Abstract
The dynamic evolution of organelle compartmentalization in eukaryotes and how strictly compartmentalization is maintained are matters of ongoing debate. While the endoplasmic reticulum (ER) is classically envisioned as the site of protein cotranslational translocation, it has recently been proposed to have pluripotent functions. Using transfected reporter constructs, organelle-specific markers, and functional enzyme assays, we now show that in an early-diverging protozoan, Giardia lamblia, endocytosis and subsequent degradation of exogenous proteins occur in the ER or in an adjacent and communicating compartment. The Giardia endomembrane system is simple compared to those of typical eukaryotes. It lacks peroxisomes, a classical Golgi apparatus, and canonical lysosomes. Giardia orthologues of mammalian lysosomal proteases function within an ER-like tubulovesicular compartment, which itself can dynamically communicate with clathrin-containing vacuoles at the periphery of the cell to receive endocytosed proteins. These primitive characteristics support Giardia's proposed early branching and could serve as a model to study the compartmentalization of endocytic and lysosomal functions into organelles distinct from the ER. This system also may have functional similarity to the retrograde transport of toxins and major histocompatibility complex class I function in the ER of mammals.
A key event in the evolution of eukaryotic cells was the compartmentalization of cellular functions into distinct organelles responsible for protein synthesis, sorting, secretion, endocytosis, and degradation (38). However, it is clear from ultrastructural and biochemical analysis of many eukaryotic cells that these functionally distinct compartments often share common aspects of biogenesis and function and, in some cases, a common tubulovesicular network (TVN) (2). For example, one current debate concerns a putative role for the endoplasmic reticulum (ER) in phagocytosis (11, 42). Gagnon et al. (11) proposed that the ER was involved in direct uptake of material from the extracellular environment via fusion with the plasma membrane. This hypothesis was based on the presence of ER markers at the initial stage of phagosome formation in mammalian macrophages. Touret et al. (42), however, found no evidence for direct ER-plasma membrane communication in either macrophages or dendritic cells. Nevertheless, the concept of pluripotent functions for the ER was left unresolved, and these studies underscore the potential for an ER function in phagocytosis or endocytosis, particularly in regard to antigen processing for major histocompatibility complex (MHC) class I presentation. Furthermore, there are intriguing examples of exogenous toxins and viruses entering mammalian cells via the ER (19, 36). Because cellular compartmentalization is a defining eukaryotic trait, clues from early-diverging eukaryotic cells could provide valuable insights into the way in which compartmentalization and discrete organelle functions evolved.
Giardia evolutionary status continues to be a matter of debate. Either Giardia lamblia is one of the earliest branches of the eukaryotic tree, with an estimated point of divergence of 1.7 to 2.1 billion years (3, 16, 17, 35, 38), or it is a very simple cell that has lost endomembrane complexity and classic organelle morphology by evolutionary reduction (6). Giardia has a simple life cycle that includes a replicating trophozoite stage and, under certain environmental conditions, an infectious, environmentally resistant cyst. The cyst form allows the replicative trophozoite to persist under conditions of desiccation outside the host and in harsh chemical environments like the host stomach. Following passage through the acidic stomach into the alkaline duodenum, the trophozoite form excysts and resides in the upper small intestine of its vertebrate host, where it opportunistically scavenges nutrients by uncharacterized endocytic pathways.
The endomembrane system of the vegetative trophozoite form of Giardia lacks complexity compared to typical eukaryotic cells. In many extant eukaryotic cells, the majority of secretory and organelle-resident proteins are delivered by cotranslational translocation to the ER lumen, prior to maturation and subsequent targeting to the Golgi apparatus (2). In Giardia, no morphological equivalent of the classic mammalian-cell Golgi apparatus has been identified, and the transient secretory pathway for cyst wall synthesis is induced only under specific conditions (13, 14, 34). Giardia does contain two nuclei, a glycogen-rich cytoplasm, acidified peripheral vacuoles (PVs), and a labyrinthine TVN, segments of which are decorated with ribosomes, consistent with rough ER (5, 21, 22). Giardia also contains a multigene family of cysteine endoproteases that are orthologous to the cathepsin L and cathepsin B found in lysosomes of higher organisms and are therefore useful markers of cell compartments where protein degradation takes place (29, 46; Giardia genome project [http://www.giardiadb.org]). Due to the limited availability of established ER/endocytic subcompartment markers and the lack of classical genetic techniques, the discrete endocytic pathway of Giardia has not been fully characterized. Despite preliminary reports, RNA interference has not been established as a reliable and consistent genetic approach (43). In spite of these limitations, we were able to use reporter gene constructs, organelle-specific markers, and functional protease cytochemistry to show that endocytosis and degradation of exogenous proteins takes place in the ER-like TVN. Such primitive characteristics of Giardia's endomembrane system support Giardia's proposed early branching and could be used as an analogous model to study the compartmentalization of endocytic/lysosomal functions into organelles distinct from the ER. Alternatively, Giardia may have had a more complex and “modern” endocytic system that has been lost by evolutionary reduction (6). These observations also have important implications for recent theories of pluripotent mammalian ER functions, including its roles in phagocytosis, entry of toxins and viruses, and MHC class I function (7, 19, 36).
MATERIALS AND METHODS
Live cell imaging.
Adherent transgenic parasites expressing G. lamblia CP2-green fluorescent protein (GlCP2::GFP) constitutively were harvested in log phase by cooling and centrifugation at 900 × g for 10 min. Cells (5 × 10e6) were resuspended in 1 ml of ice-cold culture medium, transferred to a single well in a 24-well plate kept on ice, and exposed to air for 2 h in the dark. Ten microliters of the oxygenated cells were checked for the presence of fluorescent (mature) GFP by fluorescence microscopy. ER Tracker Blue-White reagent (Molecular Probes, Eugene, OR) was added to a final concentration of 1 μM. The cells were incubated for 30 min on ice, collected by centrifugation, and washed twice with ice-cold medium. The cells were spotted on a glass slide and sealed under a coverslip with nail polish. Observation and data collection were performed on a Leica SP2 AOBS confocal laser-scanning microscope (Leica Microsystems, Wetzlar, Germany) with a 63× water objective (Leitz HCX PL APO 63×/1.2 W CORR) using the 405- and 488-nm laser lines for excitation of ER Tracker and GFP, respectively. Optical sections of adherent living trophozoites were generated for each dye, followed by a differential interference contrast (DIC) image. Appropriate detector settings and twofold line averaging were used. Cell viability was assessed and documented before and after imaging using the following criteria: morphology, attachment to the cover glass, and flagellar beating. The degree of signal overlap between ER Tracker and GlCP2::GFP from the raw image data was quantified using the colocalization function of the Imaris software suite (Bitplane AG, Zurich, Switzerland) and is shown as a scatter plot. Further processing of color merged images was done using the Huygens deconvolution software package version 2.7 (Scientific Volume Imaging, Hilversum, The Netherlands).
Imaging of fixed cells.
Adherent parasites expressing the GlCP2::GFP reporter constitutively were harvested by cooling and centrifugation at 900 × g for 10 min. Cells (5 × 10e6) were resuspended in 1 ml of ice-cold medium, transferred to a single well in a 24-well plate kept on ice, and exposed to air for 2 h in the dark. Ten microliters of the oxygenated cells was checked for the presence of fluorescent (mature) GFP, and the rest was harvested, centrifuged in the cold, and incubated with 3% formaldehyde in phosphate-buffered saline (PBS) for 40 min at 20°C. The fixative was replaced with 0.1 M glycine in PBS for 3 min to block free aldehydes.
Membranes were permeabilized by incubation with 0.2% Triton X-100 in PBS for 20 min at 20°C, and the cells were blocked for at least 2 h in 2% bovine serum albumin (BSA) in PBS. The cells were incubated with titrated monospecific mouse antibodies against the ER marker protein disulfide isomerase 2 (PDI2) (GlPDI2; Giardia Genome Database number GL50803_9413) (1:1,000 dilution), followed by Texas Red-conjugated secondary antibodies (Invitrogen, Basel, Switzerland) used at a 1:200 dilution (Invitrogen, Basel, Switzerland). All antibody incubations were done in 2% BSA-0.2% Triton X-100 in PBS at 20°C. The cells were washed between incubations with 0.5% BSA-0.05% Triton X-100 in PBS. The labeled cells were embedded for microscopy with Vectashield (Vector Laboratories) containing the DNA-intercalating agent 4′,6-diamidino-2-phenylindol dihydrochloride (DAPI) for detection of nuclear DNA. Immunofluorescence data collection was performed on a Leica SP2 AOBS confocal laser-scanning microscope (Leica Microsystems, Wetzlar, Germany) using the appropriate laser and detector settings. Image stacks of optical sections were generated by twofold oversampling, and image rendering was done using the Imaris software suite (Bitplane AG, Zurich, Switzerland). The Imaris colocalization function was used to determine the degree of signal overlap between GlCP2::GFP and PDI2. The scatter plots show the signal distribution in the three-dimensional (3D) image stack. As an internal negative control, the overlap of ER-localized PDI2 and the DAPI signal (nuclear DNA) was plotted.
Cell culture and transfection.
WBC6 isolate G. lamblia trophozoites from the American Type Culture Collection (catalog number 50803) were maintained in modified TYI-S-33 medium supplemented with 10% fetal bovine serum (Omega Scientific, Inc.), penicillin-streptomycin (UCSF Cell Culture Facility), vitamins (Gibco/Invitrogen), and Fungizone (UCSF). The pGFP.pac vector (a gift from Theodore Nash, NIH; modified by Lei Li, C. C. Wang laboratory, UCSF, by substituting the Giardia tubulin gene promoter for the Giardia giardin gene promoter) was used to transiently and episomally express C-terminal GFP fusion proteins in Giardia trophozoites. The transfection protocol used by Singer et al. (37) was followed with modifications as follows: 1 × 106 to 2 × 106 trophozoites were incubated on ice for 20 min with 50 μg of circular plasmid DNA and then electroporated (GenePulser XCell; Bio-Rad) at 0.45 kV, 950 μF. Transfectants were selected in a dose-dependent manner using puromycin dihydrochloride (Sigma Inc.) increased in 5- to 20-μg/ml increments to a final concentration of 80 to 120 μg/ml. The Giardia Genome Database (http://www.giardiadb.org/giardiadb) numbers are as follows: GlCP1, GL50803_10217; GlCP2, GL50803_14019; and GlCP3, GL50803_14019.
Microscopy.
A confocal microscope (LSM510 META; Carl Zeiss MicroImaging, Inc.) equipped with multiline (458-, 477-, 488-, and 514-nm) Ar, 543-nm HeNe, and 633-nm HeNe visible lasers and a Chameleon two-photon laser module (Coherent, Inc.) with a “Plan-Apochromat” 63×/1.40 Oil DIC oil immersion lens (Carl Zeiss MicroImaging, Inc.) was used for fluorescence and live cell imaging experiments. Microscopy was done at room temperature. The cells were pulsed with sufficient oxygen at 37°C for 1 to 3 h, fixed in 3% paraformaldehyde (Electron Microscopy Sciences) for 40 min at room temperature, and mounted with ProLong Gold mounting medium (with or without DAPI) (Molecular Probes). Live imaging was done in PBS. In the time frame of these experiments, Giardia cells were viable and active in PBS. No difference was noted in comparisons to control imaging in Giardia medium, but PBS was preferred, as background fluorescence was minimal. LSM Image Browser software (Carl Zeiss MicroImaging, Inc.) was used for confocal image acquisition. Adobe Photoshop CS (Adobe Systems, Inc.) was used for subsequent processing.
Ultrastructural tomography.
For tomography studies, a Tecnai T20 electron microscope (EM) (FEI Company) equipped with a bottom-mounted four-quadrant 4,000-by-4,000 Gatan UltraScann charge-coupled device (Gatan Inc.) was run at 200 kV, and images were collected every 2° from −60 to +60°. The camera was set at binning 2 (2,000-by-2,000) resolution, and images were collected at ×21,500 magnification. Samples were 250 nm thick. The images were processed, analyzed, and visualized using PRISM software (47). The data were analyzed further using Openlab software (Improvision Ltd., England). To obtain a pseudo-3D view, the transparent backgrounds of three selected consecutive 0.25-μm images were set to 75, 50, 35, and 0% (background layer) opacity prior to image merging.
Antibodies and reagents.
Anti-Giardia clathrin heavy chain (Giardia Genome Database number GL50803_102108) polyclonal and anti-Giardia PDI2 (GL50803_9413) polyclonal antibodies were used at 1:500 and 1:3,500, respectively. Anti-KDEL monoclonal antibody (Stressgen Bioreagents, MI) was used at the recommended dilutions. Recombinant, hemagglutinin-tagged Giardia BiP homolog (GGD; GL50803_17121) was detected by mouse monoclonal Alexa 488-conjugated anti-hemagglutinin antibody (dilution, 1:30; Roche Diagnostics GmbH, Manheim, Germany). Rhodamine-labeled anti-mouse immunoglobulin G(γ) (KPL Inc.) and fluorescein-labeled anti-mouse immunoglobulin G(γ) (KPL Inc.) were used at 1:10. For endocytosis studies, fluorescent proteins (3.75 μg/ml casein; 5.0 μg/ml albumin) or 0.04 μm biotin-conjugated Fluospheres (2.8 × 1014 particles/ml; 1.0-μl/ml final concentration; excitation/emission, 505 nm/515 nm; Molecular Probes) were incubated with trophozoites for 30 min at 37°C in medium or PBS. Sodium azide and cytochalasin D were used at 20 mM and 10 μM, respectively. Cells were fixed and visualized. Lucifer yellow lithium salt (excitation/emission, 428 nm/536 nm; Molecular Probes) was used to visualize PVs. Trophozoites were incubated in 1 ml of PBS and Lucifer yellow to a final concentration of 1 mg/ml in PBS for 15 min at 37°C. Following incubation, the cells were fixed and visualized.
Immunofluorescence localization of organelle markers.
Trophozoites were stained as previously described (26), with modifications. All experiments were carried out at room temperature unless otherwise stated. Cells were harvested following 15-min incubation on ice and a 15- to 20-min spin at 411 × g to pellet the cells. The cells were washed with cold PBS (Ca2+ and Mg2+ free) before being fixed for 40 min with fresh 3% paraformaldehyde (EMS). Following 5 min of incubation in 0.1 M glycine in PBS, the cells were permeabilized in 0.1% Triton X-100 in PBS for 30 min and blocked with 2% BSA in PBS. Trophozoites were incubated with primary and secondary antibodies (diluted in 2% BSA-0.1% Triton X-100 in PBS) for 1 h each.
Protease activity assays.
Protease activity from Giardia lysates was isolated by anion-exchange chromatography using a MonoQ column (GE Healthcare). The fluorogenic substrates Z-FR-AMC (N-carbobenzoxy-phenylalanyl-arginyl-7-amido-4-methylcoumarin) and Z-RR-AMC (N-carbobenzoxy-arginyl-arginyl-7-amido-4-methylcoumarin) (excitation/emission, 360 nm/470 nm) were incubated with Giardia lysates in citrate/dibasic sodium phosphate buffers (pH 4.0 to 8.0) containing 4 mM dithiothreitol, 5 mM Pefabloc, and 50 mM EDTA. Subsequent protease activity was measured by monitoring the increase in relative fluorescence units over time.
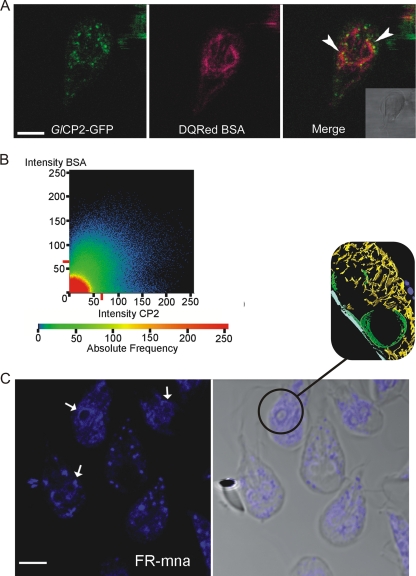
The fluorescent substrates Z-FR-MNA (N-carbobenzoxy-phenylalanyl-arginyl-4-methoxy-β-naphthylamide) and Z-RR-MNA (N-carbobenzoxy-arginyl-arginyl-4-methoxy-β-naphthylamide) (Bachem) were used in an adapted in vivo protease assay (8). Trophozoites were washed once with PBS (Ca2+ and Mg2+ free) and then incubated in PBS with 70 μM Z-FR-MNA and 2 volumes of coupling reagent (5-nitro-2-salicylaldehyde and 2 M cacodylate buffer [excitation/emission, 395 nm/575 nm]) for 30 min at 37°C. For Z-RR-MNA, trophozoites were washed in 100 mM cacodylate buffer, pH 6.8, and 5% sucrose and incubated with Z-RR-MNA at a final concentration of 500 μM in 100 mM cacodylate buffer, pH 6.8, for 1 h at 37°C, followed by 20 min of incubation at 37°C with coupling reagent. The cells were fixed and visualized.
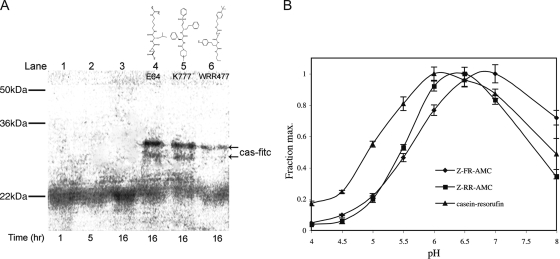
Giardia lysate was incubated for 30 min with 50 g casein-resorufin (Molecular Probes) in 200 μl citrate/dibasic sodium phosphate buffers containing 5 mM Pefabloc and 50 mM EDTA. Trichloroacetic acid (960 μl; 5% [wt/vol]) was added, and samples were incubated for 10 min and centrifuged at 16,000 × g to pellet the precipitant. The supernatant (400 μl) was added to 600 μl 0.5 M Tris-HCl, pH 8.8. Hydrolysis was quantified by measuring the fluorescence of resorufin-containing peptides (excitation/emission, 574 nm/584 nm). Giardia trophozoites (1.0 × 106) were incubated with 20 g casein-fluorescein isothiocyanate (FITC) for 30 min in vegetative medium and chased with fresh medium. The cells were incubated at 37°C for 1 h, 5 h, and 16 h. Cells were also incubated for 16 h in the presence of three known cell-permeable cysteine protease inhibitors (10 μM; dimethyl sulfoxide < 1%): E64d, K11777 (K777), and WRR477. The cells were lysed, and the proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. In-gel casein-FITC was detected using a Typhoon 8600 Variable Mode Imager (Amersham).
Localization of endocytosed albumin-gold by electron microscopy.
A trophozoite monolayer was washed and incubated with ultrasmall-gold (0.8 nm)-conjugated albumin (Electron Microscopy Sciences) or albumin-conjugated 10-nm gold (Ted Pella, Inc.; with and without sodium azide from British Biocell) at dilutions ranging from 1:10 to 1:100 in PBS at pH 7.0 for 30 min at 37°C. The cells tolerated these conditions for up to 4 h with no evidence by microscopy of deterioration. The cells were fixed (30 min in 3% glutaraldehyde-1% paraformaldehyde, 0.1 M cacodylate buffer, pH 7.4, at 4°C) and processed for LR White (EM Science) embedding. Sections were silver enhanced (HQ Silver Enhancement Kit; Nanoprobes) for 25 min at 4°C.
Localization of G6P activity by electron microscopy.
A trophozoite monolayer was fixed as described above. The glucose-6-phosphatase (G6P) reaction was performed as previously described (24). The cells were washed with 0.1 M Tris-maleate, 3 mM lead nitrate, 4 mM disodium glucose-6-phosphate, and 5% sucrose, pH 6.5, at 37°C for 60 min. The cells were postfixed with the aforementioned fixative and osmium tetroxide and embedded in Epon. Blocks were sectioned with a Leica ultracut UCT ultramicrotome. Sections were viewed with an FEI Tecnai 10 EM (FEI Company).
Endocytosis visualized by live cell imaging.
Trophozoites were allowed to adhere to a chambered coverglass (Lab-Tec) in vegetative growth medium. The medium was replaced with 400 μl PBS at 37°C. Filming began as 5 × 109 to 5 × 1010 biotin-conjugated 0.04-μm Fluospheres (excitation/emission, 505 nm/515 nm; Molecular Probes) or 250 ng of DQ Red BSA (excitation/emission, 590 nm/620 nm; Molecular Probes) were added to the chamber of adhered trophozoites.
EM tomography.
Cells incubated in the cytochemical medium designed for the localization of G6P were used (24). A single 250-nm-thick section was collected in an oyster grid, stained with uranyl acetate and lead citrate as usual, and labeled nonspecifically with colloidal gold for further alignment. The thick sections were examined at 400 keV on a Jeol 4000EX intermediate-voltage EM. The specimen was irradiated for 30 min prior to the collection of the tomographic series to avoid further shrinkage resulting from heating of the epoxy. A single-axis tilt series was obtained from the structures of interest by rotating the specimen on the y axis in equal 2° increments from 260 to 160°. Images were acquired at each tilt angle on film and were further digitized through a 1024-by-1024, 14-bit, computer-controlled Photometrics charge-coupled-device camera by flat-field transillumination of the negatives. The reconstruction was performed using IMOD (12). The automated segmentation and movie were generated using Amira (Visage Image).
RESULTS
Electron tomography defines the intracellular-membrane compartments of Giardia.
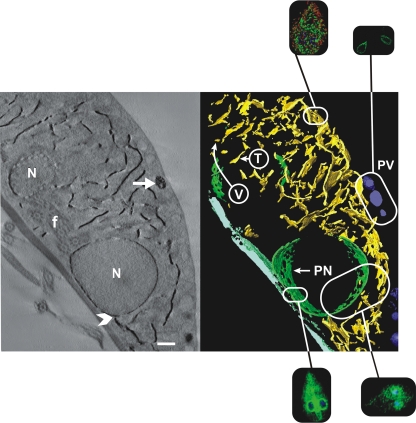
Figure 1 shows the extensive TVN that represents the ER compartment of Giardia. This compartment is recognized at the level of light microscopy by the extensive perinuclear cisternae, but it also extends to near the periphery of the cell. It is the major intracellular compartment outside of the nuclei. Tomography clearly distinguishes the PVs from the TVN. The PVs are large spherical or egg-shaped vacuoles that are situated between the TVN and the plasma membrane. The ER is referred to as a TVN because portions of the complex branching network appear vesicular when seen in cross section while in longitudinal section they appear tubular or linear. No other cellular compartment has such a distinctive appearance (5).
FIG. 1.
Electron tomography of a 250-nm section of G. lamblia labeled for G6P detection. The image on the left is from a single digital slice (2.7 nm) showing reaction product on the nuclear envelope, the TVN, and some PVs (arrow). An arrowhead (lower center below the nucleus) points to a proximity point of the TVN system and the nuclear envelope. On the right is a false-color 3D model of the same cell showing the widespread distribution of TVN tubules (yellow) and cisternae (green) that fill most of the space between the nuclei and the PVs that are positive for G6P (blue). When seen in cross section, the TVN appears vesicular (V), while in longitudinal view it appears tubular (T). Elements of the ventral disk are depicted in light blue. N, nucleus; f, flagellum. The insets show the correlated appearance of the TVN (ER), PVs (PV), and perinuclear (PN) cisternae of the TVN in confocal microscopic slices that are also shown in Fig. 2, 5, and 6. Scale bar, 500 nm.
The identification of this network by G6P and its distinction from the acidified PVs led to two models of protein endocytosis and degradation. If Giardia mirrored higher eukaryotic cells, then extracellular proteins would be endocytosed into compartments distinct from the ER and subsequently trafficked to the PVs, where lysosomal cathepsin orthologues would catalyze protein degradation. However, as documented in the figures, proteins are instead endocytosed into a TVN and ultimately into the extensive perinuclear cisternae.
Proteins and Fluospheres are endocytosed by Giardia trophozoites and rapidly traverse a TVN that extends to the perinuclear region.
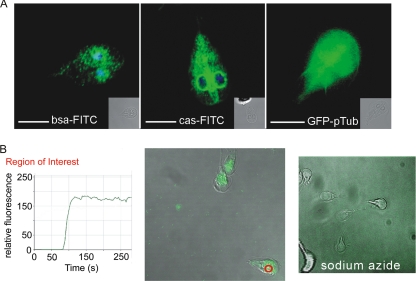
The intracellular localization of fluorescein-labeled albumin and casein was first ascertained by confocal microscopy following a 30-min incubation of labeled proteins with Giardia in culture (Fig. 2A, left and middle).
FIG. 2.
Proteins are rapidly endocytosed by trophozoites into the TVN and the perinuclear region. (A) Endocytosis of albumin (bsa-FITC) and casein (cas-FITC) resulted in both a fine “vesicular” pattern in the cytoplasm and intense perinuclear localization (see the correlates in Fig. 1). DAPI (blue) highlights the nuclei of Giardia. The perinuclear localization is more apparent with casein due to the specific optical cut that included the nuclei. Transfection with the empty GFP vector (right) resulted in dispersed signal distributed throughout the cell. Single optical sections are presented. Scale bars, 5 μm. (B) Uptake of biotin-conjugated Fluospheres. The kinetics of uptake were tracked by monitoring the increase in relative fluorescent units in the perinuclear region of a cell (red circle). The green line in the graph corresponds to fluorescence intensity over time. Sodium azide inhibited Fluosphere uptake (right).
Live cell imaging (see Movie S1 in the supplemental material) verified that uptake is extremely rapid; cells were saturated with fluorescent biotin-conjugated Fluospheres within 40 s following initial uptake (Fig. 2B). During live cell imaging, the initial Fluosphere signal was observed near the site where flagella exit the cell in the region caudal to the ventral disc (Fig. 2B) (5). This was followed by rapid distribution throughout the labyrinthine TVN with accumulation in the perinuclear region. Diffusion of the fluorochrome following protein degradation did not occur, as no difference in fluorescence localization was observed upon pretreatment with protease inhibitors (data not shown). This is in contrast to the diffuse cytosolic, noncompartmental localization seen upon expression of a GFP construct lacking a signal peptide (Fig. 2A, right). Treatment of trophozoites with sodium azide or cytochalasin D or incubation at 4°C inhibited protein uptake (Fig. 2B and data not shown).
Cathepsin B-like proteases colocalize with and degrade endocytosed proteins in the TVN.
Giardia contains a number of genes coding for clan CA cathepsin B-like cysteine proteases, of which GlCP1, GlCP2, and GlCP3 are representative members (29). Though previous Northern blot analysis did not detect GlCP1 in the vegetative trophozoites, recent studies using more sensitive reverse transcription-PCR analysis demonstrated that the genes encoding GlCP1, GlCP2, and GlCP3 are each expressed during vegetative growth (9). C-terminal fusion GFP reporter constructs indicated that each of the Giardia cathepsin B-like proteases was present in both the peripheral TVN and the perinuclear region and colocalized with endocytosed rhodamine-conjugated albumin and DQ Red BSA (Fig. 3A and B; see Fig. S4 and Movie S3 in the supplemental material).
FIG. 3.
Giardia orthologues of mammalian lysosomal proteases are present in the endocytic TVN, where they colocalize with endocytosed proteins. (A) The cathepsin B-like protease GlCP2::GFP reporter (green, Giardia orthologue of mammalian lysosomal proteases) colocalized with endocytosed albumin (red, DQ Red BSA) during live cell imaging. Yellow in the merged image shows overlap (arrowheads) in both the perinuclear region of the trophozoite and the finer network of the TVN. Validation of this overlap is shown in panel B. Bar, 5 μm. (B) Quantification of colocalization shown by plotting the pixel fluorescence intensities from channels for DQ Red BSA and GlCP2::GFP. The majority of pixels are positive for both markers. (C) Cysteine protease activity detected by fluorescent substrate cleavage (Z-FR-MNA). This activity is seen in the outer TVN but is particularly intense in the perinuclear region (arrows). The inset shows perinuclear cisternae and TVN from tomography in Fig. 1 as a correlate. Scale bar, 5 μm.
To ensure that the GFP constructs did not mislocalize due to misfolding or due to the presence of the GFP reporter protein, cathepsin activity, identified by in situ cleavage of the MNA-derivatized peptide substrate, was localized to the same region of the cell (Fig. 3C). In situ MNA peptide cleavage, indicated by coupled fluorescent activity, was completely inhibited by treatment with the cysteine protease inhibitors E64d and K11777 (data not shown). This activity correlated with the localization and degradation of endocytosed proteins. A functional assay confirmed that endocytosed casein was degraded and that this degradation was inhibited by the known clan CA cysteine protease inhibitors E64d, K11777, and WRR477 (Fig. 4A). Replacing casein-FITC with BSA-FITC in this assay yielded equivalent results (data not shown). Consistent with a proposed location and activity in the ER-like TVN, the pH optimum of the Giardia cathepsin activity was 6 to 7, notably higher than that of the mammalian cathepsin orthologues, which function in acidified lysosomes and have an acidic pH optimum of 5 to 6 (Fig. 4B) (45).
FIG. 4.
Giardia cysteine proteases degraded endocytosed proteins and were optimally active at a higher pH than their mammalian orthologues. (A) Cysteine proteases degraded endocytosed casein-FITC (cas-fitc). Cells were pulsed for 30 min with substrate and chased with fresh medium for the times indicated, and the cell lysates were examined. Lanes 1, 2, and 3 show loss of substrate in the cells following 1, 5, and 16 h of incubation. In the presence of three known cysteine protease inhibitors (10 μM) (lanes 4, 5, and 6), casein degradation was blocked (arrows). (B) pH profile of G. lamblia cathepsin B-like cysteine protease activity against the fluorogenic substrates Z-FR-AMC and Z-RR-AMC and against the macromolecular substrate casein-resorufin. The pH optimum was 6 to 7 for activities against all substrates. The error bars indicate standard errors.
The Giardia TVN corresponds to the ER and is the site of cysteine protease activity or directly contiguous to it.
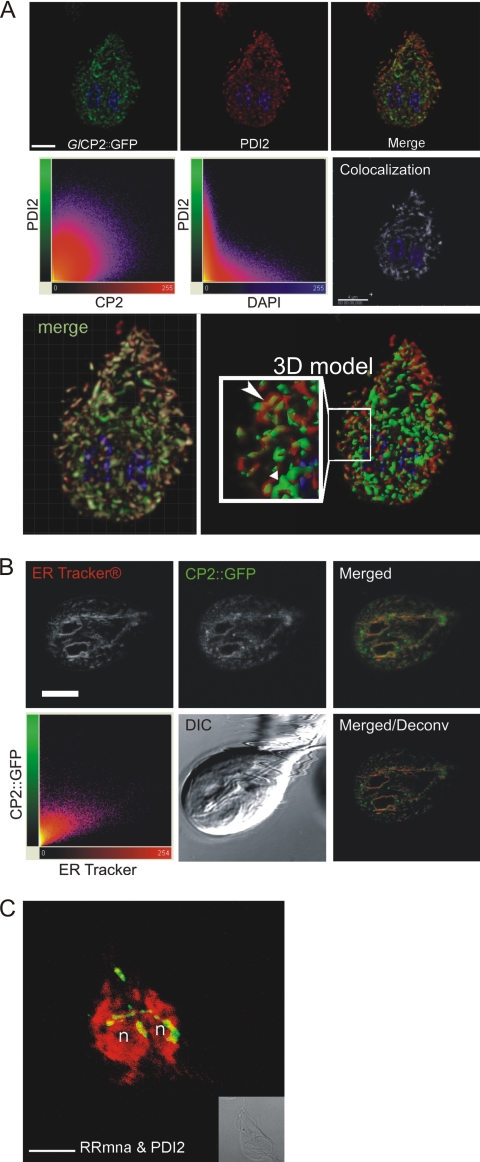
PDI2, which contains an ER retention signal and has been validated as the optimal ER marker (20, 40) (see Fig. S5 in the supplemental material), was used to visualize the TVN (Fig. 5). PDI2 colocalized with a cathepsin B-like protease in fixed cells, as shown by the GlCP2::GFP reporter (Fig. 5A, top row). Quantification of colocalization showed that the majority of GlCP2::GFP colocalized with PDI2 (Fig. 5A, bottom row). To confirm the ER localization of GlCP2, a second ER marker, ER tracker, was used in living cells, and a significant level of colocalization was detected (Fig. 5B). PDI2 also colocalized with, or was directly contiguous to, cysteine protease activity, as highlighted by cleavage of the MNA peptide substrate (Fig. 5C). Antibodies against other ER markers, Hsp70-BIP and KDEL, labeled the tubulovesicular ER network, with KDEL found predominantly in perinuclear regions (see Fig. S5 in the supplemental material).
FIG. 5.
GlCP2::GFP and GlCP protease activity colocalized with, or was immediately adjacent to, the ER marker PDI2 in the perinuclear region and the TVN. (A) (Top row) Deconvoluted single optical sections from confocal microscopy of trophozoites show that cathepsin B-like cysteine protease GlCP2::GFP (green) colocalized with PDI2 (red), extending throughout the TVN. The tubulovesicular nature of the TVN is apparent. Nuclear DNA was stained with DAPI. Scale bar, 3 μm. (Middle row) Software analysis enabled visualization of PDI2 and GlCP2::GFP colocalization. Scatter plots of the two labels show significant colocalization of CP2 and PDI2 in ER structures (left). Control: ER-localized PDI2 and DAPI-stained nuclear DNA show minimal colocalization (middle). (Right) Colocalized pixels in a separate channel colored white, with nuclear DNA in blue. Scale bar, 4 μm. (Bottom row, left) Volumetric reconstruction of confocal sections with a superimposed colocalization channel in white. (Right) Isosurface-rendered 3D model of PDI2 and GlCP2::GFP distribution. The inset shows membrane-bound PDI2 (red) and luminal CP2 (green) in the TVN (elongated arrowhead), in some instances occupying distinct subdomains (short arrowhead). GlCP2::GFP-labeled compartments lacking PDI2 staining may represent subcompartments of the ER. (B) Colocalization of ER Tracker and GlCP2::GFP in living adherent trophozoites. (Top row) Single optical sections showing the distribution of the two markers and a merged color image. (Bottom row, left) Scatter plot of the two labels. (Middle) DIC image; note the wave pattern of the ventral flagella as a hallmark of cell viability. (Right) Merged image after deconvolution (Deconv). Scale bar, 3 μm. (C) 3D projection of cysteine protease activity against the substrate Z-RR-MNA (green) colocalized with, or immediately adjacent to, PDI2 (red) in the perinuclear region and in portions of the TVN. The projection highlights the continuity of the TVN. Protease activity (green) is less prevalent than in Fig. 3C due to loss of fluorescent protease activity product during PDI2 immunolocalization washes. n, nucleus. Scale bar, 5 μm.
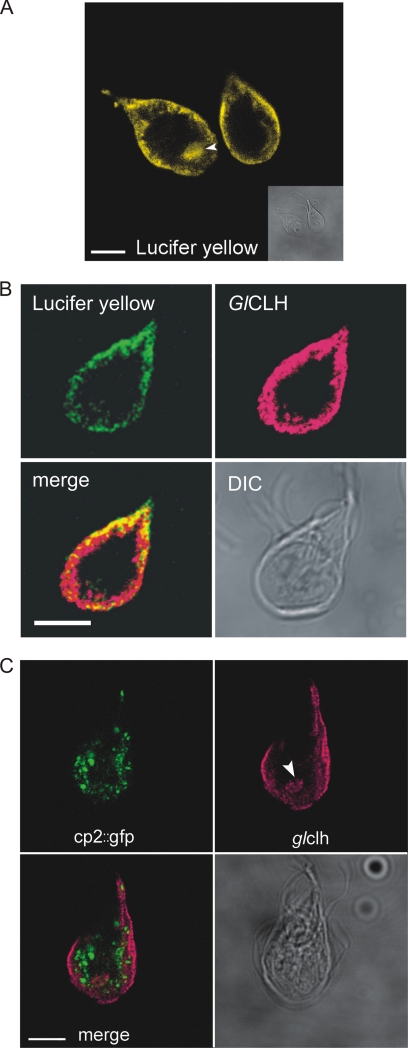
GFP-labeled cathepsins and cysteine protease activity, while predominantly in the TVN, did appear peripheral to ER markers in some optical cuts, and we therefore examined whether the cathepsins localized to the PVs. The acidified PVs were previously thought to be lysosome-like compartments where endocytosed proteins are degraded (22). The PVs were identified by accumulation of Lucifer yellow (Fig. 6A) (22) and contained abundant clathrin, which colocalized with the Lucifer yellow (Fig. 6B). A concentration of PVs was observed near the site of flagellar exit in the region of the ventral groove corresponding to the site of initial Fluosphere uptake observed in Fig. 2B (Fig. 6A and C). However, the PVs were clearly distinct from the labyrinthine TVN and the cysteine protease probe GlCP2::GFP (Fig. 6C).
FIG. 6.
Cathepsin-like cysteine proteases are not concentrated in the acidified clathrin-rich PVs of Giardia trophozoites. (A) Lucifer yellow staining (yellow) highlights the acidic nature of the PV network aside from the cell periphery. PVs are also abundant in an area of the cell near the ventral flagellar exit site (arrowhead). Scale bar, 5 μm. (B) Clathrin (red) and Lucifer yellow (green) overlap at the PV network. Colocalized pixels appear yellow on the merged image. Scale bar, 5 μm. (C) Cathepsin B-like cysteine protease GlCP2::GFP (green) marks the endocytic TVN of Giardia. This network is distinct from the clathrin-associated PVs (red). Occasional colocalization of GlCP2::GFP with clathrin was seen at the interface between the two staining patterns but was the exception. Clathrin is also abundant near the exit site of the ventral flagella (arrowhead). Scale bar, 5 μm.
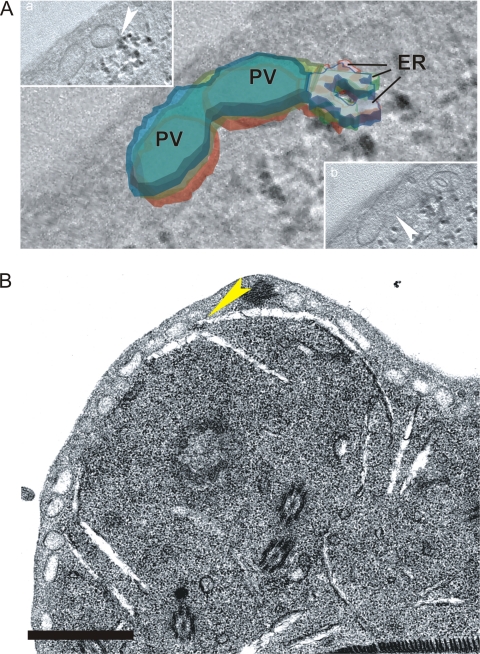
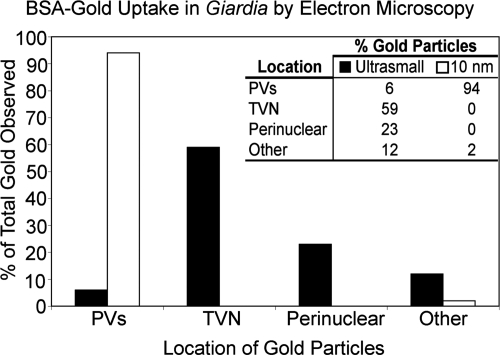
Ultrastructural analysis confirmed that the TVN has properties of the ER and identified foci of ER fusion with PVs. Sequential confocal images of the TVN along the z axis and ultrastructural analyses were both consistent with a complex, branching tubular network, appearing at times to be vesicular depending on the orientation of the optical cut (Fig. 1; see Fig. 8B; see Fig. S6 in the supplemental material). This morphology was consistent with previous reports of the Giardia ER and was confirmed by colocalization of ER markers. The ER network of Giardia appears slit-like in ultrastructural planes, as previously reported, and is highlighted by the G6P reaction (22, 23) (Fig. 1; see Fig. S6 in the supplemental material). While the PVs are differentially labeled with acid phosphatase, the G6P and acid phosphatase activities overlap, especially at focal points of ER-PV fusion (22, 23). Ultrasmall (0.8-nm) albumin-conjugated gold particles were endocytosed by Giardia and distributed throughout the TVN, including the perinuclear region (Fig. 7), consistent with the results of confocal microscopy. Larger (10-nm) gold particles entered the PVs but progressed no further (Fig. 7). While at first surprising, this result may reflect both the increased charge and relative mass of the larger gold particle. 3D reconstruction of G6P distribution provided a detailed view of the tubular network of the ER and confirmed focal fusion between the ER and PVs (Fig. 8A) (22). Figure 8B also provides evidence for a slit-like TVN extending to and contacting the PVs at the periphery of the cell (compare Fig. 1).
FIG. 8.
Identification of sites of fusion between the TVN and PVs. (A) In a stack of three consecutive EM tomography images, fusion between a large PV and the branching ER-like TVN (identified by the G6P reaction product [black]) was observed (inset a, arrowhead, inferior z-stack slice). Fusion between two adjacent PVs is also seen (inset b, arrowhead, superior z-stack slice). Each subsequent section is represented by a different color (red, green, and blue). The TVN is outlined with color, while PVs are filled. (B) Fusions between tubular extensions (slit-like spaces) of the TVN and PVs at the periphery of the cell were seen on EM sections (yellow arrowhead). Scale bar, 1 μm.
FIG. 7.
Ultrastructural analysis revealed that gold particles were endocytosed into the TVN. By EM, endocytosed albumin-coated ultrasmall (0.8-nm) gold particles localized mainly to the tubular TVN and the perinuclear region. This correlated with the endocytic pattern seen in the TVN and perinuclear region by confocal analysis in Fig. 1 to 3. Endocytosis of larger (10-nm) albumin-coated gold particles was arrested in the PVs. At least three electron microscopy images were analyzed for each gold particle size.
DISCUSSION
Electron tomography clearly defined two compartments within Giardia. The most extensive intracellular compartment is the TVN, which functions like the ER (Fig. 1). The PVs are acidified compartments originally thought to be lysosomes. If Giardia mirrored the organization of a typical eukaryotic cell, then protein endocytosis would be excluded from the ER, with proteins entering an endosome-like compartment and then delivered to lysosomes (PVs) to be degraded by lysosomal cathepsins. A very different scenario is apparent in Giardia. From video microscopy of protein endocytosis (Fig. 2) and from studies using colocalization of compartmental markers, cathepsin orthologues, and endocytosed proteins, proteins rapidly enter a TVN, even reaching the perinuclear cisternae (Fig. 2, 3, and 5). Orthologues of lysosomal cathepsin proteases are not concentrated in the acidified PVs, but rather in the TVN or in a contiguous structure that mirrors the TVN. There, they function to degrade proteins at a pH optimum higher than that of mammalian lysosomes (Fig. 4).
The ER was first identified and characterized in mammalian cells by K. R. Porter (33). Accordingly, our current concepts about the function and compartmentalization of the ER derive primarily from studies with mammalian cells and Saccharomyces cerevisiae. The ER is a labyrinthine tubular endomembrane system that coevolved with the nucleus and serves as a conduit for newly translated proteins, a subset of which are sorted by the Golgi apparatus to other membrane-bounded organelles, such as lysosomes and secretory granules (2, 10, 39). Based on the observations presented in this study, we propose that the Giardia ER is an endomembranous TVN in which the disparate functions of the ER, endosomes, and lysosomes of modern eukaryotes either have not fully diverged or have been substantially simplified (6). An alternative explanation is that a separate endosome compartment lies directly adjacent to the ER or is directly connected to it.
Regardless of the G. lamblia phylogenetic status as either an early-branching eukaryote (15, 38) or a highly simplified cell (6), its simple cellular organization can provide us with fundamental insights into the evolution and function of cell compartmentalization. The ER of Giardia was first identified through localization of BiP, an HSP70 homolog that functions as a protein-folding chaperone (39), while PDI2 has been validated as the most useful ER marker in Giardia (20, 40). This helped to distinguish the ER from the PVs of Giardia, which were thought to be secretory granules, endosomes, or lysosomes (1). In this study, the PVs were marked intensely by Lucifer yellow, which also colocalized with anti-clathrin antibodies (Fig. 6A and B). The presence of clathrin on PVs may at first seem surprising given their relatively large size (150 to 200 nm). However, a clathrin heavy-chain isoform in mammalian myocytes was also found on large vesicular compartments (44). Furthermore, recent analysis of ceramide uptake highlighted a similar clathrin-mediated pathway from peripheral vesicles to the perinuclear space (18).
By video microscopy, endocytosed proteins and Fluospheres primarily appear first in a “hot spot” corresponding to a region near the ventral flagellum exit site (4). This region appears to be abundant in clathrin-containing vacuoles (Fig. 5C and 6A). In other protozoan parasites, particularly the kinetoplastids, secretion and endocytosis are polarized, occurring only in a region known as the flagellar pocket (28). It is possible that Giardia most efficiently (though not exclusively) takes up extracellular material in a region where the action of flagellar movement may optimize sampling of the environment. The observation by electron microscopy that 10-nm gold particles only enter PVs (Fig. 7) suggests that the clathrin-rich PV network may be the initial site of uptake, followed by rapid dynamic fusion with the TVN, leading to the transfer of endocytosed material. This model is consistent with both previous ultrastructural observations that showed fusion of PVs with the plasma membrane (22) and the tomographic electron micrographs that show focal fusion of TVN with PVs in vegetative trophozoites (Fig. 1 and 8). Both studies suggest that direct delivery of endocytosed material from PVs to the TVN could occur. The recently documented pathway of ceramide uptake also supports this model of Giardia endocytosis (18). While we cannot rule out the possibility of a clathrin-coated vesicle population trafficking endocytosed material between the PVs and the TVN, we saw no direct evidence of this.
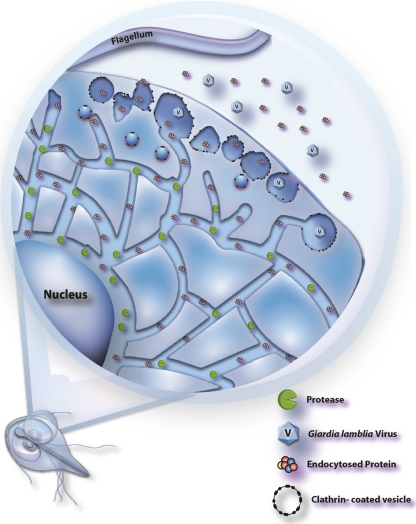
It has been proposed that the ER is an extremely dynamic organelle (10, 32); its size and shape can undergo drastic changes to meet the needs of ER-related functions. The focal fusions noted between the TVN and PVs in vegetative trophozoites and the appearance of the PVs as either spherical organelles or a fused network might both be reflections of dynamic changes occurring within the endomembranous system of Giardia (summarized in Fig. 9).
FIG. 9.
Model of the endocytic network of Giardia trophozoites. Active proteases reside primarily in the TVN, where endocytosed proteins are degraded. PVs contain clathrin and are the site of initial uptake. Membrane fusions between PVs, as well as between PVs and the TVN, are dynamic. Endocytosed proteins are passed from PVs to the TVN by these dynamic fusions and possibly by a clathrin-coated-vesicle population trafficking between the two compartments, as evidenced by Hernandez et al. (18). Giardia virus subverts this network for entry into the cell (41).
Furthermore, rather than serving as lysosomes at the end of a typical mammalian-cell endocytic pathway, the acidified clathrin-rich PVs may either denature recently endocytosed proteins, facilitating degradation by proteases in the “downstream” TVN, or neutralize the alkaline secretions of the pancreas present in the mammalian duodenum, where Giardia trophozoites reside and replicate. A previous report that cathepsin B-like cysteine protease activity resides in PVs (46) was made before the availability of ER and PV markers. Based on current observations, the vesicles observed by Ward et al. are likely the TVN in cross section, appearing larger due to diffusion of the fluorescent product (46).
Circumstantial support for this proposed pathway of protein uptake by Giardia comes from a study of the uptake of G. lamblia virus (GLV), a double-stranded RNA virus that is first translocated to the PVs after binding to the plasma membrane. GLV entry is arrested in the PVs of sodium azide-treated cells. Like some mammalian-cell viruses, GLV appears to exploit the endocytic pathway of Giardia (41). More direct support for the proposed endocytic pathway in Giardia comes from a recent publication in which the acquisition of ceramide is initiated in peripheral clathrin-coated vesicles that in turn deliver ceramide to the ER and the same perinuclear location seen for protein uptake in Fig. 1A and 2A (18).
Proteins that first enter Giardia's endocytic system through the PVs concentrate in the TVN and colocalize with GlCP::GFP fusion proteins. Although GFP reporter constructs have not been routinely used in anaerobic organisms, our studies with the pGFPpac expression vector yielded consistent results. We attribute this to the fact that Giardia is microaerophilic and cells were pulsed with sufficient oxygen levels prior to GFP visualization. Cathepsin proteolytic activity, confined to lysosomal organelles in other eukaryotes, is distributed throughout the TVN of Giardia, extending from the peripheral TVN to the perinuclear region (Fig. 2B and 4D). In keeping with the localization of Giardia cathepsins in a pH-neutral compartment, the pH optimum of the Giardia cathepsin enzymes is shifted toward neutral pH (6 to 7). The orthologous proteases of mammalian lysosomes have a strictly acidic pH optimum of 5 to 6 (45). While we saw some cathepsin activity of the GFP reporter construct peripheral to the TVN, it did not appear to be in the PVs. More likely it was either the peripheral TVN (ER) devoid of ER markers or occurred in the vesicles, like those proposed to carry ceramide from the PVs to the ER (18).
The proposal that the Giardia ER functions in endocytosis and the degradation of proteins has bearing on the recent debate concerning the role of the ER in mammalian macrophages. Gagnon et al. (11) proposed that the ER plays a pluripotent role, including direct involvement in the formation of the phagosome at the early stages of macrophage phagocytosis. Specifically, they proposed a direct fusion of the ER to the plasma membrane to initiate this process. In support of this model, they demonstrated that ER proteins are enriched in phagosomes. In contrast, however, Touret et al. (42) could not verify a physical continuity between the ER and the plasma membrane using a combination of biochemical fluorescence imaging and electron microscopy. Nevertheless, the latter authors pointed out the appeal of ER-mediated phagocytosis as an explanation of how phagocytes are able to internalize multiple large particles without large decreases in surface area. Furthermore, such a model provides a conceptual framework for understanding how antigens might be presented by MHC class I molecules when peptide loading takes place in the ER lumen. In light of this debate, two observations made in this study are noteworthy. First, the TVN of the protist G. lamblia serves as a site of protein synthesis, as well as endocytosis and degradation of material from the extracellular milieu. Second, a direct fusion of the ER in Giardia with the plasma membrane was not seen; rather, the clathrin-rich PV network appears to be the initial site of endocytosis (Fig. 8). Gagnon et al. (11) in fact suggested that the ER fuses with the plasma membrane at phagocytic cups in an area of the plasma membrane known to contain endocytic membranes, and perhaps even clathrin. Therefore, the observation that Giardia endocytoses proteins into clathrin-coated PVs that then fuse with the ER may be more analogous to the proposed model of phagocytosis than first appears.
The proposal of a multifunctional Giardia ER raises the question of how a single compartment could function as a site of protein catabolism, as well as a pathway for the import and sorting of newly translated protein. There is evidence of some polarized compartmentalization within the Giardia ER in that ribosomes are attached to the ER predominantly in the perinuclear region, where most KDEL (ER retention signal) localization is also seen (see Fig. S5 in the supplemental material) (27). The chaperones PDI2 and the Hsp70-BiP homolog, which both contain ER retrieval signals, are dispersed and colocalize throughout the TVN, including near the periphery of the cell (see Fig. S5 in the supplemental material) (40). Nevertheless, cathepsin GFP reporter constructs and functional protease activity are both present in the perinuclear region, as well as in more peripheral regions of the TVN (Fig. 1 and 2). In fine confocal sections, it appears that while GlCP2::GFP and PDI2 are dispersed throughout the TVN, single-marker compartments exist for both proteins (Fig. 4A and B). The nature of these subcompartments, including how and if they are stably maintained, remains unclear. However, this may be a mechanism by which trophozoites achieve the separation of newly synthesized proteases and degradative protease activity.
Another possible mechanism to protect newly translated proteins from the action of proteases proposed to be functioning in the ER is sequestration by chaperones (12, 30, 31). Like other eukaryotic cells, Giardia contains a number of well-characterized chaperones, including PDI2 and Hsp70-BiP (http://www.giardiadb.org). Newly translated peptide chains are rapidly enveloped and protected by chaperones as they thread into the ER lumen (10). This may allow functional separation of protein degradation and folding/transport of newly translated proteins in the same membrane-bound compartment in Giardia. Separation of anabolic and catabolic ER functions might also be aided by the fact that newly folded globular proteins should be less efficiently cleaved than endocytosed, denatured proteins that have passed sequentially through the host's acidic upper digestive tract and acidified Giardia PVs.
The “functional” compartmentalization of the TVN seen in Giardia may have parallels in modern eukaryotes, for example, in the delivery of exogenous oligopeptides to the ER of MHC class I-presenting cells (7). Day et al. found that after a brief incubation of antigen-presenting cells with fluorescein-labeled peptides in the medium, the peptides were detected within the ER bound to MHC class I molecules (7). Such ER internalization of exogenous peptides is blocked by pinocytosis inhibitors and occurs without traversing the Golgi apparatus or the cytosol. Most importantly, in cells lacking MHC class I, peptides are delivered to the ER but then are quickly removed. The authors speculated that this was due to the action of resident ER proteases. There are other examples of direct delivery of extracellular material to the ER in eukaryotic cells, including endocytosis of AB5 toxins (36) and SV40 variants (19, 25).
We conclude that the Giardia ER is a pluripotent compartment which serves as a site of protein synthesis, as well as of endocytosis and degradation of material from the extracellular milieu. We acknowledge an alternative explanation that a more classical endosome compartment lies adjacent to the ER or is in direct contact with it. In either case, such characteristics of Giardia's biology provide us with a unique model to understand how basic cell mechanisms could have operated before the complex endomembranous system of modern eukaryotes evolved.
Supplementary Material
Acknowledgments
We thank Ted Nash for his kind gift of the G. lamblia transfection vector. We thank Srini Garlapati and Lei Li for reporter constructs and many helpful comments. We thank Christopher Franklin, UCSF, for his generous assistance with graphics and overall technical support. We are very grateful to Masako Terada, from the National Center for Microscopy and Image Research at San Diego, supported by United States Public Health Service Grant NIHRR0450, and to Mark H. Ellisman for the use of the facilities to produce the G6P tomogram.
This work was supported by Tropical Disease Research Unit NIH grant AI 35707 and the Sandler Foundation. K.N.D. was supported by an NIAID training fellowship in Microbial Pathogenesis. A.H. is supported by grant 3100A0-100270 of the Swiss National Science Foundation.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adam, R. D. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts, B. 2002. Molecular biology of the cell. Garland Science, New York, NY.
- 3.Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300:1703-1706. [DOI] [PubMed] [Google Scholar]
- 4.Benchimol, M. 2004. Giardia lamblia: behavior of the nuclear envelope. Parasitol. Res. 94:254-264. [DOI] [PubMed] [Google Scholar]
- 5.Benchimol, M., B. Piva, L. Campanati, and W. de Souza. 2004. Visualization of the funis of Giardia lamblia by high-resolution field emission scanning electron microscopy—new insights. J. Struct. Biol. 147:102-115. [DOI] [PubMed] [Google Scholar]
- 6.Dacks, J. B., and M. C. Field. 2007. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci. 120:2977-2985. [DOI] [PubMed] [Google Scholar]
- 7.Day, P. M., J. W. Yewdell, A. Porgador, R. N. Germain, and J. R. Bennink. 1997. Direct delivery of exogenous MHC class I molecule-binding oligopeptides to the endoplasmic reticulum of viable cells. Proc. Natl. Acad. Sci. USA 94:8064-8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Meester, F., E. Shaw, H. Scholze, T. Stolarsky, and D. Mirelman. 1990. Specific labeling of cysteine proteinases in pathogenic and nonpathogenic Entamoeba histolytica. Infect. Immun. 58:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DuBois, K. N., M. Abodeely, J. Sakanari, C. S. Craik, M. Lee, J. H. McKerrow, and M. Sajid. 2008. Identification of the major cysteine protease of Giardia and its role in encystation. J. Biol. Chem. 283:18024-18031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Federovitch, C. M., D. Ron, and R. Y. Hampton. 2005. The dynamic ER: experimental approaches and current questions. Curr. Opin. Cell Biol. 17:409-414. [DOI] [PubMed] [Google Scholar]
- 11.Gagnon, E., S. Duclos, C. Rondeau, E. Chevet, P. H. Cameron, O. Steele-Mortimer, J. Paiement, J. J. Bergeron, and M. Desjardins. 2002. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110:119-131. [DOI] [PubMed] [Google Scholar]
- 12.Gething, M. J., and J. Sambrook. 1992. Protein folding in the cell. Nature 355:33-45. [DOI] [PubMed] [Google Scholar]
- 13.Gillin, F. D., S. E. Boucher, S. S. Rossi, and D. S. Reiner. 1989. Giardia lamblia: the roles of bile, lactic acid, and pH in the completion of the life cycle in vitro. Exp. Parasitol. 69:164-174. [DOI] [PubMed] [Google Scholar]
- 14.Gillin, F. D., D. S. Reiner, M. J. Gault, H. Douglas, S. Das, A. Wunderlich, and J. F. Sauch. 1987. Encystation and expression of cyst antigens by Giardia lamblia in vitro. Science 235:1040-1043. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, R. S., K. Aitken, M. Falah, and B. Singh. 1994. Cloning of Giardia lamblia heat shock protein HSP70 homologs: implications regarding origin of eukaryotic cells and of endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 91:2895-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto, T., Y. Nakamura, F. Nakamura, T. Shirakura, J. Adachi, N. Goto, K. Okamoto, and M. Hasegawa. 1994. Protein phylogeny gives a robust estimation for early divergences of eukaryotes: phylogenetic place of a mitochondria-lacking protozoan, Giardia lamblia. Mol. Biol. Evol. 11:65-71. [DOI] [PubMed] [Google Scholar]
- 17.Hedges, S. B. 2002. The origin and evolution of model organisms. Nat. Rev. Genet. 3:838-849. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez, Y., C. Castillo, S. Roychowdhury, A. Hehl, S. B. Aley, and S. Das. 2007. Clathrin-dependent pathways and the cytoskeleton network are involved in ceramide endocytosis by a parasitic protozoan, Giardia lamblia. Int. J. Parasitol. 37:21-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kartenbeck, J., H. Stukenbrok, and A. Helenius. 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 109:2721-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knodler, L. A., R. Noiva, K. Mehta, J. M. McCaffery, S. B. Aley, S. G. Svard, T. G. Nystul, D. S. Reiner, J. D. Silberman, and F. D. Gillin. 1999. Novel protein-disulfide isomerases from the early-diverging protist Giardia lamblia. J. Biol. Chem. 274:29805-29811. [DOI] [PubMed] [Google Scholar]
- 21.Kulda, J., and E. Nohynkova. 1996. Giardia in humans and animals, p. 225-422. In J. Kreier (ed.), Parasitic protozoa, 2nd ed. Academic Press, San Diego, CA.
- 22.Lanfredi-Rangel, A., M. Attias, T. M. de Carvalho, W. M. Kattenbach, and W. De Souza. 1998. The peripheral vesicles of trophozoites of the primitive protozoan Giardia lamblia may correspond to early and late endosomes and to lysosomes. J. Struct. Biol. 123:225-235. [DOI] [PubMed] [Google Scholar]
- 23.Lanfredi-Rangel, A., M. Attias, D. S. Reiner, F. D. Gillin, and W. De Souza. 2003. Fine structure of the biogenesis of Giardia lamblia encystation secretory vesicles. J. Struct. Biol. 143:153-163. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, P. R., and D. P. Knight. 1996. Staining methods for section matrial. Elsevier Biomedical Press, Amsterdam, The Netherlands.
- 25.Lord, J. M., and L. M. Roberts. 1998. Toxin entry: retrograde transport through the secretory pathway. J. Cell Biol. 140:733-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marti, M., A. Regos, Y. Li, E. M. Schraner, P. Wild, N. Muller, L. G. Knopf, and A. B. Hehl. 2003. An ancestral secretory apparatus in the protozoan parasite Giardia intestinalis. J. Biol. Chem. 278:24837-24848. [DOI] [PubMed] [Google Scholar]
- 27.McCaffery, J. M., and F. D. Gillin. 1994. Giardia lamblia: ultrastructural basis of protein transport during growth and encystation. Exp. Parasitol. 79:220-235. [DOI] [PubMed] [Google Scholar]
- 28.Morgan, G. W., B. S. Hall, P. W. Denny, M. Carrington, and M. C. Field. 2002. The kinetoplastida endocytic apparatus. Part I. A dynamic system for nutrition and evasion of host defences. Trends Parasitol. 18:491-496. [DOI] [PubMed] [Google Scholar]
- 29.Mottram, J., M. J. North, and M. Sajid. 2004. Trichomonad and Giardia cysteine endopeptidases, p. 1170-1173. In N. D. Rawlings, A. J. Barrett, and J. F. Woessner (ed.), Handbook of proteolytic enzymes. Academic Press, London, United Kingdom.
- 30.Nishikawa, S., J. L. Brodsky, and K. Nakatsukasa. 2005. Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). J. Biochem. 137:551-555. [DOI] [PubMed] [Google Scholar]
- 31.Paulsson, K., and P. Wang. 2003. Chaperones and folding of MHC class I molecules in the endoplasmic reticulum. Biochim. Biophys. Acta 1641:1-12. [DOI] [PubMed] [Google Scholar]
- 32.Perret, E., A. Lakkaraju, S. Deborde, R. Schreiner, and E. Rodriguez-Boulan. 2005. Evolving endosomes: how many varieties and why? Curr. Opin. Cell Biol. 17:423-434. [DOI] [PubMed] [Google Scholar]
- 33.Porter, K. R. 1953. Observations on a submicroscopic basophilic component of cytoplasm. J. Exp. Med. 97:727-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiner, D. S., H. Douglas, and F. D. Gillin. 1989. Identification and localization of cyst-specific antigens of Giardia lamblia. Infect. Immun. 57:963-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roger, A. J., H. G. Morrison, and M. L. Sogin. 1999. Primary structure and phylogenetic relationships of a malate dehydrogenase gene from Giardia lamblia. J. Mol. Evol. 48:750-755. [DOI] [PubMed] [Google Scholar]
- 36.Sandvig, K., and B. van Deurs. 2002. Transport of protein toxins into cells: pathways used by ricin, cholera toxin and Shiga toxin. FEBS Lett. 529:49-53. [DOI] [PubMed] [Google Scholar]
- 37.Singer, S. M., J. Yee, and T. E. Nash. 1998. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol. Biochem. Parasitol. 92:59-69. [DOI] [PubMed] [Google Scholar]
- 38.Sogin, M. L., J. H. Gunderson, H. J. Elwood, R. A. Alonso, and D. A. Peattie. 1989. Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science 243:75-77. [DOI] [PubMed] [Google Scholar]
- 39.Soltys, B. J., M. Falah, and R. S. Gupta. 1996. Identification of endoplasmic reticulum in the primitive eukaryote Giardia lamblia using cryoelectron microscopy and antibody to Bip. J. Cell Sci. 109:1909-1917. [DOI] [PubMed] [Google Scholar]
- 40.Stefanic, S., D. Palm, S. G. Svard, and A. B. Hehl. 2006. Organelle proteomics reveals cargo maturation mechanisms associated with Golgi-like encystation vesicles in the early-diverged protozoan Giardia lamblia. J. Biol. Chem. 281:7595-7604. [DOI] [PubMed] [Google Scholar]
- 41.Tai, J. H., S. J. Ong, S. C. Chang, and H. M. Su. 1993. Giardiavirus enters Giardia lamblia WB trophozoite via endocytosis. Exp. Parasitol. 76:165-174. [DOI] [PubMed] [Google Scholar]
- 42.Touret, N., P. Paroutis, M. Terebiznik, R. E. Harrison, S. Trombetta, M. Pypaert, A. Chow, A. Jiang, J. Shaw, C. Yip, H. P. Moore, N. van der Wel, D. Houben, P. J. Peters, C. de Chastellier, I. Mellman, and S. Grinstein. 2005. Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell 123:157-170. [DOI] [PubMed] [Google Scholar]
- 43.Touz, M. C., L. Kulakova, and T. E. Nash. 2004. Adaptor protein complex 1 mediates the transport of lysosomal proteins from a Golgi-like organelle to peripheral vacuoles in the primitive eukaryote Giardia lamblia. Mol. Biol. Cell 15:3053-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Towler, M. C., P. A. Gleeson, S. Hoshino, P. Rahkila, V. Manalo, N. Ohkoshi, C. Ordahl, R. G. Parton, and F. M. Brodsky. 2004. Clathrin isoform CHC22, a component of neuromuscular and myotendinous junctions, binds sorting nexin 5 and has increased expression during myogenesis and muscle regeneration. Mol. Biol. Cell 15:3181-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turk, V., B. Turk, and D. Turk. 2001. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 20:4629-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, W., L. Alvarado, N. D. Rawlings, J. C. Engel, C. Franklin, and J. H. McKerrow. 1997. A primitive enzyme for a primitive cell: the protease required for excystation of Giardia. Cell 89:437-444. [DOI] [PubMed] [Google Scholar]
- 47.Zheng, Q. S., M. B. Braunfeld, J. W. Sedat, and D. A. Agard. 2004. An improved strategy for automated electron microscopic tomography. J. Struct. Biol. 147:91-101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.