Abstract
Candida albicans undergoes a morphological transition from yeast to hyphae in response to a variety of stimuli and growth conditions. We previously isolated a LisH domain containing transcription factor Flo8, which is essential for hyphal development in C. albicans. To search the putative binding partner of Flo8 in C. albicans, we identified C. albicans Mss11, a functional homolog of Saccharomyces cerevisiae Mss11, which also contains a LisH motif at its N terminus. C. albicans Mss11 can interact with Flo8 via the LisH motif by in vivo coimmunoprecipitation. The results of a chromatin immunoprecipitation (ChIP) assay showed that more Mss11 and Flo8 proteins bound to the upstream activating sequence region of HWP1 promoter in hyphal cells than in yeast cells, and the increased binding of each of these two proteins responding to hyphal induction was dependent on the other. Overexpression of MSS11 enhanced filamentous growth. Deletion of MSS11 caused a profound defect in hyphal development and the induction of hypha-specific genes. Our data suggest that Mss11 functions as an activator in hyphal development of C. albicans. Furthermore, overexpression of FLO8 can bypass the requirement of Mss11 in filamentous formation, whereas overexpression of MSS11 failed to promote hyphae growth in flo8 mutants. In summary, we show that the expression level of MSS11 increases during hyphal induction, and the enhanced expression of MSS11 may contribute to cooperative binding of Mss11 and Flo8 to the HWP1 promoter.
The human fungal pathogen Candida albicans can cause superficial mucosal infections, severe surface infections and, in particular, life-threatening systemic diseases in immunocompromised patients (48). In recent years, the deaths due to C. albicans infections increased dramatically, despite the use of antifungal therapies (4). C. albicans can undergo reversible morphological transitions between yeast, pseudohyphal, and hyphal growth forms (6). Its ability to switch from yeast to hyphal growth in response to various signals is essential for its pathogenicity (20, 29, 35). Multiple signaling pathways and transcription factors are involved in regulating morphogenesis of C. albicans (13). Among them, the Efg1/Flo8-mediated cyclic AMP (cAMP)-dependent protein kinase A (PKA) pathway plays a predominant role in hyphal development and virulence (30). Many mutants in the cAMP/PKA pathway (cdc35, tpk1, tpk2, efg1, and flo8) are defective in hyphal formation and show decreased pathogenicity (8, 32).
In C. albicans, many transcriptional regulators, including Cph1 (33), Tec1 (45), and Flo8 (8), are identified based on sequence similarity and functional complementation to their homologues in S. cerevisiae. ScFlo8, a transcription factor critical for invasive growth and flocculation in haploids and pseudohyphal growth in diploids of S. cerevisiae (34), functions downstream of the cAMP/PKA pathway (44). The C. albicans FLO8 homolog was identified by functional complementation of a S. cerevisiae flo8 mutant. The C. albicans Flo8 is essential for hyphal development and hypha-specific gene expression. flo8/flo8 mutants are unable to form hyphae in all liquid-inducing media but generate increased filaments under embedded conditions at low temperature. Flo8 can interact with Efg1, which is a potential target of the cAMP/PKA pathway, and they share a common set of target genes (8). Like ScFlo8, CaFlo8 also contains a LUFS domain (LUG/LUH, Flo8, single-stranded DNA-binding protein) (12). LUFS domain containing proteins play important roles in regulating critical developmental processes. LUG (Leunig) is a key regulator of flower-specific gene expression during flower development in Arabidopsis (12). The single-stranded DNA-binding protein (Ssdp) regulates the activity of LIM-homeodomain protein complexes in Drosophila (52). Within the LUFS domain, there is a lissencephaly type 1-like homology motif (LisH) (15). The LisH motif is widespread among many proteins, some of which are implicated in human diseases (1, 17, 46). The crystal structure of the N-terminal domain of mouse LIS1 shows that the LisH motif is a thermodynamically very stable dimerization domain and forms a tightly associated homodimer with a four-helix antiparallel bundle core (25), suggesting that Flo8 might interact with another LisH domain containing protein to regulate gene expression. In fact, a previous study in Saccharomyces diastaticus showed that Flo8 and Mss11, another LisH-containing transcription factor, function as a heterodimer to activate STA1 expression (27).
MSS11 was originally isolated as a multicopy suppressor of STA10 phenotype and an activator of STA2 gene in S. cerevisiae (54). It plays a central role in the regulatory network involved in the activation of FLO11, which is essential for invasive/filamentous growth and flocculation (36). The Mss11 activity is independent of the presence of activators, including Tec1, Flo8, and Phd1, or the repressors Nrg1, Nrg2, Sok2, and Sfl1 and the signaling proteins Ras2, Kss1, and Tpk2 (19, 51). Genetic analysis shows that Mss11 is positioned downstream of Flo8 in regulating FLO11 expression. In S. diastaticus, Mss11 plays an important role in activation of the STA1 gene, which encodes an extracellular glucoamylase. The STA1 gene is activated by sequentially binding of transcriptional activators Ste12, Tec1, Flo8, and Mss11 and the Swi/Snf chromatin-remodeling complex. Furthermore, it has been shown that Mss11 and Flo8 bind cooperatively to the STA1 promoter to activate expression of the gene (27). However, the function of C. albicans Mss11 remains to be characterized.
In searching for the binding partner of Flo8 in C. albicans, we identified C. albicans Mss11 by sequence comparison and functional complementation. Like Flo8, C. albicans Mss11 acts as an activator in hyphal development and hypha-specific gene expression. We further investigated the interaction between Mss11 and Flo8 and their relationship in hyphal development.
MATERIALS AND METHODS
Strains and culture conditions.
The C. albicans and S. cerevisiae strains used in the present study are listed in Table 1. Yeast strains were routinely grown on YPD (1% yeast extract, 2% peptone, 2% glucose) medium or on synthetic complete medium with 2% glucose (SCD) for selection of prototropic strains. The pseudohyphal colony formation and flocculent phenotype of S. cerevisiae were examined as described previously (34, 38). C. albicans strains were grown in YPD at 25°C for yeast form and in Lee medium or YPD containing 10% serum (Gibco) at 37°C for hyphal induction. YPS (1% yeast extract, 2% peptone, 2% sucrose) with 1% agar was used for colony morphology assay under embedded conditions (7).
TABLE 1.
C. albicans and S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| C. albicans | ||
| SC5314 | Wild type | 18 |
| CAF2-1 | URA3/ura3::λimm434 | 18 |
| CAI4 | ura3::λimm434/ura3::λimm434 | 18 |
| CSL1 | ura3::λimm434/ura3::λimm434 MSS11/mss11::hisG-URA3-hisG | This study |
| CSL2 | ura3::λimm434/ura3::λimm434 MSS11/mss11::hisG | This study |
| CSL3 | ura3::λimm434/ura3::λimm434 mss11::hisG/mss11::hisG-URA3-hisG | This study |
| CSL4 | ura3::λimm434/ura3::λimm434 mss11::hisG/mss11::hisG | This study |
| CCF3 | ura3::λimm434/ura3::λimm434 flo8::hisG/flo8::hisG-URA3-hisG | 8 |
| CCF4 | ura3::λimm434/ura3::λimm434 flo8::hisG/flo8::hisG | 8 |
| CSL5 | ura3::λimm434/ura3::λimm434 mss11::hisG/mss11::hisG flo8::hisG/flo8::hisG-URA3-hisG | This study |
| RM1000 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG | 42 |
| CSL6 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG RP10::ACT1p-MSS11-4FLAG-HIS1 FLO8/FLO8-13MYCFLAG-URA3 | This study |
| CSL7 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG RP10::ACT1p-MSS11-4FLAG-HIS1 | This study |
| CSL8 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG RP10::ACT1p-FLO8-13MYCFLAG-HIS1 ADE2::ADH1p-MSS11-3HA-URA3 | This study |
| CSL9 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG RP10::ACT1p-FLO8-13MYCFLAG-HIS1 ADE2::ADH1p-MSS11ΔLisH-3HA-URA3 | This study |
| CSL10 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG RP10::ACT1p-MSS11-13MYCFLAG-HIS1 ADE2::ADH1p-FLO8-3HA-URA3 | This study |
| CSL11 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG RP10::ACT1p-MSS11-13MYCFLAG-HIS1 ADE2::ADH1p-FLO8ΔLisH-3HA-URA3 | This study |
| CSL12 | ura3::λimm434/ura3::λimm434 MSS11/MSS11-13MYCFLAG-URA3 | This study |
| CSL13 | ura3::λimm434/ura3::λimm434 flo8::hisG/flo8::hisG MSS11/MSS11-13MYCFLAG-URA3 | This study |
| CSL14 | ura3::λimm434/ura3::λimm434 FLO8/FLO8-13MYCFLAG-URA3 | This study |
| CSL15 | ura3::λimm434/ura3::λimm434 mss11::hisG/mss11::hisG FLO8/FLO8-13MYCFLAG-URA3 | This study |
| S. cerevisiae | ||
| MLY61 | ura3-52/ura3-52 MATa/α | 37 |
| MLY181a/α | Δmss11::G418/Δmss11::G418 ura3-52/ura3-52 MATa/α | 38 |
| BY4742 | MATα flo8-1 his3 leu2 lys2 ura3 | EUROSCARF |
| BY4742 mss11Δ | MATα flo8-1 his3 leu2 lys2 ura3 mss11Δ::KanMX4 | EUROSCARF |
| CZS1 | MATα his3 lys2 ura3 flo8-1Δ::FLO8-LEU2 | This study |
| CZS2 | MATα his3 lys2 ura3 flo8-1Δ::FLO8-LEU2 mss11Δ::KanMX4 | This study |
| EGY48 (p8op-lacZ) | MATα his3 trp1 LexAop(×6)-leu2 LexAop(×8)-lacZ flo8-1 | 23 |
Plasmid and strain construction.
SC5314 genomic DNA was used as a template for all PCR amplifications of C. albicans genes. All constructs were verified by DNA sequencing. The plasmids used in the present study are listed in Table 2. The primers used for PCR amplification are listed in Table 3.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pVT102U | S. cerevisiae URA3/2μ vector with ADH1 promoter and terminator | 53 |
| pVT102U-CaMSS11 | 1.93-kb full-length C. albicans MSS11 in pVT102U | This study |
| YIplac128 | 2μ, LEU2-based vector | 21 |
| YIplac128-ScFLO8 | 3.21-kb full-length S. cerevisiae FLO8 in YIplac128 | This study |
| pGilda | 2μ, HIS3-based vector carrying the LexA DNA-binding domain | Clontech |
| pGilda-CaMSS11 | 2.07-kb full-length C. albicans MSS11 in pGilda | This study |
| pJG4-5 | 2μ, TRP1-based vector carrying the B42 activation domain | 23 |
| YEplac195 | 2μ, URA3-based vector | 21 |
| pCUB6 | Substitution of S. cerevisiae URA3 by C. albicans URA3 in pNKY50 | 18 |
| YEplac195-MSS11Δ | 0.72-kb upstream region and 0.80-kb downstream region of MSS11 ORF and hisG-URA3-hisG in YEplac195 | This study |
| pBA1 | C. albicans ADH1 promoter in pBES116 | 8 |
| pBA1-MSS11 | 2.17-kb full-length C. albicans MSS11 in pBA1 | This study |
| pBA1-FLO8 | 2.45-kb full-length C. albicans FLO8 in pBA1 | 8 |
| pBES116 | URA3 vector, digest with AscI for integration at ADE2 | 16 |
| pBES116-MSS11 | 3.09-kb C. albicans MSS11 fragment including 1.04-kb MSS11 promoter region in pBES116 | This study |
| pPR671 | C. albicans ACT1 promoter, 13×myc-FLAG, HIS1 | 8 |
| pPR672 | 2.45-kb full-length C. albicans FLO8 in pPR671 | 8 |
| pFLAG-Act1 | C. albicans ACT1 promoter, FLAG, URA3 | 50 |
| pPR677 | 2.45-kb full-length C. albicans FLO8 in URA3 vector with 13×myc-FLAG | This study |
| pPR678 | 1.92-kb full-length C. albicans MSS11 in pPR671 | This study |
| pPR679 | 1.92-kb full-length C. albicans MSS11 in URA3 vector with 13×myc-FLAG | This study |
| pFLAG-Act1-HIS1 | C. albicans ACT1 promoter, FLAG, HIS1 | This study |
| p4FLAG-MSS11 | 1.92-kb full-length C. albicans MSS11 and 3×FLAG in pFLAG-Act1-HIS1 | This study |
| pYPB1-ADHpt | C. albicans ADH1 promoter and ADH1 terminator on a C. albicans URA3/ARS/2μ vector | 28 |
| BES-ADH | C. albicans URA3 vector with ADH1 promoter and terminator, digest with AscI for integration at ADE2 | This study |
| ADH-3HA | C. albicans ADH1 promoter, 3×HA, URA3 | This study |
| ADH-3HA-MSS11 | 1.92-kb full-length C. albicans MSS11 in ADH-3HA | This study |
| ADH-3HA-MSS11ΔN | 1.69-kb C. albicans MSS11ΔN in ADH-3HA | This study |
| ADH-3HA-FLO8 | 2.45-kb full-length C. albicans FLO8 in ADH-3HA | This study |
| ADH-3HA-FLO8ΔN | 2.10-kb C. albicans FLO8ΔN in ADH-3HA | This study |
TABLE 3.
Primers used in this study
| Primer | Sequence (5′-3′)a | Purpose and features |
|---|---|---|
| 1 | GCAGGATCCATACCACATAGATGTCTAAAC | pVT102U-CaMSS11 |
| 2 | GCAGAGCTCTTATTATTCATAACCTGGACC | |
| 3 | CTAGGATCCAATGCTGGCTCTAGTAGTAAC | YIplac128-ScFLO8 |
| 4 | CTAGAGCTCCAATGAGTGTACATCAACCAG | |
| 5 | CTAGGATCCGAATACCACATGAGATG | pGilda-CaMSS11 |
| 6 | CTGCCATGGTAGTACATAAGAAGAGC | |
| 7 | GCACTGCAGGTTGCAATATGGAGATAAGAA | YEplac195-MSS11ΔN |
| 8 | GCCTCTAGAAATAAGTTCCCTTACAACTTG | |
| 9 | GCAGGTACCGTATGCCACCTAATCAGAATC | YEplac195-MSS11ΔNΔC |
| 10 | GCAGAGCTCTTATATTAAGCGGTTGTGCGA | |
| 11 | CTAAGATCTGAGATGTCTAAACCACCACCT | pBA1-MSS11 |
| 12 | CTGATCGATATCAAGAATACCCTCTACCTC | |
| 13 | GTCCTGCAGGTATTATCGATTACAG | pBES116-MSS11 |
| 14 | CTAGGTACCAGTACATAAGAAGAGC | |
| 15 | CGGGATCCCTGAATACCACATGAGATGTCT | pPR678 and pPR679 |
| 16 | GTAGACGCGTCGTTCATAACCTGGACCTGATC | |
| 17 | ACATGCATGCCTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCTTCATAACCTGGACCTGA | p4FLAG-MSS11 |
| 18 | CTGCGCGGCCGCGATCTGTTTAGCTGGTGATTT | BES-ADH |
| 19 | TGTCTAGAGGTGTTGAAATGTTCTTGTTG | |
| 20 | CTGATCGATATGTCTAAACCACCACCTC | ADH-3HA-MSS11 |
| 21 | CTAACGCGTTTATTCATAACCTGGACCTG | |
| 22 | CTGATCGATGATAATAGTGGTCCTGATC | ADH-3HA-MSS11ΔN |
| 23 | CTGATCGATATGAATCATAAACAAGTACTACCAG | ADH-3HA-FLO8 |
| 24 | CATACGCGTTCTAATCGCCATTTTCAATTGGATC | |
| 25 | CTGATCGATAGTCAACTTCCTCTTATACAG | ADH-3HA-FLO8ΔN |
Restriction sites are underlined. The boldface sequence in primer 17 is 3×FLAG.
The pVT102U-CaMSS11 plasmid was constructed for complementation assays in S. cerevisiae mss11 mutants. A PCR fragment (primers 1 and 2) containing the CaMSS11 coding sequence was subcloned into the BamHI-SstI site of pVT102U to express CaMss11 under the control of ADH1p in S. cerevisiae. To examine the flocculation phenotype in a background with a functional ScFLO8 gene, the wild-type ScFLO8 coding sequence was amplified from a Σ strain CGx68 genomic DNA (22) with primers 3 and 4, and inserted into the BamHI-SstI site of YIplac128 to create YIplac128-ScFLO8. BglII-digested YIplac128-ScFLO8 was introduced into strains BY4742 and BY4742 mss11Δ to replace the flo8-1 mutant allele with wild-type ScFLO8, generating strains CZS1 and CZS2, respectively. pGilda-CaMSS11 for expression of LexA-CaMss11 fusion protein under the GAL1 promoter in S. cerevisiae was constructed by inserting a 2.07-kb PCR fragment (primers 5 and 6) containing the CaMSS11 coding sequence into the BamHI-NcoI site of pGilda.
The YEplac195-MSS11Δ plasmid for C. albicans MSS11 disruption was constructed as follows. Primers 7 and 8 were used to amplify the upstream region of the MSS11 open reading frame (ORF) from C. albicans genomic DNA; the PCR product was inserted into the PstI-XbaI site of YEplac195 (21), generating YEplac195-MSS11ΔN. Then, the downstream region of MSS11 ORF (primers 9 and 10) was inserted into the KpnI-SstI site of YEplac195-MSS11ΔN to generate YEplac195-MSS11ΔNΔC. A 4-kb fragment containing the HISG-URA3-HISG cassette digested from pCUB6 with BglII and BamHI was subcloned into the BamHI site of YEplac195-MSS11ΔNΔC to create YEplac195-MSS11Δ. To delete MSS11 in C. albicans, YEplac195-MSS11Δ was digested with PstI and SstI, and then transformed into CAI4 to produce MSS11/mss11 strains (CSL1). CSL1 was streaked onto a 5-fluoroorotic acid plate to select for the Ura− strain CSL2. mss11/mss11 homozygotes CSL3 (Ura+) and CSL4 (Ura−) were screened out by subsequent transformation. The disruption was confirmed by Southern blotting.
pBA1-MSS11 was constructed for expression of CaMSS11 under the control of ADH1p in C. albicans. The full-length coding sequence of MSS11 was amplified with primers 11 and 12 and inserted into the BglII-ClaI site of pBA1 to generate pBA1-MSS11. pBES116-MSS11 was for expression of CaMSS11 under its own promoter for complementation assays in C. albicans. A 3.09-kb fragment containing the entire MSS11 ORF and 1.04 kb of upstream sequence was inserted into pBES116 at the PstI-KpnI site. Primers 13 and 14 were used for the PCR amplification.
Plasmids for expression of 13×Myc fusion proteins were constructed as follows. The HIS1 fragment of pPR672 (8) (digested with HindIII) was replaced with the URA3 fragment from pFLAG-Act1 (50) to generate plasmid pPR676. pPR676 was digested with BamHI and StuI to delete the ACT1 promoter, generating pFLO8-13MYC-FLAG-URA3 (pPR677) and then digested with SpeI for targeting the integration into the FLO8 locus of CAI4 and mss11/mss11 strains under URA3 selection. The FLO8 fragments in pPR672 and pPR677 (digested with BamHI and MluI) were replaced with the MSS11 coding sequence (primers 15 and 16) to generate plasmids pPR678 and pPR679, respectively. Strains CAI4 and flo8/flo8 were transformed with SpeI-digested pPR679 to express the Mss11-13Myc fusion protein under the endogenous promoter. pPR672 and pPR678 were digested with StuI to integrate into the genomic RP10 locus for expression of 13×Myc fusion proteins under the ACT1 promoter.
pFLAG-Act1-HIS1 is a derivative of pFLAG-Act1 (50), in which the URA3 fragment in the HindIII site was substituted by HIS1 fragment from pPR671. A 2.0-kb PCR product (primers 1 and 17) containing the entire MSS11 coding region and a 3×FLAG tag was inserted into the BamHI-SphI site of pFLAG-Act1-HIS1 to create p4FLAG-MSS11. The StuI-digested p4FLAG-MSS11 DNA fragment was introduced into the RM1000 strain for 4FLAG-Mss11 expression, and then the strain was transformed with SpeI-digested pPR677 to express Flo8 under its own promoter. Expression of Mss11 and Flo8 in YPD medium at 30°C was verified by Western blot analysis using anti-FLAG-conjugated peroxidase antibody (Sigma) and anti-c-myc antibody (Santa Cruz), respectively.
To express hemagglutinin (HA) fusion proteins under the control of ADH1 promoter, a 4.5-kb PCR product (primers 18 and 19) amplified from pBES116 was digested with NotI and XbaI and then ligated with a fragment (with the same restriction sites) containing the ADH1 promoter and C. albicans URA3 gene from pYPB1-ADHpt (28) to generate BES-ADH. The synthetic 3×HA fragment (GGATCCATGTACCCATACGATGTTCCAGATTACGCTGGTTACCCATACGATGTTCCAGATTACGCTGGTTACCCATACGATGTTCCAGATTACGCTATCGATATCCCATGGAGATCTACGCGT [the introduced multiple cloning sites are underlined]) was placed into the BamHI-MluI site of the BES-ADH plasmid, generating the vector ADH-3HA. DNA fragments encoding full-length Mss11 (primers 20 and 21), C-terminal Mss11 (primers 22 and 21), full-length Flo8 (primers 23 and 24), and C-terminal Flo8 (primers 25 and 24) were inserted into the ClaI-MluI site of ADH-3HA to create expression vectors for coimmunoprecipitation assays.
Northern blotting.
RNA extraction and Northern blotting were performed as described previously (10). The HWP1 and ECE1 PCR products (41) were used for probing Northern blots. The ACT1 probe was used as previously described (31). The ImageQuant 5.2 Software is used to evaluate the relative density of different bands on the scanned image of a gel.
Immunoprecipitation.
Protein extraction was performed as described previously (8). To determine whether Mss11 interacts with Flo8 in vivo, crude extract of C. albicans cells was precleared with protein G-agarose (Roche) and then incubated with 2.0 μg of anti-c-myc antibody (Santa Cruz). After incubation for 2 h at 4°C, ∼40 μl of protein G-agarose suspension was added to precipitate the immunocomplex. The proteins were subjected to Western blotting with anti-FLAG peroxidase conjugated antibody (Sigma). For the immunoprecipitation shown in Fig. 6, 2.0 μg of anti-HA antibody (Santa Cruz) was added to the lysates, and the immunocomplex was detected by using anti-c-myc antibody.
FIG. 6.
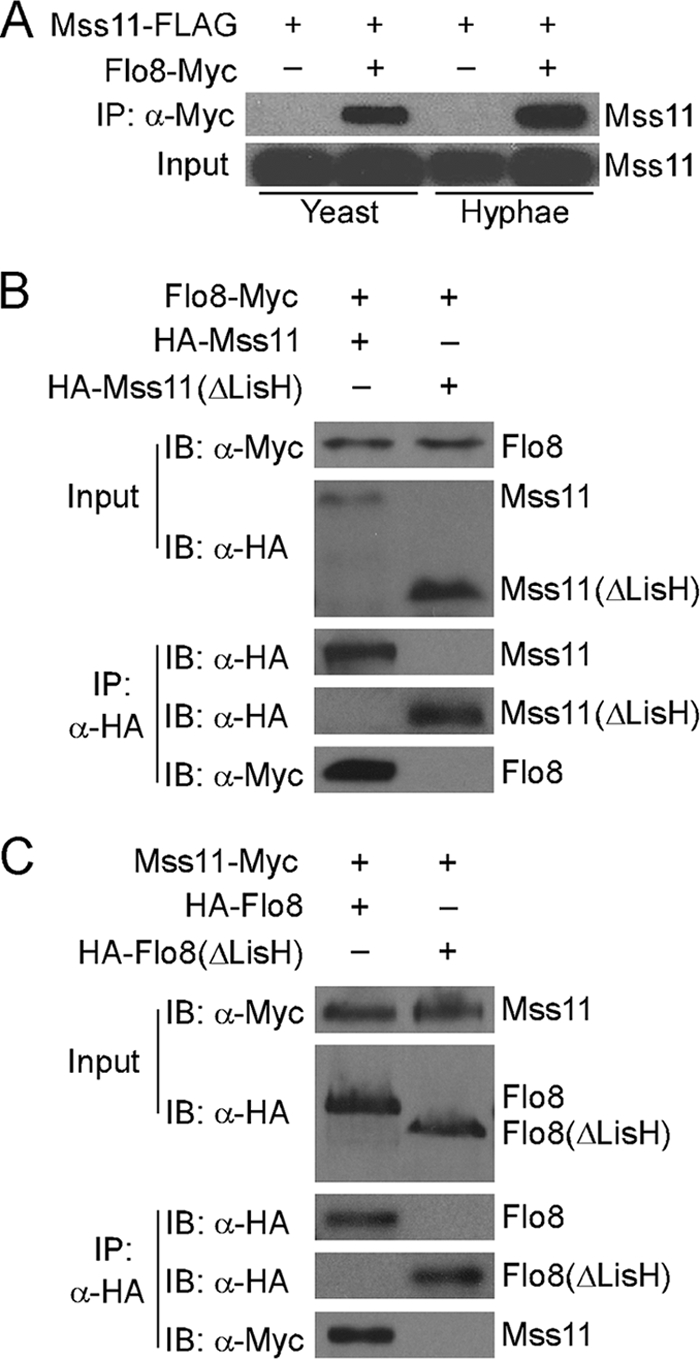
Mss11 interacts with Flo8 in vivo. (A) Mss11 interacts with Flo8 in yeast and hyphal cells. C. albicans CSL6 (Mss11-FLAG Flo8-Myc) and CSL7 (Mss11-FLAG) were grown at 25°C in YPD for yeast growth, and at 37°C in YPD + 10% serum for hyphal growth. Protein lysates were subjected to immunoprecipitation with anti-Myc antibody, and the precipitated proteins were separated by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis and probed with peroxidase-conjugated anti-FLAG. As an input control, cell lysates were analyzed by Western blotting with the peroxidase-conjugated anti-FLAG antibody. (B and C) Mss11 interacts with Flo8 via the LisH motif. Total proteins were extracted from C. albicans CSL8 (HA-Mss11 Flo8-Myc), CSL9 (HA-Mss11ΔN Flo8-Myc), CSL10 (HA-Flo8 Mss11-Myc), and CSL11 (HA-Flo8ΔN Mss11-Myc) after growth in YPD at 30°C to an optical density at 600 nm of ∼1.0. The tagged proteins were pulled down with anti-HA antibody. Western blots were probed with anti-HA antibody or anti-Myc antibody. The relative amounts of protein loaded in the input control in panels A to C were 1% of the cell extracts used for immunoprecipitation.
FIG. 2.
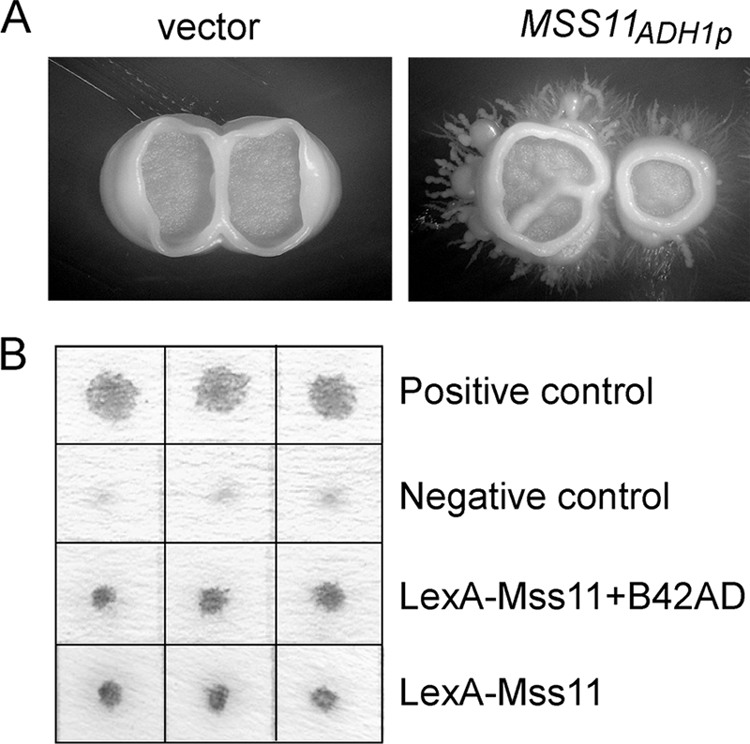
CaMss11 functions as an activator in hyphal development. (A) Overexpression of MSS11 promotes filamentous growth in C. albicans. Strain CAI4 (wild type) carrying pBA1 or pBA1-MSS11 was grown on Lee's plates at 25°C for 10 days. (B) CaMss11 exhibits strong transcriptional activity in S. cerevisiae. For the β-galactosidase assay, strain EGY48 carrying pGilda-CaMSS11 was transformed with or without vector pJG4-5. Three corresponding transformants were tested for the level of reporter gene expression using X-Gal. pSH17-4 was a positive control plasmid for LexA-AD fusion protein. pRFHM1 was a negative control plasmid for expression of a LexA-human lamin C fusion protein (43). The results are representative of three independent experiments.
ChIP analyses.
Overnight cultures were grown in YPD for 6 h at 25°C to an optical density at 600 nm of 0.8 for yeast growth or in YPD plus 10% serum for 3 h at 37°C for hyphal induction. Chromatin immunoprecipitations (ChIPs) were performed as described previously (39). Cells were formaldehyde cross linked and lysed using lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% sodium deoxycholate). Then, the DNA was sheared by sonication three times for 10 s at high power on a Bioruptor, with incubation on ice for 20 s between sonication pulses. To precipitate the Myc-tagged proteins, 8 μl of monoclonal Myc antibody (Abcam) was used in an immunoprecipitation volume of 400 μl (∼2 mg of chromatin proteins). DNA derived from the whole-cell extract, and the immunoprecipitation eluate was analyzed by PCR or quantitative PCR.
RESULTS
Identification of C. albicans MSS11.
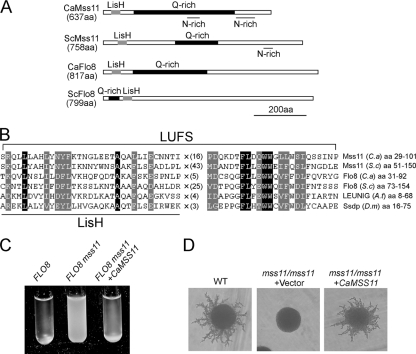
We have reported that the transcription factor Flo8 is essential for hyphal development in C. albicans (8). In S. diastaticus, Flo8 and Mss11 bind cooperatively to the STA1 promoter and function as a heterodimer in activating STA1 expression (27). Furthermore, Mss11 is a transcription factor playing important roles in flocculation, filamentous growth, starch metabolism, and activation of gene expression in S. cerevisiae (2, 19, 38, 51, 54). To determine whether a similar molecular mechanism exists in C. albicans, we searched the NCBI database (http://www.ncbi.nlm.nih.gov/BLAST/) for the S. cerevisiae Mss11 homolog and found a C. albicans protein designated Mss11 (orf19.6309) that shares similar structural features with S. cerevisiae Mss11 (ScMss11). Like ScMss11, CaMss11 contains a LisH motif at its N terminus, a glutamine-rich region in the central part and asparagine-rich regions at the C terminus. As shown in Fig. 1A, both CaFlo8 and ScFlo8 contain a LisH motif and a glutamine-rich region but lack asparagine-rich regions. In contrast to other LisH motif containing proteins in which the glutamine-rich regions are located at the right part of the motif, the glutamine-rich region of ScFlo8 is located at the left part of the LisH motif. The CaMSS11 gene encodes a putative protein of 637 amino acids, with a small region (amino acids 29 to 101) that is highly similar to a region (amino acids 51 to 150) in the ScMss11 (Fig. 1B). The conserved region, named the LUFS domain (12), exists in many regulators, including CaFlo8, ScFlo8, LEUNIG, and Ssdp. The LUFS domain consists of a conserved LisH motif in its N-terminal part and a coiled-coil region in the C terminus, and the two regions are separated by a linker varying in length. In the C. albicans genome, there are five LisH motif-containing proteins: Flo8, putative Mss11, potential SET3 histone deacetylase complex component Sif2, potential SAGA complex component Taf5, and a putative uncharacterized protein. In addition, sequence analysis shows that Mss11 and Flo8 in C. albicans contain the LUFS domain (Fig. 1B).
FIG. 1.
C. albicans Mss11 is a functional homolog of S. cerevisiae Mss11. (A) Schematic depiction of the LisH domain (gray box), glutamine-rich (Q-rich) regions (black box), and asparagine-rich (N-rich) regions (underlined areas) in the CaMss11, ScMss11, CaFlo8, and ScFlo8. aa, amino acids. (B) Sequence alignment of the LUFS domain between CaMss11 and other regulatory proteins. Identical residues are shaded in black and conserved residues are shaded in gray. The multiplication sign (×) and subscript numbers indicate spacing between the motifs in the LUFS domain. (C) Ectopically expressed CaMSS11 suppresses the nonflocculent phenotype of a haploid mss11 mutant. S. cerevisiae strains CZS1 (FLO8) carrying pVT102U and CZS2 (FLO8 mss11) carrying pVT102U or pVT102U-CaMSS11 were grown in SCD to saturation, allowed to settle for 5 min, and then photographed. (D) Ectopically expressed CaMSS11 suppresses the pseudohyphal growth defect of a diploid mss11 mutant. S. cerevisiae strains MLY61 (WT) carrying pVT102U and MLY181a/α (mss11/mss11) carrying pVT102U or pVT102U-CaMSS11 were grown on SLAD plates at 30°C for 5 days.
Although CaMss11 and ScMss11 share similar structural features, the sequence similarity between them is very low. To determine whether CaMss11 is a functional homolog (ortholog) of ScMss11, we examined the ability of CaMss11 to complement the flocculation and filamentous growth defect in Scmss11 mutants. An expression plasmid, pVT102U-CaMSS11, containing CaMSS11 ORF under the control of ADH1 promoter, was introduced into the Scmss11 mutant strains. To eliminate the effect of ScFLO8 mutation on the nonflocculent phenotype of Scmss11 mutants, a wild-type ScFLO8 gene was integrated into strains BY4742 and BY4742 mss11Δ to replace the flo8-1 mutant allele. Ectopically expressed CaMSS11 suppressed the nonflocculent phenotype of a haploid Scmss11 mutant (Fig. 1C) and the filamentous growth defect of a diploid Scmss11 mutant (Fig. 1D). The results show that CaMSS11 can functionally complement Scmss11 mutant in both flocculation and filamentous growth.
Overexpression of MSS11 promotes filaments formation in C. albicans.
ScMss11 functions as a transcriptional activator in S. cerevisiae. To examine whether CaMss11 also acts as an activator in filamentous growth of C. albicans, a single copy of CaMSS11 under the control of ADH1 promoter was introduced into the wild-type strain at the ADE2 locus. As shown in Fig. 2A, wild-type strains formed wrinkled colonies in Lee's medium at 25°C, while the CaMSS11-overexpressing strains formed wrinkled colonies surrounded by long filaments. To analyze the activating effect of CaMss11 on gene expression, we fused CaMss11 with a LexA DNA-binding domain and used lacZ as a reporter for measuring the transcriptional activity of CaMss11 in the yeast system. The β-galactosidase activity assays with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as a substrate showed that the LexA-CaMss11 fusion protein could promote expression of the lexAop-lacZ reporter with or without the existence of the B42 activation domain (Fig. 2B), a finding consistent with the result of a Gal4-ScMss11 fusion in activating lacZ expression (19). Our data suggested that C. albicans Mss11 functions as a transcriptional activator in filamentous growth and gene expression.
Deletion of MSS11 prevents hyphal formation.
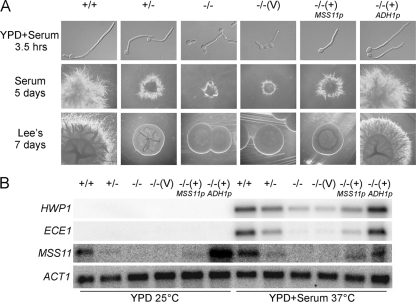
To elucidate the role of C. albicans Mss11 in hyphal development, we constructed an mss11/mss11 null mutant by sequential gene disruption using a hisG-URA3-hisG cassette in C. albicans. Successful deletion of the MSS11 was confirmed by Southern (data not shown) and Northern blot analysis (Fig. 3B). Deletion of the MSS11 caused a significant reduction of filaments formation in response to hyphal induction (Fig. 3A). In liquid serum-containing medium, wild-type cells formed true hyphae, whereas the mss11/mss11 mutant cells displayed pseudohyphalike form with a low percentage (ca. 20%) of stunted hyphae. On solid serum containing medium, the wild-type strain produced florid filamentous colonies, while the mss11/mss11 mutants formed small downy colonies without long filaments (Fig. 3A). The mss11/mss11 mutant strain displayed a more severe defective phenotype of hyphal development in Lee's medium. In contrast to the wild-type strain which developed extensive filaments, the mss11/mss11 mutant formed smooth colonies even after incubation for 7 days at 37°C on solid Lee medium. The defects of mss11/mss11 mutant strains in hyphal development could be rescued by integrating a wild-type MSS11 under the ADH1 promoter (Fig. 3A). Interestingly, MSS11 exerted its effects on hyphal development in a dosage-dependent manner. Reintroducing a single copy of MSS11 under its own promoter could not fully restore the ability of mss11/mss11 mutants to form filaments. Similarly, MSS11/mss11 heterozygotes also showed reduced filaments in solid serum-containing medium. More obviously, strains containing a single copy of MSS11 could not promote filaments formation and formed smooth-edged colonies on solid Lee's medium (Fig. 3A). Therefore, our data indicate that Mss11 is required for hyphal development in C. albicans.
FIG. 3.
Mss11 is required for hyphal development and the expression of hypha-specific genes in C. albicans. (A) Effects of MSS11 disruption on hyphal development. Cells were induced in YPD plus 10% serum for 3.5 h at 37°C (top). Colony morphology of mss11/mss11 mutants (middle and bottom rows). Strains were plated on solid serum-containing medium and solid Lee's medium, and incubated at 37°C for 5 and 7 days, respectively. (B) mss11/mss11 mutants were defective in the induction of hypha-specific genes. Cells were grown in YPD plus 10% serum at 37°C for 3.5 h or grown in YPD at 25°C for 6 h and collected for RNA extraction and Northern analysis. The image for the MSS11 hybridization was obtained after two weeks of exposure. Image for the HWP1-, ECE1-, and ACT1-probed filters were obtained after 3 h of exposure. Strains shown in panels A and B: wild type (CAF2-1), MSS11/mss11 (CSL1, +/−), mss11/mss11 (CSL3, −/−), mss11/mss11 + vector [CSL4 + pBA1, −/−(V)], mss11/mss11 + MSS11 [CSL4 + pBES116-MSS11, −/−(+)MSS11p], mss11/mss11 + ADH1p-MSS11 [CSL4 + pBA1-MSS11, −/−(+)ADH1p].
To explain the dosage effect of Mss11 on filamentous growth, we examined the transcription of MSS11 under the control of the ADH1 promoter and its own promoter. The transcription levels of MSS11 in MSS11/mss11 heterozygotes and a single-copy MSS11 revertant under its endogenous promoter were lower than that in MSS11/MSS11 wild-type cells. The transcription level of MSS11 in single copy MSS11 revertant under the ADH1 promoter was 10-fold higher than that under its own promoter in yeast growth condition, but only 2-fold higher in hyphal growth condition (Fig. 3B). This is consistent with the results reported by Swoboda et al. that the level of ADH1 mRNA fluctuates during the yeast-to-hyphal transition (49). Considering the dosage and positioning effect of URA3 in filamentation, we examined the phenotypes of MSS11/mss11 and mss11/mss11 mutants in the media containing additional uridine and observed similar phenotypes to that shown in Fig. 3A. In addition, we also compared the phenotypes between the MSS11/mss11 and mss11/mss11 mutant strains containing a single copy of URA3 at the ADE2 locus or at the MSS11 locus. The two sets of strains shared similar phenotypes in the media with or without additional uridine. Therefore, the gene dosage effect of MSS11 in affecting filamentation was not due to different expression level of URA3 or its different positions in the genome.
The expression of hypha-specific genes correlates with hyphal morphogenesis in C. albicans. HWP1, which serves as a mammalian transglutaminase substrate responsible for C. albicans-host interaction (47), was highly induced in wild-type cells under hypha-inducing conditions but was reduced 10-fold in mss11/mss11 mutant cells (Fig. 3B). As expected, the expression of ECE1, another hypha-specific gene (3), was reduced 60-fold in mss11/mss11 cells compared to that in wild-type cells. Consistent with the phenotypes, the MSS11/mss11 heterozygote as well as the single-copy MSS11 revertant under the control of the endogenous MSS11 promoter showed decreased expression of HWP1 and ECE1, whereas the MSS11 revertant under the control of the ADH1 promoter could fully restore the transcription of HWP1 and ECE1 (Fig. 3B).
To examine the expression level of MSS11 during hyphal development, wild-type (SC5314) cells were cultured overnight in YPD at 25°C and then released to liquid serum-containing medium for hyphal induction at 37°C and collected at various times up to 3 h for Northern blot analysis. The expression of MSS11, which was at a low level in yeast cells induced in response to serum, then reached the highest level in hyphal cells, which was ∼3-fold higher than that in yeast cells (Fig. 4A). To distinguish the effect of temperature switch on the expression of MSS11, we performed Northern analysis to examine the expression level of MSS11 in YPD at 37°C. Although the transcription of MSS11 was enhanced slightly (∼1.2-fold) in YPD at 37°C compared to that in YPD at 25°C but unable to reach the level that in YPD plus serum at 37°C (data not shown). Therefore, the increased expression of MSS11 responding to serum at 37°C was not due to the effect of temperature shift. Considering the existence of elongated cells in YPD at 37°C, the slightly enhanced MSS11 expression seems to correlate with the process of hyphal elongation. Consistently, deletion of Mss11 inhibited the cells from forming long filaments in serum-containing medium (Fig. 4B). The upregulated expression of MSS11 in hyphal cells reflected the regulatory activity of Mss11 in response to hyphal induction.
FIG. 4.
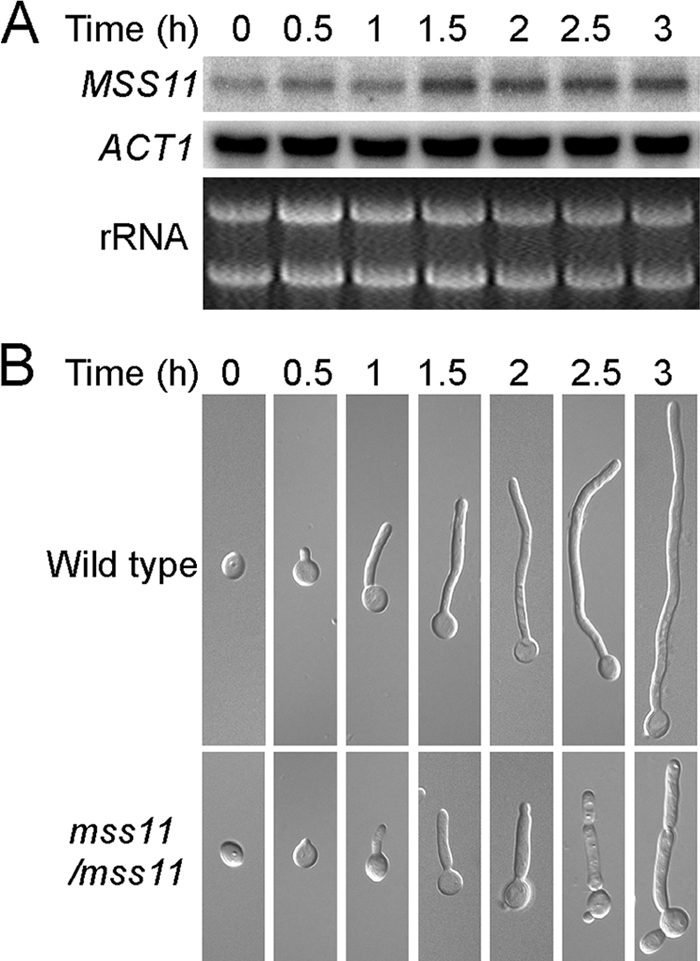
The enhanced expression of MSS11 during hyphal induction is associated with the process of cell elongation in C. albicans. (A) MSS11 transcript level increases in response to serum at 37°C. For Northern analysis, RNA was extracted from the wild-type strain (SC5314) after the indicated times of growth in liquid YPD plus 10% serum medium at 37°C. (B) Time-lapse microscopy of CAF2-1 and mss11/mss11 cells in liquid YPD containing 10% serum at 37°C.
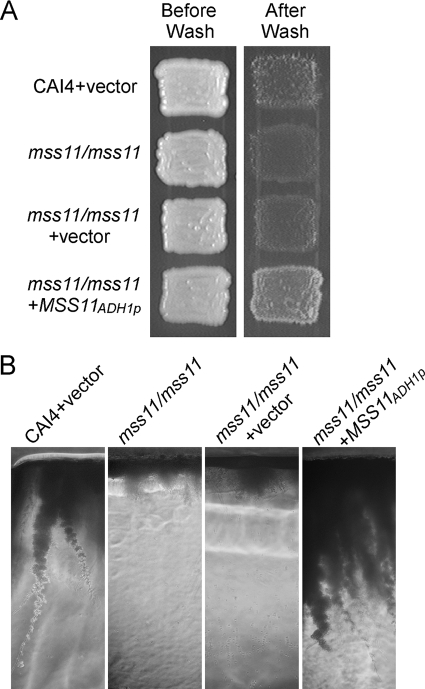
A previous study of S. cerevisiae showed that the mss11 mutant was severely defective in haploid invasive growth (51). To investigate the function of C. albicans Mss11 in the invasive growth, the strains were patched on YPD plates, incubated at 30°C for 2 days, and then washed with water for observation. Most of the wild-type cells on the surface were washed off, while some of them remained and penetrated into agar which formed invasive filaments under the agar (Fig. 5). The mss11/mss11 mutant cells were easily washed off and showed a very weak invasive growth. The invasive growth defect of mss11/mss11 mutant cells could be rescued by integrating a wild-type MSS11 under the ADH1 promoter. In fact, overexpressed Mss11 caused more cells to stick to the agar surface and promoted invasive filaments formation (Fig. 5). These observations suggest that the function of Mss11 in the invasive growth is conserved between S. cerevisiae and C. albicans.
FIG. 5.
Invasive growth assay of C. albicans. Strains of the wild type (CAI4 + pBA1), mss11/mss11 (CSL3), mss11/mss11 + vector (CSL4 + pBA1), and mss11/mss11 + ADH1p-MSS11 (CSL4 + pBA1-MSS11) were patched on solid YPD and incubated at 30°C for 2 days. (A) The plates were photographed before and after washing. (B) After washing with water, the agar containing cells was cut into slices and photographed.
Mss11 and Flo8 interact in vivo.
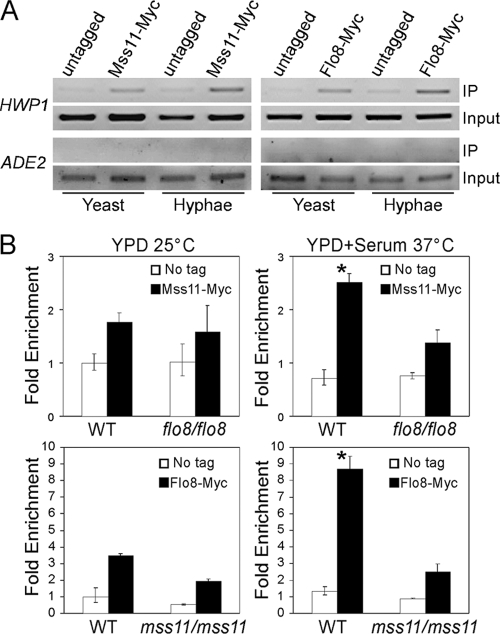
Mss11 and Flo8 are required for hyphal development and expression of hypha-specific genes in C. albicans. In addition, both Mss11 and Flo8 contain the LisH motif (amino acids 29 to 61 in Mss11 and amino acids 31 to 63 in Flo8), which is a thermodynamically stable dimerization domain. To define the relationship between Mss11 and Flo8, we examined the interaction between the two transcription factors by immunoprecipitation analysis. C-terminal 4FLAG-tagged Mss11 from ACT1 promoter at the RP10 locus was coexpressed with C-terminal 13Myc-tagged Flo8, which is under the control of its own endogenous promoter. As shown in Fig. 6A, the Mss11-4FLAG could be coimmunoprecipitated with anti-Myc antibody under both yeast and hyphal growth conditions, suggesting that Mss11 and Flo8 can interact in vivo and that the interaction is not affected by growth forms. The interaction between Mss11 and Flo8 is specific since the interaction was undetectable in the control strain that carried Mss11-4FLAG and Act1-13Myc fusion protein (data not shown).
Considering the protein level of Mss11 and Flo8 may affect their interactions, we detected the Mss11 expressing by Western blot analysis, and revealed that the expression level of MSS11 from the ACT1 promoter at the RP10 locus was almost same as that from its own promoter (data not shown). Therefore, the coimmunoprecipitation assay in Fig. 6A should represent the Mss11-Flo8 interaction in native situation. Tagging with 13Myc, 4FLAG, and 3HA may change the conformation of Mss11 and Flo8 and further affect their cellular functions. We introduced all of the N- or C-terminally tagged Flo8 and Mss11 shown in Fig. 6A, B, and C into flo8/flo8 or mss11/mss11 mutants, respectively. All of the tagged Flo8 or Mss11 can complement the defect of the mutants in filamentous growth (data not shown), indicating that the tagged Mss11 and Flo8 in Fig. 6 were all functional.
To determine whether the interaction between Mss11 and Flo8 is mediated by the LisH motif, the N-terminal 73 amino acids of Mss11 was removed. The truncated Mss11 lacking the LisH motif was fused with a 3HA tag at its N terminus (HA-Mss11ΔLisH) and subjected to the coimmunoprecipitation assay with Flo8-13Myc. The interaction between Mss11 and Flo8 was abolished by deletion of the LisH motif in Mss11 (Fig. 6B). To verify that the LisH motif in Flo8 is also required for the Mss11-Flo8 interaction, Flo8 with an N-terminal truncation of 119 amino acids was fused with a 3HA tag at its N terminus (HA-Flo8ΔLisH) and coexpressed with the Mss11-13Myc under the control of ACT1 promoter. The coimmunoprecipitation analysis with the truncated Flo8 protein indicated that the LisH motif of Flo8 is necessary for interaction with Mss11 (Fig. 6C). The results suggest that Mss11 interacts with Flo8 through the LisH motif.
Deletion of MSS11 weakens the binding of Flo8 to the HWP1 promoter in hyphal cells.
Considering the mss11/mss11 mutants are defective in the expression of HWP1, we reasoned that Mss11 might play an important role in transcriptional activation of HWP1. To test this possibility, a ChIP assay was performed to determine the presence of Myc-tagged Mss11 on the HWP1 promoter. The ADE2 promoter was served as a negative control as described in a previously published study utilizing the ChIP assay (39). As shown in Fig. 7A, Mss11-Myc expressed from its own endogenous promoter can bind to a specific region (roughly from −1381 to −1101) on the HWP1 promoter, a UAS region essential for the induction of HWP1 (26), but not to the surrounding regions of the HWP1 promoter. The promoter binding of Mss11 was detected under both yeast and hyphal growth conditions (Fig. 7A), and a significant enhancement in Mss11-Myc binding during hyphal induction was observed (P < 0.01) (Fig. 7B), a finding consistent with the phenomena of increased expression of MSS11 in response to serum at 37°C (Fig. 4A). ChIP analysis with Flo8-Myc showed that Flo8 bound to the same UAS region as Mss11 in the HWP1 promoter (Fig. 7A). Interestingly, the Flo8 binding was increased in hyphal cells compared to that in yeast cells (Fig. 7B), although the transcriptional level of FLO8 was unchanged during hyphal induction (data not shown).
FIG. 7.
Mss11 and Flo8 are present at the promoter of HWP1. (A) ChIP of Mss11 and Flo8 at a specific region of the HWP1 promoter (approximately positions −1381 to −1101) in yeast and hyphal cells. Strains CAF2-1 (no tag control), CSL12 (Mss11-Myc), and CSL14 (Flo8-Myc) were grown in either YPD at 25°C or YPD plus 10% serum at 37°C. (B) The enhanced binding of Mss11 and Flo8 proteins in the hyphal state depends on each other. For ChIP of Mss11 in the wild-type (CSL12) or flo8/flo8 (CSL13) strains (top row), wild-type (CAF2-1) and flo8/flo8 (CCF3) were used as no-tag controls, respectively. For ChIP of Flo8 in the wild-type (CSL14) or mss11/mss11 (CSL15) strains (bottom row), wild-type (CAF2-1) and mss11/mss11 (CSL3) were used as no-tag controls, respectively. DNA in the ChIP reactions was determined by quantitative PCR with primers at approximately positions −1381 to −1101 of the HWP1 promoter. The enrichment value was then presented as a ratio of immunoprecipitation versus input. Bars indicate standard deviation. *, P < 0.05 compared to the wild-type strain carrying Myc-tagged proteins under yeast growth condition. Cells were grown either in YPD at 25°C for yeast growth or in YPD plus 10% serum at 37°C for hyphal growth.
Since Mss11 can interact with Flo8 via the LisH motif, we speculated that the two proteins might cooperate to achieve the hyphal induced transcriptional activation of HWP1. Therefore, we performed a ChIP assay to examine whether Flo8 is required for Mss11-binding to the HWP1 promoter or vice versa. In the yeast growth condition, deletion of MSS11 or FLO8 has minor effect on the promoter binding of Flo8 or Mss11, respectively (Fig. 7B). However, lack of Mss11 or Flo8 has significant effect on each other's binding in hyphal growth condition, the ChIP signal of Mss11 in flo8/flo8 mutant or Flo8 in mss11/mss11 mutant remained almost unchanged during hyphal induction (Fig. 7B). The data suggested that Mss11 and Flo8 influence each other's enhanced binding to the HWP1 promoter in hyphal cells.
Overexpression of MSS11 cannot promote hyphal growth in flo8/flo8 mutant under aerobic conditions.
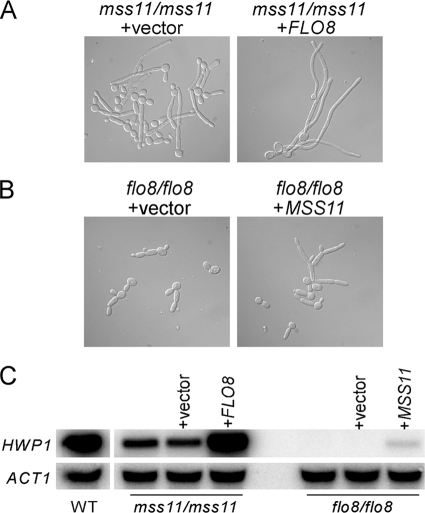
To define the functional relationship between Mss11 and Flo8, we performed reciprocal epistatic analysis. The filamentation defect in the mss11/mss11 mutant could be suppressed by overexpression of FLO8 (Fig. 8A), whereas the hyphal formation defect of the flo8/flo8 mutant was hardly suppressed by overexpression of MSS11 (Fig. 8B). The flo8/flo8 mutant carrying MSS11 under the ADH1 promoter only generated pseudohyphalike cells and exhibited a very low level of HWP1 expression when induced by serum (Fig. 8C), indicating that the presence of Flo8 is required for Mss11-mediated transcriptional activation of filamentous growth. In agreement with the phenotype, we observed that the HWP1 expression defect was restored by overexpressing FLO8 in the mss11/mss11 mutant (Fig. 8C). This result suggested that overproduction of Flo8 can activate HWP1 expression and hyphal growth in the absence of Mss11. We further constructed an mss11/mss11 flo8/flo8 double mutant for morphological observation. As expected, the double mutant strain failed to form filaments in the serum-containing medium, resulting in a phenotype similar to that of the flo8/flo8 mutant (Fig. 9A).
FIG. 8.
Mss11-stimulated hyphal development in C. albicans requires Flo8. (A) FLO8 overexpression suppresses the defect of mss11/mss11 mutant in filamentous growth. (B) Effect of MSS11 overexpression in the flo8/flo8 mutant. In panels A and B, the cells were induced in YPD plus 10% serum for 3.5 h at 37°C. (C) Northern analysis of HWP1 in the wild-type (CAF2-1), mss11/mss11 (CSL3), mss11/mss11 + vector (CSL4 + pBA1), mss11/mss11 + ADH1p-FLO8 (CSL4 + pBA1-FLO8), flo8/flo8 (CCF3), flo8/flo8 + vector (CCF4 + pBA1), and flo8/flo8 + ADH1p-MSS11 (CCF4 + pBA1-MSS11) strains. The experiment in panel C was carried out under the same conditions as in panels A and B.
FIG. 9.
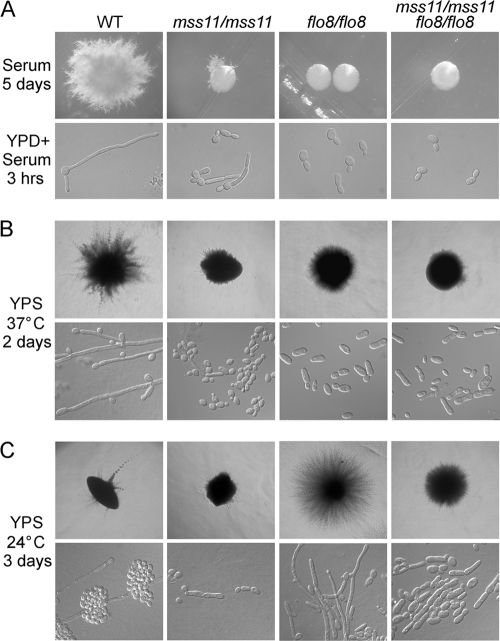
Effects of Mss11 and Flo8 on hyphal growth under embedded condition. (A) C. albicans mss11/mss11 flo8/flo8 cells are unable to form hyphae in response to serum. Strains were streaked on the solid serum-containing medium and incubated at 37°C for 5 days or grown in liquid medium (YPD plus 10% serum) at 37°C for 3 h. (B and C) Deletion of MSS11 inhibits filaments formation under microaerophilic condition. Cells were plated with molten YPS agar and grown at 37°C for 2 days (B) or at 24°C for 3 days (C). Colony and cell morphologies of the wild-type strain (CAF2-1), the mss11/mss11 mutant (CSL3), the flo8/flo8 mutant (CCF3), and the mss11/mss11 flo8/flo8 double mutant (CSL5) are shown.
Mss11 and Flo8 have different effects on hyphal growth under embedded condition at low temperature.
It has been reported that Flo8 functions as an activator at 37°C and as a repressor at 24°C under the embedded condition (8). To clarify the role of Mss11 in filamentous growth under the embedded condition, we examined the phenotype of the mss11/mss11 mutant at high and low temperatures under microaerophilic conditions. After growth in YPS agar at 37°C for 2 days, wild-type colonies generated long heterogeneous filaments, and mss11/mss11 or flo8/flo8 single mutant, as well as mss11/mss11 flo8/flo8 double mutant produced colonies surrounded with very short filaments (Fig. 9B). At 24°C, the wild type formed limited amounts of filaments after 3 days of incubation, while the mss11/mss11 single mutant formed smooth colonies surrounded with very short filaments, and the flo8/flo8 mutant produced fluffy colonies with long homogeneous filaments consisting of true hyphal cells. Although the mss11/mss11 flo8/flo8 double mutant strains formed colonies surrounded with homogeneous short filaments at 24°C, deletion of Mss11 exhibited a significant inhibitory effect on cell elongation in the flo8/flo8 mutant, the mss11/mss11 flo8/flo8 mutant cells could not develop into true hyphae (Fig. 9C). In contrast to Flo8, Mss11 functions as an activator in hyphal development under microaerophilic conditions at both temperatures.
DISCUSSION
C. albicans Mss11 is a transcriptional activator interacting with Flo8 via the LisH motif.
We identified C. albicans Mss11 as a LisH motif containing protein through a search of the NCBI database. Sequence analysis shows that a LisH motif, which is found in 114 eukaryotic proteins (15), is located at the N terminus of CaMss11, similar to that of S. cerevisiae Mss11. Previous studies suggest a central role for ScMss11 in the regulatory network that controls invasive growth in S. cerevisiae (51). By homology, CaMss11 is also important for C. albicans to invade the agar. Flo8, a transcription factor essential for hyphal development and virulence in C. albicans, also possesses a conserved LisH motif. The crystal structure of the N-terminal domain of mouse LIS1 shows that the LisH motif is a thermodynamically very stable dimerization domain (25), suggesting a common function of the LisH motif. Sif2, an integral component of the Set3 complex (SET3C), has been reported to function as a tetramer via the N-terminal LisH-containing domain in S. cerevisiae (9). Therefore, it is possible that the LisH motif in Flo8 is involved in dimerization. In fact, Flo8 and Mss11 have been found to function as a heterodimer in activating STA1 expression in S. diastaticus (27), although it remains to be defined whether the interaction is through the LisH motif. Here, we focus on the role of Mss11 in the morphological transition which is considered to be necessary for virulence of C. albicans.
Mss11 is involved in the transcriptional activation of hypha-specific genes necessary for hyphal development. Deleting MSS11 dramatically impairs hyphal formation in response to serum and the expression of hypha-specific genes, such as HWP1 and ECE1. ChIP analysis with the Mss11-Myc shows that Mss11 is located at the UAS region of the HWP1 promoter, and the Mss11 binding increases in hyphal cells. Furthermore, Mss11 functions as a transcriptional activator because the LexADB-Mss11 fusion protein has high transcriptional activity in S. cerevisiae. Taken together, our results suggest that Mss11 plays a role as a transcriptional activator in regulating the transcriptional program during hyphal induction.
The MSS11 transcript is regulated in response to serum at 37°C. The Northern blot analysis indicates that the expression level of MSS11 is enhanced during hyphal induction. In addition, constitutively overexpressed MSS11 promoted filamentation in the wild-type strain. Based on the expression pattern of MSS11, which was enhanced after 1.5 h in serum-containing medium, we infer that perhaps Mss11 is required for the elongation of hyphal cells. The regulation of MSS11 expression could explain the morphological characteristics of mss11/mss11 mutant cells which form stunted hyphae.
Changing the expression level is common for C. albicans to regulate the activity of transcription factors during hyphal development. It was previously reported that Nrg1, a repressor of filamentous growth in C. albicans, is downregulated during hyphal induction (5). Tec1, a transcription factor that regulates hyphal development in C. albicans, has been shown to be upregulated by serum-induced filamentous formation at 37°C (45). The expression level of SSN6 which encodes an important factor of morphological conversion declines significantly in response to serum (24). Our observation provides an inducible mechanism for induction of hyphal development by Mss11.
Cooperative regulation of Mss11 and Flo8 in hyphal development of C. albicans.
In this report, several lines of evidence suggest that Mss11 cooperates with Flo8 to regulate the expression of hypha-specific genes. An in vivo interaction between Mss11 and Flo8 was detected by immunoprecipitation in C. albicans, which is consistent with the notion that LisH motifs presented in both proteins contribute to dimerization. Furthermore, Mss11 and Flo8 bind to the same UAS region of the HWP1 promoter. Interestingly, there are more Mss11 and Flo8 proteins bound at this region in hyphal cells than in yeast cells, and the enhanced binding of both proteins depends on each other. Therefore, Mss11 might cooperate with Flo8 to regulate the hyphal transcriptional program in C. albicans, such as found between the Mss11 and Flo8 proteins in activation of STA1 expression in S. diastaticus. Another well-demonstrated cooperating pair in S. cerevisiae includes Ste12 and Tec1 (11). The transcription factors Ste12 and Tec1 bind cooperatively to enhancer elements called FREs (for filamentation and invasion response elements) and program transcription that is specifically responsive to the mitogen-activated protein kinase signaling components required for filamentous growth in S. cerevisiae (40). The C. albicans APSES proteins Efg1 and Efh1 cooperate to repress an alternative pathway of true hyphal formation in embedded or microaerophilic conditions (14). With coordinated activation and repression, C. albicans is able to integrate the signals from different pathways and specifically regulate the expression of target genes.
In contrast to the role of S. cerevisiae Mss11 which is genetically positioned downstream of Flo8 in regulating invasive growth and FLO11 expression (51), C. albicans Mss11 seems to act upstream of Flo8 since the CaMss11 is unable to promote hyphal development in the absence of the Flo8 in aerobic conditions. Loss of FLO8 nearly blocked the activation effect of Mss11 on hyphal formation, whereas overproducing Flo8 can suppress the hyphal growth defect of the mss11/mss11 mutant. It is possible that the increased expression of MSS11 during hyphal induction enhances the cooperative binding of Mss11 and Flo8 to the HWP1 promoter and further activates the expression of HWP1. These results, combined with the fact that flo8/flo8 mutant strains have a more severe defect in hyphal development than mss11/mss11 mutant strains, suggest that C. albicans Flo8 may play a central role in controlling the hyphal formation and activating the expression of hypha-specific genes by interacting with additional regulators. Indeed, Flo8 can interact with Efg1, a potential target of the cAMP/PKA pathway, and regulate the expression of hypha-specific genes (8). Interestingly, C. albicans Mss11 is essential for hyphal growth in microaerophilic conditions. Mss11 always functions as an activator under embedded conditions, whereas Flo8 is a dual functional regulator which acts as an activator at high temperature and as a repressor at low temperature. Loss of MSS11 in the flo8/flo8 mutant weakens the derepression effect of FLO8 disruption on hyphal formation at low temperature, indicating that the C. albicans Mss11 is required for hyphal growth of flo8/flo8 mutants in matrix.
In summary, we have identified a C. albicans transcription factor Mss11 that plays an important role in promoting hyphal development and is upregulated in response to serum. This enhanced expression of MSS11 increases the cooperative binding of Mss11 and Flo8 to the HWP1 promoter during hyphal induction, leading to transcriptional activation.
Acknowledgments
We thank Joseph Heitman and Florian F. Bauer for kindly providing S. cerevisiae mss11 strains. We thank Haoping Liu for helpful suggestions.
This study was supported by the Chinese National Natural Science Foundation (grants 30600008 and 30830003), the Chinese Academy of Sciences Foundation (KSCX2-YW-R-107), and Chinese 863 grant 2006AA02Z178.
Footnotes
Published ahead of print on 4 September 2009.
REFERENCES
- 1.Bassi, M. T., R. S. Ramesar, B. Caciotti, I. M. Winship, A. De Grandi, M. Riboni, P. L. Townes, P. Beighton, A. Ballabio, and G. Borsani. 1999. X-linked late-onset sensorineural deafness caused by a deletion involving OA1 and a novel gene containing WD-40 repeats. Am. J. Hum. Genet. 64:1604-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bester, M. C., I. S. Pretorius, and F. F. Bauer. 2006. The regulation of Saccharomyces cerevisiae FLO gene expression and Ca2+-dependent flocculation by Flo8p and Mss11p. Curr. Genet. 49:375-383. [DOI] [PubMed] [Google Scholar]
- 3.Birse, C. E., M. Y. Irwin, W. A. Fonzi, and P. S. Sypherd. 1993. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, S., P. Van Dijck, and A. Datta. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in Candida albicans, is downregulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, A. J., and N. A. Gow. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333-338. [DOI] [PubMed] [Google Scholar]
- 7.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 8.Cao, F., S. Lane, P. P. Raniga, Y. Lu, Z. Zhou, K. Ramon, J. Chen, and H. Liu. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17:295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerna, D., and D. K. Wilson. 2005. The structure of Sif2p, a WD repeat protein functioning in the SET3 corepressor complex. J. Mol. Biol. 351:923-935. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J., S. Zhou, Q. Wang, X. Chen, T. Pan, and H. Liu. 2000. Crk1, a novel Cdc2-related protein kinase, is required for hyphal development and virulence in Candida albicans. Mol. Cell. Biol. 20:8696-8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou, S., S. Lane, and H. Liu. 2006. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:4794-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conner, J., and Z. Liu. 2000. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA 97:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhillon, N. K., S. Sharma, and G. K. Khuller. 2003. Signaling through protein kinases and transcriptional regulators in Candida albicans. Crit. Rev. Microbiol. 29:259-275. [DOI] [PubMed] [Google Scholar]
- 14.Doedt, T., S. Krishnamurthy, D. P. Bockmuhl, B. Tebarth, C. Stempel, C. L. Russell, A. J. Brown, and J. F. Ernst. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emes, R. D., and C. P. Ponting. 2001. A new sequence motif linking lissencephaly, Treacher Collins, and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum. Mol. Genet. 10:2813-2820. [DOI] [PubMed] [Google Scholar]
- 16.Feng, Q., E. Summers, B. Guo, and G. Fink. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrante, M. I., G. Giorgio, S. A. Feather, A. Bulfone, V. Wright, M. Ghiani, A. Selicorni, L. Gammaro, F. Scolari, A. S. Woolf, O. Sylvie, L. Bernard, S. Malcolm, R. Winter, A. Ballabio, and B. Franco. 2001. Identification of the gene for oral-facial-digital type I syndrome. Am. J. Hum. Genet. 68:569-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagiano, M., M. Bester, D. van Dyk, J. Franken, F. F. Bauer, and I. S. Pretorius. 2003. Mss11p is a transcription factor regulating pseudohyphal differentiation, invasive growth and starch metabolism in Saccharomyces cerevisiae in response to nutrient availability. Mol. Microbiol. 47:119-134. [DOI] [PubMed] [Google Scholar]
- 20.Gale, C. A., C. M. Bendel, M. McClellan, M. Hauser, J. M. Becker, J. Berman, and M. K. Hostetter. 1998. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science 279:1355-1358. [DOI] [PubMed] [Google Scholar]
- 21.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base-pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 22.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast Saccharomyces cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 23.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 24.Hwang, C. S., J. H. Oh, W. K. Huh, H. S. Yim, and S. O. Kang. 2003. Ssn6, an important factor of morphological conversion and virulence in Candida albicans. Mol. Microbiol. 47:1029-1043. [DOI] [PubMed] [Google Scholar]
- 25.Kim, M. H., D. R. Cooper, A. Oleksy, Y. Devedjiev, U. Derewenda, O. Reiner, J. Otlewski, and Z. S. Derewenda. 2004. The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure 12:987-998. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S., M. J. Wolyniak, J. F. Staab, and P. Sundstrom. 2007. A 368-base-pair cis-acting HWP1 promoter region, HCR, of Candida albicans confers hypha-specific gene regulation and binds architectural transcription factors Nhp6 and Gcf1p. Eukaryot. Cell 6:693-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, T. S., H. Y. Kim, J. H. Yoon, and H. S. Kang. 2004. Recruitment of the Swi/Snf complex by Ste12-Tec1 promotes Flo8-Mss11-mediated activation of STA1 expression. Mol. Cell. Biol. 24:9542-9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leberer, E., D. Harcus, I. D. Broadbent, K. L. Clark, D. Dignard, K. Ziegelbauer, A. Schmidt, N. A. Gow, A. J. Brown, and D. Y. Thomas. 1996. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93:13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leberer, E., K. Ziegelbauer, A. Schmidt, D. Harcus, D. Dignard, J. Ash, L. Johnson, and D. Y. Thomas. 1997. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 7:539-546. [DOI] [PubMed] [Google Scholar]
- 30.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y., C. Su, X. Mao, F. Cao, and J. Chen. 2007. Roles of Candida albicans Sfl1 in hyphal development. Eukaryot. Cell 6:2112-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728-735. [DOI] [PubMed] [Google Scholar]
- 33.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 34.Liu, H., C. A. Styles, and G. R. Fink. 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144:967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous Candida albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 36.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenz, M. C., and J. Heitman. 1998. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics 150:1443-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, Y., C. Su, X. Mao, P. P. Raniga, H. Liu, and J. Chen. 2008. Efg1-mediated recruitment of NuA4 to promoters is required for hypha-specific Swi/Snf binding and activation in Candida albicans. Mol. Biol. Cell 19:4260-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 41.Mao, X., F. Cao, X. Nie, H. Liu, and J. Chen. 2006. The Swi/Snf chromatin remodeling complex is essential for hyphal development in Candida albicans. FEBS Lett. 580:2615-2622. [DOI] [PubMed] [Google Scholar]
- 42.Negredo, A., L. Monteoliva, C. Gil, J. Pla, and C. Nombela. 1997. Cloning, analysis, and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143(Pt. 2):297-302. [DOI] [PubMed] [Google Scholar]
- 43.Ni, J., Y. Gao, H. Liu, and J. Chen. 2004. Candida albicans Cdc37 interacts with the Crk1 kinase and is required for Crk1 production. FEBS Lett. 561:223-230. [DOI] [PubMed] [Google Scholar]
- 44.Pan, X., and J. Heitman. 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweizer, A., S. Rupp, B. N. Taylor, M. Rollinghoff, and K. Schroppel. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 46.Shoo, B. A., E. McPherson, and E. W. Jabs. 2004. Mosaicism of a TCOF1 mutation in an individual clinically unaffected with Treacher Collins syndrome. Am. J. Med. Genet. 126A:84-88. [DOI] [PubMed] [Google Scholar]
- 47.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1538. [DOI] [PubMed] [Google Scholar]
- 48.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 49.Swoboda, R. K., G. Bertram, S. Delbruck, J. F. Ernst, N. A. Gow, G. W. Gooday, and A. J. Brown. 1994. Fluctuations in glycolytic mRNA levels during morphogenesis in Candida albicans reflect underlying changes in growth and are not a response to cellular dimorphism. Mol. Microbiol. 13:663-672. [DOI] [PubMed] [Google Scholar]
- 50.Umeyama, T., Y. Nagai, M. Niimi, and Y. Uehara. 2002. Construction of FLAG tagging vectors for Candida albicans. Yeast 19:611-618. [DOI] [PubMed] [Google Scholar]
- 51.van Dyk, D., I. S. Pretorius, and F. F. Bauer. 2005. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccharomyces cerevisiae. Genetics 169:91-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Meyel, D. J., J. B. Thomas, and A. D. Agulnick. 2003. Ssdp proteins bind to LIM-interacting cofactors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development 130:1915-1925. [DOI] [PubMed] [Google Scholar]
- 53.Vernet, T., D. Dignard, and D. Y. Thomas. 1987. A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52:225-233. [DOI] [PubMed] [Google Scholar]
- 54.Webber, A. L., M. G. Lambrechts, and I. S. Pretorius. 1997. MSS11, a novel yeast gene involved in the regulation of starch metabolism. Curr. Genet. 32:260-266. [DOI] [PubMed] [Google Scholar]