Abstract
A recombinant antigen cocktail enzyme-linked immunosorbent assay (ELISA) for diagnosis of contagious bovine pleuropneumonia (CBPP) was developed after careful selection of antigens among one-third of the surface proteome proteins of the infectious agent Mycoplasma mycoides subsp. mycoides small colony (M. mycoides SC). First, a miniaturized and parallelized assay system employing antigen suspension bead array technology was used to screen 97 bovine sera for humoral immune responses toward 61 recombinant surface proteins from M. mycoides SC. Statistical analysis of the data resulted in selection of eight proteins that showed strong serologic responses in CBPP-affected sera and minimal reactivity in negative control sera, with P values of <10−6. Only minor cross-reactivity to hyperimmune sera against other mycoplasmas was observed. When applied in an ELISA, the cocktail of eight recombinant antigens allowed a fivefold signal separation between 24 CBPP-affected and 23 CBPP-free sera from different geographical origins. No false-positive results and only two false-negative results were obtained. In conclusion, the selected recombinant mycoplasma antigens qualified as highly specific markers for CBPP and could be employed in both a suspension bead array platform and a cocktail ELISA setting. This set of proteins and technologies therefore offers a powerful combination to drive and further improve serological assays toward reliable, simple, and cost-effective diagnosis of CBPP.
Contagious bovine pleuropneumonia (CBPP) is a severe infectious disease in cattle caused by Mycoplasma mycoides subsp. mycoides small colony (M. mycoides SC). Because of its potential for rapid spread with resulting massive losses of livestock and thereby severe socioeconomic consequences, an official declaration of disease to the World Organization for Animal Health is required. Vigorous and costly eradication programs involving mass slaughter, quarantine, and strict control of animal movements have been successful in eradicating CBPP from the United States, Japan, and Australia (13). In Western Europe, the disease has reemerged almost every decade in the 20th century, in spite of expensive eradication efforts, demonstrating its constant threat (13, 22). In sub-Saharan Africa, where the disease is endemic, this stamping-out procedure is not economically feasible. Vaccination is a better option for these countries, although existing vaccines so far give insufficient immunity and severe side effects (23). One of the main challenges within CBPP control is diagnosis of the subacute and chronic phases of the disease (14, 22). Without proper diagnosis, asymptomatic carriers can easily transmit the pathogen, and incubation periods of up to several months (7) hinder contact tracing. Today, there are two diagnostic tests prescribed for international trade: the Campbell and Turner complement fixation test (CFT), established in 1953 (6) and based on whole-cell antigens, and the competitive enzyme-linked immunosorbent assay (ELISA), published in 1998 (14) and based on whole-cell antigens in combination with a monoclonal antibody toward Pts-G (9). Although the two serological tests supplement each other in sensitivity, they still do not allow adequate diagnostic certainty (8).
The complete genome sequence of M. mycoides SC (26) has paved the way for new diagnostics based on subcellular components. Methods including PCR have been shown to be successful (21) but put high demands on sampling procedures. Surface lipoproteins are of high interest both for diagnostic purposes and for studies regarding the pathogenicity of the bacterium (23). To date, only a few of the surface lipoproteins from M. mycoides SC have been studied thoroughly. LppA (p72) (17, 18), LppB (25), and LppC (23) are highly conserved lipoproteins that are present in closely related species within the Mycoplasma mycoides cluster. Pts-G is a variably expressed glucose phosphotransferase system permease (9), and Vmm is a small surface protein shown to have a variable expression pattern (20). LppQ is a highly antigenic lipoprotein specific to M. mycoides SC (1). Thorough characterization studies of LppQ (1, 4) and the development of a recombinant ELISA built upon LppQ as the antigen (5) show that it is suitable as a diagnostic marker. However, of the 187 predicted surface proteins of M. mycoides SC (10), more antigens than just LppQ should trigger antibody-mediated immune responses useful in diagnostic applications. Combinations of such antigens could thereby offer a higher specificity and sensitivity than existing methods by adding discriminative power to the current LppQ-based ELISA while circumventing cross-reactivity compared to whole-cell antigen-based methods.
The aim of this study was to identify the most potent diagnostic surface antigens and to test the performance of recombinant versions in combination in an ELISA format. The selection of targets was enabled by a recently developed multiplex suspension bead array assay that allowed high-throughput screening of a large set of sera for humoral immune responses to a large number of recombinant surface proteins (10). As a result, a cocktail ELISA was developed with the selected antigens, and its capacity was evaluated.
MATERIALS AND METHODS
Recombinant surface proteins.
All proteins included in this study are listed in Table S1 in the supplemental material. The production of recombinant mycoplasma proteins was described previously (11) and included the selection of surface proteins specific to M. mycoides SC, design of full-length recombinants excluding a signal peptide and including the largest extracellular domain for transmembrane proteins, PCR and cloning into the vector pAff8c, mutagenesis to change TGA codons to TGG, expression in Escherichia coli, and, finally, purification by immobilized metal-ion chromatography. All recombinant proteins were equipped with a fusion tag consisting of a 17-kDa albumin binding protein with six N-terminal histidines (His6ABP) for enhanced solubility and purification purposes.
For the ELISA setup, relevant recombinant proteins were also produced without the fusion protein ABP. New forward PCR primers were designed to replace the N-terminal His6ABP fusion tag with a hexahistidine tag only. Sequence-specific regions were identical to those of previously used PCR primers, but the primer handles contained a 5′-terminal NcoI restriction site followed by a hexahistidine tag. A biotinylated primer complementary to this handle was used in a secondary PCR step to enable automated solid-phase restriction of the amplified mycoplasma gene fragments. Amplifications were performed as previously described, using the sequence-verified clones with His6ABP gene fusions as templates. After solid-phase restriction, clones were ligated into NcoI/AscI-digested pAff8c vector. Proteins were expressed in E. coli strain BL21(DE3) and purified using immobilized metal-ion affinity chromatography. All proteins were stored at −20°C in phosphate-buffered saline (PBS) containing 1 M urea and were named after their corresponding open reading frames in the genome sequence.
Serum collection.
The serum collection consisted of 97 bovine sera from different geographical origins as well as 17 hyperimmune sera (Table 1). Included in the collection were two CBPP-positive reference samples from Portugal, 50 CBPP-positive field samples from Spain, Namibia, and Tanzania, 24 CBPP-negative field samples from Kenya and Tanzania, 21 negative control samples from Sweden, and 17 hyperimmune sera from Denmark and Sweden. The bovine sera had well-characterized CBPP statuses, although some diagnostic test results were inconsistent. The hyperimmune sera were from four calves and 13 rabbits immunized with M. mycoides SC type strain PG1 as well as closely related Mycoplasma species to study potential cross-reactivity.
TABLE 1.
Serum collection
| Serum type (n) | Serum IDa | Diagnosisb |
Origin | Comment | |||
|---|---|---|---|---|---|---|---|
| CFT | cELISA | LAT | Gross pathology | ||||
| CBPP-positive sera (52) | B001 | + | NA | NA | NA | Portugald | CBPP reference serum 845 from OIE Reference Laboratory for CBPP |
| B002 | + | NA | NA | NA | Portugald | CBPP reference serum 840 from OIE Reference Laboratory for CBPP | |
| B003 to B008 | NA | NA | NA | NA | Spaine | Field samples from outbreaks in Spain | |
| B009 | + | + | + | − | Namibia | Field sample from CBPP outbreak (2004) | |
| B010 | − | + | + | − | Namibia | Field sample from CBPP outbreak (2004) | |
| B011 | + | − | + | + | Namibia | Field sample from CBPP outbreak (2004) | |
| B012 | − | − | + | − | Namibia | Field sample from CBPP outbreak (2004) | |
| B013 | − | + | + | − | Namibia | Field sample from CBPP outbreak (2004) | |
| B014 | + | ? | + | − | Namibia | Field sample from CBPP outbreak (2004) | |
| B015 | + | + | ? | + | Namibia | Field sample from CBPP outbreak (2004) | |
| B016 | + | + | + | + | Namibia | Field sample from CBPP outbreak (2004) | |
| B017 | + | +c | NA | NA | Tanzania | From acute CBPP case (1990); diluted 1:4 for use as reference serum | |
| B018 to B021 | + | +c | NA | NA | Tanzania | Field samples from CBPP outbreak (1990) | |
| B022 | − | −c | NA | NA | Tanzania | Field sample from CBPP outbreak (1990) | |
| B023 to B027 | − | +c | NA | NA | Tanzania | Field samples from CBPP outbreak (1990) | |
| B028 to B052 | + | NA | NA | NA | Tanzania | Field samples from CBPP outbreak | |
| CBPP-negative sera (46) | B053 to B060 | NA | − | − | NA | Kenya | CBPP-free members of vaccine trial, before start of trial (1998) |
| B061 to B076 | − | −c | NA | NA | Tanzania | From CBPP-unaffected region (1990) | |
| B077 to B092 | NA | NA | NA | NA | Sweden | From CBPP-free region (2007) | |
| B093 | NA | NA | NA | NA | Sweden | Pool of B081 to B092 | |
| B094 to B098 | NA | −c | NA | NA | Sweden | From CBPP-free region, with M. bovis infection (1988) | |
| Hyperimmune sera (17) | H099 | NA | NA | NA | NA | Denmarkf | Rabbit immunized with M. mycoides SC PG1 |
| H100 | NA | NA | NA | NA | Sweden | Rabbit immunized with M. mycoides LC Y-goat | |
| H101 | NA | NA | NA | NA | Sweden | Rabbit immunized with M. mycoides capri PG3 | |
| H102 | NA | NA | NA | NA | Sweden | Rabbit immunized with M. capricolum CK | |
| H103 | NA | NA | NA | NA | Sweden | Rabbit immunized with M. capripneumonie F38 | |
| H104 | NA | NA | NA | NA | Sweden | Rabbit immunized with Mycoplasma sp. group 7 strain PG50 | |
| H105 | NA | NA | NA | NA | Denmarkf | Rabbit immunized with M. bovis PG45 | |
| H106 | NA | NA | NA | NA | Sweden | Rabbit immunized with M. bovirhinis PG43 | |
| H107 | NA | NA | NA | NA | Sweden | Rabbit immunized with M. arginini G230 | |
| H108 | NA | NA | NA | NA | Denmarkf | Rabbit immunized with M. californicum ST-6 | |
| H109 | NA | NA | NA | NA | Denmarkf | Rabbit immunized with M. bovoculi M165/69 | |
| H110 | NA | NA | NA | NA | Denmarkf | Rabbit immunized with M. canadense 275c | |
| H111 | NA | NA | NA | NA | Sweden | Rabbit immunized with M. bovigenitalium PG11 | |
| H112 | NA | NA | NA | NA | Sweden | Calf immunized with A. laidlawii | |
| H113 | NA | NA | NA | NA | Sweden | Calf immunized with M. arginini | |
| H114 | NA | NA | NA | NA | Sweden | Calf immunized with M. fermentans | |
| H115 | NA | NA | NA | NA | Sweden | Calf immunized with M. orale | |
B, bovine sera; H, hyperimmune sera.
CFT, complement fixation test; cELISA, competitive ELISA; LAT, latex agglutination test; +, positive test result; −, negative test result; ?, inconclusive test result; NA, not available.
Indirect ELISA (3).
OIE Reference Laboratory for CBPP, Lisbon, Portugal (J. Regalla).
Laboratorio de Sanidad Animal, Santa Fe, Granada, Spain (F. Garrido Abellan).
WHO/FAO Collaborating Centre for Animal Mycoplasmas, University of Aarhus, Aarhus, Denmark (E. A. Freundt).
Screening by suspension bead array assay.
The suspension bead array assay was performed as previously described (10) on a total of 134 samples with a 62-plex surface protein array in suspension. The array consisted of 61 recombinant mycoplasma proteins and pure His6ABP covalently coupled to color-coded magnetic microspheres (MagPlex; Luminex Corp.). Coupling efficiency was confirmed, and bead collections were combined and optimized to a final concentration of 100 beads per bead ID and μl. Sera were diluted 1:3,000, and unspecific binding was reduced by preadsorption with His6ABP protein and E. coli lysate. Incubation of sera and protein-coupled beads was followed by detection of bound complex by a biotinylated secondary antibody and fluorophor-labeled streptavidin. Fluorescence intensity was registered in an Lx200 system using xPONENT software (both from Luminex Corp.). Signals were chosen from the median fluorescence intensity (MFI) for at least 50 beads per bead ID and are given in arbitrary units (AU). The obtained results were validated according to bead count, technical replicates, and background levels. Statistical analysis for the selection of the most immunogenic antigens as well as visualization of the data was performed with R, a web-based statistical computing language and environment (12). A heat map visualization was based on log10-transformed intensity values, and cluster analysis was performed with hierarchical clustering based on the Euclidean distance between data points. Subsequent selection of antigens was based on Welch's two-sample t test on log-transformed data, where antigens with a positive-to-negative signal ratio of at least 3 and a P value below 0.01 were selected for further evaluations.
Antigen cocktail ELISA.
A cocktail of the most immunogenic antigens was created by combining selected recombinant proteins in equal amounts in PBS. ELISA conditions were optimized by varying the protein coating density (10 to 1,000 ng/well), plate type (high-binding or medium-binding half-well-area 96-well plates; Greiner), reagent volume (50 to 100 μl), serum dilution (1:10 to 1:1,600), secondary antibody concentration (80 to 160 ng/ml), incubation time (10 to 60 min), and washing procedures (rounds, volumes, and addition of Tween 20 to PBS). The final assay was conducted as follows. ELISA microplates (high-binding half-well-area plates; Greiner) were coated with protein cocktail (100 μl; 1 μg/ml) and incubated overnight at 4°C. After being washed three times in 140 μl PBS, plates were blocked with 140 μl of BRE (blocking reagent for ELISA; Roche) at room temperature (RT) for 1 h, followed by three washes of 140 μl PBST (PBS with 0.1% Tween 20). Sera were diluted 1:1,000 in BRE, and 100 μl was dispensed into each well and incubated for 1 h at RT. After three washes, 100 μl anti-bovine immunoglobulin G conjugated to horseradish peroxidase (80 ng/ml; Jackson Immunoresearch) was added and incubated for 1 h at RT. After plate washing, the enzymatic color reaction was performed by the addition of 100 μl one-step ABTS [2,2′-azinobis (3-ethylbenzthiazoline-6-sulfonic acid) diammonium salt; Pierce], and the optical density was measured at 405 nm (Sunrise absorbance reader; Magellan Software, Tecan) after 30 to 60 min.
RESULTS
Recombinant production of surface proteins.
Sixty-one proteins with a predicted surface location on M. mycoides SC were produced as recombinant proteins with the fusion tag His6ABP. Protein yields from 100 ml of culture after affinity purification ranged from 0.2 to 10 mg. For nine selected antigens, ABP-free recombinants were produced for the final ELISA. Protein yields were similar to those having the ABP fusion and ranged from 0.6 to 7.3 mg. However, for recombinant R0816, expression of a tag-free version failed, and it was therefore excluded.
Screening of sera against 61 recombinant proteins.
In order to select the most immunogenic among 61 surface proteins from M. mycoides SC, a screening of sera to monitor the humoral immune response was performed. By a suspension bead array assay, a multiplex and simultaneous analysis of immunoglobulins with reactivity to the recombinant surface proteins was achieved for every serum sample. The signal intensities from all recombinant proteins formed specific and individual patterns that were shown to be reproducible for each serum in our previous work (10). Signal intensities in technical replicates varied with an average intra-assay coefficient of variation (CV) of 21%. The serum-free control, used to monitor unspecific binding of secondary antibodies to the protein-coated beads, displayed signals that peaked at 3 to 20 AU, the intensities equivalent to the intrinsic autofluorescence of the beads. The control beads carrying the tag protein His6ABP displayed a median MFI of 45 AU, reflecting binding of serum antibodies to the fusion partner of the proteins. To distinguish between noise and antibody-mycoplasma antigen signal, an MFI threshold of 142 AU was set. This threshold was based on the 95th percentile for the distribution of signal intensities from the His6ABP beads. Intensity levels exceeding this threshold value were judged as representing specific binding of serum antibodies to the recombinant mycoplasma proteins.
Distinct differences in serum patterns were seen between CBPP-affected and CBPP-free sera. For 19 proteins, the signal median for CBPP-positive sera was significantly above the threshold value of 142 AU by a Wilcoxon rank sum test, compared to 2 proteins in the CBPP-negative sera. For the majority of the proteins, however, signals were below the threshold value of 142 AU. Sera categorized as CBPP positive showed a higher overall average intensity (484 AU) than did CBPP-negative sera (106 AU). Swedish negative control sera were found to yield an even lower overall average signal intensity (64 AU) than the CBPP-free African field sera (151 AU).
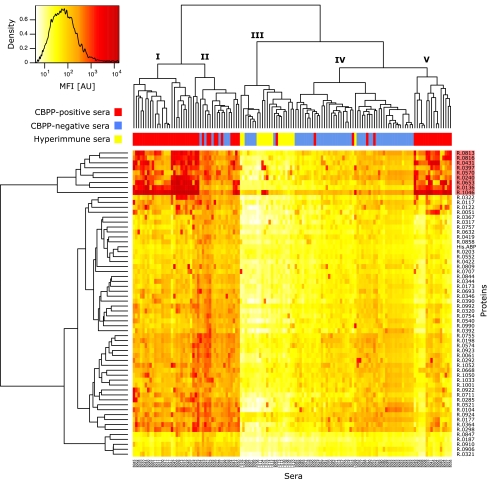
The screening data were summarized in a heat map in combination with a cluster analysis on both the serum and protein levels (Fig. 1). The sera separated into five main clusters, denoted clusters I to V. All technical replicate samples clustered together, except for one specimen of serum B079. Clusters I, II, and V consisted of 25, 20, and 16 samples, respectively, whereof 25, 13, and 16 samples, respectively, were from CBPP-positive sera. These clusters contained most of the high-signal-intensity data points. All hyperimmune sera, except for the M. mycoides SC PG1 antiserum, clustered together in cluster III, which displayed particularly low signals, together with five sera from Swedish bovines infected with Mycoplasma bovis (B094 to B098), one Swedish negative control (B084), and one CBPP-positive sample (B028). Cluster IV consisted of 50 samples, whereof 45 were CBPP negative (including 10 replicates of a negative control pool, B093), 4 were CBPP positive, and 1 was the M. mycoides SC PG1 antiserum (H099). In the vertical protein dendrogram, the top cluster, consisting of nine proteins, appeared to have potentially diagnostic importance. Large amounts of antibodies were detected in response to these proteins in CBPP-positive sera. One of them, R1046 (LppQ), yielded particularly high signals in CBPP-positive sera.
FIG. 1.
Cluster analysis of screening data. Heat map overview of log10-transformed MFI data from the bead-based screening of 134 samples (115 sera) against 61 recombinant M. mycoides SC surface proteins and His6ABP. Color intensity denotes signal intensity. Proteins against which large amounts of serum antibodies were detected in CBPP-positive sera, thus suggesting diagnostic importance, formed a cluster of nine proteins (top of vertical dendrogram; the cluster is highlighted in red). Horizontally, sera were separated into five groups. Clusters I, II, and V contained a majority of the CBPP-positive samples (red). Cluster III contained all but one hyperimmune serum (yellow), which displayed signals of low intensities, while cluster IV contained mostly CBPP-negative bovine sera (blue) as well as the positive control antiserum to M. mycoides SC PG1. All technical replicates clustered together, except for one specimen of a Swedish negative control serum.
Selection of antigens.
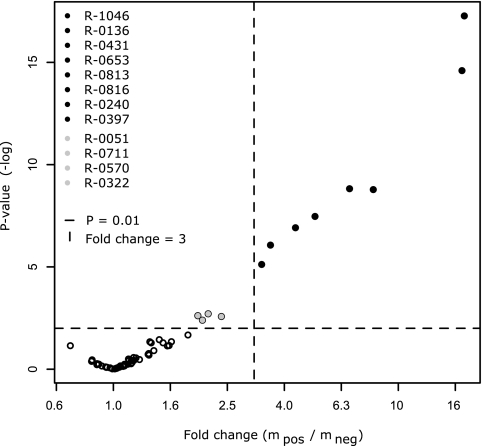
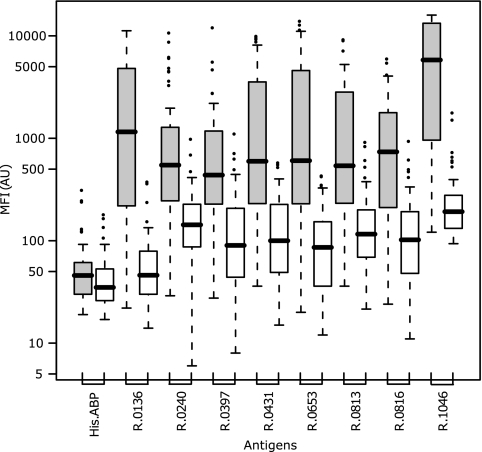
Selection of the most immunogenic antigens among the 61 recombinant proteins was based on Welch's two-sample t test. This test was performed on a log-transformed data set where blanks, replicates, hyperimmune sera, and Swedish control sera were removed. Thus, the statistical evaluation involved 24 CBPP-negative field samples, 50 CBPP-positive field samples, and 2 positive reference samples. The test revealed that among the 61 recombinant mycoplasma surface proteins, eight proteins had statistically significant positive-to-negative signal ratios of >3, with P values ranging from 10−6 to 10−18 (Fig. 2). These proteins were R1046, R0136, R0653, R0431, R0816, R0813, R0240, and R0397, all belonging to the protein cluster with apparent diagnostic importance (Fig. 1). As the theoretically most discriminative and therefore most potent diagnostic antigens among the investigated surface proteins, these eight were selected as candidates for further evaluation (Table 2). Additionally, four recombinant proteins, R0051, R0711, R0570, and R0322, appeared with P values below the significance threshold of 0.01, but their discriminating power, reflected by both the P value and the difference in antibody response, was not as good as that of the above eight proteins. An additional cluster analysis was performed on the screening data, employing all bovine sera with only the eight candidate antigens, and a distinct separation of samples into one CBPP-positive and one CBPP-negative cluster was observed (Fig. 3). Five of the seven CBPP-negative sera that previously grouped with positive sera in cluster II in Fig. 1 now clustered with the CBPP-negative group, but there were also three additional positive sera in the negative cluster. However, the cluster analysis is by no means a diagnostic test, but a visualization of signal similarities between samples. By presenting the distribution of signal intensities in the positive and negative populations in a box plot (Fig. 4), each antigen's discrepancy between CBPP-positive samples and CBPP-negative samples is shown. None of the eight proteins was discriminative enough to fully distinguish the positive from the negative sera alone, thus supporting that an antigen cocktail could offer an improved alternative for building a diagnostic system.
FIG. 2.
Statistical selection of immunogenic antigens. In a display of the results of the significance test of log10-transformed screening data, eight proteins (black) show signal changes between the CBPP-positive and CBPP-negative populations that are >3 and statistically significant, with P values ranging from 10−6 to 10−18. Additionally, four proteins (gray) displayed changes with significance exceeding the P value limit of 0.01, but with less discriminative importance than the top eight. Based on this test, the top eight proteins were selected as candidates for an antigen cocktail ELISA.
TABLE 2.
Selected recombinant mycoplasma antigens
| Recombinant protein | Native proteina | P value from suspension bead array screening | Included in cocktail ELISA | Protein size (kDa) |
|
|---|---|---|---|---|---|
| Recombinant | Native | ||||
| R1046 | LppQ | 5.4 e−18 | Yes | 49.7 | 52.1 |
| R0136 | Putative protein | 2.5 e−15 | Yes | 32.7 | 37.2 |
| R0431 | Put lipoprot | 1.5 e−9 | Yes | 38.4 | 40.8 |
| R0653 | Put lipoprot | 1.6 e−9 | Yes | 42.3 | 44.2 |
| R0813 | Put vsp | 3.4 e−8 | Yes | 58.6 | 57.9 |
| R0816 | Put vsp | 1.2 e−7 | No | 45.1 | 46.0 |
| R0240 | Put lipoprot | 8.6 e−7 | Yes | 62.5 | 63.8 |
| R0397 | Put lipoprot | 7.6 e−6 | Yes | 21.5 | 25.6 |
| R0079 | ABC transporter | Not included in screening | Yes | 49.3 | 50.1 |
Put lipoprot, putative lipoprotein; Put vsp, putative variable surface protein.
FIG. 3.
Cluster analysis based on selected proteins. Screening data for the eight selected proteins were used to categorize all bovine samples (117 samples from 97 sera). Two major clusters were formed, with one predominantly CBPP positive (dark gray) and one CBPP negative (light gray).
FIG. 4.
Discriminatory power of selected proteins. The range of signal intensities from the screening of all 52 CBPP-positive sera (gray bars) and 24 CBPP-negative sera (white bars; Swedish control sera were excluded) against the eight selected antigens shows a large discrepancy between the two groups. Potential binding to the fusion tag was monitored by the His6ABP bead. The box plot was built directly on signal intensities, and the box contains 50% of the data points (interquartile range), the black line denotes the median signal, and the whiskers extend to the furthest data points unless they are situated further away than 1.5 times the interquartile range. Outliers are presented as circles.
From the associated study (10), three additional mycoplasma surface proteins became available at a later stage and increased the number of mycoplasma proteins investigated to 64. One of these antigens, denoted R0079, displayed potential as a CBPP marker (data not shown) and was therefore included in the following evaluation and ELISA development.
Among the proteins corresponding to the nine selected recombinants, only LppQ, corresponding to R1046, had previously been described as immunodominant. The protein corresponding to R0816 is annotated as a putative variable surface protein and has been used in our previous work to study immune responses (10, 11). The protein corresponding to R0397 is a putative lipoprotein whose gene was indentified in a screening to find candidate genes for a DNA vaccine against CBPP (15), and a protective effect was shown for the protein in a mycoplasmemia mouse model. There is no information published on the remaining six proteins except for the bioinformatic annotation of the genomic sequence. The protein corresponding to R0813 is annotated as a putative variable surface protein and shows resemblance to R0816, as a ClustalW alignment gave a pairwise score of 48. Three of the proteins, corresponding to recombinants R0240, R0431, and R0653, are putative lipoproteins, while the remaining two, corresponding to R0079 and R0136, are annotated as the putative phosphonate ABC transporter PhnD and a putative protein, respectively. The MSC_0136 gene, corresponding to recombinant R0136, has previously been used in a phylogenetic study of the Mycoplasma mycoides cluster (24). The Interpro motif IPR005046, found in predicted surface proteins of many bacteria, is shared by six proteins, corresponding to R0136, R0431, R0653, R0813, R0816, and R1046. The sizes of the selected recombinant proteins, excluding His6ABP, range from 20 to 60 kDa, and those of their native counterparts range from 26 to 64 kDa. A summary of the selected antigens is presented in Table 2.
Cross-reactivity.
In the process of evaluating the recombinant proteins as antigens in a diagnostic system, the cross-reactivity to closely related Mycoplasma species was investigated. Hyperimmune sera from rabbits and calves immunized with closely related species were included in the study, and their immune responses toward all 61 proteins were analyzed. In the suspension bead array assay, these samples showed serum patterns distinctly different from those for the bovine sera. Patterns consisted of signals of 10 to 20 AU for almost all proteins, within the range of bead intrinsic autofluorescence and below the noise level threshold of 142 AU. Only a few proteins gave rise to higher signals, of 200 to 5,000 AU. Among these samples, the highest signals were noted for the positive control M. mycoides SC type strain PG1 antiserum. The hyperimmune sera from calves had the same profiles as the rabbit sera, thus excluding that results were caused by the anti-rabbit secondary antibody. Five bovines from Sweden with Mycoplasma bovis infections were included (B094 to B098) to evaluate cross-reactivity to M. bovis antigens and were more comparable to the other bovine sera than to the hyperimmune rabbit and calf sera. These samples displayed no sign of cross-reactivity, as all signals were below the noise level of 142 AU. Cross-reactivity intensities for the eight selected antigens in the hyperimmune sera are summarized in Table 3. A minor cross-reactivity was observed for some antisera from the M. mycoides cluster (H099 to H104).
TABLE 3.
Cross-reactivity of recombinant antigensa
| Serum ID | MFI (AU) with protein |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| R1046 | R0136 | R0653 | R0431 | R0816 | R0813 | R0240 | R0397 | His6ABP | |
| H099 | 5,827 | 16 | 14 | 20 | 14 | 22 | 285 | 2,074 | 20 |
| H100 | 58 | 14 | 25 | 16 | 31 | 26 | 39 | 20 | 16 |
| H101 | 2,601 | 77 | 30 | 36 | 41 | 172 | 280 | 1,523 | 31 |
| H102 | 47 | 15 | 13 | 8 | 11 | 46 | 18 | 514 | 16 |
| H103 | 1,184 | 18 | 19 | 19 | 17 | 28 | 1,465 | 119 | 18 |
| H104 | 2,812 | 23 | 24 | 1,510 | 19 | 18 | 7 | 1,446 | 20 |
| H105 | 29 | 15 | 13 | 8 | 10 | 16 | 7 | 6 | 16 |
| H106 | 101 | 16 | 20 | 15 | 116 | 29 | 16 | 23 | 17 |
| H107 | 52 | 15 | 24 | 10 | 17 | 19 | 12 | 8 | 20 |
| H108 | 33 | 13 | 14 | 10 | 11 | 17 | 30 | 6 | 15 |
| H109 | 57 | 19 | 25 | 27 | 21 | 25 | 41 | 114 | 30 |
| H110 | 50 | 22 | 42 | 21 | 19 | 25 | 10 | 11 | 19 |
| H111 | 32 | 23 | 20 | 15 | 17 | 27 | 98 | 11 | 18 |
| H112 | 69 | 15 | 15 | 16 | 11 | 17 | 6 | 12 | 17 |
| H113 | 91 | 21 | 17 | 21 | 23 | 48 | 40 | 13 | 20 |
| H114 | 94 | 32 | 24 | 27 | 16 | 33 | 10 | 12 | 26 |
| H115 | 138 | 19 | 42 | 33 | 75 | 34 | 13 | 15 | 21 |
Screening results with hyperimmune sera for different mycoplasma species (H099 to H115) to detect reactivity to the eight selected recombinant proteins and the His6ABP control. Bold values indicate intensities greater than the noise threshold of 142 AU, thus predicting cross-reactivity in a diagnostic setup. Some cross-reactivity was detected for samples within the M. mycoides cluster (H099 to H104).
Effect of ABP in suspension array assay.
A diagnostic system to monitor bacterial infection should preferably not be built on recombinant proteins with components of bacterial origin that could cause false-positive results. To verify that our screening could be relevant for tag-free recombinant proteins as well, eight proteins with and without ABP, as well as the His6ABP control, were tested with 47 sera on the suspension bead array. Hereby it was shown that the separation between CBPP-positive and CBPP-negative sera was not affected by the presence or absence of ABP (data not shown). Generally, an increase in signal intensity was observed for recombinant proteins without the fusion tag, possibly associated with bead coating properties.
Development of antigen cocktail ELISA.
A protein cocktail of the eight ABP-free proteins was used in an ELISA setup, and optimization experiments were performed to establish a robust protocol. Antigen coating concentrations and serum dilutions were varied to obtain signal intensities with the largest possible signal change between CBPP-positive and CBPP-negative sera. It was found that a protein coating concentration of 100 ng/well and a serum dilution of 1:1,000 allowed the most discriminative power. The surface and quality of the ELISA plate were crucial to obtain a robust assay. The intra-assay CV was reduced from 17% to 3% by changing the plastic from a particularly hydrophobic medium-binding-level plate to one spiked with polar groups and referred to as high binding. The most important parameter to achieve high signal intensity levels, and thus large signal changes, was the incubation time for developing the colorimetric substrate reaction. With the approach used, a signal separation between positive and negative sera was visible after 10 min, but the discrepancy increased with time. After 1 hour, signals stabilized, and this incubation time was therefore selected. The colorimetric signals were stable enough for the same relative results to be determined by remeasuring on the following day.
Performance of antigen cocktail ELISA.
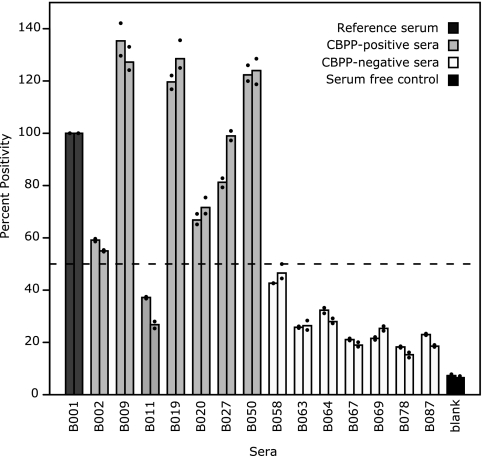
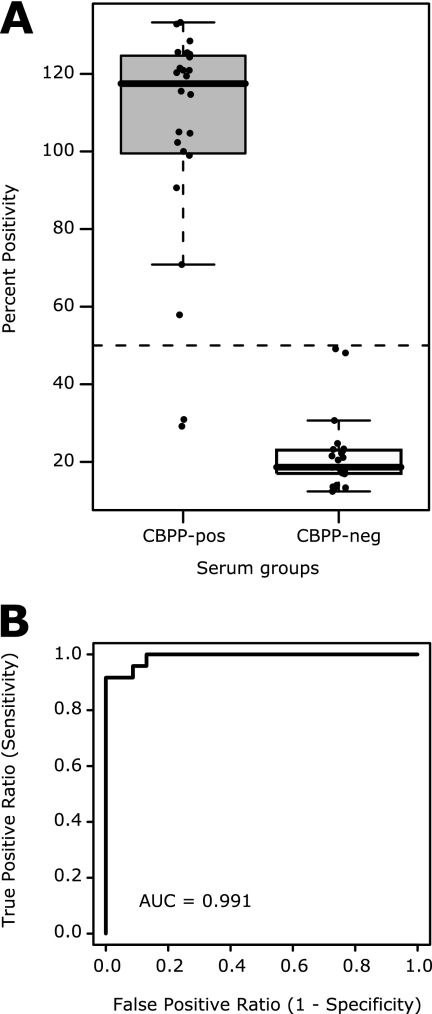
In the final setup of the eight-antigen cocktail ELISA, sera were analyzed in comparison to the reference serum B001 (16), and signals were reported in percent positivity (27). Evaluation of assay reproducibility with two experiments of triplicates of 15 sera showed an average intra-assay CV of 3% and an average interassay CV of 8% (Fig. 5). In a proof-of-concept study with 47 sera performed twice (Fig. 6), the assay allowed a fivefold signal ratio separation between CBPP-affected sera (105% average) and CBPP-free sera (22% average). The receiver operator characteristics of the assay showed a high diagnostic capability, since the area under the curve was calculated to 0.991. An initial cutoff in percent positivity of 50% resulted in no false-positive and two false-negative results, with percentages of positivity of 29% (B011) and 31% (B012). These two sera were both supplied with inconsistent results in serological tests (Table 1). Both B011 and B012 had positive results in the latex agglutination test as well as by gross pathology. B011 had a positive CFT titer, while B012 had a negative CFT titer, and both sera were negative by competitive ELISA. Two negative sera had percentages of positivity of 48% and 49%, which are close to the cutoff and would prompt additional analysis in a diagnostic situation. As observed by the bead-based array assay, Swedish and African negative sera differed in intensity level. Swedish sera had an average of 17% positivity, and African sera had an average of 24% positivity. It needs to be stated that even though origin-dependent variation in intensity levels was observed, no misclassification could be assigned to this variation.
FIG. 5.
Reproducibility of antigen cocktail ELISA. Two independent ELISA experiments with 15 sera in triplicate were performed by different persons 1 month apart to determine the reproducibility of the assay. The two bars for each sample represent the mean value for each experiment, and the dots represent the maximum and minimum values. All signals were normalized to the reference serum B001, resulting in low intra- and interassay variation (average CV, 3% and 8%, respectively). A percent positivity of 50% was suggested as the diagnostic cutoff, shown by a dashed line.
FIG. 6.
Proof-of-concept antigen cocktail ELISA. (A) Twenty-four CBPP-positive (gray) and 23 CBPP-negative (white) bovine sera were analyzed twice by the antigen cocktail ELISA. The optical density values were transformed to percent positivity with reference to sample B001 and are presented as averages for the two experiments. A suggested cutoff value of 50% would result in two false-negative and no false-positive results, assigning 96% of the tested samples correctly. The average percent positivity for CBPP-positive sera was 105%, compared to 22% for CBPP-negative sera, thus yielding a positive-to-negative signal ratio of 4.8. (B) The receiver operator characteristics of the ELISA results, with an area under the curve (AUC) of 0.991, indicate a high diagnostic capability.
DISCUSSION
In the presented study, a bead-based suspension array assay was applied to discover the most immunogenic surface antigens from M. mycoides SC among 64 recombinant surface proteins, corresponding to one-third of the bacterium's predicted surface proteome. Large-scale screening of bovine sera resulted in the selection of nine proteins to be antigen candidates for the development of enhanced serological diagnosis of CBPP. Eight of these were combined in an antigen cocktail and used to build an ELISA for diagnosis of M. mycoides SC infection.
The bead-based suspension array assay proved to be a powerful tool to monitor a multitude of humoral immune responses. Fingerprint-like immune response patterns for sera of CBPP-affected cattle appeared to reflect the individuality of humoral responses to different mycoplasma antigens and supported the idea of using multiple antigens in diagnostic tests. Importantly, objective numerical data were obtained, making it possible to apply a statistical significance test to select the antigens with the most discriminative power between CBPP-positive and -negative populations. The Swedish control samples needed to be excluded because the statistical analyses showed significant differences between the Swedish negative controls and the African negative controls. We could thereby conclude that in using the suspension bead array assay for selection of proteins that discriminate between sample populations, it is important to minimize differences between the groups and preferably to use samples with similar origins and collection procedures. In this case, the Swedish sera appeared to have lower general antibody titers than the African field sera, probably due to differences in exposure to infectious agents and other environmental factors. Still, the 21 Swedish control sera were important as technical controls to monitor assay-induced artifacts. It is noteworthy that the same top candidate diagnostic proteins were identified whether or not the Swedish samples were included.
The His6ABP tag of the recombinant proteins, used for purification and to enhance solubility, played an important role in the suspension bead array assay, as its presence made it possible to measure the coating density of the beads with an anti-ABP antibody. By including a bead carrying the fusion partner His6ABP per se, cross-reactivity to the His6ABP tag was monitored in the suspension bead array assay. An option in the data analysis was to use the signal from this negative control to normalize signals between wells, but no correlation between response toward ABP and specific antibody signals was observed. The negative control signal was instead used to define a threshold value, with intensity levels exceeding this threshold judged as signals from serum antibodies binding to the recombinant mycoplasma proteins. During optimization of the ELISA, though, the fusion protein ABP was excluded because the experimental layout would require one to additionally coat wells with His6ABP, as in other antigen ELISAs based on a fusion protein (2).
For an adequate diagnosis of CBPP, minimal cross-reactivity to related mycoplasmas is crucial. Surprisingly, the suspension bead array profiles for the 17 hyperimmune sera were found to be different from those for the African bovine sera, and signals were also obtained from antibody recognition of only a few recombinant proteins for the positive control anti-PG1 serum. However, those proteins that did appear were also among the most immunopotent surface proteins selected from the analysis of CBPP-positive sera. It therefore seems reasonable that any high signal intensity is representative for a case of infection, but “artificial” immunization appears to give a more selective immune response. Therefore, field samples from outbreaks of other mycoplasma infections would offer a preferable cohort in studies like this one. Nonetheless, the native protein corresponding to protein R1046, known as LppQ, has previously been shown to cross-react with antisera to Mycoplasma mycoides subsp. capri, Mycoplasma capricolum subsp. capripneumoniae, and Mycoplasma sp. bovine group 7 (1), and these were the same species found to cross-react with R1046 in this investigation. In a study of the antigenic specificity of LppQ (1), it was found that antisera raised against the C terminus of LppQ showed reactivity to proteins present in closely related species, while no cross-reactivity was detected for the N-terminal part of LppQ. This opens the possibility for more extended future studies that include thorough domain analysis of the selected antigens. Subcloning of the proteins could then offer a valuable strategy to adapt the target antigens to increase the specificity in the presented diagnostic application.
All serologic diagnostic methods are influenced by alterations in the immune responses of infected animals. Antibody titers are highly individual and have been reported to lack a relationship to the severity of lesions (19), and chronically infected animals may additionally not be detectable with serological tests. This is why results from serological tests are most confident on a herd level. It is a challenge to determine the specificity of a new serological test when existing methods give inconsistent results, and it is therefore required to link experimental data directly to clinical data. The serum collection available for this study was limited to include sera from bovines that had adequate diagnostic information. As observed in our analysis, several transport events and storage times might have affected sample quality, and this could affect, among other things, the discovery of false-negative results. Since none of today's serological methods are sufficiently sensitive per se, it is difficult to fully evaluate the performance of the antigen cocktail ELISA. One reason for the false-positive results discovered in our analysis could be a superior sensitivity of the bead-based array assay compared to traditional serology systems. Results from the well-characterized reference sera B001 and B002 were in clear concordance to expected and reported results (16) in both the bead-based assay and the cocktail ELISA.
The final antigen cocktail ELISA of this investigation had a discriminative power that distinguished sera from CBPP-affected and CBPP-free bovines. Thus, a promising ELISA setup has successfully evolved from the screening assay. However, to become a solid diagnostic ELISA, future evaluation on larger cohorts of preferably fresh bovine sera from herds of various geographical origins is needed to optimize positivity thresholds in order to determine the sensitivity and specificity. It is preferred to conduct future studies in facilities at the sites of livestock affected by CBPP to allow a direct comparison between the cocktail ELISA and other available tests.
In conclusion, the selected recombinant mycoplasma antigens qualified as highly specific markers for CBPP and could be employed in both a suspension bead array platform and a cocktail ELISA setting. This set of proteins and technologies therefore offers a powerful combination to drive and further improve serological assays toward reliable, simple, and cost-effective diagnostics of CBPP.
Supplementary Material
Acknowledgments
We thank the late Otto Hübschle (CVL, Namibia), Roger Ayling (VLA, United Kingdom), and John March (BigDNA Ltd., United Kingdom) for providing sera for this study. We also thank Peter Nilsson and Daniel Klevebring (KTH, Sweden) for support and for fruitful discussions on data analysis.
This study was funded by the Swedish International Development Cooperation Agency (SIDA).
Footnotes
Published ahead of print on 2 September 2009.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Abdo, E. M., J. Nicolet, and J. Frey. 2000. Antigenic and genetic characterization of lipoprotein LppQ from Mycoplasma mycoides subsp. mycoides SC. Clin. Diagn. Lab. Immunol. 7:588-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberti, A., P. Robino, B. Chessa, S. Rosati, M. F. Addis, P. Mercier, A. Mannelli, T. Cubeddu, M. Profiti, E. Bandino, R. Thiery, and M. Pittau. 2008. Characterisation of Mycoplasma capricolum P60 surface lipoprotein and its evaluation in a recombinant ELISA. Vet. Microbiol. 128:81-89. [DOI] [PubMed] [Google Scholar]
- 3.Bölske, G., H. M. Msami, A. Gunnarsson, A. M. Kapaga, and P. M. Loomu. 1995. Contagious bovine pleuropneumonia in northern Tanzania, culture confirmation and serological studies. Trop. Anim. Health Prod. 27:193-201. [DOI] [PubMed] [Google Scholar]
- 4.Bonvin-Klotz, L., E. M. Vilei, K. Kuhni-Boghenbor, N. Kapp, J. Frey, and M. H. Stoffel. 2008. Domain analysis of lipoprotein LppQ in Mycoplasma mycoides subsp. mycoides SC. Antonie van Leeuwenhoek 93:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruderer, U., J. Regalla, M. Abdo el, O. J. Huebschle, and J. Frey. 2002. Serodiagnosis and monitoring of contagious bovine pleuropneumonia (CBPP) with an indirect ELISA based on the specific lipoprotein LppQ of Mycoplasma mycoides subsp. mycoides SC. Vet. Microbiol. 84:195-205. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, A. B., and A. W. Turner. 1953. Studies on contagious bovine pleuropneumonia of cattle: an improved complement fixation test. Br. Vet. J. 29:154-163. [Google Scholar]
- 7.Egwu, G. O., R. A. J. Nicholas, J. A. Ameh, and J. B. Bashiruddin. 1996. Contagious bovine pleuropneumonia: an update. Vet. Bull. 1996:875-888. [Google Scholar]
- 8.FAO (Food and Agriculture Organization of the United Nations). 2003. Contagious bovine pleuropneumonia, Third Meeting of the Consultative Group on Contagious Bovine Pleuropneumonia, 12—14 November 2003, Rome, Italy. EMPRES Transboundary Anim. Dis. Bull. 24:2-7. [Google Scholar]
- 9.Gaurivaud, P., A. Persson, D. L. Grand, J. Westberg, M. Solsona, K. E. Johansson, and F. Poumarat. 2004. Variability of a glucose phosphotransferase system permease in Mycoplasma mycoides subsp. mycoides small colony. Microbiology 150:4009-4022. [DOI] [PubMed] [Google Scholar]
- 10.Hamsten, C., M. Neiman, J. M. Schwenk, M. Hamsten, J. B. March, and A. Persson. Recombinant surface proteomics as a tool to analyze humoral immune responses in bovines infected by Mycoplasma mycoides subsp. mycoides SC. Mol. Cell. Proteomics, in press. [DOI] [PMC free article] [PubMed]
- 11.Hamsten, C., J. Westberg, G. Bolske, R. Ayling, M. Uhlen, and A. Persson. 2008. Expression and immunogenicity of six putative variable surface proteins in Mycoplasma mycoides subsp. mycoides SC. Microbiology 154:539-549. [DOI] [PubMed] [Google Scholar]
- 12.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299-314. [Google Scholar]
- 13.Kusiluka, L. J., and F. F. Sudi. 2003. Review of successes and failures of contagious bovine pleuropneumonia control strategies in Tanzania. Prev. Vet. Med. 59:113-123. [DOI] [PubMed] [Google Scholar]
- 14.Le Goff, C., and F. Thiaucourt. 1998. A competitive ELISA for the specific diagnosis of contagious bovine pleuropneumonia (CBPP). Vet. Microbiol. 60:179-191. [DOI] [PubMed] [Google Scholar]
- 15.March, J. B., C. D. Jepson, J. R. Clark, M. Totsika, and M. J. Calcutt. 2006. Phage library screening for the rapid identification and in vivo testing of candidate genes for a DNA vaccine against Mycoplasma mycoides subsp. mycoides small colony biotype. Infect. Immun. 74:167-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martel, J. L., R. Nicholas, J. Noordhuizen, A. Pini, and J. Regalla. 2001. Diagnostic tests for contagious bovine pleuropneumonia (CBPP). Report of the Scientific Committee on Animal Health and Animal Welfare. European Commission, Brussels, Belgium.
- 17.Monnerat, M. P., F. Thiaucourt, J. Nicolet, and J. Frey. 1999. Comparative analysis of the lppA locus in Mycoplasma capricolum subsp. capricolum and Mycoplasma capricolum subsp. capripneumoniae. Vet. Microbiol. 69:157-172. [DOI] [PubMed] [Google Scholar]
- 18.Monnerat, M. P., F. Thiaucourt, J. B. Poveda, J. Nicolet, and J. Frey. 1999. Genetic and serological analysis of lipoprotein LppA in Mycoplasma mycoides subsp. mycoides LC and Mycoplasma mycoides subsp. capri. Clin. Diagn. Lab. Immunol. 6:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholas, R. A., and J. B. Bashiruddin. 1995. Mycoplasma mycoides subspecies mycoides (small colony variant): the agent of contagious bovine pleuropneumonia and member of the “Mycoplasma mycoides cluster.” J. Comp. Pathol. 113:1-27. [DOI] [PubMed] [Google Scholar]
- 20.Persson, A., K. Jacobsson, L. Frykberg, K. E. Johansson, and F. Poumarat. 2002. Variable surface protein Vmm of Mycoplasma mycoides subsp. mycoides small colony type. J. Bacteriol. 184:3712-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persson, A., B. Pettersson, G. Bolske, and K. E. Johansson. 1999. Diagnosis of contagious bovine pleuropneumonia by PCR-laser-induced fluorescence and PCR-restriction endonuclease analysis based on the 16S rRNA genes of Mycoplasma mycoides subsp. mycoides SC. J. Clin. Microbiol. 37:3815-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilo, P., J. Frey, and E. M. Vilei. 2007. Molecular mechanisms of pathogenicity of Mycoplasma mycoides subsp. mycoides SC. Vet. J. 174:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pilo, P., S. Martig, J. Frey, and E. M. Vilei. 2003. Antigenic and genetic characterisation of lipoprotein lppC from Mycoplasma mycoides subsp. mycoides SC. Vet. Res. 34:761-775. [DOI] [PubMed] [Google Scholar]
- 24.Thiaucourt, F., S. Lorenzon, A. David, and A. Breard. 2000. Phylogeny of the Mycoplasma mycoides cluster as shown by sequencing of a putative membrane protein gene. Vet. Microbiol. 72:251-268. [DOI] [PubMed] [Google Scholar]
- 25.Vilei, E. M., E. M. Abdo, J. Nicolet, A. Botelho, R. Goncalves, and J. Frey. 2000. Genomic and antigenic differences between the European and African/Australian clusters of Mycoplasma mycoides subsp. mycoides SC. Microbiology 146:477-486. [DOI] [PubMed] [Google Scholar]
- 26.Westberg, J., A. Persson, A. Holmberg, A. Goesmann, J. Lundeberg, K. E. Johansson, B. Pettersson, and M. Uhlen. 2004. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP). Genome Res. 14:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright, P. F., E. Nilsson, E. M. A. van Rooij, M. Lelenta, and M. H. Jeggo. 1993. Standardization and validation of enzyme-linked immunosorbent assay techniques for the detection of antibody in infectious disease diagnosis. Rev. Sci. Tech. 12:435-450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.