Abstract
DNA gyrase is the only topoisomerase that can introduce negative supercoils into the DNA at the cost of ATP hydrolysis. Some but not all the steps of the topoisomerization reaction are understood clearly for both eukaryotic topoII and DNA gyrase. This study is an attempt to understand whether the B subunit of DNA gyrase binds to DNA directly, which may be central to the stimulation of its ATPase activity essential for gyrase function. We have dissected the Plasmodium falciparum gyrase B (PfGyrB) subunit to identify a 45-amino-acid region in the toprim domain that is responsible for its intrinsic DNA binding activity, DNA-stimulated ATPase activity, and DNA cleavage. We find that DNA has to enter through the ATP-operated clamp of PfGyrB to gain access to the DNA binding region. Furthermore, the rate of ATP hydrolysis of PfGyrB increases significantly with increasing DNA length, suggesting a possible communication between the ATPase domain and the DNA binding region that can account for its optimal ATPase activity. These results not only highlight the mechanism of GyrB action in the deadly human parasite P. falciparum but also provide meaningful insights into the current mechanistic model of DNA transport by gyrase during the topoisomerization reaction.
DNA topoisomerases are the essential enzymes that are required for the maintenance of DNA topology and genome stability in almost all cell types. These enzymes solve a plethora of topological problems in the genome created during the processes of replication, transcription, or recombination by coupling DNA breakage and ligation with the lateral translation of single- or double-stranded DNA through the body of the enzyme (27, 38). There are two major classes of topoisomerases (type I and type II), depending upon their ability to create a nick in one or both strands of DNA (6, 8).
Bacterial gyrase as well as eukaryotic topoisomerase II (topoII) are both classified as type II topoisomerases. Gyrase is a two-subunit (subunits A and B) heterotetramer with the subunit composition A2B2, while eukaryotic topoII is a homodimer. Eukaryotic topoII is regarded as the fusion of GyrB and GyrA subunits corresponding to its N- and C-terminal regions, respectively (21). These topoII enzymes catalyze a number of reactions, including the relaxation of positive and negative supercoils, catenation and decatenation of DNA circles, and knotting and unknotting of DNA. Interestingly, DNA gyrase has the unique property of introducing negative supercoils into the DNA apart from the relaxation of supercoiled DNA (13).
Each gyrase subunit contains different functional domains. The N terminus of the gyrase B subunit has ATP hydrolysis activity (1), whereas the C-terminal domain contains the DNA binding activity (7), and it is involved in interactions with GyrA. The N terminus of gyrase A retains the cleavage and religation functions, whereas the C terminus wraps the DNA during the process of supercoiling.
During the topoisomerization reaction, the topoII enzymes capture one DNA segment, called the transport or T segment, by the ATP-operated clamp formed by the N-terminal ATPase domain. Another DNA segment known as the G segment or gate segment (coming from the same or different DNA molecules), binds to the cleavage and religation domain of the enzyme, creating a transient double-stranded nick on the phosphodiester backbone of DNA by the nucleophilic attack of a pair of catalytic active-site tyrosine residues of the enzyme. This leads to the formation of a covalent bond between the 5′ phosphate of DNA and the active-site tyrosine residue. Closing the clamp by ATP binding thereby allows the T segment to pass through the transient double-stranded break created in the G segment. The G segment is religated, and the T segment is released from the enzyme (4, 11, 15, 21, 25, 39).
Despite the individual steps of the topoisomerization reaction now being understood largely for both eukaryotic topoII and DNA gyrase, there are some fundamental questions regarding the mechanism of DNA binding by GyrB and its involvement in the stimulation of its own ATPase activity.
The human malarial parasite Plasmodium falciparum contains both subunits (subunits A and B) of gyrase (PFL1120c and PFL1915w, respectively) that are targeted exclusively to its indispensable plastid-like organelle, the apicoplast (10). Both gyrase subunits are also present in Plasmodium vivax (22). The expression of full-length P. falciparum gyrase A (PfGyrA) is difficult in the heterologous expression system due to the presence of asparagines and lysine-rich-repeat regions and the unusual codon usage in Plasmodium (10, 30). Full-length P. falciparum gyrase B (PfGyrB) shows intrinsic ATPase activity in the absence of either the gyrase A subunit or DNA. This ATP hydrolysis activity is linear and follows Michaelis-Menten kinetics (10, 30), in contrast to its full-length Escherichia coli gyrase B (GyrBEc) counterpart, which exhibits ATPase activity (nonlinear) only in the presence of the A subunit and DNA (1, 7, 36). The N-terminal ATPase domain of GyrBEc forms a dimer only in the presence ATP, whereas PfGyrB forms a dimer even in the absence of nucleotides (10). Although there may be subtle differences between PfGyrB and GyrBEc, as described above, novobiocin (a catalytic-site-specific gyrase B inhibitor) selectively blocks apicoplast DNA replication and parasite proliferation in vitro, suggesting the indispensable role of gyrase in Plasmodium (30).
DNA stimulates the intrinsic ATPase activity of PfGyrB (10, 30) and that of the gyrase A2B2 complex from E. coli (7). Gyrase A in general shows intrinsic DNA binding activity (19, 24, 32), whereas the DNA binding activity of GyrB from any organism has not yet been reported. GyrBEc binds DNA only when in a complex with GyrA (7). Therefore, it becomes difficult to study how DNA binds to GyrBEc and stimulates its ATPase activity, which in turn is responsible for other gyrase functions. To get around this problem, we used PfGyrB, which has intrinsic ATPase activity and forms a functional complex with E. coli GyrA (GyrAEc). We show that PfGyrB binds to DNA by itself in the absence of the A subunit, and this leads to the stimulation of ATP hydrolysis. Furthermore, we have dissected the PfGyrB subunit into different variants to identify a unique 45-amino-acid region in the toprim domain (a conserved catalytic domain present in DNA topoisomerases, DnaG primase, and many other proteins) (3) that is responsible for its DNA binding (see Fig. S1 in the supplemental material). We show that this region is essential for in vitro and in vivo functions of PfGyrB. We also find that this region communicates with the intrasubunit ATPase domain of PfGyrB to stimulate its ATPase activity and with the intersubunit breakage and ligation domain of GyrA to manifest the cleavage reaction.
MATERIALS AND METHODS
Generation of constructs for overexpression in bacteria.
Full-length PfGyrB (excluding the N-terminal apicoplast signal sequence) and its different deletion mutants were cloned into expression vector pET21c (Novagen) with a C-terminal His6 tag. The DNA fragments containing PfGyrB and its different variants were amplified from P. falciparum genomic DNA by PCR using Triple Master Mix PCR enzyme (Eppendorf). Primer pairs 1 and 6, 1 and 2, 1 and 3, 4 and 6, and 5 and 6 were used for the synthesis of coding regions for PfGyrB, PfGyrB74, PfGyrB64, PfGyrB47, and PfGyrB33, respectively. The sequences of all the primers used are shown in Table 1. All the amplified DNA products were cloned into NheI and XhoI restriction enzyme sites in expression vector pET21c. Site-directed mutagenesis was used to convert conserved Glu159 (E159) and Lys220 (K220) (13) in PfGyrB into Ala159 and Ala220, respectively, by PCR using primer pair 7 and 8 and primer pair 9 and 10, respectively (Table 1).
TABLE 1.
List of primers
| Primer | Sequence (5′-3′ direction) |
|---|---|
| 1 | CTAGCTAGCTATAATTATGATGCTAAAGATATTG |
| 2 | CCGCTCGAGATTGTTATTATTTTGCGTAGATTC |
| 3 | CCGCTCGAGTCTTATTAAATCCCTAGCTGCC |
| 4 | CTAGCTAGCTCAAAAAATAATCAATATTCTTCTAC |
| 5 | CTAGCTAGCAATAACAATATCTTGAATAAAAAGAAAG |
| 6 | CCGCTCGAGTTCTGATAATGAATTTGTATTTTC |
| 7 | GATTACATCAAATATTATTTGCAATTATTGATAATTCAGTAG |
| 8 | CTACTGAATTATCAATAATTGCAAATAATATTTGATGTAATC |
| 9 | CATTCAGGAGCGGCATTTTTTGATGAC |
| 10 | GTCATCAAAAAATGCCGCTCCTGAATG |
| 11 | TTAAGATATGGAAAAGTTATT |
| 12 | ATTTTTTAAATTCTTATTGTCA |
| 13 | CAAGCCGTCGACACTGGTCCCGCCA |
| 14 | CGCGAGGGATCCTTGAAGCTG |
| 15 | CGGGATCCTTCAAGAGTAATCGTTCAAC |
| 16 | TGGACCTAGGATCCAATTTATCATTAGTTATATC |
Generation of the Δ45 deletion construct.
A total of 135 bp (45 amino acids) were deleted from the toprim domain of PfGyrB by PCR using primer pairs 11 and 12 (Table 1). The deletion mutant was generated by the amplification of recombinant plasmid PfGyrB-pET21c with forward and reverse primers in tail-to-tail directions that have a corresponding gap of 135 bp between their 5′ ends (16, 20). The PCR product was treated with a large fragment of Klenow polymerase (New England Biolabs) to remove the non-template-directed nucleotides added by the Triple Master Mix PCR enzyme. The blunt-ended PCR product was self-ligated, and E. coli DH10β cells were transformed with the ligation product. The clone was subsequently sequenced to verify the desired deletion.
Protein purification.
All the proteins were overexpressed in bacterial strain BL21 Codon Plus in the presence of two antibiotics (ampicillin at 100 μg/ml and chloramphenicol at 25 μg/ml). The bacterial cultures were grown until the optical density at 600 nm reached ∼0.4, and the cells were induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 20°C for 10 h. The cells were harvested and lysed at 4°C in lysis buffer (100 mM NaHPO4 [pH 8.0], 50 mM Tris-Cl [pH 8.0], 300 mM NaCl, 10 mM β-mercaptoethanol, 10 mM imidazole, 100 μM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [Sigma]) in the presence of 1 mg/ml chicken egg lysozyme (Sigma). Three bursts of sonication (30 s) of the lysate were followed by the addition of Triton X-100 at a concentration of 0.1% and rocking for 1 h at 4°C. The supernatant was cleared by centrifugation at 10,000 × g for 30 min at 4°C and allowed to bind to Ni-nitrilotriacetic acid resin (Qiagen) for 1 h. The beads were washed three times with lysis buffer containing 500 mM NaCl and 0.1% Triton X-100. The proteins were eluted with elution buffer containing 500 mM imidazole.
The expression and purification of the GyrAEc and GyrBEc proteins were described previously (10). The proteins were further purified through a heparin-sulfate column and finally dialyzed against buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 2 mM EDTA, 10 mM β-mercaptoethanol, 100 μM phenylmethylsulfonyl fluoride, and 10% glycerol. The proteins were aliquoted, snap-frozen, and stored at −80°C until further use.
Electrophoretic mobility shift assay (EMSA).
The DNA binding reactions were carried out with binding buffer (10 mM Tris-Cl [pH 7.5], 100 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 50 μg/ml bovine serum albumin, and 6% glycerol) by using either a PCR-amplified 240-bp DNA fragment containing the strong gyrase site (SGS) from plasmid pBR322 (7) or an approximately equal-length AT-rich fragment (212 bp; ∼80% AT rich) from the gyrA gene of P. falciparum. The sequences of the primers for the amplification of SGS DNA (primers 13 and 14) and AT-rich DNA (primers 5 and 16) are shown in Table 1. The DNA fragments were end labeled using [γ-32P]ATP with T4 polynucleotide kinase (New England Biolabs) and further purified by using a PCR product purification kit (Real Genomics) according to the manufacturer's instructions. The bound and free DNA probes were resolved in a 6% native gel (6% acrylamide-bisacrylamide, 1× TGE buffer, 5% glycerol) using 1× TGE buffer (25 mM Tris base, 190 mM glycine, 1 mM EDTA) supplemented with 10 mM MgCl2 for 10 h at 150 V (4°C) (17). The gels were further dried and autoradiographed.
ATPase assay.
All the reagents used for this assay were purchased from Sigma. The substrate-dependent ATPase assay was measured by use of a coupled pyruvate kinase-lactate dehydrogenase assay system described previously (10, 26). The hydrolysis of ATP by PfGyrB and other proteins was linked to the oxidation of NADH, and the reactions were monitored spectrophotometrically (Beckman Coulter) at 340 nm. Reactions with 70-μl mixtures were carried out at 22°C in a reaction buffer containing 35 mM Tris-Cl (pH 7.5), 70 mM KCl, 5 mM MgCl2, 250 μM NADH, 1.07 mM phosphoenol pyruvate, 0.56 units of pyruvate kinase, and 0.728 units of lactate dehydrogenase and various concentrations of ATP (ATP-Mg2+). The reaction mixture was incubated at room temperature for 5 min before the addition of protein. The rate of ATP hydrolysis was calculated from the initial 5 min of continuous data collected from the decrease in absorbance at 340 nm by using the following equation:
 |
where abs is absorbance and 6.22 per mM/cm is the molar extinction coefficient of NADH at 340 nm.
The ATPase data were fitted by using nonlinear regression analysis with GraphPad Prism software, and all the kinetic parameters were calculated accordingly.
The radioisotopic ATPase assay (see Fig. 5B) was carried out as described previously (10). The hydrolyzed inorganic phosphate (Pi) was separated by thin-layer chromatography and quantified by phosphorimaging.
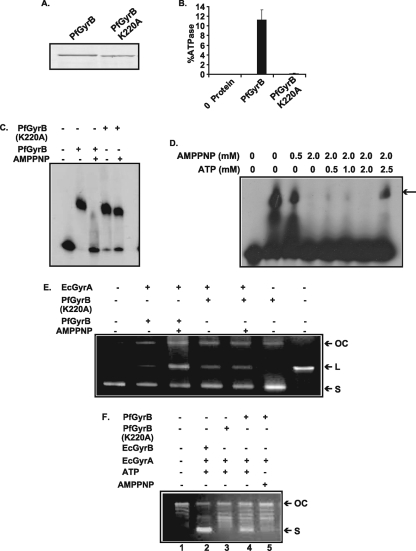
FIG. 5.
Closure of the ATP-operated clamp inhibits the DNA binding activity of PfGyrB. (A) SDS-PAGE analysis of wild-type PfGyrB and PfGyrB(K220A) with a point mutation at the ATP binding site. (B) ATPase activity of wild-type and ATP binding-site mutant proteins. The mutant protein shows a loss of ATPase activity. (C) DNA binding activity of PfGyrB (100 nM) or PfGyrB(K220A) (100 nM) in the absence or presence of 2 mM AMPPNP, a nonhydrolyzable analog of ATP. (D) DNA binding activity of PfGyrB in the presence of the various concentrations of AMPPNP and ATP indicated in figure. Competition with increasing amounts of ATP (2.5 mM) can abrogate the inhibitory effect of AMPPNP. The arrow indicates the position of the shifted band. (E) AMPPNP does not affect DNA cleavage activity. Ciprofloxacin-mediated DNA cleavage activity was determined using GyrAEc (35 nM) and PfGyrB (450 nM) or PfGyrB(K220A) (450 nM) in the absence or presence of AMPPNP. The first lane shows the DNA without any protein, and the last lane shows linearized DNA. “OC,” “L,” and “S” indicate open circular, linear, and supercoiled DNAs, respectively. (F) Supercoiling activity of PfGyrB (450 nM) and PfGyrB(K220A) (450 nM) in combination with GyrAEc (35 nM) in the presence or absence of ATP or AMPPNP as described in Materials and Methods. The results show that ATP is required for supercoiling activity. The positions of the open circular and supercoiled DNAs are marked.
DNA cleavage and supercoiling assays.
The DNA cleavage assay was carried out with cleavage buffer (35 mM Tris-Cl [pH 7.5], 24 mM KCl, 4 mM MgCl2, 5 mM dithiothreitol, 50 μg/ml bovine serum albumin, 6.5% glycerol) using plasmid pBR322 DNA as a substrate in the presence of 3 μg/ml ciprofloxacin. The samples were incubated for 1 h for the E. coli GyrAB combination and for 2 h for the GyrAEc and PfGyrB combination at room temperature. The cleavage complexes were captured by adding 0.2% sodium dodecyl sulfate (SDS). The samples were further treated with proteinase K (Sigma) at a final concentration of 166 μg/ml and incubated at 37°C for 1 h. Samples were resolved in a 1% agarose gel in 1× Tris-acetate-EDTA buffer and finally stained with ethidium bromide. A supercoiling assay was carried out according to a protocol described elsewhere previously (10). In brief, the reactions were performed using cleavage buffer (see above) supplemented with 2 mM spermidine and 2 mM ATP, and the samples were processed as described previously (10, 31).
RESULTS
The Plasmodium falciparum gyrase B subunit binds to DNA.
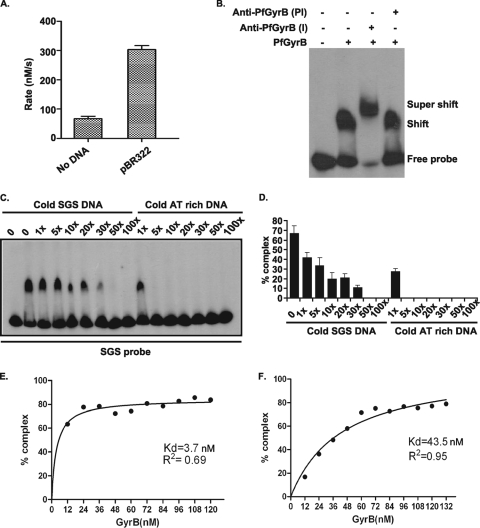
We find that the intrinsic ATPase activity of PfGyrB can be stimulated three- to fourfold in the presence of linear pBR322 DNA (Fig. 1A). These results are consistent with previously reported results (10, 30), and they raise the issue of whether PfGyrB binds to DNA directly. GyrBEc binds to DNA via the C-terminal 165-amino-acid stretch only in the presence of the GyrA subunit (see Fig. S1 in the supplemental material) (7). To investigate the DNA binding activity of PfGyrB, ∼100 nM purified protein was incubated with a 32P-radiolabeled 240-bp SGS DNA probe (see Materials and Methods) for 30 min at room temperature, and the free and protein-bound probes were separated in a native polyacrylamide gel (17). PfGyrB forms a stable DNA-protein complex, as shown in Fig. 1B. The addition of the anti-PfGyrB antibodies to the protein-DNA complex resulted in the supershift of the protein-DNA complex, while the addition of preimmune sera did not cause any supershift (Fig. 1B). These results strongly suggest that PfGyrB has an intrinsic DNA binding activity.
FIG. 1.
PfGyrB binds to DNA. (A) DNA stimulates the intrinsic ATPase activity of PfGyrB. The assay of ATPase activity was carried out with 1 μM purified PfGyrB enzyme in the absence and presence of 15 μg/ml linear pBR322 DNA. The reaction rates are plotted as shown. (B) To carry out the supershift experiments, 100 nM PfGyrB was incubated with a radiolabeled 242-bp SGS probe either in the absence or in the presence of anti-PfGyrB antibodies or preimmune antibodies. A supershift was found only in the presence of specific antibodies. (C) Affinity of PfGyrB for AT-rich DNA. The DNA binding property of PfGyrB was investigated using a [32P]ATP-labeled SGS probe in the presence of increasing amounts of cold SGS and AT-rich DNA as competitor DNA. (D) Following autoradiography, the DNA-protein complex was quantitated, and data were plotted accordingly. (E and F) Kd values for the binding of PfGyrB to AT-rich DNA (E) or SGS DNA (F) were calculated from their respective binding isotherms and by curve fittings using nonlinear regression with GraphPad Prism software.
The gyrase in the malaria parasite is targeted to the apicoplast (10), and the substrate of the enzyme is apicoplast DNA, which is ∼86% AT rich (42). To investigate whether PfGyrB has an affinity for AT-rich DNA, a gel mobility shift assay was performed using a radiolabeled SGS probe as described above, followed by competition with increasing amounts of cold SGS DNA or a similar length of AT-rich DNA from the PfGyrA coding sequence (overall, ∼79% AT-rich) (Fig. 1C). The intensity of the bands corresponding to the free probes and shifted bands under various conditions was further quantified using a phosphorimager and plotted accordingly (Fig. 1D). The results indicate that a fivefold addition of cold AT-rich DNA results in the complete loss of the protein-SGS probe complex, whereas a 50-fold excess of cold SGS DNA is required to obtain a similar effect. The gel retardation assay was repeated using the radiolabeled AT-rich DNA probe followed by competition with either cold AT-rich DNA or SGS DNA (see Fig. S2A and S2B in the supplemental material). We find that cold AT-rich DNA can compete for the radiolabeled probe-bound complex at a much lower concentration than that for cold SGS DNA. These competition experiments clearly show that AT-rich DNA is the preferred substrate for this enzyme.
To calculate the dissociation constant (Kd) of the enzyme with SGS DNA and AT-rich DNA, a gel mobility shift assay was performed with either the radiolabeled SGS probe or the AT-rich probe with various enzyme concentrations. The percentage of the protein-DNA complex was quantitated using phosphorimager analysis, and the data were directly fitted in a nonlinear regression analysis curve using GraphPad Prism software as shown in Fig. 1E and F (23). The Kd is estimated to be 43.5 nM for SGS DNA, whereas that for AT-rich DNA is estimated to be 3.7 nM. These results again confirm that PfGyrB has more affinity for AT-rich DNA.
To investigate whether PfGyrB binds to its target apicoplast DNA in vivo, we performed chromatin immunoprecipitation (CHIP) experiments. Total P. falciparum DNA extracted from the late trophozoite/early schizont stage was immunoprecipitated using anti-PfGyrB antibodies followed by PCR amplification of immunoprecipitated DNA using primer sets specific for the amplification of a region of the nuclear gene coding for the single-strand DNA binding protein (SSB) (PFE0436c) or inverted repeat region A (IRA) of apicoplast DNA (35). The CHIP experiments were repeated several times, and the results are shown in Fig. S2C and S2D in the supplemental material. The results indicate a severalfold increase in the intensity of PCR-amplified products from PfGyrB immune serum-immunoprecipitated DNA compared to that of preimmune sera for apicoplast DNA (IRA), whereas no such stimulation could be found for the nuclear SSB gene. The intensities of the control PCR products obtained from input genomic DNA were similar for both apicoplast IRA and a DNA fragment from the nuclear SSB coding region. These results strongly suggest that PfGyrB binds specifically to apicoplast DNA compared to nuclear DNA in vivo. Similar results were obtained with CHIP experiments using different primer sets specific for apicoplast DNA and nuclear DNA (data not shown).
IRA was further shown to bind to recombinant PfGyrB in vitro by EMSA using a radiolabeled IRA probe and purified PfGyrB (see Fig. S2E in the supplemental material), confirming data from the in vivo CHIP experiments.
Dissection of the PfGyrB subunit into different domains to locate the DNA binding region.
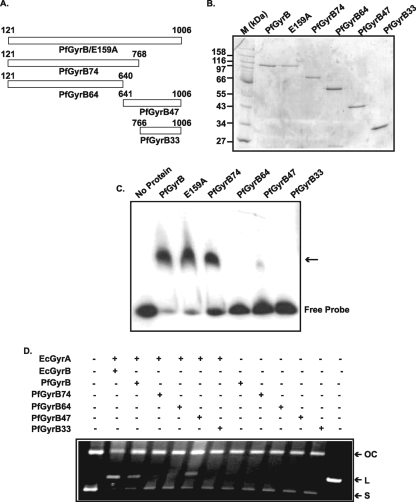
In order to map the DNA binding region of PfGyrB, several deletion mutant forms were generated from the N and C termini of GyrB. A schematic representation of all the deletion constructs is shown in Fig. 2A, and all the corresponding purified proteins are shown in Fig. 2B. Wild-type PfGyrB and a mutant that affects its ATP hydrolysis, PfGyrB(E159A) (10), were also purified under the same experimental conditions (see Materials and Methods). PfGyrB74 codes for a 74-kDa protein having the N-terminal ATPase domain and a part of the toprim domain, whereas PfGyrB64 codes for a 64-kDa N-terminal ATPase domain, which corresponds to the GyrBEc 43-kDa ATPase domain. PfGyrB47 is the 47-kDa truncated protein corresponding to the 47-kDa C-terminal domain of GyrBEc. PfGyrB33 contains the extreme C-terminal region of PfGyrB and has a molecular mass of 33 kDa. The above-described proteins were analyzed for their ability to bind DNA using a gel mobility shift assay (Fig. 2C). The results indicate that PfGyrB, PfGyrB(E159A), and PfGyrB74 bind to DNA, whereas no significant DNA binding activity was observed for PfGyrB64, PfGyrB47, and PfGyrB33. These findings indicate that the DNA binding region of PfGyrB falls within amino acid residues 641 to 768, which limit the boundaries of PfGyrB64 and PfGyrB74, respectively. Interestingly, PfGyrB47, where the above-described region is intact, does not show any DNA binding activity. Thus, the complete DNA binding domain appears to extend toward the N terminus, further upstream of residue 641.
FIG. 2.
Purification and functional analysis of PfGyrB and deletion mutant proteins. (A) Schematic representation of PfGyrB and its different deletion mutants. The amino acid coordinates are shown, and the number at the end of each mutant name indicates the molecular mass (in kilodaltons) of the respective mutant, except wild-type PfGyrB or the ATP binding mutant (E159A). (B) SDS-polyacrylamide gel electrophoresis (PAGE) analysis of the purified proteins. “M” indicates the molecular mass markers. (C) EMSA to show the DNA binding activity of wild-type PfGyrB and different deletion mutants as indicated at the top. The positions of the free probe and the shifted bands (arrow) are indicated. (D) DNA cleavage assay. Two hundred nanograms of pBR322 was incubated with GyrAEc (35 nM) in combination with either GyrBEc (55 nM) or PfGyrB (450 nM) and its different variants (450 nM each) in the presence of 3 μg/ml ciprofloxacin as described in Materials and Methods. The leftmost lane does not contain any protein. Only GyrBEc, PfGyrB, or PfGyrB47 supports E. coli GyrA-mediated DNA cleavage. “OC,” “L,” and “S” indicate open circular, linear, and supercoiled DNAs, respectively. The last lane on the right-hand side contains EcoRI-digested linearized pBR322 DNA.
During the supercoiling reaction, gyrase creates a double-stranded nick in the G segment and religates it after the passage of the T segment through the resultant gap. Ciprofloxacin, a gyrase A-specific drug, stabilizes the cleaved product and inhibits the religation activity of gyrase (7, 31). To investigate whether the DNA binding activity of PfGyrB has any role in DNA cleavage, we performed a cleavage assay using wild-type PfGyrB or its different truncated variants in combination with GyrAEc as described in Materials and Methods (Fig. 2D). The results indicate that wild-type PfGyrB and PfGyrB47 support GyrAEc-mediated DNA cleavage in the presence of ciprofloxacin, whereas PfGyrB74, PfGyrB64, or PfGyrB33 cannot perform DNA cleavage. The absence of DNA binding activity in PfGyrB47 and its involvement in cleavage suggest that the DNA binding activity of PfGyrB may not be essential for the DNA cleavage reaction. These results also suggest that the C terminus of PfGyrB including the region responsible for DNA binding (positions 641 to 768) in combination with GyrA is essential for DNA cleavage (12).
The 45-amino-acid residue insertion in the toprim domain of PfGyrB has a role in DNA binding and cleavage.
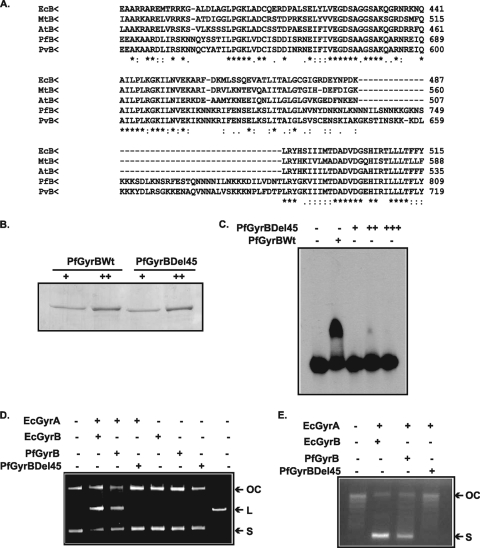
GyrBEc contains a unique stretch of 165 residues at the C terminus that is responsible for GyrBEc DNA binding activity in combination with GyrAEc (see Fig. S1 in the supplemental material) (7). A comparison of the primary sequence of PfGyrB with homologous sequences from E. coli and other organisms reveals that the above-described region is absent in PfGyrB and other organisms (see Fig. S1 in the supplemental material). Careful analysis of the toprim domain of GyrB revealed the presence of a unique 45-amino-acid-residue insertion in P. falciparum (residues 737 to 781) and P. vivax GyrB but not in other gyrase B proteins (Fig. 3A and see Fig. S1 in the supplemental material). Interestingly, this region overlaps with the DNA binding region (residues 641 to 768). To find out if this insertion in the toprim domain has a critical role in DNA binding, we deleted these 45 residues in PfGyrB using a deletion PCR method (16, 20). The resulting PfGyrBΔ45 construct was used for expression and protein purification in E. coli BL21 Codon Plus cells. The purification of wild-type and PfGyrBΔ45 proteins is shown in Fig. 3B.
FIG. 3.
Indispensable role of the 45-amino-acid insertion into the toprim domain of PfGyrB in DNA binding and cleavage. (A) Clustal W alignment of sequences of the toprim domains of GyrB subunits from different organisms (EcB, E. coli; MtB, Mycobacterium tuberculosis; AtB, Arabidopsis thaliana; PfB, Plasmodium falciparum; PvB, Plasmodium vivax) showing the insertion of the 45-amino-acid stretch in the toprim domain of Plasmodium gyrases. (B) SDS-PAGE analysis of purified wild-type PfGyrB (PfGyrBWt) and PfGyrBΔ45 at two different concentrations (“+” and “++,” respectively). (C) EMSA showing that PfGyrBΔ45 does not bind to DNA even when the protein is used in much higher concentrations (lanes +, ++, and +++) than those used for PfGyrB. (D) DNA cleavage assay as described in the legend of Fig. 2D, using GyrBEc (55 nM), wild-type PfGyrB (450 nM), or PfGyrBΔ45 (450 nM) in the presence of GyrAEc (35 nM). “OC,” “L,” and “S” indicate open circular, linear, and supercoiled DNAs, respectively. (E) Supercoiling activity of GyrBEc (55 nM), wild-type PfGyrB (450 nM), or PfGyrBΔ45 (450 nM) using PUC18 relaxed-plasmid DNA in the presence of GyrAEc (35 nM). Different bands present between the open circular and supercoiled forms correspond to different topoisomers of DNA.
To investigate if the deletion of the 45-amino-acid stretch could affect the overall structural conformation of PfGyrB, circular dichroism spectroscopy was carried out (as described in Materials and Methods) using the wild type and deletion mutants. Analysis of the absorption spectra in the far-UV region for both PfGyrB and PfGyrBΔ45 (see Fig. S2F in the supplemental material) was carried out, and the data were quantitatively analyzed using CDNN software (5) for determinations of percentages of secondary structural elements, as shown in Table 2. In conclusion, the circular dichroism data show that the deletion of the 45-amino-acid stretch results in no significant change in either the absorption spectrum or the secondary structural elements of PfGyrB (14, 26).
TABLE 2.
Comparison of the secondary structural elements of PfGyrB and PfGyrBΔ45
| Protein | % of protein with structural element |
|||||
|---|---|---|---|---|---|---|
| Helix | Antiparallel | Parallel | Beta turn | Random coil | Total | |
| PfGyrB | 26.1 | 11.3 | 10.7 | 18.5 | 39.2 | 102.8 |
| PfGyrBΔ45 | 26.3 | 11.3 | 10.5 | 18.5 | 36.7 | 102.2 |
Next, we investigated the DNA binding activities of PfGyrB and PfGyrBΔ45 using a gel retardation assay as mentioned above (Fig. 3C). No significant DNA binding was detected even at high concentrations of the PfGyrBΔ45 protein, whereas the wild-type protein showed a strong band shift, suggesting that the deletion of the 45 amino acid residues affects the DNA binding of PfGyrB completely.
Furthermore, the role of PfGyrBΔ45 in DNA cleavage and supercoiling activity was assessed in the presence of GyrAEc as described above in Materials and Methods. This protein could not support either DNA cleavage (Fig. 3D) or DNA supercoiling (Fig. 3E) in combination with GyrAEc, whereas the wild-type enzyme can perform both functions. The GyrABEc complex was used as a positive control for both gyrase activities.
It is important that the region containing the 45 amino acid residues is necessary but not sufficient to sustain DNA cleavage activity. This is demonstrated by the lack of cleavage with the GyrAEc-PfGyrB74 complex (Fig. 2D).
We showed previously that PfGyrB can functionally complement an E. coli GyrB temperature-sensitive mutant strain at the restrictive temperature (10). To investigate whether the deletion of the 45 amino acid residues would also affect the in vivo function of PfGyrB, a complementation experiment was carried out using an E. coli gyrB temperature-sensitive strain (N4177) (7) transformed with plasmid pAD2 or pAD2Δ45 expressing either wild-type or deletion mutant PfGyrB proteins, respectively. We find that the wild type but not the deletion mutant can rescue the temperature-sensitive phenotype at the restrictive temperature (42°C) (see Fig. S2G in the supplemental material). These results show the indispensable role of the 45-amino-acid insertion for the in vitro DNA binding and in vivo function of PfGyrB.
Analysis of the effect of DNA on the intrinsic ATPase activity of PfGyrB.
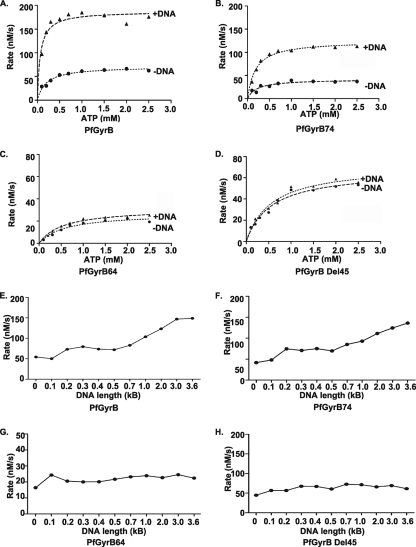
We have shown above that the intrinsic ATPase activity of PfGyrB is stimulated in the presence of DNA (10, 30). DNA-stimulated ATP hydrolysis by PfGyrB was measured by a lactate dehydrogenase-pyruvate kinase-linked assay as described in Materials and Methods (1, 10, 26). The rates of ATP hydrolysis at a fixed enzyme concentration (1 μM) were calculated with various concentrations of AT-rich DNA (15, 18, 29), and 15 μg/ml DNA was found to be optimal for maximal ATPase activity (data not shown). In order to confirm whether the 45-amino-acid region has a role in the DNA-dependent modulation of the ATPase activity of PfGyrB, ATP hydrolysis rates at a fixed enzyme concentration (1 μM) and at various substrate concentrations (0.1 mM to 2.5 mM) were measured spectrophotometrically for different proteins (wild-type PfGyrB, PfGyrB74, PfGyrB64, and PfGyrBΔ45) in the absence and presence of DNA (15 μg/ml AT-rich DNA). The kinetic parameters (Km, Kcat, and Vmax) for these ATPase reactions are listed in Table 3. The addition of DNA resulted in an increase in the rate of ATP hydrolysis for PfGyrB and PfGyrB74 to approximately threefold with respect to their DNA-independent rates, as shown in Fig. 4A and B and Table 3. These results are consistent with the DNA binding activity of PfGyrB and PfGyrB74 (with intact DNA binding regions). The Km values sharply decreased upon the addition of DNA in both cases, suggesting that DNA binding increases the affinity of the enzyme for ATP. The analysis of kinetic parameters of ATP hydrolysis reactions of PfGyrB64 and PfGyrBΔ45 (lacking a DNA binding region) revealed no significant change in the Km or Kcat in the absence or presence of DNA, confirming the critical role of the DNA binding region in modulating ATP hydrolysis rates by DNA (Fig. 4C and D and Table 3).
TABLE 3.
Kinetic parameters of hydrolysis of ATP
| Protein |
Km (mM) |
Vmax (nM/s) |
Kcat (s−1) |
|||
|---|---|---|---|---|---|---|
| Without DNA | With DNA | Without DNA | With DNA | Without DNA | With DNA | |
| PfGyrB | 0.19 | 0.06 | 71.0 | 187.4 | 0.07 | 0.18 |
| PfGyrB74 | 0.21 | 0.11 | 41.7 | 124.8 | 0.04 | 0.12 |
| PfGyrB64 | 0.58 | 0.55 | 27.0 | 31.3 | 0.02 | 0.03 |
| PfGyrBΔ45 | 0.55 | 0.56 | 66.8 | 71.9 | 0.06 | 0.07 |
FIG. 4.
DNA-stimulated ATPase activity of PfGyrB and effect of DNA lengths on the ATPase rate. (A to D) ATPase assays were carried out by incubating 1 μM PfGyrB (A), PfGyrB74 (B), PfGyrB64 (C), and PfGyrBΔ45 (D) in the absence or presence of 15 μg/ml AT-rich DNA at various substrate (Mg2+-ATP) concentrations, and the reaction rates were plotted accordingly. The individual curve in each graph was drawn by fitting the average of ATPase rates of three individual experiments using GraphPad Prism software. (E to H) Effect of the length of DNA on the ATPase activity of PfGyrB, PfGyrB74, PfGyrB64, and PfGyrBΔ45. The ATP hydrolysis rates of different proteins (1 μM) were determined (as described above) in the presence of equal concentrations (15 μg/ml) of AT-rich DNA fragments of various lengths, as indicated.
PfGyrB(E159A), PfGyrB47, and PfGyrB33 did not show any ATPase activity in the presence or absence of DNA (data not shown).
DNA length influences the ATP hydrolysis rate of PfGyrB.
The stimulation of the PfGyrB ATPase activity in the presence of DNA suggests that there may be a coordination between the ATPase domain and the DNA binding region. The binding of DNA may induce a conformational change in the ATPase domain in PfGyrB, resulting in the stimulation of ATPase activity.
To investigate if DNA has a role in coordinating the ATPase domain and DNA binding region during the DNA-stimulated ATPase reaction, we produced AT-rich DNA fragments of various lengths, from 100 bp to ∼3.7 kb, by means of PCR using coding regions of PfGyrA or PfGyrB as templates. The overall AT richnesses of PfGyrA and PfGyrB were ∼79% and 76%, respectively. Details of the primers, the length of the DNA fragments, and their coordinates are shown in Table S1 in the supplemental material. Each DNA fragment (15 μg/ml) was used to investigate its effect on the ATP hydrolysis rates of PfGyrB, PfGyrB74, PfGyrB64, and PfGyrBΔ45. The addition of DNA of various lengths showed an increase in the ATP hydrolysis rate with increasing DNA fragment lengths for PfGyrB and PfGyrB74 (Fig. 4E and F). Interestingly, both the PfGyrB64 and PfGyrBΔ45 proteins lacking the DNA binding region did not show an overall increase in the ATP hydrolysis rate with an increasing length of DNA fragments (Fig. 4G and H). These results strongly suggest that communication between the DNA binding region and the ATPase domain of PfGyrB may depend on the length of the DNA, which may be important for the optimal ATPase activity of PfGyrB. A similar coordination between the ATPase domain and DNA binding domain was reported previously for human and yeast topoII (15, 25). PfGyrB64 and PfGyrBΔ45 did not show any DNA length-dependent ATPase stimulation since these proteins do not have a DNA binding region.
The ATP-operated clamp of the ATPase domain of PfGyrB acts as an entry gate for DNA binding.
The DNA binding activity of PfGyrB and the increase in the ATP hydrolysis rate of PfGyrB in a DNA length-dependent manner raise the issue of whether the DNA molecule needs to pass through the ATP-operated clamp of PfGyrB. AMP-PNP (adenylyl-imidodiphosphate, tetralithium salt; nonhydrolyzable analog of ATP) is used to lock the ATP-operated clamp of topoisomerases. The preincubation of yeast topoII with AMPPNP does not allow the enzyme to bind to circular DNA (34), whereas the preincubation of E. coli gyrase with the ATP analog reduced but did not abolish its DNA binding activity (41). We were interested to analyze the DNA binding activity of PfGyrB in the presence of AMPPNP. For this purpose, we also purified a point mutant, PfGyrB(K220A) (Fig. 5A), which is equivalent to the GyrBEc(K103A) protein (see Fig. S1 in the supplemental material), which does not bind to ATP (28). We find that the ATP hydrolysis activity is completely lost for the PfGyrB(K220A) mutant compared to that of wild-type PfGyrB (Fig. 5B). The preincubation of PfGyrB and PfGyrB(K220A) with AMPPNP and subsequent incubation with a linear 212-bp radiolabeled AT-rich DNA probe abolished the DNA binding activity of the wild-type protein but did not affect the DNA binding activity of PfGyrB(K220A) (Fig. 5C). The loss of DNA binding activity of PfGyrB in the presence of AMPPNP indicates that the closure of the ATP-operated clamp limits the passage of DNA through it. This is similar to the function of the ATP-operated clamp described previously for yeast topoII (34).
The simultaneous preincubation of PfGyrB with 2 mM AMPPNP and a range of ATP concentrations resulted in the appearance of protein-DNA complexes when the ATP concentration (2.5 mM) exceeded the AMPPNP concentration (2 mM), as shown in Fig. 5D. These results suggest that the clamp of the enzyme remains closed when bound to AMPPNP, and it is further not available for the passage of DNA, whereas the hydrolysis of ATP allows the clamp to open up and the passage of DNA through it.
To assess the effect of the closure of the ATP-operated clamp with AMPPNP on the DNA cleavage and supercoiling reactions, we used PfGyrB and PfGyrB(K220A) in combination with GyrAEc. The intensity of the linear band, characteristic of DNA cleavage, increased in the presence of AMPPNP in the case of PfGyrB (Fig. 5E), suggesting that nucleotide binding shifts the equilibrium between uncleaved and cleaved states toward the cleaved state (11). On the other hand, there was no significant change in the level of cleavage in the case of PfGyrB(K220A) when used in the absence or presence of AMPPNP (Fig. 5E). These observations resemble the results reported previously for yeast topoII, where DNA cleavage occurred in the absence of ATP and the activity was greatly stimulated by the presence of AMPPNP (11).
We find that PfGyrB can introduce supercoils into relaxed pUC18 DNA in combination with GyrAEc only in the presence of ATP but not in the presence of AMPPNP (Fig. 5F, lanes 4 and 5). However, PfGyrB(K220A) cannot introduce the supercoils into relaxed pUC18 DNA because of its inability to bind to nucleotide (Fig. 5F, lane 3). GyrBEc along with GyrAEc were used as a positive control for supercoiling activity (Fig. 5F, lane 2).
The above-described results suggest that the passage of DNA through the ATP-operated clamp is not essential for the DNA cleavage reaction since DNA cleavage in the G segment is the direct manifestation of the DNA breakage and religation domain of GyrA in association with the C-terminal domain of PfGyrB (including the DNA binding region). DNA capture by the ATP-operated clamp is needed when the T segment has to be passed through the gate created in the G segment by the breakage and religation domain during the process of supercoiling.
DISCUSSION
The communication between the individual domains (ATPase domain and DNA cleavage domain) of eukaryotic topoII during the DNA topoisomerization reaction is largely understood (11, 25). This study is an attempt to understand the interdomain communications in the DNA gyrase, especially GyrB. GyrBEc possesses DNA binding activity (via the 165-amino-acid stretch) only in the presence of the A subunit (7), which shows a DNA binding property by itself (19, 32). The ATPase activity of GyrBEc is dependent on its association with GyrA and DNA (1, 13), making it difficult to study intra- and intersubunit domain communication during the various steps of the topoisomerization reaction. To address these issues, we have used PfGyrB with intrinsic ATPase activity (10, 30), which is stimulated in the presence of DNA and forms a functional gyrase complex with GyrAEc (10).
PfGyrB shows more affinity toward AT-rich DNA (Kd = 3.7 nM) than SGS DNA (Kd = 43.5 nM) (Fig. 1E and F). This finding is consistent with the overall AT richness of the apicoplast genome (∼86%), the target of P. falciparum gyrase (10, 42). PfGyrB contains a unique 45-amino-acid insertion region within the toprim domain (Fig. 3A) (3) that is essential for its in vitro (DNA binding, DNA cleavage, and DNA supercoiling) and in vivo (complementation of the E. coli gyrB temperature-sensitive mutant) functions. The apicoplast genome undergoes several rounds of rapid DNA replication during the asexual blood stage, liver stage, and sexual mosquito stage within a short span of time (35). The unique 45-amino-acid insertion accounting for its intrinsic DNA binding leading to the proficient stimulation of ATPase activity may provide better efficiency to Plasmodium gyrase to accommodate and relieve the tensions in circular apicoplast DNA molecules that may arise due to the rapid replication rate.
It is interesting that C-terminal PfGyrB47 does not show significant DNA binding activity, although it contains the toprim domain, including the 45-amino-acid insertion. It is possible that the core 45-amino-acid insertion region, along with the upstream region (absent in PfGyrB47), is required for the DNA binding of PfGyrB. This may explain the DNA binding activity of PfGyrB74 (containing the core DNA binding region and ATPase domain) and the inability of PfGyrB64 to bind to DNA (lacking the DNA binding region).
DNA binding of PfGyrB may not be required for the DNA cleavage reaction since cleavage was observed with wild-type PfGyrB and PfGyrB47 (which does not bind DNA) but not with PfGyrB74, PfGyrB64, and PfGyrB33 (Fig. 2D). These results suggest that the C terminus containing the toprim domain (3, 9) (including the core 45-residue insertion) of PfGyrB is the minimum requirement for the cleavage reaction in the presence of the GyrA subunit. This may also explain the inability of PfGyrBΔ45 to show cleavage activity.
DNA significantly increases the Vmax and Kcat values for PfGyrB and PfGyrB74 (both have their DNA binding regions intact) and decreases the Km values in both the cases (Table 3). There was a partial increase in the Vmax and Kcat values in the case of PfGyrB64 and PfGyrBΔ45 (both lacking a DNA binding region) but no significant change in their Km values in the presence of DNA. The partial increase in the ATPase activity of PfGyrB64 and PfGyrΔB45 in the presence of DNA suggests that the ATPase domain might have transient affinity for DNA. A mutation in Arg286 in the ATPase domain of GyrBEc abolishes the DNA-stimulated ATPase activity of the E. coli GyrAB complex (37, 40). The presence of equivalent residue K536 in the PfGyrB ATPase domain may be responsible for its transient affinity for DNA.
We found an unusual DNA length-dependent increase in the ATP hydrolysis rates of PfGyrB and PfGyrB74 where the core DNA binding region is intact (Fig. 4E and F), compared to PfGyrB64 and PfGyrBΔ45 lacking the DNA binding region (Fig. 4G and H). These results suggest possible cross talk between the ATPase domain and the DNA binding region in the presence of long and continuous DNA fragments that can span both the ATPase domain and the DNA binding region (15, 25). This possible cross talk would be absent in PfGyrB64 and PfGyrBΔ45 due to the absence of a core DNA binding region. It is also possible that the DNA length-dependent increase in the ATP hydrolysis rate of PfGyrB is due to the cooperative binding of PfGyrB to longer DNA fragments. However, it is less likely since increasing protein concentrations for a DNA probe with a fixed length do not result in multiple bands in gel shift experiments (data not shown).
Closing the ATP-operated clamp in the ATPase domain of PfGyrB with AMPPNP does not allow the protein to bind DNA (Fig. 5C). It seems that DNA has to enter through the ATP-operated clamp, slide down through the dimeric interface of the PfGyrB enzyme, and settle at the DNA binding region. A mutant form of PfGyrB, PfGyrB(K220), that is unable to bind ATP still binds to DNA in the presence of AMPPNP (Fig. 5C), and competition with ATP can override the inhibitory effect of AMPPNP on the DNA binding activity of wild-type PfGyrB (Fig. 5D). These results validate our hypothesis of the entry of the DNA through the ATP-operated clamp to facilitate the DNA binding activity of PfGyrB.
Based on our observations, we propose the following model for gyrase activity, as shown in Fig. S3 in the supplemental material. We believe that during the topoisomerization reaction, the ATP-operated clamp has to be open to capture the T segment of DNA to facilitate its binding with the DNA binding region and passage through cleaved gate or G-segment DNA held by GyrA to complete the reaction. For DNA cleavage activity in the G segment, the C-terminal domain including the DNA binding region of PfGyrB along with the breakage and religation domain of GyrA are essential (33). It seems that the DNA cleavage domain of topoII of higher organisms (11) has been evolutionarily distributed between the A and B subunits of gyrase to prevent GyrA from an unnecessary tearing of the genome. The finding of a unique DNA binding region in PfGyrB, its intrinsic ATPase activity, and the stimulation of ATPase activity in the presence of DNA validates the above-described model. Rapid and huge amounts of DNA replication (both nuclear and apicoplast) take place during various stages of the Plasmodium life cycle (2). While the eukaryote-type nuclear topoI and topoII (PFE0520c and PF14_0316, respectively) present in Plasmodium take care of nuclear DNA, unique features of PfGyrB may be central to maintain the topology of the apicoplast DNA to cope with the rapid DNA replication of this organelle during development. These results may have far-reaching effects, with the potential of using PfGyrB as an effective drug target in the light of growing incidences of conventional drug-resistant strains in malaria.
Supplementary Material
Acknowledgments
This work is supported by the Wellcome Trust London, the Swarnajayanti Fellowship (Department of Science and Technology, Government of India), and the MALSIG project funded by the European Union. S.K.D. is a Wellcome Trust International Senior Research Fellow. A.D. and D.P. acknowledge University Grant Commission, India, and CSIR, India, for fellowships. N.M. acknowledges financial support from the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases.
We greatly acknowledge the circular dichroism facility at the Advanced Instrument Facility at Jawaharlal Nehru University. We thank V. Nagaraja (IISc, Bangalore, India) for his valuable suggestions.
Footnotes
Published ahead of print on 21 August 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Ali, J. A., A. P. Jackson, A. J. Howells, and A. Maxwell. 1993. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyses ATP and binds coumarin drugs. Biochemistry 32:2717-2724. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., L. M. Iyer, T. E. Wellems, and L. H. Miller. 2003. Plasmodium biology: genomic gleanings. Cell 115:771-785. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., D. D. Leipe, and E. V. Koonin. 1998. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjergbaek, L., P. Kingma, I. S. Nielsen, Y. Wang, O. Westergaard, N. Osheroff, and A. H. Andersen. 2000. Communication between ATPase and cleavage/religation domain of human topoisomerase IIα. J. Biol. Chem. 17:13041-13048. [DOI] [PubMed] [Google Scholar]
- 5.Bohm, G., R. Muhr, and R. Jaenicke. 1992. Quantitative analysis of proteins in far UV circular dichroism spectra by neural networks. Protein Eng. 5:191-195. [DOI] [PubMed] [Google Scholar]
- 6.Champoux, J. J. 2001. DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 7.Chatterji, M., S. Unniraman, A. Maxwell, and V. Nagaraja. 2000. The additional 165 amino acids in the B protein of Escherichia coli gyrase have an important role in DNA binding. J. Biol. Chem. 275:22888-22894. [DOI] [PubMed] [Google Scholar]
- 8.Corbett, K. D., and J. M. Berger. 2004. Structure, molecular mechanisms and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33:95-118. [DOI] [PubMed] [Google Scholar]
- 9.Costenaro, L., J. G. Grossmann, C. Ebel, and A. Maxwell. 2007. Modular structure of full length gyrase B subunit revealed by small-angle-X-ray scattering. Structure 15:329-339. [DOI] [PubMed] [Google Scholar]
- 10.Dar, M. A., A. Sharma, N. Mondal, and S. K. Dhar. 2007. Molecular cloning of apicoplast-targeted Plasmodium falciparum DNA gyrase genes: unique intrinsic ATPase activity and ATP-independent dimerization of PfGyrB subunit. Eukaryot. Cell 6:398-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong, K. C., and J. M. Berger. 2007. Structural basis for gate-DNA recognition and bending by type II topoisomerases. Nature 450:1201-1206. [DOI] [PubMed] [Google Scholar]
- 12.Gellert, M., L. M. Fisher, and M. H. O'Dea. 1979. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc. Natl. Acad. Sci. USA 76:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellert, M., K. Mizuuchi, M. H. O'Dea, and H. A. Nash. 1976. DNA gyrase: an enzyme that introduces negative supercoils into DNA. Proc. Natl. Acad. Sci. USA 73:3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenfield, N. J. 2007. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1:2876-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammonds, T. R., and A. Maxwell. 1997. The DNA dependence of ATPase activity of human topoisomerase IIα. J. Biol. Chem. 272:32696-32703. [DOI] [PubMed] [Google Scholar]
- 16.Heckman, K. L., and L. R. Pease. 2007. Gene splicing and mutagenesis by PCR driven overlap extension. Nat. Protoc. 2:924-932. [DOI] [PubMed] [Google Scholar]
- 17.Hellman, L. M., and M. G. Fried. 2007. Electrophoretic mobility shift assay (EMSA) for detecting protein nucleic acid interactions. Nat. Protoc. 2:1849-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, T., H. Sage, and T. Hsieh. 2002. ATPase domain of eukaryotic DNA topoisomerase II. J. Biol. Chem. 277:5944-5951. [DOI] [PubMed] [Google Scholar]
- 19.Huang, Y., J. Deng, J. Gu, Z. Zhang, A. Maxwell, L. Bi, Y. Chen, Y. Zhou, Z. Yu, and X. Zhang. 2006. The key DNA-binding residues in the C-terminal domain of Mycobacterium tuberculosis DNA gyrase A subunit (GyrA). Nucleic Acids Res. 34:5650-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai, Y., Y. Matsushima, T. Sugimura, and M. Terada. 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kampranis, S. C., and A. Maxwell. 1996. Conversion of gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci. USA 93:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khor, V., C. Yowell, J. B. Dame, and T. C. Rowe. 2005. Expression and characterization of the ATP binding domain of a malarial Plasmodium vivax gene homologous to the B subunit of bacterial topoisomerase DNA gyrase. Mol. Biochem. Parasitol. 140:107-117. [DOI] [PubMed] [Google Scholar]
- 23.Kim, C. S., S. H. Lee, R. Y. Kim, B. J. Kim, S. Z. Li, I. H. Lee, E. J. Lee, S. K. Lim, Y. S. Bae, W. Lee, and J. H. Baik. 2002. Identification of domains directing specificity of coupling to G-proteins for the melanocortin MC3 and MC4. J. Biol. Chem. 277:31310-31317. [DOI] [PubMed] [Google Scholar]
- 24.Lother, H., and R. Lurz. 1984. DNA binding and antigenic specifications of DNA gyrase. Nucleic Acids Res. 12:901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller-Planitz, F., and D. Herschlag. 2006. Interdomain communication in DNA topoisomerase II. J. Biol. Chem. 281:23395-23404. [DOI] [PubMed] [Google Scholar]
- 26.Nitharwal, R. G., S. Paul, A. Dar, N. R. Choudhury, R. K. Soni, D. Prusty, S. Sinha, T. Kashav, G. Mukhopadhyay, T. K. Chaudhuri, S. Gourinath, and S. K. Dhar. 2007. The domain structure of Helicobacter pylori DnaB helicase: the N-terminal domain can be dispensable for helicase activity whereas the extreme C-terminal region is essential for its function. Nucleic Acids Res. 35:2861-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nöllmann, M., M. D. Stone, Z. Bryant, J. Gore, N. J. Crisona, S.-C. Hong, S. Mitelheiser, A. Maxwell, C. Bustamante, and N. R. Cozzarelli. 2007. Multiple modes of E. coli DNA gyrase activity revealed by force and torque. Nat. Struct. Mol. Biol. 14:264-271. [DOI] [PubMed] [Google Scholar]
- 28.O'Dea, M. H., J. K. Tamura, and M. Gellert. 1996. Mutations in the B subunit of Escherichia coli DNA gyrase that affect ATP-dependent reactions. J. Biol. Chem. 271:9723-9729. [DOI] [PubMed] [Google Scholar]
- 29.Olland, S., and J. C. Wang. 1999. Catalysis of ATP hydrolysis by two NH2-terminal fragments of yeast DNA topoisomerase II. J. Biol. Chem. 31:21688-21694. [DOI] [PubMed] [Google Scholar]
- 30.Raghu, R. E. V., A. Kumar, S. Biswas, A. Kumar, S. Chaubey, M. I. Siddiqi, and S. Habib. 2007. Nuclear encoded gyrB encodes a functional subunit of Plasmodium falciparum gyrase that is involved in DNA replication. Mol. Biochem. Parasitol. 154:30-39. [DOI] [PubMed] [Google Scholar]
- 31.Reece, R. J., and A. Maxwell. 1989. Tryptic fragments of Escherichia coli DNA gyrase A protein. J. Biol. Chem. 264:19648-19653. [PubMed] [Google Scholar]
- 32.Reece, R. J., and A. Maxwell. 1991. The C-terminal domain of Escherichia coli DNA gyrase A subunit is a DNA binding protein. Nucleic Acids Res. 19:1399-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roca, J. 2008. Topoisomerase II: a fitted mechanism for the chromatin landscape. Nucleic Acids Res. 37:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roca, J., and J. C. Wang. 1992. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell 71:833-840. [DOI] [PubMed] [Google Scholar]
- 35.Singh, D., S. Chaubey, and S. Habib. 2003. Replication of the Plasmodium falciparum apicoplast DNA initiates within the inverted repeat region. Mol. Biochem. Parasitol. 126:9-14. [DOI] [PubMed] [Google Scholar]
- 36.Sugino, A., and N. Cozzarelli. 1980. The intrinsic ATPase of DNA gyrase. J. Biol. Chem. 225:6299-6303. [PubMed] [Google Scholar]
- 37.Tingey, A. P., and A. Maxwell. 1996. Probing the role of ATP operated clamp in the strand passage reaction of DNA gyrase. Nucleic Acids Res. 24:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, J. C. 1991. DNA topoisomerases: why so many? J. Biol. Chem. 266:6659-6662. [PubMed] [Google Scholar]
- 39.Wang, J. C. 2007. Unlocking and opening a DNA gate. Proc. Natl. Acad. Sci. USA 104:4773-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wigley, D. B., G. J. Davie, E. J. Dodson, A. Maxwell, and G. Dodson. 1991. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351:624-629. [DOI] [PubMed] [Google Scholar]
- 41.Williams, N. L., A. J. Howells, and A. Maxwell. 2001. Locking the ATP operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 306:969-984. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, R. J. M., P. W. Denny, P. R. Preiser, K. Rangachari, K. Roberts, A. Roy, A. Whyte, M. Strath, D. J. Moore, P. W. Moore, and D. H. Williamson. 1996. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 261:155-172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.