Abstract
Toll-like receptors (TLRs) participate in detecting microbial pattern molecules for activation of the host immune response. We investigated possible roles of TLRs in the chicken response to Clostridium perfringens infection by examining the expression of TLR genes and other genes involved in TLR-mediated signaling within the spleens and ilea of C. perfringens-challenged broilers. Upregulation of a tumor necrosis factor alpha-inducing factor homolog in challenged chickens compared to naïve chickens was observed, regardless of the incidence of necrotic enteritis. In addition, the members of the TLR2 subfamily were found to be most strongly involved in the host response to C. perfringens challenge, although the expression of TLR4 and TLR7 was also upregulated in spleen tissues. While the combination of TLR1.2, TLR2.1, and TLR15 appeared to play a major role in the splenic response, the expression of TLR2.2 and TLR1.1 was positively correlated to the expression of adaptor molecules MyD88, TRAF6, TRIF, and receptor interacting protein 1 in the ileal tissues, demonstrating a dynamic spatial and temporal innate host response to C. perfringens.
Necrotic enteritis (NE) is an enteric disease in chickens caused by Clostridium perfringens (23, 36, 40). Despite the fact that C. perfringens is part of the normal intestinal microbiota, flock management changes, such as changes in diet or exposure to additional pathogens such as coccidia, can change the population of C. perfringens in the small intestine. Overgrowth of C. perfringens often leads to NE disease through the accumulation of multiple extracellular toxins that damage the intestinal wall (40). Alpha-toxin has historically been considered to be the major disease determinant (41). However, this notion has been challenged by the newly discovered NetB (NE toxin B-like) toxin, which has been shown to be critical for the production of NE (15). The acute form of the disease leads to high mortality in broiler flocks, while the damaged intestinal mucosa observed in the subclinical form of C. perfringens infection can result in decreased production output due to decreased digestion and nutrient absorption, reduced weight gain, and increased feed conversion ratios (22).
C. perfringens has been well controlled in the past through the prophylactic use of antibiotics in commercial chicken production. However, this practice of feeding growth-promoting antibiotics has been banned in the European Union countries (8) and is under review by other countries due to public concerns over widespread antibiotic resistance among bacterial pathogens. Since the reduction of dietary antibiotic use, the incidence of NE disease in chicken production has significantly increased (23). Therefore, effective alternatives to dietary antibiotics, including options that can improve the overall host immune response, are urgently required. A full understanding of C. perfringens infection and its related pathogenesis and immunology is essential for developing effective control strategies other than antibiotic therapies. Recently, there has been significant progress in understanding the molecular mechanisms for the pathogenesis of C. perfringens infection in chickens. Specifically, NE toxin B-like (NetB) toxin and bacterium-secreted collagenases were described as major determinants for NE lesions, in addition to alpha-toxin, which had been considered to cause the disease (15, 16, 28). In contrast to pathogenesis, the immunology relating to C. perfringens infection, including immune recognition of the pathogen, is still poorly understood.
The Toll-like receptor (TLR) family is a highly conserved group of proteins that participate in pathogen detection and in the initiation and regulation of innate and adaptive immune responses (18, 39). In avian species, 10 TLRs, including orthologs of mammalian TLR3, TLR4, TLR5, and TLR7, have been discovered to date. In humans, TLR1 and TLR6 have been shown to form a heterodimer with TLR2 to recognize bacterial lipoproteins and peptidoglycan. In chickens, TLR1.1 and TLR1.2, two members of the TLR1/TLR6/TLR10 family, demonstrate the same function, recognizing lipoproteins and peptidoglycan, respectively (3, 10). The chicken genome also contains duplicated TLR2 (TLR2.1 and TLR2.2) genes and genes for unique TLRs (TLR15 and TLR21) that may function in association with the TLR2 subfamily (31). In mammals, many of the TLR signaling pathways have been defined. Typically, TLRs activate innate and adaptive responses through a series of intracellular signaling cascades that are MyD88 dependent or independent and TRIF dependent (18). Following MyD88 or TRIF activation, several adaptor molecules, such as IRAK molecules, are recruited to initiate transcription factor activation and subsequently induce cytokine (e.g., interleukin-6 [IL-6]) and chemokine (e.g., IL-8) production (18). Although the genes that encode some of these adaptor molecules, transcription factors, and immune mediators in chickens have been identified, little is known about chicken TLR-regulated pathogen recognition and the subsequently initiated systemic host responses to C. perfringens infection. As a first step to identify the relationship among key molecules in TLR signaling during C. perfringens infection, the present study sought to examine gene expression profiles for TLR-mediated signaling molecules in the spleen and ileal tissues of broiler chickens challenged with C. perfringens.
MATERIALS AND METHODS
Bacterium.
A type A strain of C. perfringens was grown in Mueller-Hinton broth or on Mueller-Hinton agar containing 5% (vol/vol) sheep blood at 37°C under an anaerobic atmosphere (85% N2, 10% CO2, and 5% H2). The bacterium has routinely been used to induce NE in broiler chickens at Nutreco Canada Agresearch as described previously (4).
Chicken trial.
The chickens used for the present study were the same birds used for our previous reports of alpha-toxin gene expression in the chicken intestine (35) and the host response to C. perfringens infection as evaluated by both a low-density chicken immune cDNA microarray and a 44,000-slide-format whole-chicken-genome Agilent microarray (34, 42). The chickens were cared for under the 1993 guidelines of the Canadian Council on Animal Care. Briefly, 600 1-day-old chicks (Ross × Ross) were originally used and were randomly allocated to 12 pens and fed an all-vegetable starter diet (Shur-Gain; Nutreco Canada). Three hundred of these birds (in six pens) were fed an antibiotic-medicated diet and showed no NE disease and low-level C. perfringens colonization in the ileum (35) and thus were not subjected to further analysis in the present study. On day 18 posthatching, birds were challenged with the type A strain of C. perfringens (4) in the stationary phase at 107 CFU per g of feed in 40 g of feed per chicken for 16 h after 8 h of starvation. The day of C. perfringens challenge was designated day zero (D0). On D0, two birds from each pen were randomly selected for downstream molecular analyses to examine the host responses and euthanized with CO2 before the C. perfringens challenge. The remaining birds in each pen were originally used for the statistical analysis of animal performance that was reported earlier (35). The same sampling procedure was repeated for 4 days after the challenge (on D1, D2, D3, and D4 postinfection [p.i.]). Spleen and ileal tissues from each bird were collected for total-RNA isolation. The 5-cm-long sections of ileal tissues were collected 1 cm away from the end of the cecal junction, digesta were removed, and the tissues were rinsed with 3 to 5 ml saline. Tissues were snap-frozen until RNA extraction. RNA extracted from frozen samples collected on D0 and D1, D2, and D4 p.i. was used for quantitative reverse transcription-PCR (QRT-PCR) assays.
RNA isolation.
Spleen or ileal tissues were homogenized using a PRO200 homogenizer (DiaMed, Mississauga, ON, Canada). Total RNA was isolated from each homogenized tissue sample by the Trizol extraction method as described by the Trizol manufacturer (Invitrogen, Carlsbad, CA). The RNA samples were treated to remove DNA by using the TURBO DNA-free kit according to the protocol of the manufacturer (Ambion, Austin, TX). The RNA quantity was determined by using an ND-1000 spectrophotometer at 260 nm/280 nm (NanoDrop Technologies, Wilmington, DE). cDNA for QRT-PCR assays was synthesized from 1 μg of purified RNA by using 50 ng of random hexamers and the SuperScript II first-strand cDNA synthesis kit according to the instructions of the manufacturer (Invitrogen, Mississauga, ON, Canada).
QRT-PCR.
QRT-PCR primers were either synthesized based on published sequences or designed for this experiment. PCR primers for the genes encoding a tumor necrosis factor alpha (TNF-α)-inducing factor homolog, IL-6, and β-actin were those described previously by Mohammed et al. (25), Hong et al. (12), and Abdul-Careem et al. (1), respectively. PCR primers for the IL-8 (CXCLi2) gene were obtained from S. Sharif's laboratory (unpublished data). PCR primers for the remaining target genes were developed through the present study. The primers were designed using the Vector NTI 8 software program (Invitrogen Corporation) according to sequences acquired from GenBank by employing keyword searches. The specificity of the designed primers was firstly examined by comparison of the primers with sequences in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) through BLAST analysis and then experimentally verified through PCR assays. The PCR conditions for each target gene were optimized, and the optimal conditions are listed in Table 1.
TABLE 1.
PCR primers
| Primera | Sequence | Product size (bp) | GenBank accession no. for target gene | Annealing temperature (°C) | Source or reference |
|---|---|---|---|---|---|
| TLR1-1 F | CTGTCTTGCCAATCTGTC | 194 | AY633574 | 56 | Present studyb |
| TLR1-1 R | GTGAAGGCTCCGTGTATT | ||||
| TLR1-2 F | AGCTGCAGGACTTCCTGCGC | 264 | NM_001098854 | 56 | Present study |
| TLR1-2 R | TTGTCTGCGTCCACTGCCAC | ||||
| TLR2-1 F | TTAAAAGGGTGTGCCAGGAG | 271 | AB050005 | 56 | Present study |
| TLR2-1 R | GTCCAAACCCATGAAAGAGC | ||||
| TLR2-2 F | AGGCACTTGAGATGGAGCAC | 314 | AB046533 | 56 | Present study |
| TLR2-2 R | CCTGTTATGGGCCAGGTTTA | ||||
| TLR4 F | GTCTCTCCTTCCTTACCTGCTGTTC | 187 | AY064697 | 56 | Present study |
| TLR4 R | AGGAGGAGAAAGACAGGGTAGGTG | ||||
| TLR7 F | GGAAACGCTACTAACCTG | 282 | DQ780342 | 56 | Present study |
| TLR7 R | GTTTGCTTCCAGACTCAG | ||||
| TLR15 F | GTTCTCTCTCCCAGTTTTGTAAATAGC | 262 | NM_001037835 | 56 | Present study |
| TLR15 R | GTGGTTCATTGGTTGTTTTTAGGAC | ||||
| TLR21 F | ATGATGGAGACAGCGGAGAAGG | 172 | NM_001030558 | 62 | Present study |
| TLR21 R | GGATGCAGCGGAAGTACAAAGG | ||||
| MyD88 F | AGAAGGTGTCGGAGGATGGTG | 365 | NM_001030962 | 57 | Present study |
| MyD88 R | GGGCTCCAAATGCTGACTGC | ||||
| RIP1 F | AGTGCTCCAAAAAGTCCCAGTACC | 211 | AB108485 | 56 | Present study |
| RIP1 R | GGTCTCTTCTTTGGTCAGCCG | ||||
| TRAF6 F | GAGTGTCCAAGGCGTCAAGTCTG | 243 | XM_421089 | 57 | Present study |
| TRAF6 R | GTGTCGTGCCAGTTCATTCCTC | ||||
| IRF3 F | CGTATCTTCCGCATCCCTTGG | 206 | U20338 | 56 | Present study |
| IRF3 R | TCGTCGTTGCACTTGGAGCG | ||||
| TRIF F | TCAGCCATTCTCCGTCCTCTTC | 339 | EF025853 | 57 | Present study |
| TRIF R | GGTCAGCAGAAGGATAAGGAAAGC | ||||
| MAL F | CTCATAGCACCACCAGCCACTC | 472 | DQ019929 | 56 | Present study |
| MAL R | GGGTAATCCTTCCTGTCAATGTCC | ||||
| IFN-β F | ACCACAGCCCTCTCCATCAAC | 156 | X14455 | 56 | Present study |
| IFN-β R | CTGCATCTTCTCCGTCATCTCC | ||||
| TNF-like Fc | TGCTGTTCTATGACCGCC | 174 | AY765397 | 57 | 18 |
| TNF-like R | CTTTCAGAGCATCAACGCA | ||||
| IL-6 F | CAAGGTGACGGAGGAGGAC | 254 | AJ309540 | 56 | 19 |
| IL-6 R | TGGCGAGGAGGGATTTCT | ||||
| IL-8 F | ATGAACGGCAAGCTTGGAGCTG | 233 | AJ009800 | 57 | S. Sharifd |
| IL-8 R | TCCAAGCACACCTCTCTTCCATCC | ||||
| β-Actin F | CAACACAGTGCTGTCTGGTGG | 205 | X00182 | 55 | 20 |
| β-Actin R | ATCGTACTCCTGCTTGCTGAT |
F, forward primer; R, reverse primer.
Primers were developed for the present study.
Primer for chicken TNF-α-inducing factor gene homolog.
Primers were obtained from S. Sharif's laboratory (unpublished data).
QRT-PCR assays were performed on a Stratagene MX3005 thermal cycler with brilliant SYBR green QRT-PCR master mix (Stratagene, La Jolla, CA). cDNA was diluted 10-fold, and 1 μl of each diluted sample was added to a 25-μl reaction solution containing 12.5 μl of 2× master mix, 150 nM (each) primers, and 30 nM ROX reference dye. Cycling parameters were as follows: 10 min at 95°C; 35 to 40 cycles of 30 s at 95°C, 30 s at the annealing temperature, and 30 s at 72°C; and extension for 2 min at 72°C.
For relative quantitative measurement, a standard curve for each target gene was acquired. The standard curves were established through amplification and cloning of individual target genes (including the β-actin gene that served as a reference) into a plasmid (Topo 10 vector from Invitrogen, Burlington, ON, Canada) followed by log10 serial dilution of the plasmid harboring cloned genes. Each experiment was performed in duplicate with the same plasmid dilution used as a calibrator. The amplification efficiency (E) was calculated as 10−1/slope, and the relative expression was calculated with efficiency correction as follows: ET[CpT(c) − CpT(s)] × ER[CpR(s) − CpR(c)], where ET is the efficiency for the target gene as determined by the standard curve, ER is the efficiency for the reference gene as determined by the standard curve, CpT and CpR are the crossing points (threshold cycles) for the target and reference genes, respectively, s indicates the value for the sample, and c indicates the value for the calibrator (the plasmid) (30). Degrees of change (n-fold) were then calculated by taking ratios of the relative expression in C. perfringens-challenged birds to that in naïve birds at D0.
Statistical analysis.
The relative expression value for each gene was log2 transformed before the statistical analysis. Statistical computations were performed using the Statistical Analysis System (version 9.0; SAS Institute Inc., Cary, NC). The gene expression data were subjected to analysis of variance using the general linear model (GLM) procedure. Least-square means of the relative gene expression levels were calculated using the LSMEANS option, and statistical differences between results for the treatments were identified at P values of <0.05 by using the PDIFF option. Spearman correlation analysis was used to examine relationships among genes.
RESULTS
Challenge results.
The response to C. perfringens challenge was described previously (35) and therefore will not be elaborated on within this study. However, it is important that birds challenged with C. perfringens became infected beginning at D1 p.i. and showed ileal C. perfringens counts and lesion scores beginning on D1 and D2 p.i., respectively. Furthermore, spleen tissues from this study were examined and tested positive for bacterial presence by Q-PCR analysis of alpha-toxin gene expression across all time points (data not shown).
Generation of QRT-PCR standard curves.
Standard curves for relative quantification of target and housekeeping gene expression were generated using the Stratagene MX3005 thermal cycler with three technical replicates and 10 serial dilutions. The PCR efficiency (E) values for standard curves ranged from 1.89 to 2.03, with the majority of efficiencies ranging from 1.97 to 2.02. R2 values for the standard curve reactions ranged from 0.991 to 0.999, with the majority of values above 0.995.
Profiles of TLR gene expression during C. perfringens challenge.
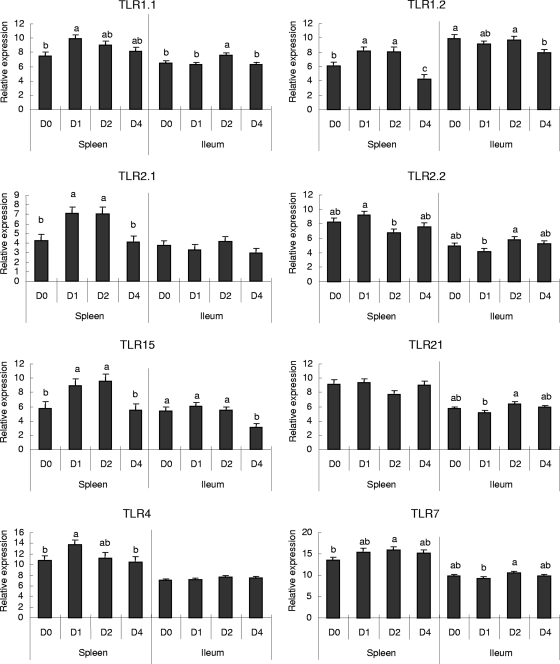
QRT-PCR analysis was performed to further define and compare profiles of TLR-mediated signaling gene expression in both ileal and splenic tissues. TLR expression data acquired from QRT-PCR are presented in Fig. 1, which demonstrates that the expression of TLR7 was increased on D2 p.i. in spleens and that increases in TLR1.2, TLR2.1, and TLR15 expression on D1 and D2 p.i. were followed by decreases on D4 p.i., which were significant only for TLR1.2 (Fig. 1). While TLR1.1 and TLR4 showed upregulation in spleen tissues only on D1 p.i., TLR2.2 expression was not significantly affected by C. perfringens infection at any time point. In the ileal tissues, significant differences in expression were observed only for some members of the TLR2 subfamily. TLR1.1 was upregulated on D2 p.i., and TLR1.2 and TLR15 were downregulated on D4 p.i.
FIG. 1.
Changes (n-fold) in the expression of TLRs in the spleen and ileal tissues of C. perfringens-challenged chickens as determined by QRT-PCR assays. Relative expression of TLRs in the spleens and ilea of chickens was measured by QRT-PCR. Relative expression data were log2 transformed to acquire normal distribution after normalization with respect to β-actin expression and a constant concentration of a plasmid calibrator by using GLM statistical analysis of values for the same tissue type (splenic or ileal) (n = 8). Data are presented as changes (n-fold) calculated by comparing expression levels on D1, D2, and D4 p.i. with those on D0. A value of >1 represents upregulation; a value of <−1 represents downregulation. The bars denoted with different letters represent significant differences before and after challenge (P < 0.05).
Expression of adaptor and signaling molecule genes involved in the TLR signaling pathway.
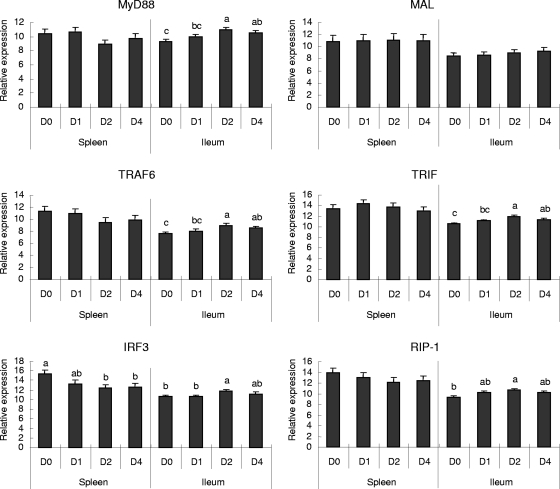
Several adaptor and signaling molecules involved in the TLR signaling pathway have been shown to play important roles in bacterial infections of mammals. In this study, the expression of TLR pathway-associated adaptors in chickens before and after C. perfringens challenge was investigated (Fig. 2). Compared with expression on D0, no significant differences in the expression of most adaptor or signaling molecules in spleen tissues on later days were observed, except for decreases in IRF3 (interferon [IFN] regulatory factor 3) expression on D2 and D4 p.i. In ileal tissues, the levels of expression of MyD88, TRAF6 (TNF receptor-associated factor 6), and TRIF (TIR domain-containing adapter-inducing IFN-β) all increased from D2 to D4 after C. perfringens challenge. However, IRF3 and RIP-1 (receptor interacting protein 1) genes were also upregulated on D2 p.i. MAL adaptor expression in spleen or ileal tissues was not affected by C. perfringens challenge at any time point.
FIG. 2.
Changes (n-fold) in the expression of adaptor molecules involved in the TLR signaling pathway in the spleen and ileal tissues of C. perfringens-challenged chickens as determined by QRT-PCR assays. Relative expression of adaptors from the TLR signaling pathway in the spleens and ilea of chickens was measured by QRT-PCR. Relative expression data were log2 transformed to acquire normal distribution after normalization with respect to β-actin expression and a constant concentration of a plasmid calibrator by using GLM statistical analysis of values for the same tissue type (splenic or ileal) (n = 8). Data are presented as changes (n-fold) calculated by comparing expression levels on D1, D2, and D4 p.i. with those on D0. A value of >1 represents upregulation; a value of <−1 represents downregulation. The bars denoted with different letters represent significant differences before and after challenge (P < 0.05).
Expression of cytokines.
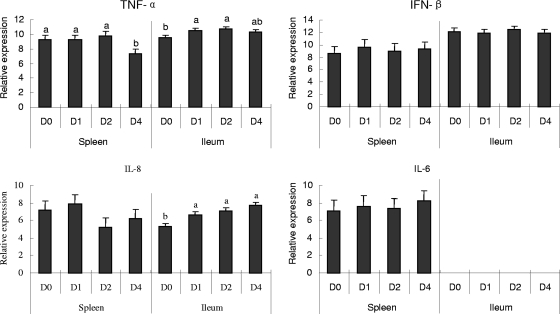
The expression of genes for four cytokines, including the TNF-α-inducing factor homolog, IFN-β, IL-6, and CXCLi2 (IL-8), that can be produced as a result of TLR activation was assessed by QRT-PCR assays. Splenic and ileal tissues demonstrated different patterns of expression of the cytokines in response to clostridial infection (Fig. 3). Expression of IL-6 was not detected in ileal samples. The cytokine was expressed in the spleen but showed no significant changes in response to C. perfringens challenge. The IFN-β gene also exhibited no significant differences in expression before and after C. perfringens challenge. In contrast, chicken TNF-α-inducing factor homolog expression was decreased significantly on D4 p.i. in the spleen tissues but increased in the ileal tissues after C. perfringens challenge (on D1 and D2 p.i.). CXCLi2 (IL-8) expression in the ileal tissues increased significantly at all time points after C. perfringens challenge, but this pattern was not observed in the spleen tissues.
FIG. 3.
Changes (n-fold) in the expression of cytokines in the spleen and ileal tissues of C. perfringens-challenged chickens as determined by QRT-PCR assays. Relative expression of cytokines in the spleens and ilea of chickens was measured by QRT-PCR. Relative expression data were log2 transformed to acquire normal distribution after normalization with respect to β-actin expression and a constant concentration of a plasmid calibrator by using GLM statistical analysis of values for the same tissue type (splenic or ileal) (n = 8). Data are presented as changes (n-fold) calculated by comparing expression levels on D1, D2, and D4 p.i. with those on D0. A value of >1 represents upregulation; a value of <−1 represents downregulation. The bars denoted with different letters represent significant differences before and after challenge (P < 0.05).
Correlation of members of the TLR2 subfamily and other genes.
To investigate the relationships among expressed TLR2 subfamily members, the correlation coefficients (CCs) for pairs of TLR signaling pathway genes after C. perfringens challenge (D1, D2, and D4 p.i.) have been estimated. A significant correlation is defined by a CC with a P value of less than 0.05. Positive correlations between TLR2 subfamily gene expression levels in the spleen after C. perfringens challenge were observed (Table 2), with the levels of TLR1.2 and TLR15 gene expression being the most highly correlated. In the ileum, correlations among the TLR2 subfamily genes were also observed. The greatest correlation found was between the TLR21 and TLR2.2 genes. Among the findings for the four members of the chicken TLR1/TLR2 subfamily (TLR1.1, TLR1.2, TLR2.1, and TLR2.2), the greatest correlations were those for TLR1.2 and TLR2.1 in the spleen and TLR1.1 and TLR2.2 in the ileum (Table 2). In the present study, TLR15 in the spleen and TLR21 in the ileum were the TLRs most frequently observed to have significant correlations with members of the chicken TLR2 subfamily. When the expression profiles of TLR genes were compared to those of adaptor and cytokine genes, positive correlations were also observed. Table 3 outlines the correlations between TLR genes and genes with expression levels significantly changed after C. perfringens challenge, with the gene for the TNF-α-inducing factor homolog being the only gene significantly correlated with the TLR family members in spleen tissues. In contrast, the expression of more than one adaptor molecule gene and the CXCLi2 (IL-8) gene was correlated to the expression of multiple TLR genes. Expression of the TLR15 gene in the ileum demonstrated no correlation with the expression of genes encoding adaptor molecules and CXCLi2 (IL-8).
TABLE 2.
Spearman's rank CCs for TLR expression levels within splenic and ileal tissues from C. perfringens-challenged chickensa
| TLR | CC for splenic expression of: |
CC for ileal expression of: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TLR1.1 | TLR1.2 | TLR2.1 | TLR2.2 | TLR15 | TLR1.1 | TLR1.2 | TLR2.1 | TLR2.2 | TLR15 | |
| TLR1.2 | 0.65 | 0.32 | ||||||||
| TLR2.1 | 0.32 | 0.67 | 0.29 | 0.11 | ||||||
| TLR2.2 | 0.42 | 0.21 | 0.05 | 0.55 | 0.31 | 0.35 | ||||
| TLR15 | 0.54 | 0.71 | 0.68 | 0.21 | −0.01 | 0.56 | 0.21 | −0.22 | ||
| TLR21 | 0.13 | −0.11 | −0.06 | 0.38 | −0.04 | 0.56 | 0.24 | 0.43 | 0.75 | −0.05 |
CCs were calculated in a pairwise manner, with high CC values (e.g., approaching 1.00 or −1.00) indicating a high degree of correlation. Positive CC values indicate positive correlation between TLR expression levels, and negative CC values indicate negative correlation between TLR expression levels. Values indicating significant correlations (P < 0.05) are in bold.
TABLE 3.
Spearman's rank CCs for TLR expression and adaptor molecule and cytokine expression within splenic and ileal tissues from C. perfringens-challenged chickensa
| Tissue | Protein | CC for expression of: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| TLR1.1 | TLR1.2 | TLR2.1 | TLR2.2 | TLR15 | TLR21 | TLR7 | TLR4 | ||
| Spleen | TNF-like factor | 0.38 | 0.69 | 0.66 | −0.01 | 0.90 | −0.16 | 0.48 | 0.49 |
| Ileum | MyD88 | 0.53 | 0.41 | 0.29 | 0.79 | 0.01 | 0.85 | 0.71 | 0.58 |
| TRAF6 | 0.54 | 0.09 | 0.25 | 0.49 | −0.25 | 0.61 | 0.47 | 0.39 | |
| TRIF | 0.36 | 0.17 | 0.19 | 0.55 | −0.05 | 0.67 | 0.43 | 0.58 | |
| IRF3 | 0.49 | 0.08 | 0.46 | 0.63 | −0.02 | 0.70 | 0.59 | 0.28 | |
| IL-8 | 0.14 | 0.19 | 0.45 | 0.43 | 0.12 | 0.48 | 0.37 | 0.57 | |
CCs were calculated in a pairwise manner, with high CC values (e.g., approaching 1.00 or −1.00) indicating a high degree of correlation. Positive CC values indicate positive correlation between expression levels, and negative CC values indicate negative correlation between expression levels. Values indicating significant correlations (P < 0.05) are in bold.
DISCUSSION
The TLR family is a highly conserved group of proteins that participate in pathogen recognition and in the initiation and regulation of the immune response (5). The avian TLR repertoire comprises both orthologous and distinct TLR genes, since avian and mammalian lineages diverged from a common ancestor about 300 million years ago. For example, the chicken genome contains genes with homology to mammalian TLR3, TLR4, TLR5, and TLR7 genes and fish/amphibian TLR21 genes, as well as the novel chicken TLR15 gene (3, 37). In the present study, several TLR genes appeared to be involved in the host response to C. perfringens infection, with emphasis on the upregulation of the TLR2 subfamily, although the expression of both TLR4 and TLR7 in the spleen also increased. These results are consistent with observations for mammals, in which gram-positive bacteria present a tendency for signaling through the TLR2 pathway. Signaling via TLR4 was also observed, however, to a lesser extent (38).
C. perfringens infection occurs in the small intestines of chickens. Previous studies relating to the host response to C. perfringens have involved mainly the examination of pathological effects on the intestinal tissues and the identification of NE disease-causing bacterial components. Nonetheless, the pathogen has also been detected systemically, in the spleens of NE-positive chickens, by Collier et al. (7) and our group (unpublished data). Similarly, a systemic response was indicated by elevated levels of antibodies to C. perfringens in serum samples from approximately 2 to 4 weeks p.i. (20, 21). Given these reports of a systemic host response to C. perfringens and our previous findings of differential TLR gene expression patterns in the spleens of C. perfringens-infected chickens (34), our aim was to evaluate TLR pathway signaling in association with host recognition of C. perfringens and the local and systemic host responses.
C. perfringens challenge had a robust effect on gene expression profiles. In many cases, expression of several TLR genes was upregulated after challenge; however, at certain time points, only significant downregulation was observed (Fig. 1 to 3). These results show the variability of the TLR response to C. perfringens antigens in both the spleen and ileum, where one TLR signaling pathway does not appear to be dominant over the other in terms of the host response to C. perfringens infection. This pattern is also true of the adaptor and cytokine genes analyzed throughout the study, suggesting that the innate host response to C. perfringens can be characterized only by a complex combination of responses varying based on temporal as well as spatial influences. Of the TLRs affected by challenge, some have no previously identified functions in chickens and the others have been described only in the context of their responses to bacterial pathogens other than C. perfringens. Specifically, TLR15 gene expression has been previously noted only to be upregulated in the ceca of Salmonella enterica serovar Typhimurium-infected chickens and in heterophils stimulated with Salmonella serovar Typhimurium and thus, until this study, has been considered to be specific to chicken host responses to Salmonella pathogenesis alone (9, 27). Similar to TLR15 expression, TLR21 expression in chickens has been described only briefly, with respect to the response to Salmonella and Campylobacter infection (24). Furthering previous observations, our results indicate that following C. perfringens exposure, TLR15 and TLR21 expression patterns are differentially regulated, which may be considered central to host responses to intestinal bacterial infection in chickens.
Upon examination of the gene expression profiles in ileal and splenic tissues, large differences between tissue responses in regard to the specific gene affected or the direction of the change (i.e., up- or downregulation in comparison with expression in naïve chickens) were detected (Fig. 1 to 3). Primarily, we observed that C. perfringens infection caused a change in the expression of most TLRs within the spleen. Since the expression of TLR1.2, TLR2.1, and TLR15 was upregulated on D1 and D2 p.i. and the expression of TLR4 and TLR7 was upregulated on D1 and D2 p.i., respectively, we would have predicted upregulation of MyD88-dependent downstream signaling events leading to proinflammatory cytokine production, which is typical in mammalian counterparts. However, no significant changes in MyD88 or TRAF6 pathway expression were observed, except for the decrease in TNF-α-inducing factor homolog expression. Similarly, TRIF and its downstream signaling molecules, which are involved in mammalian MyD88-independent TLR3 and TLR4 activation, also showed no significant changes in the spleen except for the decrease in IRF3 expression. The relationship between TLR4 activation and IRF3 upregulation has been fairly well investigated. In concert with our results, Sakaguchi et al. (33) showed that stimulation with bacterial lipopolysaccharide activates TLR4 in mice in an IRF3-dependent response, which extends to IFN-β dependence when IRF7 is also activated. To interpret the failure of upregulation of MyD88 pathway-dependent adaptors, one could hypothesize that these TLRs may not signal through the MyD88 pathway as TLR2 molecules do in mammals. Alternatively, our sampling may have missed the time window to capture transcription; also, we did not assess protein mechanisms that may be more indicative of MyD88 activation, such as docking or phosphorylation. Lastly, in the case of TLR4, overstimulation of TLR4 by both endogenous ligands and chronic exposure to exogenous ligands following bacterial lysis has been reported to cause systemic inflammation, leading to death (32). Whether the typical TLR4 signaling pathway involving MyD88 is activated during this exacerbated response is yet to be determined, and the answer may explain the upregulation of TLR4 that is not necessarily linked to MyD88 gene expression.
Seemingly important to the host response to C. perfringens, the chicken TNF-α-inducing factor homolog has been suggested previously to be involved in the initiation of TNF-α production. TNF-α in mammals is a member of a group of NF-κB-activated signaling cytokines that stimulate systemic inflammation. In chickens, the production of a TNF-α-modulating factor has been shown to increase, along with the production of inflammatory cytokines, after exposure to Eimeria and Salmonella species (6, 11, 17, 29). Given the inflammatory nature of NE disease and the predominance of innate and inflammatory responses observed in the present study, TNF-α-inducing factor homolog expression may be indicative of the initiation of a TNF-α-type response in chickens infected with C. perfringens (2, 13).
In the ileum, the expression of TLR4 and the adaptor MAL was unchanged. In addition, there was no IL-6 expression. Previously, C. perfringens-derived β-toxin failed to induce the production of IL-6 in mice, despite the typical role of IL-6 as a proinflammatory cytokine produced following TLR pathway activation (26), suggesting that this cytokine is not involved in TLR regulation of C. perfringens-induced NE. In contrast to the expression of IL-6, the expression of MyD88, TRAF6, TRIF, and RIP-1 in the ileum was increased after C. perfringens challenge in the present study. Considering that the activation of adaptor molecules, including MyD88, is typically followed by the production of inflammatory immune mediators such as IL-6, IL-1β, and TNF-α in mammals (14, 18), one could expect that the corresponding genes would be upregulated in the ileal tissues of C. perfringens-infected chickens. The present data show that the expression of both the TNF-α-inducing factor homolog and CXCLi2 (IL-8) was upregulated in the ileum, in contrast to the spleen, where the only significant change was decreased TNF-α-inducing factor homolog expression.
In addition to the statistical significance of gene expression changes, correlations between gene expression patterns were investigated to further infer functional relationships of TLR-mediated genes following C. perfringens challenge (Tables 2 and 3). In particular, data for genes were highlighted when the gene expression profile was correlated with significant changes after C. perfringens challenge. In the spleen, the cooperative effects of TLR1.2, TLR2.1, and TLR15 in the initiation of the immune response on D1 or D2 p.i. were confirmed by positive Pearson's correlation among TLR1.2, TLR2.1, TLR15, and TNF-α-inducing factor homolog gene expression levels. However, the IRF3 gene was the only gene with significantly decreased expression after C. perfringens challenge, yet its expression was not significantly correlated with TLR expression. In contrast, ileal gene expression profiles showed no changes for these genes on D1 or D2 p.i. Yet similar to those in the spleen, the levels of expression of TLR1.2, TLR2.1, and TLR15 genes in the ileum were decreased on D4 p.i. and all expression profiles were positively correlated, suggesting an overlap in the functionalities of these TLR genes among immune-related tissues at the later time points.
In summary, the present study has revealed the possible role of the TLR2 subfamily and less studied, novel chicken TLRs, TLR15 and TLR21, in innate chicken responses to C. perfringens infection. Chickens seem to differ in part from mammals in TLR-mediated signaling, particularly based on the divergence of the TLR2 subfamily and potential functional differences. These observations warrant further functional studies to evaluate the TLR signaling pathway in chickens, which may lead to a discovery of potential targets in innate immunity for the control of NE disease.
Acknowledgments
This research was supported by Agriculture & Agri-Food Canada through the A-base program. The chicken trial with sample collection was made possible through support from Nutreco Canada Agresearch and the Poultry Industry Council. Y.L. was a visiting graduate student in the laboratory of J.G. and was supported by the China Scholar Council through the MOE-AAFC Ph.D. research program. A.J.S. was an NSERC visiting fellow to Canadian federal government laboratories.
Footnotes
Published ahead of print on 23 September 2009.
REFERENCES
- 1.Abdul-Careem, M. F., B. D. Hunter, A. J. Sarson, A. Mayameei, H. Zhou, and S. Sharif. 2006. Marek's disease virus-induced transient paralysis is associated with cytokine gene expression in the nervous system. Viral Immunol. 19:167-176. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, J. W., and P. S. Holt. 1996. Cytotoxicity in chicken alimentary secretions as measured by a derivative of the tumor necrosis factor assay. Poult. Sci. 75:329-334. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, A., V. J. Philbin, and A. L. Smith. 2007. Conserved and distinct aspects of the avian Toll-like receptor (TLR) system: implications for transmission and control of bird-borne zoonoses. Biochem. Soc. Trans. 35:1504-1507. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, J., R. Bagg, D. Barnum, J. Wilson, and P. Dick. 2001. Efficacy of narasin in the prevention of necrotic enteritis in broiler chickens. Avian Dis. 45:210-214. [PubMed] [Google Scholar]
- 5.Brisbin, J. T., J. Gong, and S. Sharif. 2008. Interactions between commensal bacteria and the gut-associated immune system of the chicken. Anim. Health Res. Rev. 9:101-110. [DOI] [PubMed] [Google Scholar]
- 6.Carvajal, B. G., U. Methner, J. Pieper, and A. Berndt. 2008. Effects of Salmonella enterica serovar Enteritidis on cellular recruitment and cytokine gene expression in caecum of vaccinated chickens. Vaccine 26:5423-5433. [DOI] [PubMed] [Google Scholar]
- 7.Collier, C. T., C. L. Hofacre, A. M. Payne, D. B. Anderson, P. Kaiser, R. I. Mackie, and H. R. Gaskins. 2008. Coccidia-induced mucogenesis promotes the onset of necrotic enteritis by supporting Clostridium perfringens growth. Vet. Immunol. Immunopathol. 122:104-115. [DOI] [PubMed] [Google Scholar]
- 8.European Commission. May 2001, posting date. 2nd opinion on anti-microbial resistance. European Commission, Brussels, Belgium. http://ec.europa.eu/food/fs/sc/ssc/out203_en.pdf.
- 9.Higgs, R., P. Cormican, S. Cahalane, B. Allan, A. T. Lloyd, K. Meade, T. James, D. J. Lynn, L. A. Babiuk, and C. O'Farrelly. 2006. Induction of a novel chicken Toll-like receptor following Salmonella enterica serovar Typhimurium infection. Infect. Immun. 74:1692-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi, M., A. Matsuo, M. Shingai, K. Shida, A. Ishii, K. Funami, Y. Suzuki, H. Oshiumi, M. Matsumoto, and T. Seya. 2008. Combinational recognition of bacterial lipoproteins and peptidoglycan by chicken Toll-like receptor 2 subfamily. Dev. Comp. Immunol. 32:147-155. [DOI] [PubMed] [Google Scholar]
- 11.Hong, Y. H., H. S. Lillehoj, S. H. Lee, D. W. Park, and E. P. Lillehoj. 2006. Molecular cloning and characterization of chicken lipopolysaccharide-induced TNF-alpha factor (LITAF). Dev. Comp. Immunol. 30:919-929. [DOI] [PubMed] [Google Scholar]
- 12.Hong, Y. H., H. S. Lillehoj, S. H. Lee, R. A. Dalloul, and E. P. Lillehoj. 2006. Analysis of chicken cytokine and chemokine gene expression following Eimeria acervulina and Eimeria tenella infections. Vet. Immunol. Immunopathol. 114:209-223. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser, G. C., F. Yan, and D. B. Polk. 1999. Mesalamine blocks tumor necrosis factor growth inhibition and nuclear factor κB activation in mouse colonocytes. Gastroenterology 116:602-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai, T., and S. Akira. 2006. TLR signaling. Cell Death Differ. 13:816-825. [DOI] [PubMed] [Google Scholar]
- 15.Keyburn, A. L., J. D. Boyce, P. Vaz, T. L. Bannam, M. E. Ford, D. Parker, A. Di Rubbo, J. I. Rood, and R. J. Moore. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keyburn, A. L., S. A. Sheedy, M. E. Ford, M. M. Williamson, M. M. Awad, J. I. Rood, and R. J. Moore. 2006. Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect. Immun. 74:6496-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, D. K., Y. H. Hong, D. W. Park, S. J. Lamont, and H. S. Lillehoj. 2008. Differential immune-related gene expression in two genetically disparate chicken lines during infection by Eimeria maxima. Dev. Biol. (Basel) 132:131-140. [DOI] [PubMed] [Google Scholar]
- 18.Krishnan, J., K. Selvarajoo, M. Tsuchiya, G. Lee, and S. Choi. 2007. Toll-like receptor signal transduction. Exp. Mol. Med. 39:421-438. [DOI] [PubMed] [Google Scholar]
- 19.Kucharzik, T., and I. R. Williams. 2002. Neutrophil migration across the intestinal epithelial barrier—summary of in vitro data and description of a new transgenic mouse model with doxycycline-inducible interleukin-8 expression in intestinal epithelial cells. Pathobiology 70:143-149. [DOI] [PubMed] [Google Scholar]
- 20.Kulkarni, R. R., V. R. Parreira, S. Sharif, and J. F. Prescott. 2007. Immunization of broiler chickens against Clostridium perfringens-induced necrotic enteritis. Clin. Vaccine Immunol. 14:1070-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkarni, R. R., V. R. Parreira, S. Sharif, and J. F. Prescott. 2008. Oral immunization of broiler chickens against necrotic enteritis with an attenuated Salmonella vaccine vector expressing Clostridium perfringens antigens. Vaccine 26:4194-4203. [DOI] [PubMed] [Google Scholar]
- 22.Lovland, A., and M. Kaldhusdal. 2001. Severely impaired production performance in broiler flocks with high incidence of Clostridium perfringens-associated hepatitis. Avian Pathol. 30:73-81. [DOI] [PubMed] [Google Scholar]
- 23.McDevitt, R. M., J. D. Brooker, T. Acamovic, and N. H. C. Sparks. 2006. Necrotic enteritis: a continuing challenge for the poultry industry. World's Poult. Sci. J. 62:221-247. [Google Scholar]
- 24.Meade, K. G., F. Narciandi, S. Cahalane, C. Reiman, B. Allan, and C. O'Farrelly. 2009. Comparative in vivo infection models yield insights on early host immune response to Campylobacter in chickens. Immunogenetics 61:101-110. [DOI] [PubMed] [Google Scholar]
- 25.Mohammed, J., S. Frasca, Jr., K. Cecchini, D. Rood, A. C. Nyaoke, S. J. Geary, and L. K. Silbart. 2007. Chemokine and cytokine gene expression profiles in chickens inoculated with Mycoplasma gallisepticum strains Rlow or GT5. Vaccine 25:8611-8621. [DOI] [PubMed] [Google Scholar]
- 26.Nagahama, M., A. Kihara, H. Kintoh, M. Oda, and J. Sakurai. 2008. Involvement of tumour necrosis factor-alpha in Clostridium perfringens beta-toxin-induced plasma extravasation in mice. Br. J. Pharmacol. 153:1296-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nerren, J. R., C. L. Swaggerty, K. M. MacKinnon, K. J. Genovese, H. He, I. Pevzner, and M. H. Kogut. 2009. Differential mRNA expression of the avian-specific toll-like receptor 15 between heterophils from Salmonella-susceptible and -resistant chickens. Immunogenetics 61:71-77. [DOI] [PubMed] [Google Scholar]
- 28.Olkowski, A. A., C. Wojnarowicz, M. Chirino-Trejo, B. Laarveld, and G. Sawicki. 2008. Sub-clinical necrotic enteritis in broiler chickens: novel etiological consideration based on ultra-structural and molecular changes in the intestinal tissue. Res. Vet. Sci. 85:543-553. [DOI] [PubMed] [Google Scholar]
- 29.Park, S. S., H. S. Lillehoj, P. C. Allen, D. W. Park, S. FitzCoy, D. A. Bautista, and E. P. Lillehoj. 2008. Immunopathology and cytokine responses in broiler chickens coinfected with Eimeria maxima and Clostridium perfringens with the use of an animal model of necrotic enteritis. Avian Dis. 52:14-22. [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl, M. W., T. M. Georgieva, I. P. Georgiev, E. Ontsouka, M. Hageleit, and J. W. Blum. 2002. Real-time RT-PCR quantification of insulin-like growth factor (IGF)-1, IGF-1 receptor, IGF-2, IGF-2 receptor, insulin receptor, growth hormone receptor, IGF-binding proteins 1, 2 and 3 in the bovine species. Domest. Anim. Endocrinol. 22:91-102. [DOI] [PubMed] [Google Scholar]
- 31.Roach, J. C., G. Glusman, L. Rowen, A. Kaur, M. K. Purcell, K. D. Smith, L. E. Hood, and A. Aderem. 2005. The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. USA 102:9577-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossignol, D. P., and M. Lynn. 2005. TLR4 antagonists for endotoxemia and beyond. Curr. Opin. Investig. Drugs 6:496-502. [PubMed] [Google Scholar]
- 33.Sakaguchi, S., H. Negishi, M. Asagiri, C. Nakajima, T. Mizutani, A. Takaoka, K. Honda, and T. Taniguchi. 2003. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem. Biophys. Res. Commun. 306:860-866. [DOI] [PubMed] [Google Scholar]
- 34.Sarson, A. J., Y. Wang, Z. Kang, S. E. Dowd, Y. Lu, H. Yu, Y. Han, H. Zhou, and J. Gong. 2009. Gene expression profiling within the spleen of Clostridium perfringens-challenged broilers fed antibiotic-medicated and non-medicated diets. BMC Genomics 10:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Si, W., J. Gong, Y. Han, H. Yu, J. Brennan, H. Zhou, and S. Chen. 2007. Quantification of cell proliferation and alpha-toxin gene expression of Clostridium perfringens in the development of necrotic enteritis in broiler chickens. Appl. Environ. Microbiol. 73:7110-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Songer, J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Temperley, N. D., S. Berlin, I. R. Paton, D. K. Griffin, and D. W. Burt. 2008. Evolution of the chicken Toll-like receptor gene family: a story of gene gain and gene loss. BMC Genomics 9:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tietze, K., A. Dalpke, S. Morath, R. Mutters, K. Heeg, and C. Nonnenmacher. 2006. Differences in innate immune responses upon stimulation with gram-positive and gram-negative bacteria. J. Periodontal Res. 41:447-454. [DOI] [PubMed] [Google Scholar]
- 39.Underhill, D. M., and A. Ozinsky. 2002. Toll-like receptors: key mediators of microbe detection. Curr. Opin. Immunol. 14:103-110. [DOI] [PubMed] [Google Scholar]
- 40.Van Immerseel, F., J. De Buck, F. Pasmans, G. Huyghebaert, F. Haesebrouck, and R. Ducatelle. 2004. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 33:537-549. [DOI] [PubMed] [Google Scholar]
- 41.Van Immerseel, F., J. I. Rood, R. J. Moore, and R. W. Titball. 2009. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. 17:32-36. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, H., J. Gong, J. Brisbin, H. Yu, A. J. Sarson, W. Si, S. Sharif, and Y. Han. 2009. Transcriptional profiling analysis of host response to Clostridium perfringens infection in broilers. Poult. Sci. 88:1023-1032. [DOI] [PubMed] [Google Scholar]