Abstract
Using a Tn7 transposon library of Candida albicans, we have identified a mutant that exhibited sensitivity in drop plate assays to oxidants such as menadione and hydrogen peroxide. To verify the role of the mutated gene in stress adaptation, null mutants were constructed and phenotypically characterized. Because of its apparent functions in growth and oxidant adaptation, we have named the gene GOA1. Goa1p appears to be unique to the CTG subclade of the Saccharomycotina, including C. albicans. Mutants of C. albicans lacking goa1 (strain GOA31) were more sensitive to 6 mM H2O2 and 0.125 mM menadione than the wild type (wt) or a gene-reconstituted (GOA32) strain. The sensitivity to oxidants correlated with reduced survival of the GOA31 mutant in human neutrophils and avirulence compared to control strains. Other phenotypes of GOA31 include reduced growth and filamentation in 10% serum, Spider, and SLAD agar media and an inability to form chlamydospores. Since Goa1p has an N-terminal mitochondrion localization site, we also show that green fluorescent protein-tagged Goa1p shows a mitochondrionlike distribution during oxidant or osmotic stress. Further, the inability of GOA31 to grow in medium containing lactate, ethanol, or glycerol as the sole carbon source indicates that the mitochondria are defective in the mutant. To determine how Goa1p contributes to mitochondrial function, we compared the wt, GOA32, and GOA31 strains for mitochondrial electrical membrane potential, respiration, and oxidative phosphorylation. We now show that GOA31, but not the wt or GOA32, had decreased respiration and mitochondrial membrane potential such that mutant cells are unable to drive oxidative phosphorylation. This is the first report in C. albicans of a respiratory defect caused by a loss of mitochondrial membrane potential.
Antioxidant proteins are part of the fungal cellular response to reactive oxidant species (ROS) in Candida albicans, both in vivo when the organism encounters phagocytic cells or competitors on the mucosal surfaces of humans or in vitro when confronted with external or internal metabolically generated ROS. Interestingly, increased ROS production in C. albicans, and probably fungi in general, occurs following treatment with antifungal compounds, such as diallyl disulfide (the active ingredient in garlic) and with miconazole, observations that may explain in part the activity of these compounds (35, 39).
To counter the stress induced by ROS, a number of antioxidant proteins are induced in C. albicans, including catalases (43, 58), superoxide dismutases (30, 40), and enzymes that require the cofactor glutathione (13). Biosynthesis of trehalose also provides protection against oxidant stress (4, 27, 41). Of the mitogen-activated protein kinase (MAPK) signal pathways that regulate new gene transcription during stress, the MKC1 MAPK and HOG1 MAPK (for hyperosmotic glycerol) pathways are prominent in a number of yeasts and filamentous fungi in regard to adaptation to hyperosmotic and/or oxidant stress (2, 3, 5, 6, 11, 12, 18-21, 29, 44, 50, 52, 55). Transcription factors such as Cap1p are also critical to oxidant resistance (7, 20, 21, 60). The Cek2 MAPK is not apparently required for stress adaptation (14).
Resistance to stress in C. albicans is complex but, in general, genes can be assigned to two categories based upon transcriptional profiling: (i) genes that make up a general response (or core) profile to common stress conditions such as osmotic, oxidative, and heavy metal and (ii) genes that are specific for each stress condition (20, 21, 52). In C. albicans a core transcriptional response to all stress conditions included genes that encode redox regulation, mitochondrial activity, carbohydrate metabolism, and cell wall biogenesis, among others (20). Interestingly, the core stress response set of genes in C. albicans is much smaller than that of Saccharomyces cerevisiae or Schizosaccharomyces pombe, suggesting that C. albicans relies more upon diversity in gene transcription to counter stress conditions (20, 52). A common theme is that most stress conditions result in an increase in ROS during cellular respiration. Thus, resistance to osmostress, for example, requires restoration of redox potential, reduction of ROS, and increased mitochondrial function.
Adaptation to stress in C. albicans is also most likely critical to interactions of the organism during encounters with host defenses. For example, innate immunity including phagocytic killing by human polymorphonuclear cells (PMNs) and activated macrophages is critical to protection against blood-borne Candida species. Unactivated macrophages provide some protection although less so than neutrophils. This important observation has been demonstrated both clinically (neutropenic patients are susceptible to blood-borne candidiasis) and experimentally (25, 26, 47). In the latter instance, a sevenfold-greater killing of C. albicans by PMNs was reported compared to blood monocytes; additionally, germination of the organism occurred in monocytes. Similar data have been previously reported by another group (38). After phagocytosis, in both PMNs and monocytes, the expression of many fungal genes associated with oxidative stress responses was especially prominent (25). However, it would appear that resistance to oxidant stress, and probably other anti-Candida factors, was achieved only in infected monocytes. Thus, in neutropenic patients, the paradigm is that mononuclear phagocytes are less able to kill the organism that, in turn, favors spread of the organism in tissues.
We have identified a gene (GOA1) from C. albicans whose deletion results in oxidative sensitivity. One of the features of this protein is that it becomes localized to the mitochondria during oxidant stress. In the absence of GOA1, we show that mitochondrial functions are compromised as revealed by a decrease in respiration and membrane potential. We hypothesize that Goa1p provides functions that relate to its role in mitochondria energetics especially during stress.
MATERIALS AND METHODS
Strains, strain maintenance, and plasmids.
All strains used in the present study are listed in Table 1 and were maintained as frozen stocks and propagated on yeast extract-peptone-dextrose (YPD) agar when needed (1% yeast extract, 2% peptone, 2% glucose, 2% agar). Plasmids pGEM-URA3, pRS-ARG4, and pGEM-HIS1 were provided by Aaron Mitchell (56, 57). To create pGEM-HIS1-GOA1, GOA1 was amplified by using primers listed in Table 2, which contained flanking BamHI restriction sequences. The PCR product was purified and digested with BamHI restriction enzymes and ligated into pGEM-HIS1, which had been digested using BamHI.
TABLE 1.
Strains of C. albicans used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| DAY286 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG,ARG4,URA3 | 56 |
| SC5314 | Clinical isolate; Ura+ parent of CAI4 | 23 |
| CAI4 | ura3::imm434/ura3::imm434 | 23 |
| DAY185 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/HIS1::his1::hisG arg4::hisG/URA3::ARG4::arg4::hisG | 16 |
| BWP17 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 56 |
| SN148 | arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δura3Δ::imm434/ura3::imm434 iro1Δ::imm434/iro1Δ::imm434 | 46 |
| GOA21 | goa1::UAU1/goa1::URA3 ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | This study |
| GOA31 | goa1::URA3/goa1::ARG4 arg4/arg4leu2/leu2 his1/his1ura3Δ::imm434/ura3::imm434 iro1::imm434 | This study |
| GOA41 | goa1::URA3/goa1::ARG4 ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | This study |
| GOA22 | goa1::UAU1/goa1::URA3::GOA1-HIS1 | This study |
| GOA32 | goa1::URA3/goa1::ARG4::GOA1-HIS1 | This study |
| GOA42 | goa1::URA3/goa1::ARG4::GOA1-HIS1 | This study |
| GOA-GFP | GOA1/GOA1-GFP-URA3 ura3::imm434/ura3::imm434 | This study |
TABLE 2.
Primer sets used in this study
| Primer | Sequence | Application |
|---|---|---|
| AnuKOF | CAGCCATTAAGAGATTGCCAATAAAATAACTAAGAATCACAAACCCTGGTGCTAAATTGCGTTTTCCCAGTCACGACGTT | URA3/ARG4 disruption cassette: 5′ region of GOA1 |
| AnuKOR | CCAAGTAAAACACCCCTTGAATTATCAGTATGCCAACATGTTTCATTGTTTGTGTTAGTAATTGTGGAATTGTGAGCGGATA | URA3/ARG4 disruption cassette: 3′ region of GOA1 |
| ABA-REV-BamHIF | GGATCCGGCATTCAATGTCTGCGTGC | Upstream region of GOA1 with BamHI restriction sequence to create GOA32 and GOA42 |
| ABA-REV-BamHIR | GGATCCCTCCCATGACGAAATGACG | Downstream region of GOA1 with BamHI restriction sequence to create GOA32 and GOA42 |
| 19.3818F | ACGTGCACCCACGGTATTACATCA | GOA1 specific |
| 19.3818R | ACTTCTGATCTCCAATTCGGGCAA | GOA1 specific |
| AnuDIAGF | GGGTCAGGCAGGGGTTACCTA | Confirm intergration at the GOA1 locus |
| AnuDIAGR | CACGGTGTTTTGGGTACCTCTC | Confirm intergration at the GOA1 locus |
| SouthernF | ACTTCTGAATTTGATGGATCTCGGA | Generate probe for Southern blot |
| SouthernR | AGCAATCCAACAGGTGGACCTAGA | Generate probe for Southern blot |
| GOA-GFPF | CACAAAGCTTGAAAGTAAAGCATTAATGCAAAGTGACATATACAAATCTCTCAACAAAGAAAATCAGAATGGTGGTGGTTCTAAAGGTGAAGAATTATT | GFP cassette, 3′ region of GOA1 excluding stop codon |
| GOA-GFPR | TATCAGTATGCCAACATGTTTCATTGTTTGTGTTAGTAATTTTTGAGCTTGCATTCCTAGTCGGAATACGTCTAGAAGGACCACCTTTGATTG | GFP cassette, downstream of GOA1 |
Construction of GOA1 deletion mutants.
GOA21 was obtained from a transposon mutagenized library (16). In addition to GOA21, we constructed two other GOA1 deletion mutants, GOA31 and GOA41. To do this, cassettes for transformation were designed by amplifying ARG4 and URA3 from plasmids pRS-ARG4 and pGEM1-URA3 using the primers described in Table 2. Primers containing the GOA1 flanking sequences were also used for PCR amplification, and URA3 or ARG4 was inserted within the GOA1 flanking sequences (46). Cassettes were then transformed into SN148 to generate GOA31 or into BWP17 to generate GOA41 (see Fig. 1A [only GOA31 is shown]).
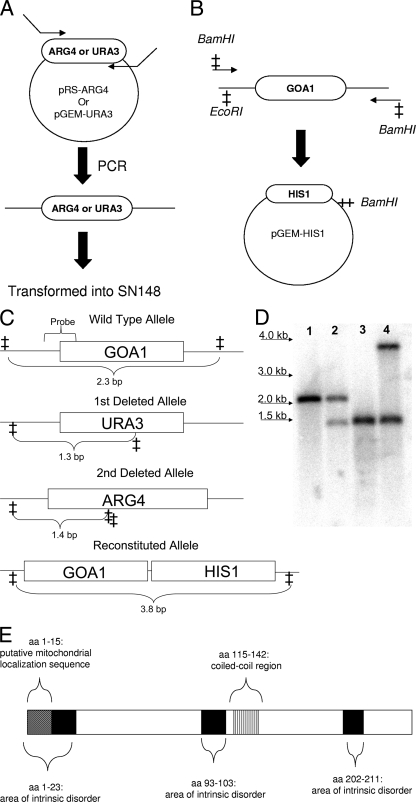
FIG. 1.
Construction of the goa1Δ deletion mutant (GOA31) and its gene-reconstituted derivative GOA32. (A) Cassettes for transformation were constructed by amplifying ARG4 and URA3 from plasmids pRS-ARG4 and pGEM-URA3 using primers homologous to sequences upstream and downstream of GOA1 (Table 2). These cassettes were then transformed into SN148 to create GOA31 and into BWP17 to generate GOA41 (data not shown). (B) GOA1 was amplified using primers containing BamHI restriction sites. pGEM-HIS1 and the amplified GOA1 fragment were digested with BamHI and the GOA1 fragment inserted into pGEM-HIS1. The plasmid was digested with EcoRI and transformed into GOA31 to create GOA32. (C) Restriction maps of GOA1 and derived constructs. (D) Southern hybridization. Strains were digested with NdeI (sites are indicated with a cross). Lanes 1 to 4: 1, SC5314; 2, heterozygote; 3, GOA31; 4, GOA32. The probe for the blot is shown in Fig. 1C. (E) The Goa1p of C. albicans. Goa1p is 279 aa in length. The cross-hatched box (aa 1 to 15) indicates a putative mitochondrial localization signal. The black boxes (aa 1 to 23, 93 to 103, and 202 to 211) indicate areas of intrinsic disorder. The vertical-lined box (aa 115 to 142) indicates a coiled-coil region.
Construction of the gene reconstituted strains GOA22, GOA32, and GOA42.
GOA22 was constructed as follows. GOA1 was amplified by using primers listed in Table 2. The PCR product was purified and digested with NdeI and ligated into pGEM-HIS1, which had been digested by using NdeI. The pGEM-HIS1-GOA1 plasmid was linearized by using SalI and subsequently transformed into GOA21 (Fig. 1B). The integration and orientation of GOA1 were confirmed by PCR using primers listed in Table 2. GOA32 and GOA42 were constructed as follows. GOA1 was amplified using primers listed in Table 2, which contained flanking BamHI restriction sequences. The PCR product was purified, digested with BamHI, and ligated into pGEM-HIS1, which had been digested by using BamHI. The pGEM-HIS1-GOA1 plasmid was linearized using EcoRI and subsequently transformed to yield strains GOA32 and GOA42.
Southern hybridization.
We utilized standard methods for doing Southern blot hybridization (8, 23). Purified C. albicans genomic DNA was isolated and digested with NdeI and separated by gel electrophoresis and transferred to a nylon membrane. The reaction proceeded overnight in a solution containing 0.4 M NaOH and 1 M NaCl. The membrane was then placed in a neutralization buffer (0.5 M Tris, 1 M NaCl [pH 7.2]), UV cross-linked and placed in a 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) solution for hybridization. A 600-bp probe was generated by PCR with primers (Table 2) containing sequences upstream of and within GOA1. The probe (Fig. 1C) was purified and labeled with [32P]dCTP. All blots were hybridized at 68°C in a solution containing 0.5 M NaH2PO4 (pH 7.2), 7% sodium dodecyl sulfate, and 1 mM EDTA. After hybridization, the membranes were washed with decreasing concentrations of SSC and sodium dodecyl sulfate. All membranes were imaged by using a Storm phosphorimager (Amersham Biosciences) (Fig. 1D).
Screening of the transposon mutant library for oxidant sensitivity.
Approximately 400 transposon insertion mutants were evaluated for oxidant sensitivity by plating each strain on YPD agar medium containing 0, 4, 6, or 8 mM H2O2 using a pin replicator. Plates were incubated at 30°C for 48 h and growth was evaluated. Strains that failed to grow or exhibited reduced growth from at least three experiments were screened further by plating known numbers of cells onto YPD medium containing either hydrogen peroxide or menadione as described below.
Determination of generation time and growth measurements.
To determine generation times of all strains, yeast cells from overnight cultures grown at 30°C were diluted to a starting optical density at 600 nm (OD600) of 0.10 in 50 ml of YPD broth containing leucine, histidine, arginine, and uridine. Cultures were incubated at 30°C, the OD600 was recorded hourly for 12 h or 20 h (see Results for details), and the generation times were calculated by using the method of Walia and Calderone (55).
Drop plate sensitivity assays.
According to the method of Chauhan et al. (11), the sensitivity of all strains to oxidant and osmotic stress was tested by plating serial dilutions of 5 × 101 to 5 × 105 cells (each in a total of 5 μl) onto YPD agar plates containing 6 mM H2O2 or 0.125 mM menadione. Yeast cells were obtained from overnight cultures grown in YPD broth at 30°C, washed with saline, and standardized by hemacytometer counts. The growth of each strain was evaluated for sensitivities after 48 h of incubation at 30°C.
Strains were also evaluated for their growth on YNB (yeast nitrogen base) agar supplemented with histidine (20 mg/ml) and leucine (30 mg/ml) containing 2% glucose, 4% glycerol, 4% lactate, or 6% ethanol. Drop plates were incubated at 30°C for 5 days and then photographed.
PMN killing and phagocytosis assays.
Neutrophil killing assays were performed as previously described (18, 49). Briefly, PMNs were isolated from the peripheral blood of healthy human volunteers by dextran sedimentation and centrifuged through the lymphocyte separation medium Polymorphprep (Axis-Shield). PMNs were enriched in number by a brief hypotonic lysis of erythrocytes and then suspended in RPMI 1640 medium containing 10% fetal bovine serum (FBS). Cells were judged as >99% viable by trypan blue dye exclusion. Strains were grown in YPD broth overnight as described above, washed with phosphate-buffered saline (PBS; pH 7.4), and then opsonized with 50% human serum for 30 min at 37°C. PMNs and opsonized yeasts (collected by centrifugation) were suspended in RPMI medium containing 10% FBS at a ratio of 10:1 (107 PMNs to 106 yeasts). Cultures were incubated at 37°C for 3 h, centrifuged, and suspended in water to lyse the neutrophils. Serial dilutions were performed of the yeast cells, and 100 μl was plated onto YPD agar. Plates were incubated for 48 h at 30°C, at which time CFU were counted. The percent killing was calculated by using the following formula for each strain: [(CFU without PMNs) − (CFU with PMNs/CFU without PMNs)] × 100.
To determine the percent phagocytosis of all strains, we used the method of Klippel and Bilitewski (34). Human neutrophils were prepared as described above. For fluorescence labeling, 107 yeasts were harvested by centrifugation (13,000 rpm, 5 min, 4°C), washed twice with PBS, and stained with 1 ml of carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Leiden, The Netherlands; 500 μM in PBS-0.1% dimethyl sulfoxide) for 1.5 h at 37°C. Stained cells are green in color. Yeasts were washed three times in PBS to remove the residual dye. For phagocytosis assays, in brief, 6 × 105 PMNs/ml/well were incubated with CFSE-labeled Candida (1:2) for 30 min at 30°C in a final volume of 400 μl. Samples were kept on ice until immediately before analysis. To quench the fluorescence of yeasts that were not internalized, trypan blue (0.2%, final concentration) was added. The images were analyzed and captured with a Zeiss Axiovert 200M microscope, coupled with a digital camera (magnification, ×400). Thus, intracellular (phagocytized) cells appear green, whereas nonphagocytized yeasts are red. We determined the percentage of PMN that contained phagocytized yeasts of each strain, as well as the number of yeast cells per PMN per strain, from 20 fields of mixed suspensions.
Morphogenesis assays.
For these experiments, cells from an overnight culture grown in YPD were washed, and 250 cells of each strain were plated on YPD, 10% serum, Spider, and SLAD agar media and incubated at 37°C for 5 days according to standard procedures (8, 15). Representative colonies were then photographed.
Chlamydospore formation was evaluated by spotting 10 μl of an overnight yeast culture (105 cells) of each strain onto cornmeal agar, placing a coverslip over the cells, and incubating the cultures for 5 days in the dark at room temperature. Subsequently, chlamydospore formation was evaluated for each strain by light microscopy. Cultures were photographed by using an Olympus DP12 microscope equipped with a digital camera.
Murine model of hematogenously disseminated candidiasis.
We followed the method of Calera et al. (8) to evaluate the virulence of a set of wild-type (wt), null, and gene-reconstituted strains. All strains were grown overnight in YPD at 30°C. Cells were washed in PBS (pH 7.2), counted by using a hemacytometer, and suspended in PBS. Three groups of 13 female BALB/c mice (Harlan) were injected intravenously via the lateral tail vein with 106 yeast cells in 50 μl of SC5314, GOA31, and GOA32 samples. At 24, 48, and 72 h postinfection, one mouse per group was euthanized. One kidney from each animal was removed, fixed in 10% formalin, and prepared for histological examination using the periodic acid-Schiff stain. The other kidney was removed, weighed, and homogenized in PBS. The homogenates were diluted and plated on YPD agar containing 50 μg per ml of streptomycin to inhibit bacterial growth. The plates were incubated at 30°C and, after 48 h, the number of colonies was counted to determine fungal load per gram of kidney. All remaining mice were observed for signs of morbidity twice per day. If moribund, animals were euthanized by cervical dislocation.
Construction of GOA1-GFP.
Using primers (Table 2) containing sequences from the 3′ end of GOA1 (excluding the stop codon) and downstream of GOA1, as well as containing sequences of the green fluorescent protein (GFP)-URA3 cassette from plasmid pGFP-URA3, a GOA-GFP cassette was amplified by PCR. This cassette was transformed into CAI4. The resultant strain was verified by PCR (data not shown).
Mitochondrial colocalization.
Strain GOA-GFP was grown at 30°C overnight and then diluted to a starting OD600 of 0.10 in 50 ml of YPD broth. Cells were incubated at 30°C and upon reaching log-phase were stained with 250 nM MitoTracker Red (Molecular Probes) for 45 min. Cells were washed and stressed with 1 M NaCl or 5 mM H2O2 for 15 min, or grown in glycerol-YP for 15 min, and then imaged by using an Olympus Fluoview-FV300 laser scanning confocal system.
Spheroplast formation.
Cultured GOA31, GOA32, or SC5314 cells in exponential phase were harvested by centrifugation from 350-ml of cultures from YPD medium, and the cells were washed with cold water, followed by a wash using buffer A (1 M sorbitol, 10 mM MgCl2, 50 mM Tris-HCl [pH 7.5]). Cells were suspended in buffer A (3 ml/g of cells) containing 30 mM dithiothreitol. After 15 min of incubation at 30°C with shaking, the cells were harvested by centrifugation, suspended in buffer A containing lyticase (1 mg/g of cells) and 1 mM dithiothreitol, and incubated at 30°C until ca. 90% of the cells converted to spheroplasts (60 to 80 min). The digestion was stopped by the addition of an equal volume of ice-cold buffer A, and spheroplasts were washed twice with the same buffer. Protein concentration of the final suspension was determined using the biuret assay (28) in the presence of 0.2% deoxycholate.
Mitochondrial membrane potential.
The mitochondrial membrane potential (ΔΨm) of permeabilized cells (spheroplasts) was monitored by measuring the fluorescence spectrum of Safranine O on a RF5301PC Shimadzu spectrofluorophotometer (Kyoto, Tokyo, Japan) operating at excitation and emission wavelengths of 495 and 586 nm, respectively, and with slit widths of 5 nm (24). All experiments were performed at 28°C with 1 mg of spheroplasts in 2 ml containing 5 μM Safranine O, 5 mM succinate, and 0.05% bovine serum albumin. Relative changes in membrane potential were expressed as arbitrary fluorescence units.
Oxygen uptake.
Oxygen uptake was measured polarographically at 28°C using a Clark-type electrode (Hansatech Instruments, Ltd., England) with 1 mg of spheroplasts in 1 ml of standard incubation medium containing 125 mM sucrose, 65 mM KCl, 10 mM HEPES (pH 7.2), 2.5 mM KH2PO4, and 1 mM MgCl2 with 1 mg of spheroplasts.
Statistical analysis.
The Student t test was used for all analyses. Differences were considered significant when P was <0.05.
RESULTS
Isolation of GOA21 from a UAU insertion mutant library.
A library of ∼400 transposon insertion mutants was used to screen for oxidant-sensitive strains on YPD agar plates containing 0, 4, 6, and 8 mM H2O2. From this primary screen, 34 strains exhibited a peroxide sensitive phenotype and were further evaluated for sensitivity to both peroxide and menadione in drop-plate assays. Of these strains, a single mutant designated strain GOA21, named for growth and oxidant adaptation, was chosen for further study based upon its sensitivity to both peroxide and menadione and its slower growth rate (see below). The DNA sequence corresponding to the transposon insertion site was provided by Aaron Mitchell. The open reading frame (19.3818) encodes a protein of 279 amino acids (aa) in length. SMART (Simple Modular Architecture Research Tool) analysis indicated protein domains that include a putative mitochondrial localization signal (aa 1 to 15), areas of intrinsic disorder (aa 1 to 23, 93 to 103, and 202 to 211), and a coiled-coil domain (aa 115 to 142) (Fig. 1E).
Orthologs of Goa1p were identified by BLAST comparison with fungal, plant, and animal genomes in the NCBI database and genomics provided by the Fungal Genome Initiative at the Broad Institute. The presence of orthologs was confined to species within the CTG subclade of the Saccharomycotina, so named because the CTG codon of most of the species within this subclade has been reassigned from leucine to serine (17, 22). An apparent ortholog was present in each of the available genome sequences of species within this subclade, even those that have lost the codon reassignment. There are no known homologues in Saccharomyces cerevisiae or other human pathogens except the Candida species in this subclade.
Construction of null strains GOA31 and GOA41.
To address the concern of secondary mutations due to the use of the TN7 transposon insertion cassette in the original library, we constructed new deletion mutants in the SN148 and BWP17 backgrounds (Fig. 1A). Cassettes for transformation were constructed by amplifying ARG4 and URA3 from plasmids pRS-ARG4 and pGEM-URA3 using primers containing sequences homologous to regions upstream and downstream of GOA1. The null mutant GOA31 was constructed by using these cassettes in the transformation of strain SN148 (46). The same cassettes were used to construct a second null mutant (GOA41) using BWP17 as the parental strain (not shown). Here, we describe only data from GOA31, although GOA41 exhibited similar phenotypes in assays described below.
To reconstitute GOA1 into its native locus, the gene was amplified using primers containing BamHI restriction sites and subcloned into the BamHI site of pGEM-HIS1 (Fig. 1B). pGEM-HIS1 and amplified GOA1 were digested with BamHI, ligated to create pGEM-HIS-GOA1, and evaluated by PCR to ensure that GOA1 was inserted. After digestion with EcoRI, the cassette was transformed into GOA31 to generate strains GOA32 and GOA42 from GOA41 (only GOA31 and 32 are shown). All strains were confirmed by Southern Blot (Fig. 1C and D). Subsequent results in the present study will focus on strains GOA31 and GOA32.
A strain lacking GOA1 has a longer generation time.
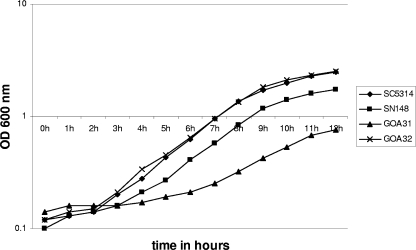
To determine the generation time for each strain, overnight grown cells in YPD (30°C) were diluted to a starting OD600 of 0.10, and growth was monitored hourly spectrophotometrically (OD600) for 12 h at 30°C (Fig. 2). The data represent averages from three independent experiments for the strains indicated and three replicates per time point. The wt strains SN148 and SC5314 doubled at 2.09 and 1.88 h, respectively (P = 0.009). In contrast, GOA31 (goa1/goa1) doubled significantly more slowly at 2.9 h (P = 0.0005 versus SN148). The GOA1 reconstituted strain (GOA32) had a doubling time equal to SC5134 of 1.86 h (P = not significant versus SC5314; P = 0.003 versus SN148; and P = 0.0002 versus GOA31). We also found that GOA31 had a longer lag phase than the parental or gene-reconstituted strains and did not reach the same density at stationary phase (Fig. 2), even if incubation was carried out for 20 h (not shown). Therefore, GOA1 is required for optimum growth and did not reach the same stationary phase by 12 h as wt cells, or even at 20 h (data not shown).
FIG. 2.
Deletion of GOA1 results in a longer generation time. The growth curves of strains SC5314, SN148, GOA31, and GOA32 are shown. Strains were grown overnight in YPD, transferred to fresh YPD at an OD600 of 0.1, and incubated at 30°C. The absorbance was measured every 1 h for 12 h, and the generation times were calculated. The growth curves represent the averages of three experiments and three replicates per time point per strain. Generation times: GOA31, 2.9 h; GOA32, 1.8 h; SC5314, 1.8 h; SN148, 2.09 h.
Goa1p protects cells against some oxidants.
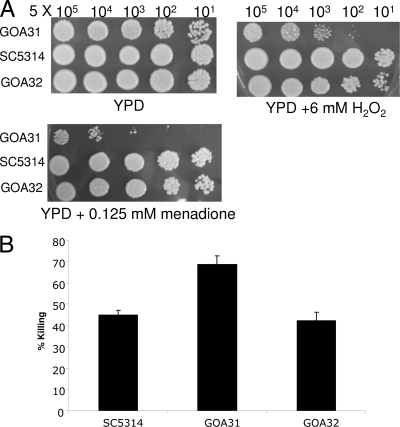
We tested the sensitivity of the strains to H2O2, tert-butyl hydrogen peroxide, menadione, and potassium superoxide. Cells were grown overnight, dilutions were plated on YPD agar containing each oxidant mentioned above, and the plates were incubated at 30°C for 48 h. Compared to the wt, GOA31 showed increased sensitivity to 6 mM H2O2 and to 0.125 mM menadione (Fig. 3A). Resistance to these oxidants was partially or completely restored in GOA32. There was no difference in growth among strains on YPD agar containing tert-butyl hydrogen peroxide and potassium superoxide (data not shown). Thus, GOA1 is required for protection to some but not all types of oxidants, a result we have observed with the C. albicans ssk2Δ null mutant (54). We also evaluated the killing of each strain (107 cells/strain) in the presence of 10 mM H2O2 by incubating strains with H2O2 for 6 h (Fig. 3B). The concentration chosen for this assay was greater than the drop plate assays since we also used a greater number of yeast cells (107 per strain). We found that GOA31 was killed to a significantly greater extent than wt or GOA32 by H2O2 (P = 0.03, GOA31 versus GOA32).
FIG. 3.
The GOA31 strain is sensitive to H2O2 and menadione and is killed significantly more by H2O2. (A) Drop plate assays of strains GOA31, GOA32, and wt (SC5314) cells in the presence of H2O2 or menadione. Strains were grown overnight and standardized as described in Materials and Methods. Pictured is the growth of strains on YPD or YPD containing H2O2 (6 mM) or menadione (0.125 mM) at yeast cell concentrations of 5 × 105, 104, 103, 102, and 50 cells. Plates were incubated at 30°C for 48 h. (B) Killing of strains was determined in vitro by incubating 107 yeasts with 10 mM H2O2. Samples were removed after 6 h of incubation at 30°C, and dilutions were plated on YPD agar.
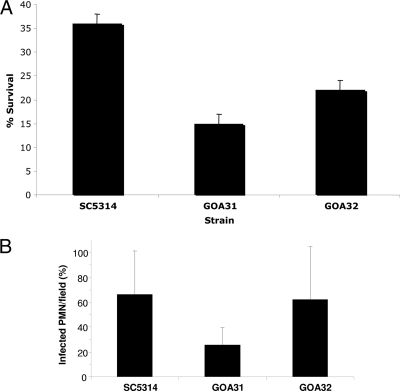
The GOA-null strains are more sensitive to killing by neutrophils.
Since GOA31 is sensitive to oxidant stress imposed by incubating cells in medium containing menadione or H2O2, we hypothesized that this strain would be more sensitive to killing by PMNs. To measure killing of all strains, PMNs were isolated and mixed with yeasts previously opsonized with 50% human serum at a 10:1 (PMN-to-yeast) ratio in RPMI medium containing 10% FBS for 2 h at 37°C. PMNs were then lysed with water, the yeast cells were plated on YPD agar, and cultures were incubated for 48 h at 30°C. Colonies were counted from three independent experiments, and the averages are shown in Fig. 4A. The survival of GOA31 cells was significantly less than that of wt cells (P = 0.018) and lower for GOA32 than GOA31 (P = 0.04), while there was a significant difference observed between wt and GOA32 strains (P = 0.05). Killing of the original transposon mutant GOA21 versus its parental strain (DAY286) by PMNs was significantly greater also, similar to the results with GOA31 (data not shown). Thus, these experiments indicate a correlation between the sensitivity of the goa1Δ null mutant to hydrogen peroxide and menadione and survival in human PMNs.
FIG. 4.
GOA31 is killed more by human PMNs than wt or GOA32 but is phagocytized less. (A) GOA31 is more susceptible to killing by PMNs. Strains were grown overnight at 30°C, standardized, opsonized with human serum, mixed with human PMNs at a ratio of 10:1 (PMNs to yeast), and incubated at 37°C for 3 h, after which PMNs were lysed, and the yeast cell suspensions were plated on YPD plates (30°C for 48 h). The colonies were counted, and the percent survival was calculated. GOA31 versus SC5314, P = 0.018. (B) Percent phagocytosis by human PMNs of wt, GOA31, and GOA32 strains. Phagocytosis was determined by mixing CFSE-treated, green fluorescing yeast cells of each strain with PMNs. Subsequently, after phagocytosis for 30 min, extracellular, green fluorescent yeasts were quenched with trypan blue, whereas internalized yeasts remained green. The percentage of PMN cells containing green fluorescing yeasts was greater with wt and GOA32 cells than with GOA31 cells.
Reduced survival of the GOA31 null mutant than the wt or the reconstituted strains might be associated with a higher amount of phagocytosis by neutrophils. To address this issue, we determined the percentage of neutrophils that phagocytized each strain by observing 20 microscope fields of PMN-yeast mixtures. We found that the percentage of phagocytosis was unexpectedly lower for GOA31 than the wt or reconstituted strains (ca. 67% for SC5314, 26% for GOA31, and 63% GOA32 (P = 0.0001, wt versus GOA31; P = 0.01, GOA31 versus GOA32, and wt = GOA32) (Fig. 4B). We also determined the number of yeast cells of each strain per PMN, which was, on average, wt = 3.4, GOA31 = 1.1, and GOA32 = 1.8 (data not shown). The differences between GOA31 and both the wt and GOA32 strains were significant (P = 0.0001, wt versus GOA31; P = 0.005, GOA31 versus GOA32; wt versus GOA32, P = not significant). Thus, these data indicate that the increased killing (reduced survival) of GOA31 is not associated with greater phagocytosis or a greater number of GOA31 cells per PMN. In fact, we found less phagocytosis of GOA31, suggesting that killing occurs by a different mechanism.
GOA1 is required for morphogenesis and chlamydospore formation.
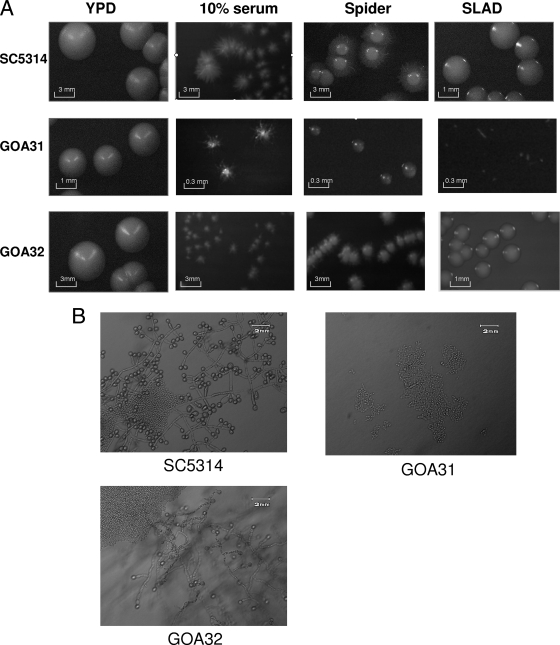
The role of C. albicans genes in nutrient stress can be evaluated in medium lacking specific carbon sources or low nitrogen (15). Most often, wt cells under these conditions convert from yeast to hyphal growth. To determine whether GOA1 is required for the yeast-to-hyphal transition, we evaluated the morphogenesis and growth of all strains on YPD (nutrient rich), 10% serum, spider, and SLAD agar at 37°C for a minimum of 5 days (Fig. 5A). In a comparison of the strains, there were 3- to 10-fold differences in the colony diameters. On YPD agar, GOA31 colonies were smaller than those of wt and GOA32 strains, a finding consistent with the slower growth rate and longer lag phase (Fig. 2). On 10% serum agar, GOA31 colonies were smaller that those of the wt and GOA32, and all strains formed filaments, although GOA31 hyphae were considerably shorter in length than those of the wt but the same as GOA32 (Fig. 5A). On Spider and especially SLAD agar media, GOA31 colonies appeared minute and lacked filamentation. Each of these phenotypes was partially restored in GOA32, indicating a gene dosage-related phenotype. Thus, GOA1 is required for morphogenesis under the conditions tested.
FIG. 5.
GOA31 is compromised in growth on hypha-inducing media and lacks chlamydospore formation compared to other strains. (A) GOA31 colonies are smaller than those of wt and GOA32 and fail to filament or filament less extensively on 10% serum, Spider, or SLAD medium than other strains. Strains SC5314, GOA31, and GOA32 (indicated to the left of the figure) were grown overnight in YPD at 30°C and counted, and an equal number were incubated at 37°C on YPD, 10% serum, Spider, and SLAD agars for 48 h. Representative colonies are indicated. Note that the differences in the colony sizes of the strains on filamentation or YPD medium. A bar in each photograph indicates 0.3, 1, or 3 mm. (B) GOA31 fails to form chlamydospores compared to wt and GOA32 strains, which is intermediate in spore formation compared to wt and GOA31 strains. Cells of all strains were plated on cornmeal-Tween agar and incubated in the dark at room temperature for 5 days. After 5 days, chlamydospore formation was viewed by using light microscopy.
Chlamydospore formation by all strains was also evaluated. To do this, 10-μl portions of overnight grown YPD cultures were standardized to cell number, spotted onto cornmeal Tween agar plates, and then covered with a glass coverslip. The plates were then allowed to incubate at room temperature for up to 5 days in the dark. GOA31 was unable to form chlamydospores compared to wt cells, while chlamydospore production in GOA32 was intermediate to that of wt and GOA31, indicating that GOA1 is required for chlamydospore formation (Fig. 5B).
GOA1 is required for virulence in a hematogenously disseminated murine model of candidiasis.
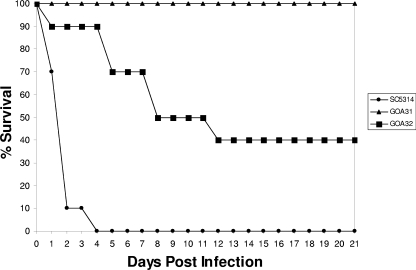
To investigate the role of Goa1p in virulence, wt, GOA31 and GOA32 strains were inoculated intravenously into immunocompetent mice. The survival of mice over a 21-day period of time was determined for each strain, as shown in Fig. 6, and the colonization of their kidneys determined from mice infected for 24 to 72 h. We found that mice infected with wt cells rapidly succumbed to the infection during 4 days, whereas mice infected with the null strain GOA31 survived for at least 3 weeks. Mice infected with the gene-reconstituted strain (GOA32) showed attenuation of virulence, such that 40% survived for 21 days. Tissue loads of the organism, expressed as CFU/g of kidney, were relatively constant for all strains over the course of 72 h but much lower in strain GOA31 (Table 3). Hyphae were visible in kidneys of infected animals, although much more difficult to find in mice infected with GOA31 (data not shown). These data clearly demonstrate that Goa1p is required for virulence in this murine model.
FIG. 6.
GOA31 is avirulent in a murine model of hematogenously disseminated candidiasis. Mice were infected with 106 cells of each strain (wt, GOA31, and GOA32) in a murine model of hematogenously disseminated candidiasis. The survival of mice at 0 to 21 days postinfection is shown for each strain.
TABLE 3.
CFU counts in mice infected with strains of C. albicansa
| C. albicans strain | Log10 CFU/g of tissue (mean ± SD) in kidneys at: |
||
|---|---|---|---|
| 24 h | 48 h | 72 h | |
| SC5314 | 6.0 ± 0.3 | 6.6 ± 0.2 | 6.6 ± 0.2 |
| GOA31 | 3.7 ± 0.3 | 3.2 ± 0.1 | 3.0 ± 0.2 |
| GOA32 | 5.8 ± 0.4 | 5.5 ± 0.3 | 5.2 ± 0.2 |
At 24, 48, and 72 h postinfection, mice were sacrificed, and the CFU of each strain were determined by plating homogenates of kidneys on YPD agar. Cultures were incubated at 30°C, and colonies were counted after a 48-h incubation.
Goa1p is translocated from the cytoplasm to mitochondria during oxidant and osmotic stress.
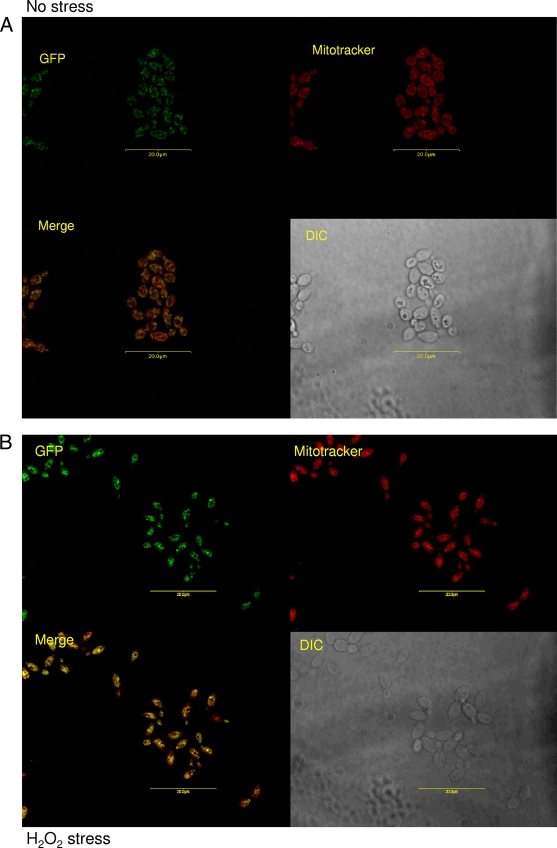
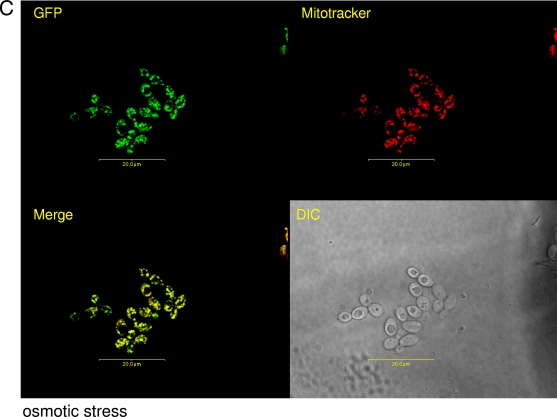
Since Goa1p has a putative mitochondrial localization signal, we tagged Goa1p with GFP and performed fluorescence microscopy. Cells were grown to early log phase in YPD and then stained with MitoTracker (Molecular Probes) in order to visualize mitochondria. Cells were either treated or not with 5 mM H2O2 or 1.0 M NaCl or had glycerol substituted for glucose. We observed that, in unstressed cells, Goa1p was located in a punctuate pattern in the cytoplasm; the fluorescent signal was not uniformly coincident with the MitoTracker signal, indicating that Goa1p is not generally found in the mitochondria (Fig. 7A). However, upon either peroxide or osmotic stress, Goa1p partially relocalizes to the mitochondria as evidenced by merged photographs that indicate superimposition of GFP and MitoTracker (Fig. 7B, peroxide, and Fig. 7C, osmotic; see also Fig. S1 in the supplemental material). Goa1p also shows a mitochondrionlike distribution when cells were incubated in glycerol as a carbon source (data not shown). These data indicate that during peroxide or osmotic stress, the function of Goa1p is probably related to its ability to translocate to mitochondria.
FIG. 7.
Translocation of Goa1p to mitochondria is dependent upon peroxide or osmotic stress and a nonfermentative (glycerol) carbon source. A Goa1p-GFP fusion was incubated at 30°C, grown to early log phase, stained with MitoTracker Red (Molecular Probes/Invitrogen) for 45 min, washed, and subjected to treatment regimes for 20 min in YPD. Cells were immediately viewed by using fluorescence confocal microscopy. (A) Unstressed; (B) 5 mM hydrogen peroxide; (C) 1.0 M NaCl.
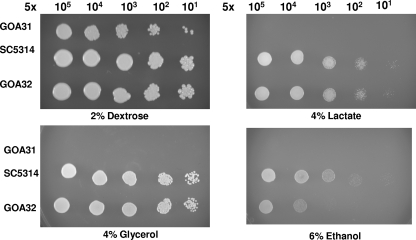
To determine whether Goa1p is critical for mitochondria function, we evaluated growth of all strains on YNB medium supplemented with 2% glucose or nonfermentable carbon sources such as glycerol, lactate, and ethanol. We observed that while wt and GOA32 grew on all carbon sources, GOA31 was unable to utilize glycerol, lactate, or ethanol (Fig. 8). We surmise that mitochondria function in the mutant was compromised as the nonfermentable carbon sources were not utilized via mitochondrial oxidation.
FIG. 8.
The GOA31 mutant is unable to utilize nonfermentable carbon sources. Drop plate assays of wt, GOA32, and GOA31 strains on YNB agar medium supplemented with histidine, leucine, and either 2% glucose, 4% glycerol, 4% lactate, or 6% ethanol. GOA31 is able to grow only on YNB plus 2% glucose.
Mitochondria of GOA31 have defective membrane potential and reduced respiration.
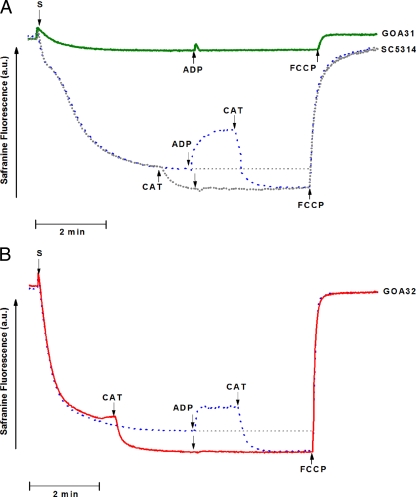
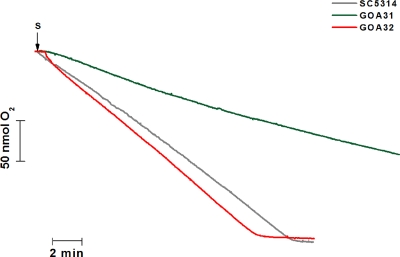
Safranine O is a dye whose fluorescence spectrum shifts as it binds to and stacks upon increasingly polarized inner mitochondrial membranes (1). The spectral shift is related to a developed mitochondrial membrane potential (ΔΨm) up to at least 170 mV (1) and can be monitored by using a spectrofluorophotometer operating at excitation and emission wavelengths of 495 and 586 nm, respectively (9). Figure 9A shows that spheroplasts suspended in the standard buffer containing 5 mM succinate as a substrate and 5 μM safranine was followed by a large decrease in fluorescence, which reached a steady state after a period of about 2 min. The blue line (Fig. 9A) shows that the addition of ADP to the experiment with the SC5314 spheroplasts, after the fluorescence had stabilized, was followed by an increase in fluorescence compatible with the utilization of the electrochemical proton potential to drive ADP phosphorylation by the FoF1-ATP synthase, as previously observed with Candida parapsilosis (9) and C. albicans (32). As expected, this fluorescence increase was reversed by carboxyatractyloside (CAT), an inhibitor of the adenine nucleotide translocase (Fig. 9A, black line). After CAT addition, the ΔΨm returned to values slightly higher than the initial value, suggesting that, even in the absence of exogenous ADP, the mitochondria were using the membrane potential to phosphorylate endogenous ADP. The addition of FCCP, a protonophore, promoted a fast downward deflection of the trace compatible with depolarization of the inner mitochondrial membrane and return of safranine to the water phase. In contrast to the SC5314 spheroplasts, the GOA31 spheroplasts developed a very small ΔΨm that was not modified by the addition of ADP, strongly indicating that this strain is not able to oxidatively phosphorylate ADP (Fig. 9A, green line). Figure 9B shows that GOA32 spheroplasts develop a ΔΨm similar to SC5314 spheroplasts, allowing ADP phosphorylation consistent with the development of a membrane potential. Compatible with the ΔΨm experiments of both SC5314 and GOA32 strains, respiration supported by endogenous substrates was similar and much faster than the GOA31 respiration (Fig. 10). The respiratory rates for strains SC5314, GOA32, and GOA31 were 13.62, 15.06, and 5.56 nM O2/min/mg of protein, respectively.
FIG. 9.
Electrical membrane potential (ΔΨm) in different strains of C. albicans. GOA31 is unable to drive the phosphorylation of ADP. Spheroplasts (S, 1 mg/ml) were added to a reaction medium containing 125 mM sucrose, 65 mM KCl, 10 mM HEPES (pH 7.2), 2.5 mM KH2PO4,1 mM MgCl2, 5 μM Safranine O, and 5 mM succinate in a total volume of 2 ml. Additions—200 μM ADP, 0.5 μM CAT, and 1 μM FCCP—are indicated by the arrows. (A) The dark green line represents GOA31 spheroplasts; the blue and black lines represent experiments performed with SC5314 spheroplasts in the absence (blue) or presence (black) of CAT (dashed line), respectively. (B) Experiments performed with GOA32 spheroplasts. The results shown are representative of four independent experiments performed in duplicate. a.u., arbitrary units.
FIG. 10.
The GOA31 strain shows a significantly reduced rate of respiration compared to wt and GOA32 strains, as measured by oxygen consumption. The test system (final volume, 1 ml, 28°C) contained 125 mM sucrose, 65 mM KCl, 10 mM HEPES (pH 7.2), 2.5 mM KH2PO4, and 1 mM MgCl2. The addition of C. albicans spheroplasts (1 mg/ml) is indicated by an arrow. The results shown are a representative trace from four independent experiments performed in duplicate. Respiratory rates for each strain are listed in the text.
DISCUSSION
From a transposon mutant library of C. albicans, a gene was identified that provided adaptation to 6 mM H2O2 (16). Subsequently, null mutants were constructed in two strain backgrounds, and both exhibited sensitivity to peroxide and menadione. Since mutants lacking this gene also had growth defects, the gene was named GOA1 to indicate its functions in both growth and oxidant adaptation. A thorough search of the literature and genome databases indicated its absence in S. cerevisiae as well as non-Candida human pathogenic fungi but a similarity to proteins only from the CTG subclade of the Saccharomycotina that includes, in addition to C. albicans, five other human pathogenic Candida species (17, 22). SMART analysis of the protein revealed an N-terminal mitochondrial localization domain as well as a coiled-coil and intrinsic disorder domains, the latter two of which indicate that it may functionally be important in its interactions with other proteins. Interestingly, Goa1p is required for optimal growth in the presence of H2O2 and menadione in vitro as well as optimum survival in human PMNs. Presumably, the latter function is associated with the ability to partially withstand the ROS of these phagocytes. Our data on killing of strains by PMNs was not related to a difference in the percent phagocytosis since wt cells were phagocytized more than GOA31. Our interpretation of this observation is that GOA31 is more susceptible to extracellular killing by PMN, which has been shown to be part of the PMN killing repertoire of C. albicans (25). Alternatively, while the percent PMN phagocytosis is lower in GOA31, the intracellular killing is still greater. Of importance, Goa1p is also required for virulence in a hematogenously disseminated murine model of candidiasis. The null strain GOA31 was totally avirulent in this model and, based upon tissue counts, the organism colonized (or persisted) in kidney tissue to a much lower extent than wt and GOA32.
One of the more striking phenotypes of strain GOA31 was its minimal growth on media that induce filamentation such as 10% serum, SLAD, and Spider agar media. This observation demonstrates the importance of respiratory metabolism in filamentation of C. albicans. Interestingly, there remains a controversy over the role of metabolic status associated with morphogenesis since both a need for and a lack of an intact respiratory pathway have been both suggested as important (10, 36, 37, 42, 54). Our data support the former conclusion.
Because of its mitochondrial localization signal, we integrated a GOA1-GFP cassette in wt cells and monitored translocation of the protein with or without stress to determine whether in fact the protein shuttled to the mitochondria during stress. In stressed cells or those grown in glycerol, the protein did partially relocalize to the mitochondria from the cytoplasm, whereas in unstressed cells, Goa1p was not associated with mitochondria. It is clear that adaptation to these stress conditions requires extensive energy production and mitochondria activity (see below). Cellular ROS accumulation is an outcome of increased cell metabolism and in addition, as cells sense increased external ROS, a stress response is induced to adapt cells. The relationship between stress adaptation and energy metabolism has been shown by others. During treatment with peroxide or high salinity, genes associated with energy production and mitochondrial functions are upregulated in S. cerevisiae (48, 59) and the halotolerant black yeast, Hortaea werneckii (53). In S. cerevisiae, respiration-associated transcripts (metabolism and energy functions) were especially strongly induced within 30 min of osmotic stress and persisted throughout exposure (59). The authors of that study showed that after treatment with high salinity, ca. 10% of the open reading frames were changed transcriptionally, and ca. 9% of the altered transcripts included open reading frames functionally important in providing energy via mitochondrial activities. In addition, a number of genes referred to as “cell defense” transcripts, including those for glycerol and trehalose production and cell detoxification (glutaredoxin and metallothionein) also increased significantly. Posas et al. (48) showed that a number of functional gene families are induced early during saline stress, including carbohydrate metabolism (as represented by increased glycerol, glycogen, and trehalose content), sugar transporters, protein biosynthesis, ion homeostasis, and signal transduction. Clearly, osmotic stress can cause the loss of mitochondrial functions and apoptotic programmed cell death, as evidenced by mitochondrial swelling, reduction of mitochondrial inner membranes, and increased production of ROS (51). Growth of H. werneckii in a hypersaline medium resulted in increased ATP synthesis and oxidative damage protection (53). In the same study, the authors identified the 14-3-3 protein-encoding gene HwBMH1 that localized to the mitochondria during hypersaline stress but not during growth of cells in physiological saline. In S. cerevisiae, the Bmh1/2 proteins are thought to be involved in the regulation of the mitochondrial proteome, especially in regard to carbohydrate metabolism (31). Transcriptional studies of stress responses have been carried out in C. albicans, although there were differences reported in regard to whether or not this organism has a core stress response (20, 21, 52). If a core stress response to adapt cells to oxidative, osmotic, or heavy metal (Cd2+) stress does exist in C. albicans, it apparently upregulates a smaller set of genes compared to S. cerevisiae or S. pombe. The core stress response includes genes involved in redox regulation, cell wall biogenesis, protein folding and degradation, and carbohydrate metabolism, as well as genes encoding mitochondrial activities. In summary, it appears that an increase in energy metabolism is part of a stress response in a variety of yeasts and that adaptation to most stress conditions includes regulation of redox conditions and ROS detoxification.
Little is known about the coordination of events that shuttle cytoplasmic proteins to the mitochondria during stress in C. albicans although the literature is abundant with studies in other eukaryotes (reviewed in reference 45). About 10 to 15% of nuclear genes encode mitochondrial proteins, and they are recognized by translocase receptors that are of two general groups of proteins: transmembrane outer proteins and transmembrane inner proteins. Goa1p, by nature of its intrinsic and coiled-coil domains, may have functions related to protein escorting that are critical to translocation.
There are no other reports of proteins in C. albicans that are translocated to the mitochondria that play a role in membrane potential and ATP synthesis. C. albicans Hmi1p, like that of S. cerevisiae, has a C-terminal mitochondria targeting signal. Deletion of HMI1 causes fragmentation of mtDNA, reduction of mtDNA mass, and loss of nucleoid distribution, but the deletion is not lethal, and the deleted strain is able to grow on nonfermentable carbon sources, unlike the goa1Δ null mutant (33). Recently, a hog1Δ null mutant was shown to be sensitive to inhibitors of the respiratory chain, with an enhanced respiratory basal rate and increased ROS production, although oxidant-detoxifying enzymes were elevated (2). However, mutant cells were able to still undergo oxidative phosphorylation even though the mitochondrial membrane potential was lower (2). In our studies, the functional activity of Goa1p is clearly related to its ability to mediate membrane integrity and, in the presence of peroxide, this event is lethal in the GOA31 strain. The studies described here focused upon the suggested protein escort function in which Goa1p may participate, as well as the identity of mitochondrial proteins lacking in GOA31.
Supplementary Material
Acknowledgments
We thank Aaron Mitchell for providing the transposon mutant library and Alexander Johnson for C. albicans SN148. The fluorescence microscopy was done in the Lombardi Core microscopy facility. We thank Guillermo Palchik of the Lombardi Comprehensive Cancer Center Microscopy (LCCC) for his technical assistance and Luzia Lyra for help with the C. albicans cultures in the Laboratory of Clinical Pathology at the State University of Campinas. We also thank Cheryl Gale (The University of Minnesota) for providing the GFP-URA3 plasmid.
Our study was supported by a National Institutes of Health grant (NIH-NIAID-43465) to R.C. This study was supported in part by the LCCC, Imaging Shared Resource, and U.S. Public Health Service grants 2P30-CA-51008 and 1S10 RR15768-01. O.A.A. was supported by the National Counsel of Technological and Scientific Development (CNPq/Brazil). N.C. is the recipient of an NIH-NIAID grant (AI076084) and an American Heart Association National Scientist Development Grant (0635108N).
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Åkerman, K. E. O., and K. Wikstrom. 1976. Safranine as a probe of the mitochondrial membrane potential. FEBS Lett. 68:191-197. [DOI] [PubMed] [Google Scholar]
- 2.Alonso-Monge, R., S. Cavalheiro, C. Nombela, E. Rial, and J. Pla. 2009. The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology 155:413-423. [DOI] [PubMed] [Google Scholar]
- 3.Alonso-Monge, R., E. Navarro-Garcia, A. Romain, B. Negredo, C. Eisman, C. Nombela, and J. Pla. 2003. The HOG1 MAP Kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez-Peral, F., O. Zaragoza, Y. Pedreno, and J. Arguelles. 2002. Protective role of trehalose during severe oxidative stress caused by hydrogen peroxide and the adaptive oxidative stress response in Candida albicans. Microbiology 148:2599-2606. [DOI] [PubMed] [Google Scholar]
- 5.Arana, D., R. Alonso-Monge, C. Du, R. Calderone, and J. Pla. 2007. Differential susceptibility of mitogen-activated protein kinase pathways to oxidative-mediated killing by phagocytes in the fungal pathogen Candida albicans. Cell. Microbiol. 9:1647-1659. [DOI] [PubMed] [Google Scholar]
- 6.Arana, D., C. Nombela, R. Alonso, and J. Pla. 2005. The Pbs2 MAP kinase kinase is essential for the oxidative stress response in the fungal pathogen Candida albicans. Microbiology 151:1033-1049. [DOI] [PubMed] [Google Scholar]
- 7.Bahn, V. S., and P. Sundstrom. 2001. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transition, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 183:3211-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calera, J. A., X.-J. Zhao, and R. Calderone. 2000. Defective hyphal formation and avirulence caused by a deletion of the CSSK1 response regulator gene in Candida albicans. Infect. Immun. 68:518-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavalheiro, R. A., F. Fortes, J. Borecky, V. C. Faustinoni, A. Z. Schreiber, and A. E. Vercesi. 2004. Respiration, oxidative phosphorylation, and uncoupling protein in Candida albicans. Braz. J. Med. Biol. Res. 37:1455-1461. [DOI] [PubMed] [Google Scholar]
- 10.Chattaway, F., R. Bishop, R. Holmes, F. Odds, and A. Barlow. 1973. Enzyme activities associated with carbohydrate synthesis and breakdown in yeast and mycelial forms of Candida albicans. Microbiology 75:97-109. [DOI] [PubMed] [Google Scholar]
- 11.Chauhan, N., D. Inglis, J. Pla, D. Li, J. A. Calera, and R. Calderone. 2003. The SSK1 of Candida albicans is associated with oxidative stress adaptation and cell wall biosynthesis. Eukaryot. Cell 2:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chauhan, N., J.-P. Latge, and R. Calderone. 2006. Signaling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Microbiol. Rev. 4:435-444. [DOI] [PubMed] [Google Scholar]
- 13.Chaves, G. M., S. Bates, D. MacCallum, and F. Odds. 2007. Candida albicans GRX2, encoding a putative glutaredoxin, is required for virulence in a murine model. Genet. Mol. Res. 6:1051-1063. [PubMed] [Google Scholar]
- 14.Chen, J., J. Chen, S. Lan, and H. Liu. 2002. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol. Microbiol. 46:1335-1344. [DOI] [PubMed] [Google Scholar]
- 15.Csank, C., K. Schroppel, E. Lebrerer, D. Harcus, O. Mohamed, S. Melocha, D. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homologue Cek1 in hyphal development and systemic candidiasis. Infect. Immun. 66:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis, D. A., V. M. Bruno, L. Loza, S. G. Filler, and A. P. Mitchell. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diezmann, S., C. Cox, G. Schonian, R. Vilgalys, and T. G. Mitchell. 2004. Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J. Clin. Microbiol. 42:5624-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du, C., R. Calderone, J. Richert, and D. Li. 2005. Deletion of the SSK1 response regulator gene of Candida albicans contributes to enhanced killing by human polymorphonuclear neutrophils. Infect. Immun. 73:865-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisman, B., R. Alonso Monge, D. Arana, E. Roman, C. Nombela, and J. Pla. 2006. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot. Cell 5:347-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enjalbert, B., D. Smith, M. Cornell, I. Alam, S. Nicholls, A. J. P. Brown, and J. Quinn. 2006. Role of the Hog1 stress activated protein kinase in the global transcriptional response to stress in the fungal pathogen. Candida albicans. Mol. Biol. Cell 17:1018-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enjalbert, B., A. Nantel, and M. Whiteway. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzpatrick, D. A., M. E. Logue, J. Stajich, and G. Butler. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6:99-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fonzi, W., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fortes, F., R. Castilho, R. Catisti, R. E. Carnieri, and A. E. Vercesi. 2001. Ca2+ induces a cyclosporin A-insensitive permeability transition pore in isolated potato tuber mitochondria mediated by reactive oxygen species. J. Bioenerg. Biomembr. 33:43-51. [DOI] [PubMed] [Google Scholar]
- 25.Fradin, C., P. de Groot, D. MacCallum, M. Schaller, F. Klis, F. Odds, and B. Hube. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56:397-415. [DOI] [PubMed] [Google Scholar]
- 26.Fradin, C., M. Kretschmar, T. Nichterlein, C. Gaillarding, C. d'Enfert, and B. Hube. 2003. Stage-specific gene expression of Candida albicans in blood. Mol. Microbiol. 47:1523-1543. [DOI] [PubMed] [Google Scholar]
- 27.Gonzales-Parraga, J. Hernandez, and J. C. Arguelles. 2003. Role of anti-oxidant enzymatic defenses against oxidative stress H2O2 and the acquisition of oxidative tolerance in Candida albicans. Yeast 20:161-169. [DOI] [PubMed] [Google Scholar]
- 28.Gornall, A., C. Bardawill, and M. Davis. 1949. Determinations of serum proteins by means of the biuret reaction. J. Biol. Chem. 177:751-766. [PubMed] [Google Scholar]
- 29.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang, C., Y. Baek, H. Yim, and S. Kang. 2003. Protective roles of mitochondrial manganese-containing superoxide dismutase against various stresses in Candida albicans. Yeast 29:929-941. [DOI] [PubMed] [Google Scholar]
- 31.Ichimura, T., H. Kubota, T. Goma, N. Mizushima, Y. Ohsumi, M. Iwago, K. Kakiuchi, H. Shekhar, T. Shinkawa, M. Taoka, T. Ito, and T. Isobe. 2004. Transcriptomic and proteomic analysis of a 14-3-3 gene-deficient yeast. Biochemistry 43:6149-6158. [DOI] [PubMed] [Google Scholar]
- 32.Jarmuszkiewicz, W., G. Milani, F. Fortes, A. Z. Schreiber, F. E. Sluse, and A. E. Vercesi. 2000. First evidence and characterization of an uncoupling protein in fungi kingdom: CpUCP of Candida parapsilosis. FEBS Lett. 467:145-149. [DOI] [PubMed] [Google Scholar]
- 33.Joers, P., J. Gerhold, T. Sedman, S. Kuusk, and J. Sedman. 2007. The helicase CaHmi1p is required for wild-type mitochondrial DNA organization in Candida albicans. FEMS Yeast Res. 7:118-130. [DOI] [PubMed] [Google Scholar]
- 34.Klippel, N., and U. Bilitewski. 2007. Phagocytosis assay based on living Candida albicans for the detection of effects on macrophage function. Anal. Lett. 40:1400-1411. [Google Scholar]
- 35.Kobayashi, D., K. Kondo, N. Uehara, S. Otokozawa, N. Tsuji, A. Yagihashi, and N. Watanabe. 2002. Endogenous reactive oxygen species is an important mediator of the miconazole effect. Antimicrob. Agents Chemother. 46:3113-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Land, G., W. McDonald, R. Stjernholm, and L. Friedman. 1975. Factors affecting filamentation in Candida albicans: changes in respiratory activity of Candida albicans during filamentation. Infect. Immun. 12:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Land, G., W. McDonald, R. Stjernholm, and L. Friedman. 1975. Factors affecting filamentation in Candida albicans: relationship of the uptake and distribution of proline to morphogenesis. Infect. Immun. 11:1014-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehrer, R. I., and M. J. Cline. 1969. Interaction of Candida albicans with human leukocytes and serum. J. Bacteriol. 98:996-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemar, K., M. Aon, S. Cortassa, B. O'Rouke, C. Muller, and D. Lloyd. 2007. Diallyl disulphide depletes glutathione in Candida albicans. Yeast 24:695-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martchenko, M., A.-M. Alarco, D. Harcus, and M. Whiteway. 2004. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15:456-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Esparza, M., A. Aguniaga, P. Gonzalez-Parraga, P. Garcia-Penarrubia, T. Jouault, and J. C. Arguelles. 2007. Role of trehalose in resistance to macrophages killing: study with a tps/tps trehalose-deficient mutant of Candida albicans. Clin. Microbiol. Infect. 13:384-394. [DOI] [PubMed] [Google Scholar]
- 42.McDonough, J., V. Bhattacherjee, T. Sadlon, and M. Hostetter. 2002. Involvement of Candida albicans NADH dehydrogenase complex I in filamentation. Fungal Genet. Biol. 36:117-127. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa, Y., T. Kanbe, and I. Mizugchi. 2003. Disruption of the human pathogenic yeast Candida albicans catalase gene decreases survival in mouse-model infection and elevates susceptibility to higher temperature and to detergents. Microbiol. Immunol. 47:395-403. [DOI] [PubMed] [Google Scholar]
- 44.Navarro-Garcia, F., R. Alonso-Monge, H. Rico, J. Pla, R. Sentandreu, and C. Nombela. 1998. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology 144:411-421. [DOI] [PubMed] [Google Scholar]
- 45.Neupert, W., and J. M. Hermann. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723-749. [DOI] [PubMed] [Google Scholar]
- 46.Noble, S., and A. D. Johnson. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Posas, F., J. Chambers, J. Heyman, J. Hoeffler, E. de Nadal, and J. Arino. 2000. The transcriptional response of yeast to saline stress. J. Biol. Chem. 275:17249-17255. [DOI] [PubMed] [Google Scholar]
- 49.Roilides, E., K. Uhlig, D. Venzon, P. Pizzo, and T. Walsh. 1992. Neutrophil oxidative burst in response to blastoconidia and pseudohyphae of Candida albicans: augmentation by granulocyte colony-stimulating factor and interferon-gamma. J. Infect. Dis. 166:668-673. [DOI] [PubMed] [Google Scholar]
- 50.San Jose, C., R. Alonso-Monge, R. Perez-Diaz, J. Pla, and C. Nombela. 1996. The mitogen-activated protein kinase HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 178:5850-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva, R., R. Sotoca, B. Johansson, P. Ludovico, F. Sansonetty, M. Silva, J. Peinado, and M. Corte-Real. 2005. Hyperosmotic stress induces metacaspase-and mitochondria-dependent apoptosis in Saccharomyces cerevisiae. Mol. Microbiol. 58:824-834. [DOI] [PubMed] [Google Scholar]
- 52.Smith, D. A., S. Nicholls, B. Morgan, A. J. P. Brown, and J. Quinn. 2004. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell 15:4179-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaupotic, T., P. Veranic, P. Jenoe, and A. Plemenitas. 2008. Mitochondrial mediation of environmental osmolytes discrimination during osmoadaptation in the extremely halotolerant black yeast Hortaea werneckii. Fungal Genet. Biol. 45:994-1007. [DOI] [PubMed] [Google Scholar]
- 54.Vellucci, V., S. Gygax, and M. Hostetter. 2007. Involvement of Candida albicans pyruvate dehydrogenase complex protein X (Pdx1) in filamentation. Fungal Genet. Biol. 44:979-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walia, A., and R. Calderone. 2007. The SSK2 MAPKKK of Candida albicans is required for oxidant adaptation in vitro. FEMS Yeast Res. 8:287-299. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, R., D. Davis, B. Enloe, and A. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR through gene disruption with short homology. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson, R., D. Davis, B. Enloe, and A. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR-product-directed gene disruption. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 58.Wysong, D. R., L. Christin, A. Sugar, P. W. Robbins, and R. D. Diamond. 1998. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 66:1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yale, J., and H. J. Bohnert. 2001. Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 276:15996-16007. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, X., M. De Micheli, S. Coleman, D. Sanglard, and W. S. Moye-Rowley. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36:618-629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.