALLY ENCODED RESISTANCE
This review has highlighted the impressive ability of P. aeruginosa to develop antibacterial resistance through mutational changes in the function and/or production of chromosomally encoded resistance mechanisms. Furthermore, the most difficult challenge with this pathogen is the ability of P. aeruginosa to become resistant during treatment of an infection. Unfortunately, the prospect for bringing new, and specifically novel, antipseudomonal drugs to clinical use in the near future is not promising. Therefore, the challenge facing us today is to slow the emergence of resistance through optimizing therapy with currently available drugs.
The two most common strategies considered to address this need are (i) optimizing therapy through our understanding of basic antibacterial pharmacodynamic principles and (ii) treating P. aeruginosa with a combination of antibacterial drugs. Although our understanding of antibacterial pharmacodynamics can help in the selection of the best antibiotic and/or dosing strategy to optimize therapy, preventing the emergence of resistance requires the inclusion of potential resistant subpopulations into the equation. Unfortunately, this is not always straightforward, since the shifts in susceptibility associated with mutations can vary widely depending upon the resistance mechanism involved, the differential impact of resistance mechanisms on specific antibiotics, and the potential cooperation of multiple resistance mechanisms. Therefore, pharmacodynamic optimization based upon the susceptibility of the original clinical isolate does not always address the risk of resistance emerging during therapy. Some investigators have promoted replacing the MIC with the mutation prevention concentration, but this approach has not been adopted by clinical laboratories.
A more accepted approach is to treat serious P. aeruginosa infections with a combination of antibacterial agents. Although synergistic interactions are an important aspect for some drug combinations (e.g., trimethoprim-sulfamethoxazole), the primary focus of combination therapy against P. aeruginosa is preventing the emergence of resistance. The combination of an antipseudomonal β-lactam with an aminoglycoside has often been the treatment of choice for this pathogen. However, this combination does not always prevent the emergence of AmpC-mediated resistance to the β-lactams, and clinical failures are still a risk (97, 118, 147, 226, 228, 235). Therefore, the search for more effective combinations must be a priority. The combination of levofloxacin-imipenem has been shown to prevent the emergence of resistance during “therapy” of P. aeruginosa in a two-compartment pharmacodynamic model (129, 130). In these in vitro studies, the levofloxacin-imipenem combination prevented emergence of resistance in susceptible clinical isolates, as well as in strains with characterized mechanisms that provided resistance to one or both drugs in the combination. Although these two studies focused specifically on levofloxacin-imipenem, it is expected that ciprofloxacin, meropenem, and doripenem could be used as alternatives in this combination without compromising efficacy. At this time, clinical evaluation of the combination is needed.
CONCLUDING COMMENTS
Treatment of infectious diseases becomes more challenging with each passing year. Continued increases in immunosuppressed/compromised patient populations and the evolutionary advantage of bacteria to rapidly mutate and adapt to antibacterial/biocide threats in their environment make the treatment of infectious diseases a serious challenge. This is especially true for the opportunistic pathogen P. aeruginosa and its ability to develop a multidrug-resistant phenotype. Although the potential import of resistance mechanisms on mobile genetic elements is a continuing threat, perhaps the most difficult challenge we face with P. aeruginosa is its ability to rapidly develop resistance to multiple classes of antibiotics during the course of treating a patient. The chromosomal AmpC cephalosporinase, the outer membrane porin OprD, and the multitude of efflux pumps are particularly relevant to this therapeutic challenge, and the discussion presented in this review highlights the complex mechanisms and pathways by which P. aeruginosa regulates and/or coregulates their expression. As the pipeline of new drugs achieving FDA approval continues to diminish, it is critical that we look for novel strategies to combat the threat of antibacterial resistance.
One potential strategy is to target the regulation of bacterial resistance mechanisms as a pathway to enhance the potency of available drugs and, perhaps, restore the efficacy of available drugs. In addition to the development of direct inhibitors of resistance mechanisms, i.e., β-lactamase inhibitors, another strategy is to target the regulation of gene expression. Although a great deal of knowledge has been gained toward understanding the mechanisms by which P. aeruginosa regulates AmpC, OprD, and efflux pumps, it is clear that we have a long road ahead and have much to learn about how this resourceful pathogen coregulates different resistance mechanisms to meet the antibacterial challenges it faces.
Acknowledgments
Daniel Wolter created Fig. 2 through 8. We acknowledge Patrick Lane for his enhancement of the figures.
Philip Lister received research grant support from the National Institutes of Health (5R21AI070188-02), Ortho-McNeil, Merck, Wyeth-Ayerst, and Astra-Zeneca. Nancy Hanson received research grant support from the National Institutes of Health (5R21AI070188-02), Merck, Astra-Zeneca, Johnson & Johnson, and Becton-Dickinson.
Biography
 Philip Lister is a Professor of Medical Microbiology and Immunology and Associate Director of the Center for Research in Anti-Infectives and Biotechnology at Creighton University. Dr. Lister received his B.S. in microbiology from Kansas State University (1986) and his Ph.D. in medical microbiology from Creighton University (1992). After a postdoctoral fellowship, Dr. Lister joined the faculty at Creighton University in 1994 and was promoted to Professor of Medical Microbiology and Immunology in 2007, with a secondary appointment for the School of Pharmacy Science. Dr. Lister has been involved in antibacterial pharmacodynamics and resistance research for 15 years and currently serves as an Editor for the Journal of Antimicrobial Chemotherapy and on the editorial boards of other journals. Dr. Lister's research has focused on clinically important gram-positive and gram-negative pathogens, but it has been the therapeutic challenge of P. aeruginosa that has been a primary focus.
Philip Lister is a Professor of Medical Microbiology and Immunology and Associate Director of the Center for Research in Anti-Infectives and Biotechnology at Creighton University. Dr. Lister received his B.S. in microbiology from Kansas State University (1986) and his Ph.D. in medical microbiology from Creighton University (1992). After a postdoctoral fellowship, Dr. Lister joined the faculty at Creighton University in 1994 and was promoted to Professor of Medical Microbiology and Immunology in 2007, with a secondary appointment for the School of Pharmacy Science. Dr. Lister has been involved in antibacterial pharmacodynamics and resistance research for 15 years and currently serves as an Editor for the Journal of Antimicrobial Chemotherapy and on the editorial boards of other journals. Dr. Lister's research has focused on clinically important gram-positive and gram-negative pathogens, but it has been the therapeutic challenge of P. aeruginosa that has been a primary focus.
 Nancy Hanson is a Professor of Medical Microbiology and Immunology and Director of Molecular Biology for the Center for Research in Anti-Infectives and Biotechnology at Creighton University. Dr. Hanson received her Ph.D. in medical microbiology from the University of Nebraska Medical Center in 1991 and joined the faculty of Creighton University in 1995. Her area of expertise involves the study of molecular mechanisms of antibiotic resistance in gram-negative organisms, such as E. coli, K. pneumoniae, Salmonella spp., and Pseudomonas aeruginosa. Her research explores the following two aspects of antibiotic resistance mechanisms: (i) the regulation of the genes involved in resistance and (ii) the development of PCR-based diagnostic tests that can be used by clinical laboratories to detect resistance genes in clinical isolates. In 2007, Dr. Hanson was awarded researcher of the year by the Nebraska Chapter of the Cystic Fibrosis Foundation for her work on P. aeruginosa.
Nancy Hanson is a Professor of Medical Microbiology and Immunology and Director of Molecular Biology for the Center for Research in Anti-Infectives and Biotechnology at Creighton University. Dr. Hanson received her Ph.D. in medical microbiology from the University of Nebraska Medical Center in 1991 and joined the faculty of Creighton University in 1995. Her area of expertise involves the study of molecular mechanisms of antibiotic resistance in gram-negative organisms, such as E. coli, K. pneumoniae, Salmonella spp., and Pseudomonas aeruginosa. Her research explores the following two aspects of antibiotic resistance mechanisms: (i) the regulation of the genes involved in resistance and (ii) the development of PCR-based diagnostic tests that can be used by clinical laboratories to detect resistance genes in clinical isolates. In 2007, Dr. Hanson was awarded researcher of the year by the Nebraska Chapter of the Cystic Fibrosis Foundation for her work on P. aeruginosa.
 Daniel Wolter received his B.S. in biology from Gonzaga University in 1997 and his Ph.D. in medical microbiology from Creighton University in 2004. Dr. Wolter continued his training as a postdoctoral fellow in medical microbiology at Creighton University until 2008 and has since joined the Department of Pediatrics at the University of Washington as a Senior Fellow. A majority of his research has focused on the antimicrobial resistance mechanisms of Pseudomonas aeruginosa, with particular emphasis on efflux-mediated drug resistance. His current research at the University of Washington explores the genotypic and phenotypic adaptations of P. aeruginosa isolated from patients with cystic fibrosis (CF) and the polymicrobial interactions of this organism with other CF lung microbiota, such as Staphylococcus aureus.
Daniel Wolter received his B.S. in biology from Gonzaga University in 1997 and his Ph.D. in medical microbiology from Creighton University in 2004. Dr. Wolter continued his training as a postdoctoral fellow in medical microbiology at Creighton University until 2008 and has since joined the Department of Pediatrics at the University of Washington as a Senior Fellow. A majority of his research has focused on the antimicrobial resistance mechanisms of Pseudomonas aeruginosa, with particular emphasis on efflux-mediated drug resistance. His current research at the University of Washington explores the genotypic and phenotypic adaptations of P. aeruginosa isolated from patients with cystic fibrosis (CF) and the polymicrobial interactions of this organism with other CF lung microbiota, such as Staphylococcus aureus.
REFERENCES
- 1.Abdalhamid, B., P. A. Wickman, and N. D. Hanson. 2005. Correlation of ampC induction with PBP binding in Enterobacter cloacae, abstr. C1-2211. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., Washington, DC.
- 2.Adewoye, L., A. Sutherland, R. Srikumar, and K. Poole. 2002. The mexR repressor of the mexAB-oprM multidrug efflux operon in Pseudomonas aeruginosa: characterization of mutations compromising activity. J. Bacteriol. 184:4308-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aendekerk, S., S. P. Diggle, Z. Song, N. Hoiby, P. Cornelis, P. Williams, and M. Camara. 2005. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 151:1113-1125. [DOI] [PubMed] [Google Scholar]
- 4.Aendekerk, S., B. Ghysels, P. Cornelis, and C. Baysse. 2002. Characterization of a new efflux pump, MexGHI-OpmD from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148:2371-2381. [DOI] [PubMed] [Google Scholar]
- 5.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akasaka, J. R., M. Tanaka, A. Yamaguchi, and K. Sato. 2001. Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob. Agents Chemother. 45:2263-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aloush, V., S. Navon-Venezia, Y. Seigman-Igra, S. Cabili, and Y. Carmeli. 2006. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob. Agents Chemother. 50:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avison, M. B., P. Niumpsup, K. Nurmahomed, T. R. Walsh, and P. M. Bennett. 2004. Role of the ‘cre/blr-tag’ DNA sequence in regulation of gene expression by the Aeromonas hydrophila beta-lactamase regulator, BlrA. J. Antimicrob. Chemother. 53:197-202. [DOI] [PubMed] [Google Scholar]
- 9.Bagge, N., O. Ciofu, M. Hentzer, J. I. Cambell, M. Givskov, and N. Hoiby. 2002. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnaud, G., R. Labia, L. Raskine, M. J. Sanon-Le Pors, A. Philippon, and G. Arlet. 2001. Extension of resistance to cefepime and cefpirome associated to a six amino acid deletion in the H-10 helix of the cephalosporinase of an Enterobacter cloacae clinical isolate. FEMS Microbiol. Lett. 195:185-190. [DOI] [PubMed] [Google Scholar]
- 11.Bartowsky, E., and S. Normark. 1993. Interactions of wild-type and mutant AmpR of Citrobacter freundii with target DNA. Mol. Microbiol. 10:555-565. [DOI] [PubMed] [Google Scholar]
- 12.Bartowsky, E., and S. Normark. 1991. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol. Microbiol. 5:1715-1725. [DOI] [PubMed] [Google Scholar]
- 13.Bellido, F., J. C. Pechere, and R. E. W. Hancock. 1991. Reevaluation of the factors involved in the efficacy of new beta-lactams against Enterobacter cloacae. Antimicrob. Agents Chemother. 35:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bert, F., C. Branger, and N. Lambert-Zechovsky. 2002. Identification of PSE and OXA beta-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J. Antimicrob. Chemother. 50:11-18. [DOI] [PubMed] [Google Scholar]
- 15.Bisbe, J., J. M. Gatell, and J. Puig. 1988. Pseudomonas aeruginosa bacteremia: univariate and multivariate analyses of factors influencing the prognosis in 133 episodes. Rev. Infect. Dis. 10:629-635. [DOI] [PubMed] [Google Scholar]
- 16.Bishop, R. E., and J. H. Weiner. 1993. Overproduction, solubilization, purification, and DNA-binding properties of AmpR from Citrobacter freundii. Eur. J. Biochem. 213:405-412. [DOI] [PubMed] [Google Scholar]
- 17.Blanc, D. S., C. Petignat, B. Janin, J. Bille, and P. Fancioli. 1998. Frequency and molecular diversity of Pseudomonas aeruginosa upon admission and during hospitalization: a prospective epidemiologic study. Clin. Microbiol. Infect. 4:242-247. [DOI] [PubMed] [Google Scholar]
- 18.Bonten, M. J., D. C. Bergmans, H. Speijer, and E. E. Stobberingh. 1999. Characteristics of polyclonal endemicity of Pseudomonas aeruginosa colonization in intensive care units. Implications for infection control. Am. J. Crit. Care Med. 160:1212-1219. [DOI] [PubMed] [Google Scholar]
- 19.Boutoille, D., S. Corvec, N. Caroff, C. Giraudeau, E. Espaze, J. Caillon, P. Plesiat, and A. Reynaud. 2004. Detection of an IS21 insertion sequence in the mexR gene of Pseudomonas aeruginosa increasing beta-lactam resistance. FEMS Microbiol. Lett. 230:143-146. [DOI] [PubMed] [Google Scholar]
- 20.Breidenstein, E. B. M., B. K. Khaira, I. Wiegand, J. Overhage, and R. E. Hancock. 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown, S. D., and M. M. Traczewski. 2005. Comparative in vitro antimicrobial activity of a new carbapenem, doripenem: tentative disc diffusion criteria and quality control. J. Antimicrob. Chemother. 55:944-949. [DOI] [PubMed] [Google Scholar]
- 22.Caille, O., C. Rossier, and K. Perron. 2007. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 189:4561-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell, J. I. A., O. Cioufu, and N. Hoiby. 1997. Pseudomonas aeruginosa isolates from patients with cystic fibrosis have different beta-lactamase expression phenotypes but are homogenous in the ampC-ampR genetic region. Antimicrob. Agents Chemother. 41:1380-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423-1436. [DOI] [PubMed] [Google Scholar]
- 25.Carmeli, Y., N. Troillet, A. W. Karchmer, and M. H. Samore. 1999. Health and economic outcomes of antibiotic resistant Pseudomonas aeruginosa. Arch. Intern. Med. 159:1127-1132. [DOI] [PubMed] [Google Scholar]
- 26.Castang, S., H. R. McManus, K. H. Turner, and S. L. Dove. 2008. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. USA 105:18947-18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castanheira, M., R. E. Mendes, T. R. Walsh, A. C. Gales, and R. N. Jones. 2004. Emergence of the extended-spectrum beta-lactamase GES-1 in a Pseudomonas aeruginosa strain from Brazil: report from the Sentry antimicrobial surveillance program. Antimicrob. Agents Chemother. 48:2344-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castanheira, M., M. A. Toleman, R. N. Jones, F. J. Schmidt, and T. R. Walsh. 2004. Molecular characterization of a beta-lactamase gene, blaGIM-1, encoding a new subclass of metallo-beta-lactamase. Antimicrob. Agents Chemother. 48:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, H., J. Hu, P. R. Chen, L. Lan, Z. Li, L. M. Hicks, A. R. Dinner, and C. He. 2008. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc. Natl. Acad. Sci. USA 105:13586-13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng, Q., and J. T. Park. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184:6434-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuanchuen, R., J. B. Gaynor, R. Karkhoff-Schweizer, and H. P. Schweizer. 2005. Molecular characterization of MexL, the transcriptional repressor of the mexJK multidrug efflux operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:1844-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chuanchuen, R., T. Murata, N. Gotoh, and H. P. Schweizer. 2005. Substrate-dependent utilization of OprM and OpmH by the Pseudomonas aeruginosa MexJK efflux pump. Antimicrob. Agents Chemother. 49:2133-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colom, K., A. Fdz-Aranguiz, E. Suinaga, and R. Cisterna. 1995. Emergence of resistance to beta-lactam agents in Pseudomonas aeruginosa with group 1 beta-lactamases in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 14:964-971. [DOI] [PubMed] [Google Scholar]
- 35.Daigle, D. M., L. Cao, S. Fraud, M. S. Wilke, A. Pacey, R. Klinoski, N. C. Strynadka, C. R. Dean, and K. Poole. 2007. Protein modulator of multidrug efflux gene expression in Pseudomonas aeruginosa. J. Bacteriol. 189:5441-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danel, F., L. M. Hall, D. Gur, H. E. Akalin, and D. M. Livermore. 1995. Transferable production of PER-1 beta-lactamase in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 35:281-294. [DOI] [PubMed] [Google Scholar]
- 37.Dean, C. R., M. A. Visalli, S. J. Projan, P. E. Sum, and P. A. Bradford. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:972-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Kievit, T. R., M. D. Parkins, R. J. Gillis, R. Srikumar, H. Ceri, K. Poole, B. H. Iglewski, and D. G. Storey. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 45:1761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietz, H., D. Pfeifle, and B. Wiedemann. 1996. Location of N-acetylmuramyl-l-alanyl-d-glutamylmesodiaminopimelic acid, presumed signal molecule for beta-lactamase induction, in the bacterial cell. Antimicrob. Agents Chemother. 40:2173-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dietz, H., D. Pfeifle, and B. Wiedemann. 1997. The signal molecule for beta-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 41:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimatatac, E. L., M. M. Alejandria, C. Montalban, C. Pineda, C. Ang, and R. Delino. 2003. Clinical outcomes and costs of care of antibiotic resistant Pseudomonas aeruginosa infections. Philipp. J. Microbiol. Infect. Dis. 32:159-167. [Google Scholar]
- 42.Doi, Y., D. de Oliveira Garcia, J. Adams, and D. Paterson. 2007. Coproduction of novel 16S rRNA methylase RmtD and metallo-beta-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob. Agents Chemother. 51:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doi, Y., K. Yokoyama, K. Yamane, J. Wachino, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dotsch, A., T. Becker, C. Pommerenke, Z. Magnowska, L. Jansch, and S. Haussler. 2009. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehrhardt, A. F., and C. C. Sanders. 1993. Beta-lactam resistance amongst Enterobacter species. J. Antimicrob. Chemother. 32(Suppl. B):1-11. [DOI] [PubMed] [Google Scholar]
- 46.Ehrhardt, A. F., C. C. Sanders, J. R. Romero, and J. S. Leser. 1996. Sequencing and analysis of four new Enterobacter ampD alleles. Antimicrob. Agents Chemother. 40:1953-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El Amin, N., C. G. Giske, S. Jalal, B. Keijser, G. Kronvall, and B. Wretlind. 2005. Carbapenem resistance mechanisms in Pseudomonas aeruginosa: alterations of porin OprD and efflux proteins do not fully explain resistance patterns observe in clinical isolates. APMIS 113:187-196. [DOI] [PubMed] [Google Scholar]
- 48.Emori, T. G., and R. P. Gaynes. 1993. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erol, S., U. Altoparlak, M. N. Akcay, F. Celebi, and M. Parlak. 2004. Changes of microbial flora and wound colonization in burned patients. Burns 30:357-361. [DOI] [PubMed] [Google Scholar]
- 50.Eron, L. J., R. I. Goldenberg, D. M. Poretz, and C. H. Park. 1983. Piperacillin therapy for Pseudomonas infections. South. Med. J. 76:859-862. [DOI] [PubMed] [Google Scholar]
- 51.Evans, J. C., and H. Segal. 2007. A novel insertion sequence, ISPA26, in oprD of Pseudomonas aeruginosa is associated with carbapenem resistance. Antimicrob. Agents Chemother. 51:3776-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans, K., and K. Poole. 1999. The MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa is growth-phase regulated. FEMS Microbiol. Lett. 173:35-39. [DOI] [PubMed] [Google Scholar]
- 54.Flamm, R. K., M. K. Weaver, C. Thornsberry, M. E. Jones, J. A. Karlowsky, and D. F. Sahm. 2004. Factors associated with relative rates of antibiotic resistance in Pseudomonas aeruginosa isolates tested in clinical laboratories in the United States from 1999 to 2002. Antimicrob. Agents Chemother. 48:2431-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraud, S., A. J. Campigotto, Z. Chen, and K. Poole. 2008. MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa: involvement in chlorhexidine resistance and induction by membrane-damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob. Agents Chemother. 52:4478-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freeman, L. 1916. Chronic general infection with the Bacillus pyocyaneus. Ann. Surg. 64:195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fritsche, T. R., H. S. Sader, M. A. Toleman, T. R. Walsh, and R. N. Jones. 2005. Emerging metallo-beta-lactamase-mediated resistances: a summary report from the worldwide SENTRY antimicrobial surveillance program. Clin. Infect. Dis. 41(Suppl. 4):S276-S278. [DOI] [PubMed] [Google Scholar]
- 58.Fukuda, H., M. Hosaka, S. Iyobe, N. Gotoh, T. Nishino, and K. Hirai. 1995. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:790-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gales, A. C., L. C. Menezes, S. Silbert, and H. S. Sader. 2003. Dissemination in distinct Brazilian regions of an epidemic carbapenem-resistant Pseudomonas aeruginosa producing SPM metallo-beta-lactamase. J. Antimicrob. Chemother. 52:699-702. [DOI] [PubMed] [Google Scholar]
- 60.Galimand, M., P. Courvalin, and T. Lambert. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galimand, M., P. Sabtcheva, P. Courvalin, and T. Lambert. 2005. Worldwide disseminated armA aminoglycoside resistance methylase gene borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 49:2949-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gasink, L. B., N. O. Fishman, M. G. Weiner, I. Nachamkin, W. B. Bilker, and E. Lautenbach. 2006. Fluoroquinolone-resistant Pseudomonas aeruginosa: assessment of risk factors and clinical impact. Am. J. Med. 119:526e19-526e25. [DOI] [PubMed] [Google Scholar]
- 63.Gates, M. L., C. C. Sanders, R. V. Goering, and W. E. Sanders. 1986. Evidence for multiple forms of type I chromosomal beta-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 30:453-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaynes, R., and J. R. Edwards. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41:848-854. [DOI] [PubMed] [Google Scholar]
- 65.Gessard, C. 1984. Classics in infectious diseases. On the blue and green coloration that appears on bandages. Rev. Infect. Dis. 6(Suppl. 3):S775-S776. [PubMed] [Google Scholar]
- 66.Gillis, R. J., K. G. White, K. H. Choi, V. E. Wagner, H. P. Schweizer, and B. H. Iglewski. 2005. Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 49:3858-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Girlich, D., T. Naas, and P. Nordmann. 2004. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:2043-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez-Zorn, B., A. Catalan, J. A. Escudero, L. Dominguez, T. Teshager, C. Porrero, and M. A. Moreno. 2005. Genetic basis for dissemination of armA. J. Antimicrob. Chemother. 56:583-585. [DOI] [PubMed] [Google Scholar]
- 69.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gotoh, N., H. Tsujimoto, M. Tsuda, K. Okamoto, A. Nomura, T. Wada, M. Nakahashi, and T. Nishino. 1998. Characterization of the MexC-MexD-OprJ multidrug efflux system in delta-mexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gribble, M. J., A. W. Chow, S. C. Naiman, J. A. Smith, W. R. Bowie, S. L. Sacks, L. Grossman, N. Buskard, G. H. Growe, and L. H. Plenderleith. 1983. Prospective randomized trial of piperacillin monotherapy versus carboxypenicillin-aminoglycoside combination regimens in the empirical treatment of serious bacterial infections. Antimicrob. Agents Chemother. 24:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grundstrom, T., and S. Normark. 1985. Initiation of translation makes attenuation of ampC in E. coli dependent on growth rate. Mol. Gen. Genet. 198:411-415. [DOI] [PubMed] [Google Scholar]
- 73.Gutierrez, O., C. Juan, E. Cercenado, F. Navarro, E. Bouza, P. Coll, J. L. Perez, and A. Oliver. 2007. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob. Agents Chemother. 51:4329-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hancock, R. E. W. 1998. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis. 27(Suppl. 1):S93-S99. [DOI] [PubMed] [Google Scholar]
- 75.Hancock, R. E. W., and F. S. Brinkman. 2002. Function of pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56:17-38. [DOI] [PubMed] [Google Scholar]
- 76.Hanson, N. D., and C. C. Sanders. 1999. Regulation of inducible AmpC beta-lactamase expression among Enterobacteriaceae. Curr. Pharm. Des. 5:881-894. [PubMed] [Google Scholar]
- 77.Harris, A. A., L. Goodman, and S. Levin. 1984. Community-acquired Pseudomonas aeruginosa pneumonia associated with the use of a home humidifier. West. J. Med. 141:521-523. [PMC free article] [PubMed] [Google Scholar]
- 78.Hiraoka, M., S. Masuyoshi, S. Mitsuhashi, K. Tomatsu, and M. Inoue. 1988. Cephalosporinase interactions and antimicrobial activity of BMY-28142, ceftazidime, and cefotaxime. J. Antibiot. 41:86-93. [DOI] [PubMed] [Google Scholar]
- 79.Hocquet, D., P. Nordmann, F. El Garch, L. Cabanne, and P. Plesiat. 2006. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holtje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holtje, J. V., U. Kopp, A. Ursinus, and B. Wiedemann. 1994. The negative regulator of beta-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol. Lett. 122:159-164. [DOI] [PubMed] [Google Scholar]
- 82.Honore, N., M. H. Nicolas, and S. T. Cole. 1986. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 20:3709-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Honore, N., M. H. Nicolas, and S. T. Cole. 1989. Regulation of enterobacterial cephalosporinase production: the role of a membrane-bound sensory transducer. Mol. Microbiol. 3:1121-1130. [DOI] [PubMed] [Google Scholar]
- 84.Hooper, D. C. 2000. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31:S24-S28. [DOI] [PubMed] [Google Scholar]
- 85.Islam, S., S. Jalal, and B. Wretlind. 2004. Expression of MexXY efflux pump in amikacin-resistant isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 10:877-883. [DOI] [PubMed] [Google Scholar]
- 86.Jacobs, C., J. M. Frere, and S. Normark. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible beta-lactam resistance in gram-negative bacteria. Cell 88:823-832. [DOI] [PubMed] [Google Scholar]
- 87.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreuix, J. van Heijenoort, J. T. Park, S. Normark, and J.-M. Frere. 1995. AmpD, essential for both beta-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:533-539. [DOI] [PubMed] [Google Scholar]
- 89.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47:559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jalal, S., O. Ciofu, N. Hoiby, N. Gotoh, and B. Wretlind. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 44:710-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jalal, S., and B. Wretlind. 1998. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microb. Drug Resist. 4:257-261. [DOI] [PubMed] [Google Scholar]
- 92.Jeannot, K., S. Elsen, T. Kohler, I. Attree, C. van Delden, and P. Plesiat. 2008. Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob. Agents Chemother. 52:2455-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jeannot, K., M. L. Sobel, F. El Garch, K. Poole, and P. Plesiat. 2005. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 187:5341-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jimenez-Lucho, V. E., L. D. Saravolatz, A. A. Meideros, and D. Pohlod. 1986. Failure of therapy in Pseudomonas endocarditis: selection of resistant mutant. J. Infect. Dis. 154:64-68. [DOI] [PubMed] [Google Scholar]
- 95.Jones, M. E., D. C. Draghi, C. Thornsberry, J. A. Karlowsky, D. F. Sahm, and R. P. Wenzel. 2004. Emerging resistance among bacterial pathogens in the intensive care unit—a European and North American surveillance study (2000-2002). Ann. Clin. Microbiol. Antimicrob. 3:14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jones, R. N., D. J. Biedenbach, H. S. Sader, T. R. Fritsche, M. A. Toleman, and T. R. Walsh. 2005. Emerging epidemic of metallo-beta-lactamase-mediated resistances. Diagn. Microbiol. Infect. Dis. 51:77-84. [DOI] [PubMed] [Google Scholar]
- 97.Juan, C., M. D. Macia, O. Gutierrez, C. Vidal, J. L. Perez, and A. Oliver. 2005. Molecular mechanisms of beta-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 49:4733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Juan, C., B. Moya, J. L. Perez, and A. Oliver. 2006. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level beta-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 50:1780-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karlowsky, J. A., D. C. Draghi, M. E. Jones, C. Thornsberry, I. R. Friedland, and D. F. Sahm. 2003. Surveillance for antimicrobial susceptibility among clinical isolates of Pseudomonas aeruginosa and Acinetobacter baumannii from hospitalized patients in the United States, 1998-2001. Antimicrob. Agents Chemother. 47:1681-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karlowsky, J. A., L. J. Kelly, C. Thornsberry, M. E. Jones, A. T. Evangelista, I. A. Critchley, and D. F. Sahm. 2002. Susceptibility to fluoroquinolones among commonly isolated gram-negative bacilli in 2000: TRUST and TSN data for the United States. Int. J. Antimicrob. Agents 19:21-31. [DOI] [PubMed] [Google Scholar]
- 101.Kohler, T., S. F. Epp, L. K. Curty, and J. C. Pechere. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181:6300-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kohler, T., M. Kok, M. Michea-Hamzehpour, P. Plesiat, T. Gotoh, T. Nishino, L. K. Curty, and J. C. Pechere. 1996. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:2288-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kohler, T., M. Michea-Hamzehpour, S. F. Epp, and J. C. Pechere. 1999. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob. Agents Chemother. 43:424-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kohler, T., M. Michea-Hamzehpour, U. Henz, N. Gotoh, L. K. Curty, and J. C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 105.Kohler, T., C. van Delden, L. K. Curty, M. M. Hamzehpour, and J. C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kolleff, M. H., A. Shorr, Y. P. Tabak, V. Gupta, L. Z. Liu, and R. S. Johannes. 2005. Epidemiology and outcomes of health-care-associated pneumonia: results from a large US database of culture-positive pneumonia. Chest 128:3854-3862. [DOI] [PubMed] [Google Scholar]
- 107.Kong, K.-F., S. R. Jayawardena, S. D. Indulkar, A. del Puerto, C.-L. Koh, N. Hoiby, and K. Mathee. 2005. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 49:4567-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kopp, U., B. Wiedemann, S. Linquist, and S. Normark. 1993. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob. Agents Chemother. 37:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Korfmann, G., and C. C. Sanders. 1989. ampG is essential for high-level expression of AmpC beta-lactamase activity in Enterobacter cloacae. Antimicrob. Agents Chemother. 33:1946-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Korfmann, G., C. C. Sanders, and E. S. Moland. 1991. Altered phenotypes associated with ampD mutations in Enterobacter cloacae. Antimicrob. Agents Chemother. 35:358-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuga, A., R. Okamoto, and M. Inoue. 2000. ampR gene mutations that greatly increase class C beta-lactamase activity in Enterobacter cloacae. Antimicrob. Agents Chemother. 44:561-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Labia, R., A. Morand, and J. Peduzzi. 1986. Timentin and beta-lactamases. Antimicrob. Agents Chemother. 17(Suppl. C):17-26. [DOI] [PubMed] [Google Scholar]
- 113.Landman, D., S. Bratu, S. Kochar, M. Panwar, M. Trehan, M. Doymaz, and J. Quale. 2007. Evolution of antimicrobial resistance among Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae in Brooklyn, N.Y. J. Antimicrob. Chemother. 60:78-82. [DOI] [PubMed] [Google Scholar]
- 114.Langaee, T. Y., M. Dargis, and A. Huletsky. 1998. An ampD gene in Pseudomonas aeruginosa encodes a negative regulator of AmpC beta-lactamase expression. Antimicrob. Agents Chemother. 42:3296-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Langaee, T. Y., L. Gagnon, and A. Huletsky. 2000. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC beta-lactamase expression. Antimicrob. Agents Chemother. 44:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee, J. K., Y. S. Lee, Y. K. Park, and B. S. Kim. 2005. Alterations in the GyrA and GyrB subunits of topoisomerase II and the ParC and ParE subunits of topoisomerase IV in ciprofloxacin-resistant clinical isolates of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 25:290-295. [DOI] [PubMed] [Google Scholar]
- 117.Lee, K., G. Y. Ha, B. M. Shin, J. J. Kim, J. O. Kang, S. J. Jang, D. Yong, and Y. Chong. 2004. Metallo-beta-lactamase-producing gram-negative bacilli in Korean Nationwide Surveillance of Antimicrobial Resistance group hospitals 2003: continued prevalence of VIM-producing Pseudomonas spp. and increase of IMP-producing Acinetobacter spp. Diagn. Microbiol. Infect. Dis. 50:51-58. [DOI] [PubMed] [Google Scholar]
- 118.Letendre, E. D., R. Mantha, and P. L. Turgeon. 1988. Selection of resistance by piperacillin during Pseudomonas aeruginosa endocarditis. J. Antimicrob. Chemother. 22:557-562. [DOI] [PubMed] [Google Scholar]
- 119.Li, X. Z., N. Barre, and K. Poole. 2000. Influence of MexA-MexB-OprM multidrug efflux system on expression of the MexC-MexD-OprJ and MexE-MexF-OprN multidrug efflux systems in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 46:885-893. [DOI] [PubMed] [Google Scholar]
- 120.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li, X. Z., L. Zhang, R. Srikumar, and K. Poole. 1998. Beta-lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li, Y., T. Mima, Y. Komori, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. A new member of the tripartite multidrug efflux pumps, MexVW-OprM, in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 52:572-575. [DOI] [PubMed] [Google Scholar]
- 123.Lindberg, F., S. Lindquist, and S. Normark. 1988. Genetic basis of induction and overproduction of chromosomal class I beta-lactamase in nonfastidious gram-negative bacilli. Rev. Infect. Dis. 10:782-785. [DOI] [PubMed] [Google Scholar]
- 124.Lindberg, F., S. Lindquist, and S. Normark. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii beta-lactamase. J. Bacteriol. 169:1923-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lindberg, F., L. Westman, and S. Normark. 1985. Regulatory components in Citrobacter freundii ampC beta-lactamase induction. Proc. Natl. Acad. Sci. USA 82:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lindquist, S., F. Lindberg, and S. Normark. 1989. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible AmpC beta-lactamase. J. Bacteriol. 171:3746-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lindquist, S., K. Weston-Hafer, H. Schmidt, C. Pul, G. Korfmann, J. Erickson, C. C. Sanders, H. H. Martin, and S. Normark. 1993. AmpG, a signal transducer in chromosomal beta-lactamase induction. Mol. Microbiol. 9:703-715. [DOI] [PubMed] [Google Scholar]
- 128.Lister, P. D., V. M. Gardner, and C. C. Sanders. 1999. Clavulanate induces expression of the Pseudomonas aeruginosa AmpC cephalosporinase at physiologically relevant concentrations and antagonizes the antibacterial activity of ticarcillin. Antimicrob. Agents Chemother. 43:882-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lister, P. D., and D. J. Wolter. 2005. Levofloxacin-imipenem combination prevents the emergence of resistance among clinical isolates of Pseudomonas aeruginosa. Clin. Infect. Dis. 40:S105-S114. [DOI] [PubMed] [Google Scholar]
- 130.Lister, P. D., D. J. Wolter, P. A. Wickman, and M. D. Reisbig. 2006. Levofloxacin/imipenem prevents the emergence of high-level resistance among Pseudomonas aeruginosa strains already lacking susceptibility to one or both drugs. J. Antimicrob. Chemother. 57:999-1003. [DOI] [PubMed] [Google Scholar]
- 131.Livermore, D. M. 1992. Interplay of impermeability and chromosomal beta-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:2046-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare. Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 133.Livermore, D. M. 2003. The threat from the pink corner. Ann. Med. 35:226-234. [DOI] [PubMed] [Google Scholar]
- 134.Livermore, D. M., and N. Woodford. 2006. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas, and Acinetobacter. Trends Microbiol. 14:413-420. [DOI] [PubMed] [Google Scholar]
- 135.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 136.Livermore, D. M., and Y.-J. Yang. 1987. Beta-lactamase lability and inducer power of newer beta-lactam antibiotics in relation to their activity against beta-lactamase-inducibility mutants of Pseudomonas aeruginosa. J. Infect. Dis. 4:775-782. [DOI] [PubMed] [Google Scholar]
- 137.Llanes, C., D. Hocquet, C. Vogne, D. Benali-Baitich, C. Neuwirth, and P. Plesiat. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lodge, J. M., S. J. W. Busby, and L. J. Piddock. 1993. Investigation of the Pseudomonas aeruginosa ampR gene and its role at the chromosomal ampC beta-lactamase promoter. FEMS Microbiol. Lett. 111:315-320. [DOI] [PubMed] [Google Scholar]
- 139.Lodge, J. M., S. D. Minchin, L. J. Piddock, and S. J. W. Busby. 1990. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC beta-lactamase. Biochem. J. 272:627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lynch, M. J., G. L. Drusano, and H. L. Mobley. 1987. Emergence of resistance to imipenem in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 31:1892-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mammeri, H., M. Galleni, and P. Nordmann. 2009. Role of the Ser-287-Asn replacement in the hydrolysis spectrum extension of AmpC beta-lactamases in Escherichia coli. Antimicrob. Agents Chemother. 53:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mammeri, H., P. Nordmann, A. Berkani, and F. Eb. 2008. Contribution of extended-spectrum AmpC (ESAC) beta-lactamases to carbapenem resistance in Escherichia coli. FEMS Microbiol. Lett. 282:238-240. [DOI] [PubMed] [Google Scholar]
- 143.Mammeri, H., L. Poirel, P. Bemer, H. Drugeon, and P. Nordmann. 2004. Resistance to cefepime and cefpirome due to a 4-amino-acid deletion in the chromosome-encoded AmpC beta-lactamase of a Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 48:716-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Margaret, B. S., G. L. Drusano, and H. C. Standiford. 1989. Emergence of resistance to carbapenem antibiotics in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 24(Suppl. A):161-167. [DOI] [PubMed] [Google Scholar]
- 145.Maseda, H., K. Saito, A. Nakajima, and T. Nakae. 2000. Variation of the mexT gene, a regulator of the MexEF-OprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 192:107-112. [DOI] [PubMed] [Google Scholar]
- 146.Maseda, H., I. Sawada, K. Saito, H. Uchiyama, T. Nakae, and N. Nomura. 2004. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:1320-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Masterton, R. G., P. J. Garner, N. A. Harrison, and D. J. Rainford. 1987. Timentin resistance. Lancet ii:975-976. [DOI] [PubMed] [Google Scholar]
- 148.Masuda, N., N. Gotoh, C. Ishii, E. Sakagawa, S. Ohya, and T. Nishino. 1999. Interplay between chromosomal beta-lactamase and the MexAB-OprM efflux system in intrinsic resistance to beta-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Masuda, N., N. Gotoh, S. Ohya, and T. Nishino. 1996. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:909-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, and T. Nishino. 2001. Hypersusceptibility of the Pseudomonas aeruginosa nfxB mutant to beta-lactams due to reduced expression of the AmpC beta-lactamase. Antimicrob. Agents Chemother. 45:1284-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Matsumura, N., S. Minami, and S. Mitsuhashi. 1998. Sequences of homologous beta-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob. Agents Chemother. 42:176-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Matsuo, Y., S. Eda, N. Gotoh, E. Yoshihara, and T. Nakae. 2004. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol. Lett. 238:23-28. [DOI] [PubMed] [Google Scholar]
- 155.Mavroidi, A., E. Tzelepi, A. Tsakris, V. Miriagou, D. Sofianou, and L. S. Tzouvelekis. 2001. An integron associated beta-lactamase (IBC-2) from Pseudomonas aeruginosa is a variant of the extended-spectrum beta-lactamase IBC-1. J. Antimicrob. Chemother. 48:627-630. [DOI] [PubMed] [Google Scholar]
- 156.Micek, S. T., A. E. Lloyd, D. J. Ritchie, R. M. Reichley, V. J. Fraser, and M. H. Kollef. 2005. Pseudomonas aeruginosa bloodstream infection: importance of appropriate initial antimicrobial treatment. Antimicrob. Agents Chemother. 49:1306-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mima, T., S. Joshi, M. Gomez-Escalada, and H. P. Schweizer. 2007. Identification and characterization of TriABC-OpmH, a triclosan efflux pump of Pseudomonas aeruginosa requiring two membrane fusion proteins. J. Bacteriol. 189:7600-7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mima, T., H. Sekiya, T. Mizushima, T. Kuroda, and T. Tsuchiya. 2005. Gene cloning and properties of the RND-type multidrug efflux pump, MexPQ-OpmE and MexMN-OprM from Pseudomonas aeruginosa. Microbiol. Immunol. 49:999-1002. [DOI] [PubMed] [Google Scholar]
- 159.Mine, T., Y. Morita, T. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Morita, Y., L. Cao, V. C. Gould, M. B. Avison, and K. Poole. 2006. nalD encodes a second repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J. Bacteriol. 188:8649-8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Morita, Y., N. Kimura, T. Mima, T. Mizushima, and T. Tsuchiya. 2001. Roles of MexXY- and MexAB-multidrug efflux pumps in intrinsic multidrug resistance of Pseudomonas aeruginosa. J. Gen. Appl. Microbiol. 47:27-32. [DOI] [PubMed] [Google Scholar]
- 162.Morita, Y., Y. Komori, T. Mima, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2001. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol. Lett. 202:139-143. [DOI] [PubMed] [Google Scholar]
- 163.Morita, Y., M. L. Sobel, and K. Poole. 2006. Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J. Bacteriol. 188:1847-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Morrison, A. J., and R. P. Wenzel. 1984. Epidemiology of infections due to Pseudomonas aeruginosa. Rev. Infect. Dis. 6(Suppl. 3):S627-S642. [DOI] [PubMed] [Google Scholar]
- 165.Mouneimne, H., J. Robert, V. Jarlier, and E. Cambau. 1999. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Moya, B., A. Dotsch, C. Juan, J. Blazquez, L. Zamorano, S. Haussler, and A. Oliver. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 5:e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Moya, B., C. Juan, S. Alberti, J. L. Perez, and A. Oliver. 2008. Benefit of having multiple ampD genes for acquiring beta-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:3694-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Murata, T., N. Gotoh, and T. Nishino. 2002. Characterization of outer membrane efflux proteins OpmE, OpmD and OpmB of Pseudomonas aeruginosa: molecular cloning and development of specific antisera. FEMS Microbiol. Lett. 217:57-63. [DOI] [PubMed] [Google Scholar]
- 169.Mushtaq, S., Y. Ge, and D. M. Livermore. 2004. Doripenem versus Pseudomonas aeruginosa in vitro: activity against characterized isolates, mutants, and transconjugants and resistance selection potential. Antimicrob. Agents Chemother. 48:3086-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Naas, T., and P. Nordmann. 1999. OXA-type beta-lactamases. Curr. Pharm. Des. 5:865-879. [PubMed] [Google Scholar]
- 171.National Nosocomial Infections Surveillance System. 1998. National Nosocomial Infections Surveillance (NNIS) System report, data summary from October 1986 to April 1998, issued June 1998. Am. J. Infect. Control 26:522-533. [DOI] [PubMed] [Google Scholar]
- 172.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nikaido, H. 1989. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob. Agents Chemother. 33:1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Niumsup, P., A. M. Simm, K. Nurmahomed, T. R. Walsh, P. M. Bennett, and M. B. Avison. 2003. Genetic linkage of the penicillinase gene, amp, and blrAB, encoding the regulator of beta-lactamase expression in Aeromonas spp. J. Antimicrob. Chemother. 51:1351-1358. [DOI] [PubMed] [Google Scholar]
- 175.Nordmann, P., and T. Nass. 1994. Sequence analysis of PER-1 extended-spectrum beta-lactamase from Pseudomonas aeruginosa and comparison with class A beta-lactamases. Antimicrob. Agents Chemother. 38:104-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 177.Normark, S., T. Edlund, T. Grundstrom, S. Bergstrom, and H. Wolf-Watz. 1977. Escherichia coli K-12 mutants hyperproducing chromosomal beta-lactamase by gene repetitions. J. Bacteriol. 132:912-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Obritsch, M. D., D. N. Fish, R. MacLaren, and R. Jung. 2004. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob. Agents Chemother. 48:4606-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ochs, M. M., C.-D. Lu, R. E. W. Hancock, and A. T. Abdelal. 1999. Amino acid-mediated induction of the basic amino acid-specific outer membrane porin OprD from Pseudomonas aeruginosa. J. Bacteriol. 181:5426-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ochs, M. M., M. P. McCusker, M. Bains, and R. E. Hancock. 1999. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob. Agents Chemother. 43:1085-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Oh, H., J. Stenhoff, S. Jalal, and B. Wretlind. 2003. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb. Drug Resist. 9:323-328. [DOI] [PubMed] [Google Scholar]
- 182.Ohara, T., and K. Itoh. 2003. Significance of Pseudomonas aeruginosa colonization of the gastrointestinal tract. Intern. Med. 42:1072-1076. [DOI] [PubMed] [Google Scholar]
- 183.Oliva, B., P. M. Bennett, and I. Chopra. 1989. Penicillin-binding protein 2 is required for induction of the Citrobacter freundii class I chromosomal beta-lactamase in Escherichia coli. Antimicrob. Agents Chemother. 33:1116-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pai, H., J. Kim, J. H. Lee, K. W. Choe, and N. Gotoh. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 45:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Palma, M., J. Zurita, J. A. Ferraras, S. Worgall, D. H. Larone, L. Shi, F. Campagne, and L. E. Quadri. 2005. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect. Immun. 73:2958-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)-Ib-cr, encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Peleg, A. Y., C. Franklin, J. Bell, and D. W. Spelman. 2004. Emergence of IMP-4 metallo-beta-lactamase in a clinical isolate from Australia. J. Antimicrob. Chemother. 54:699-700. [DOI] [PubMed] [Google Scholar]
- 188.Perez, F. J., C. Gimeno, D. Navarro, and J. Garcia-de-Lomas. 1996. Meropenem permeation through the outer membrane of Pseudomonas aeruginosa can involve pathways other than OprD porin channel. Chemotherapy 42:210-214. [DOI] [PubMed] [Google Scholar]
- 189.Perron, K., O. Caille, C. Rossier, C. Van Delden, J. L. Dumas, and T. Kohler. 2004. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 279:8761-8768. [DOI] [PubMed] [Google Scholar]
- 190.Pfeifle, D., E. Janas, and B. Wiedemann. 2000. Role of penicillin-binding proteins in the initiation of AmpC beta-lactamase expression in Enterobacter cloacae. Antimicrob. Agents Chemother. 44:169-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Philippon, L. N., T. Naas, A.-T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Picao, R. C., L. Poirel, A. C. Gales, and P. Nordmann. 2009. Further identification of CTX-M-2 extended-spectrum beta-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:2225-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4:629-636. [DOI] [PubMed] [Google Scholar]
- 194.Piddock, L. J., and D. J. Griggs. 1991. Selection and characterization of cefepime-resistant gram-negative bacteria. J. Antimicrob. Chemother. 28:669-676. [DOI] [PubMed] [Google Scholar]
- 195.Pirnay, J.-P., D. De Vos, D. Mossialos, A. Vanderkelen, P. Cornelis, and M. Zizi. 2002. Analysis of the Pseudomonas aeruginosa oprD gene from clinical and environmental isolates. Environ. Microbiol. 4:872-882. [DOI] [PubMed] [Google Scholar]
- 196.Pitt, T. L. 1998. Pseudomonas, Burkholderia, and related genera, p. 1109-1138. In B. I. Duerden (ed.), Microbiology and microbial infections, vol. 2. Oxford University Press Inc., New York, NY. [Google Scholar]
- 197.Poirel, L., V. O. Rotimi, and E. M. Mokaddas. 2001. VEB-1-like extended spectrum beta-lactamases in Pseudomonas aeruginosa, Kuwait. Emerg. Infect. Dis. 17:468-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Poirel, L., M. Van De Loo, H. Mammeri, and P. Nordmann. 2005. Association of plasmid-mediated quinolone resistance with extended-spectrum beta-lactamase VEB-1. Antimicrob. Agents Chemother. 49:3091-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pollack, M. 1995. Pseudomonas aeruginosa, p. 1820-2003. In G. L. Mandell, R. Dolan, and J. E. Bennett (ed.), Principles and practices of infectious diseases. Churchill Livingstone, New York, NY.
- 200.Poole, K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Poole, K. 2002. Outer membranes and efflux: the path to multidrug resistance in gram-negative bacteria. Curr. Pharm. Biotechnol. 3:77-98. [DOI] [PubMed] [Google Scholar]
- 203.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J. Yamagishi, X. Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 204.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 205.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms, and clinical significance. Curr. Top. Med. Chem. 1:59-71. [DOI] [PubMed] [Google Scholar]
- 206.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Pumbwe, L., and L. J. V. Piddock. 2000. Two efflux systems expressed simultaneously in multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2861-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Queenan, A. M., W. Shang, M. Kania, M. G. P. Page, and K. Bush. 2007. Interactions of ceftobiprole with beta-lactamases from molecular classes A to D. Antimicrob. Agents Chemother. 51:3089-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Quinn, J. P., E. J. Dudek, C. A. DiVincenzo, D. A. Lucks, and S. A. Lerner. 1986. Emergence of resistance to imipenem during therapy for Pseudomonas aeruginosa infections. J. Infect. Dis. 154:289-294. [DOI] [PubMed] [Google Scholar]
- 210.Quinn, J. P., A. E. Studemeister, C. A. DiVincenzo, and S. A. Lerner. 1988. Resistance to imipenem in Pseudomonas aeruginosa: clinical experience and biochemical mechanisms. Rev. Infect. Dis. 10:892-898. [DOI] [PubMed] [Google Scholar]
- 211.Rhomberg, P. R., L. M. Deshpande, J. T. Kirby, and R. N. Jones. 2007. Activity of meropenem as serine carbapenemases evolve in US medical centers: monitoring report from the MYSTIC program (2006). Diagn. Microbiol. Infect. Dis. 59:425-432. [DOI] [PubMed] [Google Scholar]
- 212.Rhomberg, P. R., and R. N. Jones. 2007. Contemporary activity of meropenem and comparator broad-spectrum agents: MYSTIC program report from the United States component (2005). Diagn. Microbiol. Infect. Dis. 57:207-215. [DOI] [PubMed] [Google Scholar]
- 213.Richards, M. J., J. R. Edwards, D. H. Culver, R. P. Gaynes, et al. 1999. Nosocomial infections in medical intensive care units in the United States. Crit. Care Med. 5:887-892. [DOI] [PubMed] [Google Scholar]
- 214.Richards, M. J., J. R. Edwards, D. H. Culver, R. P. Gaynes, et al. 1999. Nosocomial infections in pediatric intensive care units in the United States. Pediatrics 103:e39. [DOI] [PubMed] [Google Scholar]
- 215.Robicsek, A., J. Strahilevitz, D. Sahm, G. A. Jacoby, and D. C. Hooper. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50:2872-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rodriquez-Martinez, J.-M., L. Poirel, and P. Nordmann. 2009. Extended-spectrum cephalosporinases in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:1766-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Roh, I. K., I. J. Kim, J. H. Chung, and S. M. Byun. 1996. Affinity purification and binding characteristics of Citrobacter freundii AmpR, the transcriptional regulator of the ampC beta-lactamase gene. Biotechnol. Appl. Biochem. 23:149-154. [PubMed] [Google Scholar]
- 218.Saier, M. H., I. T. Paulsen, M. K. Sliwinski, S. S. Pao, R. A. Skurray, and H. Nikaido. 1998. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 12:265-274. [DOI] [PubMed] [Google Scholar]
- 219.Saito, K., H. Akama, E. Yoshihara, and T. Nakae. 2003. Mutations affecting DNA-binding activity of the MexR repressor of mexR-mexA-mexB-oprM operon expression. J. Bacteriol. 185:6195-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Saito, K., S. Eda, H. Maseda, and T. Nakae. 2001. Molecular mechanism of MexR-mediated regulation of MexAB-OprM efflux pump expression in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 195:23-28. [DOI] [PubMed] [Google Scholar]
- 221.Sakyo, S., H. Tomita, K. Tanimoto, S. Fujimoto, and Y. Ike. 2006. Potency of carbapenems for the prevention of carbapenem-resistant mutants of Pseudomonas aeruginosa. J. Antibiot. 59:220-228. [DOI] [PubMed] [Google Scholar]
- 222.Sanders, C. C. 1992. Beta-lactamases of gram-negative bacteria: new challenges for new drugs. Clin. Infect. Dis. 14:1089-1099. [DOI] [PubMed] [Google Scholar]
- 223.Sanders, C. C. 1993. Cefepime: the next generation? Clin. Infect. Dis. 17:369-379. [PubMed] [Google Scholar]
- 224.Sanders, C. C., P. A. Bradford, A. F. Ehrhardt, K. Bush, K. D. Young, T. A. Henderson, and W. E. Sanders. 1997. Penicillin-binding proteins and induction of AmpC beta-lactamase. Antimicrob. Agents Chemother. 41:2013-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sanders, C. C., M. L. Gates, and W. E. Sanders. 1988. Heterogeneity of class I beta-lactamase expression in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 32:1893-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sanders, C. C., and E. C. Sanders. 1983. Emergence of resistance during therapy with newer beta-lactam antibiotics: role of inducible beta-lactamases and implications for the future. Rev. Infect. Dis. 5:639-648. [DOI] [PubMed] [Google Scholar]
- 227.Sanders, C. C., and W. E. Sanders. 1986. Type I beta-lactamases of gram-negative bacteria: interactions with beta-lactam antibiotics. J. Infect. Dis. 154:792-800. [DOI] [PubMed] [Google Scholar]
- 228.Sanders, W. E., and C. C. Sanders. 1988. Inducible beta-lactamases: clinical and epidemiological implications for use of newer cephalosporins. Rev. Infect. Dis. 10:830-838. [DOI] [PubMed] [Google Scholar]
- 229.Satake, S., H. Yoneyama, and T. Nakae. 1991. Role of OmpD2 and chromosomal beta-lactamase in carbapenem resistance in clinical isolates of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 28:199-207. [DOI] [PubMed] [Google Scholar]
- 230.Sawada, I., H. Maseda, T. Nakae, H. Uchiyama, and N. Nomura. 2004. A quorum-sensing autoinducer enhances the mexAB-oprM efflux pump expression without the MexR-mediated regulation of Pseudomonas aeruginosa. Microbiol. Immunol. 48:435-439. [DOI] [PubMed] [Google Scholar]
- 231.Schmidtke, A. J., and N. D. Hanson. 2006. Model system to evaluate the effect of ampD mutations on AmpC-mediated beta-lactam resistance. Antimicrob. Agents Chemother. 50:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schmidtke, A. J., and N. D. Hanson. 2008. Role of ampD homologs in overproduction of AmpC in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:3922-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Schurek, K. N., A. K. Marr, P. K. Taylor, I. Wiegand, L. Semenec, B. K. Khaira, and R. E. W. Hancock. 2008. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:4213-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Schweizer, H. P. 2003. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet. Mol. Res. 2:48-62. [PubMed] [Google Scholar]
- 235.Scully, B. E., C. N. Ores, A. S. Prince, and H. C. Neu. 1985. Treatment of lower respiratory tract infections due to Pseudomonas aeruginosa in patients with cystic fibrosis. Rev. Infect. Dis. 7(Suppl.):S669-S674. [DOI] [PubMed] [Google Scholar]
- 236.Sekiya, H., T. Mima, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Functional cloning and characterization of a multidrug efflux pump, mexHI-opmD, from a Pseudomonas aeruginosa mutant. Antimicrob. Agents Chemother. 47:2990-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Sevillano, E., L. Gallego, and J. M. Garcia-Lobo. 26 June 2008, posting date. First detection of the OXA-40 carbapenemase in P. aeruginosa isolates, located on a plasmid also found in A. baumannii. Pathol. Biol. (Paris) [Epub ahead of print.] doi: 10.1016/j.patbio.2008.05.002. [DOI] [PubMed]
- 238.Shiba, T., K. Ishiguro, N. Takemoto, H. Koibuchi, and K. Sugimoto. 1995. Purification and characterization of the Pseudomonas aeruginosa NfxB protein, the negative regulator of the nfxB gene. J. Bacteriol. 177:5872-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sobel, M. L., D. Hocquet, L. Cao, P. Plesiat, and K. Poole. 2005. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:1782-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 48:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sobel, M. L., S. Neshat, and K. Poole. 2005. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 187:1246-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Spencer, R. C. 1996. Predominant pathogens found in the European prevalence of infection in intensive care study. Eur. J. Clin. Microbiol. Infect. Dis. 15:281-285. [DOI] [PubMed] [Google Scholar]
- 243.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive E. coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Srikumar, R., X. Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Stapleton, P., K. Shannon, and I. Phillips. 1995. DNA sequence differences of ampD mutants of Citrobacter freundii. Antimicrob. Agents Chemother. 39:2494-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Stobberingh, E. E. 1988. Induction of chromosomal beta-lactamase by different concentrations of clavulanic acid in combination with ticarcillin. Antimicrob. Agents Chemother. 21:9-16. [DOI] [PubMed] [Google Scholar]
- 248.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, and Y. Yuan. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 249.Takenouchi, T., E. Sakagawa, and M. Sugawara. 1999. Detection of gyrA mutations among 335 Pseudomonas aeruginosa strains isolated in Japan and their susceptibilities to fluoroquinolones. Antimicrob. Agents Chemother. 43:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Takesue, Y., T. Yokoyama, S. Akagi, H. Ohge, Y. Imamura, Y. Murakami, and T. Sueda. 2002. Changes in the intestinal flora after the administration of prophylactic antibiotics to patients undergoing a gastrectomy. Surg. Today 32:581-586. [DOI] [PubMed] [Google Scholar]
- 251.Tam, V. M., K. T. Chang, A. N. Schilling, M. T. LaRocco, L. O. Gentry, and K. W. Garey. 2009. Impact of AmpC overexpression on outcomes of patients with Pseudomonas aeruginosa bacteremia. Diagn. Microbiol. Infect. Dis. 63:279-285. [DOI] [PubMed] [Google Scholar]
- 252.Thuong, M., K. Arvaniti, R. Ruimy, P. de la Salmoniere, A. Scanvic-Hameg, J. C. Lucet, and B. Regnier. 2003. Epidemiology of Pseudomonas aeruginosa and risk factors for carriage acquisition in an intensive care unit. J. Hosp. Infect. 53:274-282. [DOI] [PubMed] [Google Scholar]
- 253.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, M. E. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-beta-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance program. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 254.Trias, J., and H. Nikaido. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Trias, J., and H. Nikaido. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 265:15680-15684. [PubMed] [Google Scholar]
- 256.Tsuji, M., Y. Ishii, A. Ohno, S. Miyazaki, and K. Yamaguchi. 1998. In vitro and in vivo antibacterial activities of S-4661, a new carbapenem. Antimicrob. Agents Chemother. 42:94-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Valles, J., D. Mariscal, P. Cortes, P. Coll, A. Villagra, E. Diaz, A. Artigas, and J. Rello. 2004. Patterns of colonization by Pseudomonas aeruginosa in intubated patients: a 3-year prospective study of 1607 isolates using pulsed-field gel electrophoresis with implications for prevention of ventilator-associated pneumonia. Intensive Care Med. 30:1768-1775. [DOI] [PubMed] [Google Scholar]
- 258.Vallet, I., S. P. Diggle, R. E. Stacey, M. Camara, I. Ventre, S. Lory, A. Lazdunski, P. Williams, and A. Filloux. 2004. Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol. 186:2880-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Van Bambeke, R., E. Balzi, and P. M. Tulkens. 2000. Antibiotic efflux pumps. Biochem. Pharmacol. 60:457-470. [DOI] [PubMed] [Google Scholar]
- 260.Van Delden, C., and B. H. Iglewski. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Villavicencio, R. T. 1998. The history of blue pus. J. Am. Coll. Surg. 187:212-216. [DOI] [PubMed] [Google Scholar]
- 262.Villegas, M. V., K. Lolans, A. Correa, J. N. Kattan, J. A. Lopez, J. P. Quinn, and the Colombian Nosocomial Resistance Study Group. 2007. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob. Agents Chemother. 51:1553-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Vogne, C., J. R. Aires, C. Bailly, D. Hocquet, and P. Plesiat. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 48:1676-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wachino, J., K. Yamane, K. Kimura, N. Shibata, S. Suzuki, Y. Ike, and Y. Arakawa. 2006. Mode of transposition and expression of 16S rRNA methyltransferase gene rmtC accompanied by ISEcp1. Antimicrob. Agents Chemother. 50:3212-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wachino, J., K. Yamane, K. Shibayama, H. Kurokawa, N. Shibata, S. Suzuki, Y. Doi, N. Kimura, Y. Ike, and Y. Arakawa. 2006. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob. Agents Chemother. 50:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Watanabe, M., S. Iyobe, M. Inoue, and S. Mitsuhashi. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Weber, D. A., and C. C. Sanders. 1990. Diverse potential of beta-lactamase inhibitors to induce class I enzymes. Antimicrob. Agents Chemother. 34:156-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Weldhagen, G. F., L. Poirel, and P. Nordmann. 2003. Ambler class A extended-spectrum beta-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob. Agents Chemother. 47:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Westfall, L. W., N. L. Carty, N. Layland, P. Kuan, J. A. Colmer-Hamood, and A. N. Hamood. 2006. mvaT mutation modifies the expression of the Pseudomonas aeruginosa multidrug efflux operon mexEF-oprN. FEMS Microbiol. Lett. 255:247-254. [DOI] [PubMed] [Google Scholar]
- 271.Wiegand, I., A. K. Marr, E. B. M. Breidenstein, K. N. Schurek, P. Taylor, and R. E. W. Hancock. 2008. Mutator genes giving rise to decreased antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:3810-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wilke, M. S., M. Heller, A. L. Creagh, C. A. Haynes, L. P. McIntosh, K. Poole, and N. C. Strynadka. 2008. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc. Natl. Acad. Sci. USA 105:14832-14837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Wolter, D. J., D. Acquazzino, R. V. Goering, P. Sammut, N. Khalaf, and N. D. Hanson. 2008. Emergence of carbapenem resistance in Pseudomonas aeruginosa isolates from a patient with cystic fibrosis in the absence of carbapenem therapy. Clin. Infect. Dis. 46:137-141. [DOI] [PubMed] [Google Scholar]
- 274.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2005. AmpC and OprD are not involved in the mechanism of imipenem hypersusceptibility among Pseudomonas aeruginosa isolates overexpressing the mexCD-oprJ efflux pump. Antimicrob. Agents Chemother. 49:4763-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2008. Discordance between imipenem resistance and oprD expression in a Pseudomonas aeruginosa overexpressing mexEF-oprN, abstr. C1-1061. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., Washington, DC.
- 276.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2002. Efflux-mediated resistance among Pseudomonas aeruginosa: complex regulation of a complex resistance mechanism, abstr. A-132. Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., Salt Lake City, UT.
- 277.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2004. Insertional inactivation of oprD in clinical isolates of Pseudomonas aeruginosa leading to carbapenem resistance. FEMS Microbiol. Lett. 236:137-143. [DOI] [PubMed] [Google Scholar]
- 278.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2008. MexT-associated downregulation of oprD in Pseudomonas aeruginosa involves inhibition of transcription initiation, abstr. C1-1056. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., Washington, DC.
- 279.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2008. Novel mechanism of mexEF-oprN efflux pump overexpression in Pseudomonas aeruginosa without coregulation of oprD expression, abstr. C1-1058. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., Washington, DC.
- 280.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2005. Phenotypic reversion of imipenem hypersusceptibility among mexCD-oprJ-overexpressing P. aeruginosa: a complex system becomes even more complex, abstr. C1-2214. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., Washington, DC. [DOI] [PMC free article] [PubMed]
- 281.Wolter, D. J., N. Khalaf, I. E. Robledo, G. J. Vazquez, M. I. Sante, E. E. Aquino, R. V. Goering, and N. D. Hanson. 2009. Surveillance of carbapenem-resistant Pseudomonas aeruginosa isolates from Puerto Rican medical center hospitals: dissemination of KPC and IMP-18 beta-lactamases. Antimicrob. Agents Chemother. 53:1660-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wolter, D. J., A. J. Schmidtke, N. D. Hanson, and P. D. Lister. 2007. Increased expression of ampC in Pseudomonas aeruginosa mutants selected with ciprofloxacin. Antimicrob. Agents Chemother. 51:2997-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Wolter, D. J., E. Smith-Moland, R. V. Goering, N. D. Hanson, and P. D. Lister. 2004. Multidrug resistance associated with mexXY expression in clinical isolates of Pseudomonas aeruginosa from a Texas hospital. Diagn. Microbiol. Infect. Dis. 50:43-50. [DOI] [PubMed] [Google Scholar]
- 284.Woodford, N., J. Zhang, M. E. Kaufmann, S. Yarde, M. D. M. Tomas, C. Faris, M. S. Vardhan, S. Dawson, S. L. Cotterill, and D. M. Livermore. 2008. Detection of Pseudomonas aeruginosa isolates producing VEB-type extended-spectrum beta-lactamases in the United Kingdom. J. Antimicrob. Chemother. 62:1265-1268. [DOI] [PubMed] [Google Scholar]
- 285.Yamane, K., Y. Doi, K. Yokoyama, T. Yagi, H. Kurokawa, N. Shibata, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Genetic environments of the rmtA gene in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 48:2069-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Yamane, K., J. Wachino, Y. Doi, H. Kurokawa, and Y. Arakawa. 2005. Global spread of multiple aminoglycoside resistance genes. Emerg. Infect. Dis. 11:951-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Yokoyama, K., Y. Doi, K. Yamane, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888-1893. [DOI] [PubMed] [Google Scholar]
- 288.Yoneyama, H., and T. Nakae. 1993. Mechanism of efficient elimination of protein D2 in outer membrane of imipenem-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Yoshimura, F., and H. Nikaido. 1985. Diffusion of beta-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob. Agents Chemother. 27:84-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zhao, Q., X. Z. Li, R. Srikumar, and K. Poole. 1998. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob. Agents Chemother. 42:1682-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]